SUMMARY
Previous genetic association studies of the fibrinogen gene cluster have identified associations with plasma fibrinogen levels. These studies are typically limited to plasma fibrinogen measured among European-descent populations. We sought to replicate previous well-known associations with fibrinogen variants and plasma fibrinogen. We then sought to identify and characterise novel associations with fibrinogen variants with plasma fibrinogen and several haematological traits in three racial/ethnic populations. We genotyped 25 single nucleotide polymorphisms (SNPs) in the fibrinogen gene cluster in 2,631 non-Hispanic whites, 2,108 non-Hispanic blacks, and 2,073 Mexican Americans from the Third National Health and Nutrition Examination Survey (NHANES). We performed single SNP tests of association for plasma fibrinogen, mean platelet volume, platelet distribution width, platelet count, white blood cell count, and serum triglycerides. Five previously identified associations with plasma fibrinogen replicated in our study in non-Hispanic whites and blacks. We identified two novel associations between genetic variants and decreased plasma fibrinogen: rs2227395 (p=0.0007; non-Hispanic whites) and rs2070022 (p=0.001; Mexican Americans). Several fibrinogen SNPs were also associated with haematological traits: rs6050 with decreased platelet distribution width in non-Hispanic whites; rs6050 and rs2066879 with decreased and increased platelet distribution width, respectively, in non-Hispanic whites; rs2227409 with increased mean platelet volume, rs2070017 with decreased platelet count, and rs6063 with increased platelet distribution width in non-Hispanic blacks; and rs4220 and rs2227395 with decreased white blood cell count, rs2227409 with increased platelet distribution width, rs2066860 and rs1800792 with increased and decreased triglyceride levels, respectively, and rs1800792 with decreased platelet counts in Mexican Americans. We successfully replicated and identified novel associations with fibrinogen variants and plasma fibrinogen. These data confirm the importance of the fibrinogen gene cluster for plasma fibrinogen levels as well as suggest this gene cluster may have pleiotropic effects on haematological traits.
Keywords: fibrinogen, pleiotropy, replication
INTRODUCTION
Circulating levels of haemostatic factors, such as plasma fibrinogen, and cellular components of the blood such as the concentration of haemoglobin (Hb), the numbers of white blood cells (WBC), red blood cells (RBC) and platelets (PLT), and the volumes of red blood cells (mean corpuscular volume, MCV) and platelets (mean platelet volume, MPV) are tightly regulated [1;2]. Abnormal levels of Hb, MPV, PLT, plasma fibrinogen, serum triglycerides, and WBC are associated with myocardial infarction (MI), coronary artery disease (CAD), and stroke [3–8]. Plasma fibrinogen is a key player in the pathogenesis of cardiovascular disease, specifically arterial thrombosis. In the coagulation pathway, fibrinogen is converted to fibrin, the main protein component of an arterial clot [9;10]. The roles of genetic factors for plasma fibrinogen and other coagulation factors have been well studied over the past 30 years; however, little is known about the genetics of other common haematological measurements and their relationship with cardiovascular related diseases.
Genetic factors are reported to account for up to 50% of the variability observed in plasma fibrinogen levels [10]. Heritability estimates for haematological measurements such as Hb, WBC, and PLT range from 0.37–0.89 [1;2]. Several genetic association studies have been conducted to assess the relationship between candidate genes of the coagulation pathway and fibrinogen, coagulation factors, and other common haemostatic measurements. Candidate gene studies have demonstrated that variation in fibrinogen genes (FBA, FGB and FGG) are associated with plasma fibrinogen levels [11–15]. More recently, genome-wide association (GWAS) studies have been performed for haematological traits. Meisinger, et al. conducted a GWAS and identified three common loci that were associated with mean platelet volume [16]. Another GWAS conducted by the Haem Gen Consortium identified 22 loci associated with eight haematological traits: Hb, RBC, WBC, PLT, MPV, MCV, mean corpuscular hemoglobin concentration (MCHC) and mean corpuscular content (MCH) [17].
While these studies have identified genetic variation associated with these quantitative traits, the discovery effort has been limited to European-descent populations. It is well known that genetic backgrounds vary among populations, both in allele frequency and in linkage disequilibrium patterns for any diseases including venous thrombosis [18]. As an example of allele frequency differences, one study performed in diverse populations demonstrated that Factor V Leiden (rs6025) and the prothrombin G20210A (rs1799963) variants differ across populations [19]. The prevalence of these variants in African Americans and Hispanics is 1–2% and <0.04%, respectively compared to 5% and 2–4% observed in Europeans [20–22]. Since these well-known variants are less common in African Americans and Hispanics, it is possible that other variants have yet to be identified in these populations that explain more of the variance in haemostatic and/or haematological traits.
Using a candidate gene approach we investigated the role of several genetic variants in the fibrinogen gene cluster with plasma fibrinogen and several haematological parameters. We characterised the association of 25 single nucleotide polymorphisms (SNPs) and plasma fibrinogen levels in >7,000 DNA samples of non-Hispanic whites, non-Hispanic blacks, and Mexican Americans from the Third National Health and Nutrition Examination Survey (NHANES). We also tested for an association with these genetic variants and mean platelet volume, white blood cell count, platelet count, platelet distribution width, and serum triglycerides. Overall, we replicated well-known associations between the fibrinogen cluster and plasma fibrinogen levels in a European-descent population, generalized these associations to populations of non-European descent, and identified novel associations with the gene cluster and haematological traits, expanding our knowledge the role the fibrinogen gene cluster plays on plasma fibrinogen levels and haematologic traits in diverse populations.
MATERIALS AND METHODS
Study population
All procedures were approved by the CDC’s Ethics Review Board and written informed consent was obtained from all participants. This candidate gene association study was approved by the CDC’s Ethics Review Board (protocols #2003-08 and #2006-11) and the University of Washington’s Institutional Review Board (IRB #23667; HSRC D committee). Because no identifying information was accessed by the investigators, this study was considered exempt from Human Subjects by Vanderbilt University’s Institutional Review Board (IRB #061062; HS2 committee).
Ascertainment of NHANES III and method of DNA collection have been previously described [23–25]. The National Health and Nutritional Examination Surveys are cross-sectional surveys conducted across the United States by the National Center for Health Statistics (NCHS) at the Centers for Disease Control and Prevention (CDC). NHANES III was conducted between 1988–1990 (phase 1) and 1991–1994 (phase 2) [26;27] as a complex survey design that over-sampled minorities (non-Hispanic blacks and Mexican Americans), the young, and the elderly. All NHANES have interviews that collect demographic, socioeconomic, dietary, and health-related data. Also, all NHANES study participants undergo a detailed medical examination at a central location known as the Mobile Examination Center (MEC). The medical examination includes the collection of physiological measurements by CDC medical personnel and blood and urine samples for laboratory tests. Beginning with phase 2 of NHANES III, DNA samples were collected from study participants aged 12 years and older.
Haematological measurements
Blood was collected from all participants with the exception of those who reported having haemophilia or chemotherapy in the past four weeks [28]. Complete blood counts (CBCs) were performed on qualifying NHANES participants using the Beckman Coulter method, a method that sizes and counts particles by using measurable changes in electrical resistance produced by nonconductive particles suspended in an electrolyte [29]. A quantitative, automated, differential cell counter was used to calculate exact values for haematological measurements [28]. Plasma fibrinogen was measured from blood plasma on participants 40 years and older using the Clauss clotting method [29]. Serum triglycerides were measured using standard enzymatic methods.
SNP selection and genotyping
DNA was extracted from crude cell lysates from lymphoblastoid cell lines established for NHANES III participants aged 12 and over as part of Genetic NHANES [25]. TagSNPs were selected using LDSelect [30] for multiple populations at r2 >0.80 for common variants (minor allele frequency (MAF) >5%) in three candidate genes (FGA, FGB, and FGG) based on data available for European Americans and African Americans in SeattleSNPs [31]. A total of 25 SNPs were genotyped using the Illumina GoldenGate assay (as part of a custom 384 oligonucleotide pool assay (OPA) by the Center for Inherited Disease Research (CIDR) through the National Heart Lung and Blood Institute’s Resequencing and Genotyping Service. A total of 7,159 samples were genotyped, including 2,631 non-Hispanic whites, 2,108 non-Hispanic blacks and 2,073 Mexican Americans. Quality control measurements were calculated locally using the Platform for the Analysis, Translation, and Organization of large-scale data [32]. The average genotyping call rate was 96%. We flagged SNPs that deviated from Hardy Weinberg Equilibrium expectations (p-value <0.001), MAF<0.05, and SNP call rates <95% for each subpopulation. In addition to these quality control metrics, we genotyped blinded duplicates as required by CDC, and all SNPs reported here passed quality control metrics required by CDC. All genotype data reported here were deposited into the NHANES III Genetic database and are available for secondary analysis through CDC.
Statistical methods
All analyses were performed using the Statistical Analysis Software (SAS v.9.2; SAS Institute, Cary, NC) either locally or via the Analytic Data Research by Email (ANDRE) portal of the CDC Research Data Center (RDC) in Hyattsville, MD. Analyses were limited to participants 18 year of age and older. Participants with measurements outside of the normal range for any trait were excluded from the analysis, as extreme measurements for these traits can indicate inflammation or recent trauma. Pair-wise correlations for each trait were calculated using Pearson’s correlation coefficient. None of the traits were correlated (defined as r>0.50; data not shown). Descriptive statistics for all traits are reported in Table 1. All traits followed a normal distribution with the exception of triglycerides, which was uniformly transformed by using the natural logarithm.
Table 1. Study population characteristics and haematological trait descriptive statistics for NHANES III participants.
Unweighted means (± standard deviations) or percentages are given for demographic, plasma fibrinogen, and haematologic traits, by subpopulation for adults in phase 2 of NHANES III. Abbreviations: non-Hispanic white (NHW), non-Hispanic black (NHB), and Mexican American (MA).
| Variable | Mean or % | Standard Deviation | ||||
|---|---|---|---|---|---|---|
| NHW | NHB | MA | NHW | NHB | MA | |
| Age (yrs) | 53.4 | 40.7 | 41 | ±20 | ±16 | ±17 |
| Female (%) | 60 | 58 | 50 | - | - | - |
| Current Smokers (%) | 26 | 37 | 24 | - | - | - |
| Body Mass Index (kg/m2) | 26.6 | 28.2 | 27.7 | ±5.56 | ±6.67 | ±5.42 |
| Plasma Fibrinogen (g/L) | 2.93 | 2.95 | 2.97 | ±0.50 | ±0.51 | ±0.50 |
| Platelet Count | 252 | 271 | 266 | ±58 | ±64 | ±62 |
| Platelet Distribution Width (mean %) | 16.5 | 16.3 | 16.4 | ±0.47 | ± 0.57 | ±0.54 |
| Mean Platelet Volume (fL) | 8.43 | 8.56 | 8.57 | ± 0.92 | ±0.98 | ±0.98 |
| Ln Triglycerides (mg/dL) | 4.51 | 4.38 | 4.52 | ± 0.32 | ±0.35 | ±0.31 |
| White Blood Cell Count | 7.38 | 6.69 | 7.73 | ±2.46 | ±2.11 | ±2.08 |
Pairwise linkage disequilibrium was calculated for all 25 SNPs using Haploview [33]. Single-locus tests of association were performed using linear regression for each fibrinogen SNP and each haematological trait: plasma fibrinogen, mean platelet volume, white blood cell count, platelet distribution width, log-transformed serum triglycerides, and platelet count. For each test of association, we assumed an additive genetic model, and we coded the same risk allele for each population (Supplemental Table 1). Analyses were performed unweighted and unadjusted and adjusted for age, sex, body mass index (BMI) and current smoking status (yes/no), and results were plotted using Synthesis View [34]. Analyses were repeated to include time to last meal (in hours) as a covariate (data not shown). Current smoking status was determined by the question “Do you smoke cigarettes now?” and cotinine levels (>15 ng/ml). All analyses were stratified by self-reported race/ethnicity.
RESULTS
Plasma fibrinogen and replicated associations
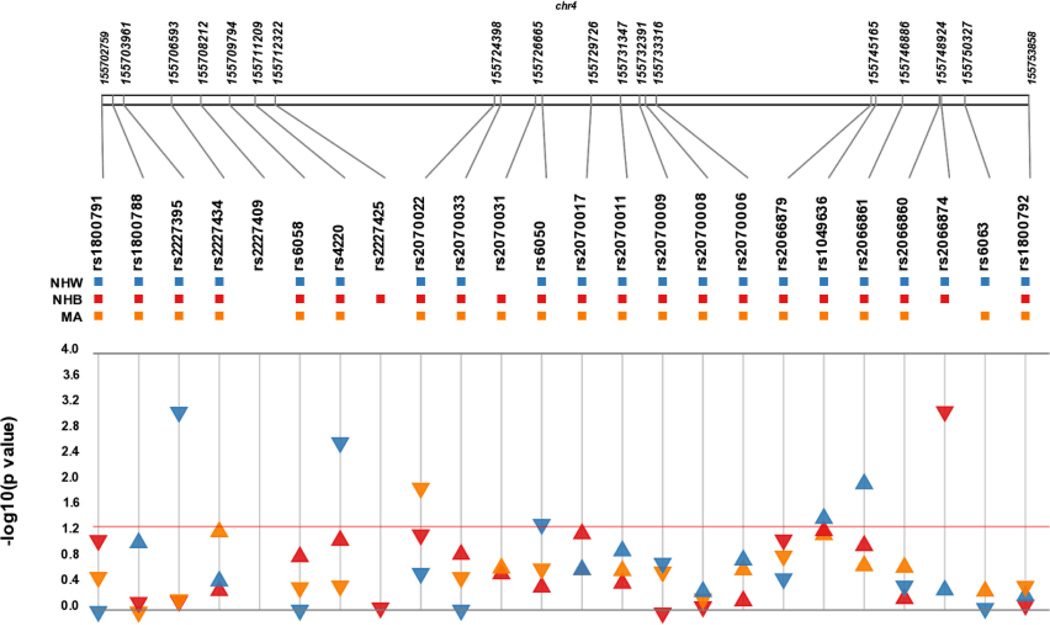
We replicated several single SNP associations from previous studies (Figure 1; Table 2). Replication was defined by statistical significance (p <0.05) and consistent direction of effect with the same SNP in the same ancestral population across studies. In non-Hispanic whites, four of the six tests of association replicated at p<0.05 and, after accounting for the coded allele, all tests trended in the same direction as previous reports regardless of significance (Table 2). In non-Hispanic whites, we replicated two associations previously reported in a candidate gene study [35]: rs6050 (Thr312Ala; p = 0.03) and intronic rs1049636 (p =0.04) were associated with decreased and increased plasma fibrinogen, respectively. We also replicated two associations reported from recent GWA studies [36;37]: FGG intronic rs2066861 (β = 0.05; p=0.01) and FGB nonsynonymous rs4220 (β = −0.08; p=0.002) (Table 2).
Figure 1. Synthesis view plot comparing results from tests of association across three NHANES subpopulations for plasma fibrinogen.
The –log10 of the p-value and the direction of the effect (indicated by arrow direction) are on the y-axis and SNPs are located on the x-axis. Each colour represents a different NHANES III population: blue (non-Hispanic white; NHW), red (non-Hispanic black; NHB), and yellow (Mexican American; MA). Arrows above the red line represent a significant associat ion (p<0.05).
Table 2. Replication of results from previous studies in non-Hispanic whites and blacks for plasma fibrinogen levels.
We compared results from our tests of association with plasma fibrinogen and all SNPs tested with the results from previous studies. Note that the coded allele may be different across studies. Abbreviations: Candidate gene Association Resource (CARe), Coronary Artery Risk Development in Young Adults (CARDIA), Rotterdam Study (RS), Framingham Heart Study (FHS), Cardiovascular Health Study (CHS), Atherosclerosis Rick in Communities Study (ARIC), Monitoring of Trends and Determinants of Cardiovascular Disease/Cooperative Health Research in the Region Augsburg (MONICA/KORA).
| Non-Hispanic White | |||||||||||||
| NHANES III |
CARe Wassel et al |
CARDIA Reiner et al |
RS/FHS/CHSs/ARIC / MONICA Wassel et al |
||||||||||
| SNP | Alleles | Coded Allele | β | P-value | Coded Allele | β | P-value | Coded Allele | β | P-value | Coded Allele | β | P-value |
| rs4220 | A/G | G | −0.08 | 2.10E-3 | A | 0.14 | 1.04E-34 | -- | -- | -- | G | −0.08 | 2.14E-27 |
| rs2070011 | A/G | G | 0.02 | 0.14 | -- | -- | -- | A | −3.82 | 0.005 | -- | -- | -- |
| rs6050 | A/G | G | −0.04 | 0.03 | -- | -- | -- | G | −7.04 | 0.001 | -- | -- | -- |
| rs1800788 | C/T | C | 0.04 | 0.10 | -- | -- | -- | T | −6.41 | 0.008 | -- | -- | -- |
| rs1049636 | C/T | C | 0.04 | 0.04 | -- | -- | -- | C | 6.16 | 0.003 | -- | -- | -- |
| rs2066861 | A/G | G | 0.05 | 0.01 | T | −0.06 | 4.18E-10 | -- | -- | -- | -- | -- | -- |
| Non-Hispanic White | |||||||||||||
| NHANES III |
CARe Wassel et al |
CARDIA Reiner et al |
|||||||||||
| SNP | Alleles | Coded Allele | β | P-value | Coded Allele | β | P-value | Coded Allele | β | P-value | |||
| rs2070017 | A/G | G | 0.07 | 0.07 | T | −0.16 | 4.82E-9 | A | decreased | NS | |||
| rs2066874 | A/G | G | −0.28 | 7.00 E-4 | C | −0.33 | 2.86E-11 | G | decreased | NS | |||
| rs6050 | A/G | G | 0.01 | 0.52 | -- | -- | -- | G | decreased | NS | |||
| rs6058 | G/T | G | 0.06 | 0.18 | T | −0.17 | 9.52E-7 | -- | -- | -- | |||
Similar to that observed in non-Hispanic whites, we were able to replicate reported associations in non-Hispanic blacks (Table 2)[35;36]. Intronic FGG rs2066874 (MAF = 0.02) was associated with decreased plasma fibrinogen in non-Hispanic blacks (β = −0.28; p=0.0007; Table 2). Although not significant, intronic FGA rs2070017, which is rare in non-Hispanic whites (MAF=0.0012), trended towards an association with increased plasma fibrinogen in non-Hispanic blacks (p=0.07; β = 0.07). Overall, regardless of significance, all but one test in non-Hispanic blacks (FGA rs6050) trended in the same direction as the previous report after accounting for the coded allele (Figure 1; Table 2).
We identified two novel associations between genetic variants and plasma fibrinogen. In non-Hispanic whites, intronic FGB rs2227395 (β = −0.08, p=0.0007) was associated with decreased plasma fibrinogen levels. FGB rs2227395 is in strong linkage disequilibrium (LD) (r2= 0.86; Supplemental Figure 1) with rs4220, and thus likely represents the same effect. FGA rs2070022, located in the 3′ untranslated region of the gene, was associated with decreased plasma fibrinogen levels in Mexican Americans (β = −0.07) and trended towards significance in non-Hispanic blacks (p=0.057; β = −0.07; Figure 1 and Table 3).
Table 3. Novel significant associations between the fibrinogen cluster and haematological traits in NHANES III.
Linear regressions adjusting for age, sex, BMI and smoking status were performed for 25 SNPs, assuming an additive genetic model for five hematological traits. Significant associations (p < 0.05) between the fibrinogen cluster variants and any one haematologic traits in any NHANES III population are shown.
| SNP | Phenotype | Location/Gene | Coded Allele |
Race/Ethnicity | N | BETA (95% CI) |
SE | p- value |
|---|---|---|---|---|---|---|---|---|
| rs2227395 | Plasma fibrinogen | Intron/ FGB | G | Non-Hispanic White | 1391 | −0.08 (−0.13, −0.03) | 0.02 | 7.00E-04 |
| rs2070022 | Plassma fibrinogen | 3′ UTR/ FGA | G | Mexican American | 659 | −0.07 (−0.13, −0.02) | 0.03 | 1.06E-02 |
| rs2066860 | LN Triglycerides | Intron/ FGG | G | Mexican American | 1005 | 0.13 (0.02, 0.24) | 0.06 | 2.27E-02 |
| rs1800792 | LN Triglycerides | 5′ flanking/FGG | G | Mexican American | 1005 | −0.03 (−0.06, −0.003) | 0.01 | 2.84E-02 |
| rs6050 | Platelet Distribution Width | Thr312Ala/FGA | G | Non-Hispanic White | 2270 | −0.03 (−0.06, −0.001) | 0.02 | 4.06E-02 |
| rs2070017 | Platelet Count | Intron/ FGA | G | Non-Hispanic Black | 1624 | −8.12 (−14.93, −1.31) | 3.47 | 1.95E-02 |
| rs1800792 | Platelet Count | Promoter/ FGG | G | Mexican American | 1663 | −5.32 (−9.83, −0.81) | 2.30 | 2.07E-02 |
| rs4220 | White blood cell count | Arg478Lys/FGB | G | Mexican American | 1682 | −0.28 (−0.48, −0.08) | 0.10 | 6.20E-03 |
| rs2227395 | White blood cell count | Intron/ FBG | G | Mexican American | 1682 | −0.23 (−0.43, −0.04) | 0.10 | 2.00E-02 |
| rs2066879 | Platelet Distribution Width | 3’ near FGG | G | Non-Hispanic White | 2,268 | 0.77 (0.15, 1.39) | 0.32 | 2.00E-02 |
| rs2227409 | Mean Platelet Volume | Asn170His/FGB | C | Non-Hispanic Black | 1,657 | 1.11 (0, 2.22) | 0.57 | 5.00E-02 |
| rs6063 | Platelet Distribution Width | Gly191Arg/ FGG | G | Non-Hispanic Black | 1,628 | 0.61 (0.06, 1.15) | 0.28 | 3.00E-02 |
| rs2227425 | Platelet Distribution Width | 3’UTR/FGB | G | Mexican Americans | 1,661 | 1.14 (0.08, 2.21) | 0.54 | 3.00E-02 |
Any one SNP (replicated and novel) significantly associated with plasma fibrinogen in non-Hispanic whites explained 0.3–0.7% of the trait variability. Collectively, the five associations we identified account for 1.6% of the trait variability in plasma fibrinogen in non-Hispanic whites. The two SNPs (replicated and novel) associated with decreased plasma fibrinogen in non-Hispanic blacks collectively account for 1.7% of the trait variability. With the inclusion of SNP rs2070017, which trended towards significance, these three SNPs accounted for 2.0% of the trait variability of plasma fibrinogen in non-Hispanic blacks. FGA rs2070022, associated with decreased plasma fibrinogen in Mexican Americans, accounted for 1.0% of the trait variability.
Associations with haematological traits
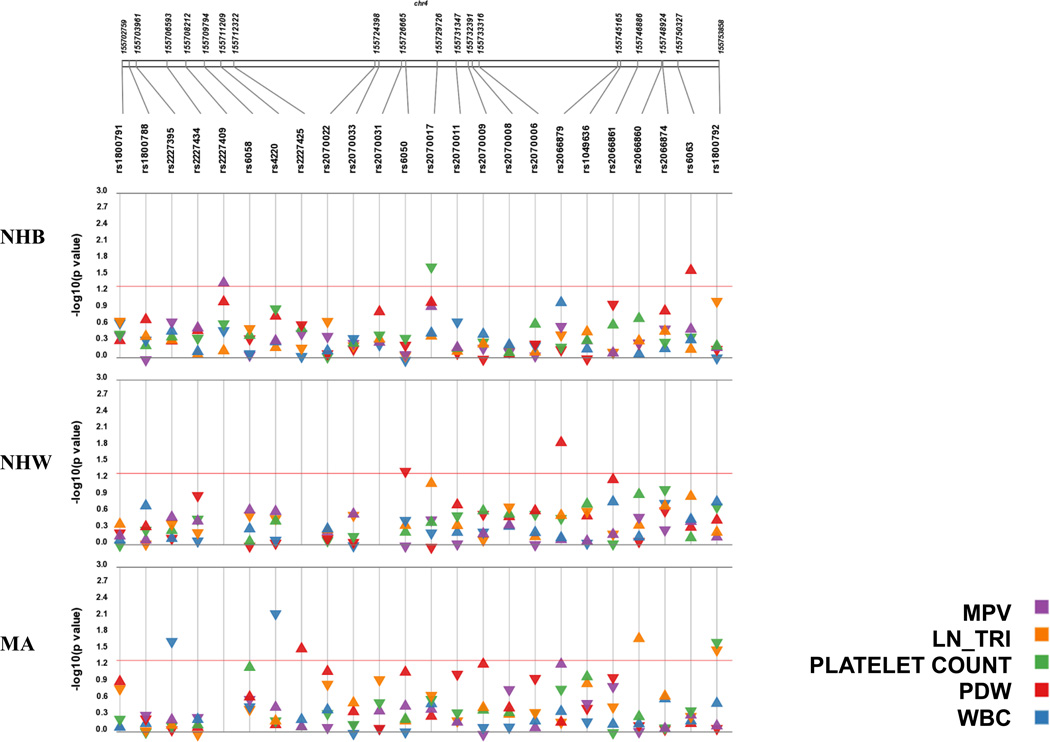
In addition to testing for an association with plasma fibrinogen levels, we tested for associations between the fibrinogen gene cluster variation and traits measured in the CBC: platelet count, mean platelet volume, platelet distribution width, and white blood cell count (Figure 2). We identified nine significant associations with fibrinogen SNPs and other haematological traits (Figure 2; Table 3). FGA rs6050 (Thr312Ala) and FGG rs2066879 were associated with decreased (β = −0.03; p = 0.04) and increased (β = 0.77; p = 0.02) platelet distribution width in non-Hispanic whites, respectively. Increased platelet distribution width was associated with rs2227425 in Mexican Americans and rs6063 in non-Hispanic blacks. Non-synonymous FGG rs4220 (Arg478Lys) and FGB intronic rs2227395 were both associated with decreased white blood cell count in Mexican Americans (β = −0.28, p = 0.006 and β = −0.23, p = 0.02, respectively). We identified two significant associations with decreased platelet count: intronic FGA rs2070017 associated with decreased platelet count in non-Hispanic blacks, and 5′ flanking FGG rs1800792 associated with decreased platelet count in Mexican Americans (Figure 2; Table 3). There was one association between FGB rs2227409 and mean platelet volume in non-Hispanic blacks (β = 1.11; p=0.05).
Figure 2. Synthesis view plot comparing significant results from tests of association for five hematological traits across three NHANES subpopulations.
The –log10 of the p-value and the direction of the effect (indicated by arrow direction) are on the y-axis and SNPs are located on the x-axis. Each panel represents a different population from the NHANES III dataset and each colour represents the haematological traits tested for each SNP. Points above the red line represent a significant association (p<0.05). Abbreviations: non-Hispanic black (NHB), non-Hispanic white (NHW), Mexican American (MA), mean platelet volume (MPV), transformed triglycerides (ln_TRI), white blood cell count (WBC), and platelet distribution width (PDW).
Associations with triglycerides
There were two significant associations with serum triglycerides in Mexican Americans. Intronic FGG rs2066860 was associated with increased serum triglycerides and 5′ flanking FGG rs1800792 was associated with decreased triglyceride levels. Inclusion of fasting status (time to last meal) as a covariate in the models did not alter the results substantially (data not shown).
Associations across populations
There were no associations with any trait that were significant across all three populations. While there were no significant associations observed in all three NHANES populations, four of the seven associations we identified with plasma fibrinogen have a consistent direction of effect across all three populations (Figure 1; Supplement Table 2). There was also a consistent direction of effect with rs2066860 across all three populations with serum triglycerides (Figure 2). For the other associations we identified with platelet distribution width, platelet count, and white blood cell count, the direction of effect was different across populations (Figure 2; Supplement Table 2).
Sex-specific associations
To determine if sex acts as a modifier of these associations, we also performed tests of association stratified by sex for the significant associations we report (replicated and novel). Among non-Hispanic whites, of the seven SNPs tested for sex effects, both rs2066861 and rs6050 were significantly associated in males (p = 0.002 and 0.006, respectively) but not in females (p = 0.50 and 0.69, respectively) despite the larger sample size in females compared with males (Supplementary Table 3). Among non-Hispanic blacks, four SNPs were examined for sex effects and two SNPs (rs2070017 and rs6058) were associated among females (p = 0.02 and 0.02, respectively) but not males. For these female-specific associations, the genetic effect size was larger (β = 0.14 versus 0.006 for rs2070017) in females compared with males, and, in the case of rs6058, in the opposite direction (β = 0.15 versus −0.05). Large genetic effect sizes among females compared with males were also observed for platelet counts in non-Hispanic blacks (rs2070017; β = −12.78 versus −0.52) and Mexican Americans (rs1800792; β = −9.65 versus −0.32; Supplementary Table 3). It is important to note that small sample size could be a consequence of the large genetic effects we report.
DISCUSSION
Using a large, diverse study population, we were able to replicate five associations as well as identify two novel associations between SNPs located in the fibrinogen gene cluster and levels of plasma fibrinogen. Additionally, we were able to detect associations with fibrinogen SNPs and platelet count, white blood cell count and platelet distribution width, indicative of pleiotropic effects. This is also the first genetic association performed in Mexican Americans with variants within the fibrinogen gene cluster.
While we successfully replicated genetic associations reported for plasma fibrinogen in non-Hispanic whites and blacks (Table 2), three variants did not replicate the findings of previous studies (Table 2) [35–37]. We were unable to replicate the association with rs2070011 and rs1800788 in non-Hispanic whites. In comparison to Reiner et al, we were under-powered to detect the same effects with our sample size in non-Hispanic whites. We were also unable to replicate the association with rs6058 and plasma fibrinogen in non-Hispanic blacks, again most likely due to power (Table 2).
We identified two novel associations with plasma fibrinogen in FGB and FGA. The novel association we identify between plasma fibrinogen and FGA is significant in Mexican Americans and trends towards significant in non-Hispanic blacks.
Several genetic variants are associated with increased plasma fibrinogen in all three genes in the fibrinogen cluster. Most of these genetic variants are in non-coding regions of FGB, the rate limiting gene for the production of the fibrinogen poly peptide [38]. Little is known about function of genetic variation in the other genes, FGA and FGG, in the fibrinogen gene cluster. Of the SNPs targeted for replication for plasma fibrinogen levels, three were putatively functional SNPs based on gene location. For example, nonsynonymous variant, rs6050 in the coding region of the FGA gene (Thr331Ala), was significantly associated with plasma fibrinogen in non-Hispanic whites (β=0.04, p = 0.03). Interestingly, the G allele was associated with decreased plasma fibrinogen levels in European Americans and trended in the same direction in African Americans from CARDIA (Table 2) [35]; however, in NHANES III, the same allele replicated in whites but in the opposite direction. None of the novel variants we identified are in the coding regions of the fibrinogen cluster. Novel variant rs2070022, associated with decreased plasma fibrinogen, is located in the 3’ untranslated region of the FGA gene, which could potentially affect polyadenylation and mRNA stability. Functional studies are needed to confirm role of rs2070022 in fibrinogen production.
This study, which accesses the diverse NHANES III, has many strengths. One strength is the breadth of laboratory measurements available for tests of association. Until recently, most genetic association studies have ignored pleiotropy and have limited tests of association to a single trait or pathway. Pleiotropy is defined here and elsewhere as a locus affecting multiple traits [39]. In a statistical setting, pleiotropy is detected as a single genetic variant associated with multiple traits, but this setting does not establish causality without further experiments. Epidemiologic studies such as NHANES make statistical tests for pleiotropy possible, which may ultimately identify novel genotype-phenotype relationships and pathways [40]. Our study is unique in that we also investigated the pleiotropic effects of the fibrinogen cluster with haematological traits. This is important given the complex pathophysiology of cardiovascular disease and the role haematological traits play in the development of disease. Several studies have been published on genetic variation with levels of plasma fibrinogen but little is known about how these same variants impact haematological traits. It is known that elevated levels of plasma fibrinogen are observed for several disease states such as inflammation, deep venous thrombosis, and arterial thrombosis [41]. However, it is still unclear as to whether elevated levels of plasma fibrinogen are a result of fibrinogen itself or other intermediate phenotypes underlying the pathophysiology. For example, arterial thrombosis, a common characteristic and intermediate phenotype for cardiovascular disease, is very complex and is likely to involve the fibrinogen gene cluster. However, unlike venous thrombosis, which is mainly due to the accumulation fibrin clots, arterial thrombosis is primarily due to platelet adhesion and it is likely involve haematological traits tested in this study [42–45]. We identified genetic associations with fibrinogen SNPs and white blood cell count, platelet distribution width, and platelet count suggesting possible pleiotropic effects. These findings support the idea that variants in the fibrinogen gene cluster may also affect the regulation of haematological traits.
Another major strength of our study is its diversity. Although there have been studies performed in European and African descent populations, none have included Mexican Americans. The characterization of variants in non-European descent populations has considerable scientific benefits. For example in Mexican Americans, we identified a novel association between plasma fibrinogen levels and rs2070022 that also trended towards significance in non-Hispanic blacks. We also were able to report a consistent direction of effect for rs1049636 across all the three NHANES populations despite sample size. In addition to plasma fibrinogen levels, we identified significant associations with serum triglycerides in Mexican Americans.
A weakness of the present study is power and sample size. While NHANES III contains over 33,000 study participants, our study population sample size was limited for several reasons. The Centers for Disease Control and Prevention (CDC) only collected DNA samples from a subset of the entire dataset, which includes 7,159 participants from phase 2 of NHANES III. DNA was not collected from participants who reported having haemophilia or chemotherapy within four weeks of collection [28]. Of these samples, plasma fibrinogen levels in NHANES III was only measured for participants > 40 years of age [28], which drastically reduces our sample size with plasma fibrinogen compared to the haematological traits. Despite the small sample sizes for plasma fibrinogen, we were able to successfully replicate several known associations at a liberal significance threshold of p<0.05. We also identified potentially novel associations at the same significance threshold, but because of multiple testing and the increased risk of false positive findings, these novel associations will require replication in other studies. Similarly, the sex differences reported here with this limited sample size will require replication.
In summary, we have identified both previous and novel associations within the fibrinogen gene cluster with plasma fibrinogen levels in the diverse population-based cohort. We also characterise these variants in Mexican Americans. Additionally, we show that these variants are also associated with haematological traits including serum triglyceride levels. Identifying these associations will provide important information about intermediate phenotypes with the ultimate goal of predicting cardiovascular disease outcome.
Supplementary Material
Extra Table.
| What is known on this topic? | What this paper adds? |
|---|---|
| • To date there are several genetic association studies that identify genetic risk factors that explain a portion of the inter-individual variability of plasma fibrinogen levels. | • We replicate previous well-known associations as well as identify three novel associations with variants in this cluster with plasma fibrinogen in a population-based setting, NHANES. |
| • Genetic variants in the fibrinogen gene cluster are associated with variable levels of plasma fibrinogen in African Americans and European descent populations. | • To date there are no genetic association studies performed with fibrinogen variants in Mexican Americans. Our study tests previous associations initially described in African Americans and Europeans in Mexican Americans. |
| • Most genetic association studies test for an association with variants in the fibrinogen gene cluster and plasma fibrinogen; however, the possibility of pleiotropic effects with these variants have yet to be explored. | • We are the first to test and/or report an association with these fibrinogen variants and hematological traits: mean platelet volume, platelet count, platelet distribution width, white blood cell count, and serum triglycerides. We report nine novel associations with variants in the fibrinogen cluster and these haematological traits. |
ACKNOWLEGEMENTS
Genotyping services were provided by the Johns Hopkins University under federal contract number (N01-HV-48195) from the National Heart, Lung, and Blood Institute (NHLBI). We would like to thank Dr. Geraldine McQuillan and Jody McLean for their help in accessing the Genetic NHANES III data. The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention. The Vanderbilt University Center for Human Genetics Research, Computational Genomics Core provided computational and/or analytical support for this work.
References
- 1.Pennington J, Garner SF, Sutherland J, Williamson LM. Residual subset population analysis in WBC-reduced blood components using real-time PCR quantitation of specific mRNA. Transfusion. 2001 Dec;41(12):1591–1600. doi: 10.1046/j.1537-2995.2001.41121591.x. [DOI] [PubMed] [Google Scholar]
- 2.Evans DM, Frazer IH, Martin NG. Genetic and environmental causes of variation in basal levels of blood cells. Twin Res. 1999 Dec;2(4):250–257. doi: 10.1375/136905299320565735. [DOI] [PubMed] [Google Scholar]
- 3.Meade TW, Mellows S, Brozovic M, Miller GJ, Chakrabarti RR, North WR, et al. Haemostatic function and ischaemic heart disease: principal results of the Northwick Park Heart Study. Lancet. 1986 Sep 6;2(8506):533–537. doi: 10.1016/s0140-6736(86)90111-x. [DOI] [PubMed] [Google Scholar]
- 4.Tosetto A, Prati P, Baracchini C, Manara R, Rodeghiero F. Association of plasma fibrinogen, C-reactive protein and G-455>A polymorphism with early atherosclerosis in the VITA Project cohort. Thromb Haemost. 2011 Feb 1;105(2):329–335. doi: 10.1160/TH10-08-0522. [DOI] [PubMed] [Google Scholar]
- 5.Boos CJ, Lip GY. Platelet activation and cardiovascular outcomes in acute coronary syndromes. J Thromb Haemost. 2006 Dec;4(12):2542–2543. doi: 10.1111/j.1538-7836.2006.02250.x. [DOI] [PubMed] [Google Scholar]
- 6.Danesh J, Lewington S. Plasma homocysteine and coronary heart disease: systematic review of published epidemiological studies. J Cardiovasc Risk. 1998 Aug;5(4):229–232. [PubMed] [Google Scholar]
- 7.Ensrud K, Grimm RH., Jr The white blood cell count and risk for coronary heart disease. Am Heart J. 1992 Jul;124(1):207–213. doi: 10.1016/0002-8703(92)90942-o. [DOI] [PubMed] [Google Scholar]
- 8.Kannel WB, Wolf PA, Castelli WP, D'Agostino RB. Fibrinogen and risk of cardiovascular disease. The Framingham Study. JAMA. 1987 Sep 4;258(9):1183–1186. [PubMed] [Google Scholar]
- 9.Collet JP, Soria J, Mirshahi M, Hirsch M, Dagonnet FB, Caen J, et al. Dusart syndrome: a new concept of the relationship between fibrin clot architecture and fibrin clot degradability: hypofibrinolysis related to an abnormal clot structure. Blood. 1993 Oct 15;82(8):2462–2469. [PubMed] [Google Scholar]
- 10.Voetsch B, Loscalzo J. Genetic Determinants of Arterial Thrombosis. Arterioscler Thromb Vasc Biol. 2003;24:216–229. doi: 10.1161/01.ATV.0000107402.79771.fc. [DOI] [PubMed] [Google Scholar]
- 11.Siegerink B, Rosendaal FR, Algra A. Genetic variation in fibrinogen; its relationship to fibrinogen levels and the risk of myocardial infarction and ischemic stroke. J Thromb Haemost. 2009 Mar;7(3):385–390. doi: 10.1111/j.1538-7836.2008.03266.x. [DOI] [PubMed] [Google Scholar]
- 12.Boekholdt SM, Bijsterveld NR, Moons AH, Levi M, Buller HR, Peters RJ. Genetic variation in coagulation and fibrinolytic proteins and their relation with acute myocardial infarction: a systematic review. Circulation. 2001 Dec 18;104(25):3063–3068. doi: 10.1161/hc5001.100793. [DOI] [PubMed] [Google Scholar]
- 13.Folsom AR, Aleksic N, Ahn C, Boerwinkle E, Wu KK. Beta-fibrinogen gene-455G/A polymorphism and coronary heart disease incidence: the Atherosclerosis Risk in Communities (ARIC) Study. Ann Epidemiol. 2001 Apr;11(3):166–170. doi: 10.1016/s1047-2797(00)00203-9. [DOI] [PubMed] [Google Scholar]
- 14.Koch W, Hoppmann P, Biele J, Mueller JC, Schomig A, Kastrati A. Fibrinogen genes and myocardial infarction: a haplotype analysis. Arterioscler Thromb Vasc Biol. 2008 Apr;28(4):758–763. doi: 10.1161/ATVBAHA.107.157842. [DOI] [PubMed] [Google Scholar]
- 15.Hamsten A, Iselius L, de FU, Blomback M. Genetic and cultural inheritance of plasma fibrinogen concentration. Lancet. 1987 Oct 31;2(8566):988–991. doi: 10.1016/s0140-6736(87)92557-8. [DOI] [PubMed] [Google Scholar]
- 16.Meisinger C, Prokisch H, Gieger C, Soranzo N, Mehta D, Rosskopf D, et al. A genome-wide association study identifies three loci associated with mean platelet volume. Am J Hum Genet. 2009 Jan;84(1):66–71. doi: 10.1016/j.ajhg.2008.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Soranzo N, Spector TD, Mangino M, Kuhnel B, Rendon A, Teumer A, et al. A genome-wide meta-analysis identifies 22 loci associated with eight hematological parameters in the HaemGen consortium. Nat Genet. 2009 Nov;41(11):1182–1190. doi: 10.1038/ng.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Margaglione M, Grandone E. Population genetics of venous thromboembolism. A narrative review. Thromb Haemost. 2011 Feb 1;105(2):221–231. doi: 10.1160/TH10-08-0510. [DOI] [PubMed] [Google Scholar]
- 19.Rosendaal FR, Doggen CJ, Zivelin A, Arruda VR, Aiach M, Siscovick DS, et al. Geographic distribution of the 20210 G to A prothrombin variant. Thromb Haemost. 1998 Apr;79(4):706–708. [PubMed] [Google Scholar]
- 20.Grody WW, Griffin JH, Taylor AK, Korf BR, Heit JA. American College of Medical Genetics consensus statement on factor V Leiden mutation testing. Genet Med. 2001 Mar;3(2):139–148. doi: 10.1097/00125817-200103000-00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grody WW. Molecular genetic risk screening. Annu Rev Med. 2003;54:473–490. doi: 10.1146/annurev.med.54.101601.152127. [DOI] [PubMed] [Google Scholar]
- 22.McGlennen RC, Key NS. Clinical and laboratory management of the prothrombin G20210A mutation. Arch Pathol Lab Med. 2002 Nov;126(11):1319–1325. doi: 10.5858/2002-126-1319-CALMOT. [DOI] [PubMed] [Google Scholar]
- 23.Crawford DC, Sanders CL, Qin X, Smith JD, Shephard C, Wong M, et al. Genetic variation is associated with C-reactive protein levels in the Third National Health and Nutrition Examination Survey. Circulation. 2006 Dec 5;114(23):2458–2465. doi: 10.1161/CIRCULATIONAHA.106.615740. [DOI] [PubMed] [Google Scholar]
- 24.Chang MH, Lindegren ML, Butler MA, Chanock SJ, Dowling NF, Gallagher M, et al. Prevalence in the United States of selected candidate gene variants: Third National Health and Nutrition Examination Survey, 1991–1994. Am J Epidemiol. 2009 Jan 1;169(1):54–66. doi: 10.1093/aje/kwn286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Steinberg KK, Sanderlin KC, Ou CY, Hannon WH, McQuillan GM, Sampson EJ. DNA banking in epidemiologic studies. Epidemiol Rev. 1997;19(1):156–162. doi: 10.1093/oxfordjournals.epirev.a017938. [DOI] [PubMed] [Google Scholar]
- 26.Centers for Disease Control and Prevention Third National Health and Nutrition Examination Survey: 1988–94, Plan and Operations Procedures Manual. 1996 [Google Scholar]
- 27.Centers for Disease Control and Prevention Plan and Operation of the Third National Health and Nutrition Examination Survey: 1988–94. Bethesda, MD: 2004. [Google Scholar]
- 28.Elaine W, Lewis B, Koncikowski S. Laboratory Procedures Used for the Third National Health and Nutrition Examination Survey (NHANES III), 1988–1994. 1996 [Google Scholar]
- 29.Daum P, Estergreen J, Wener M. Laboratory Procedure Manual:Rate of Clot Formation on the STA-Compact. 2000 [Google Scholar]
- 30.Carlson CS, Eberle MA, Rieder MJ, Yi Q, Kruglyak L, Nickerson DA. Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. Am J Hum Genet. 2004 Jan;74(1):106–120. doi: 10.1086/381000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Crawford DC, Akey DT, Nickerson DA. The patterns of natural variation in human genes. Annu Rev Genomics Hum Genet. 2005;6:287–312. doi: 10.1146/annurev.genom.6.080604.162309. [DOI] [PubMed] [Google Scholar]
- 32.Grady BJ, Torstenson E, Dudek SM, Giles J, Sexton D, Ritchie MD. Finding unique filter sets in plato: a precursor to efficient interaction analysis in gwas data. Pac Symp Biocomput. 2010:315–326. [PMC free article] [PubMed] [Google Scholar]
- 33.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005 Jan 15;21(2):263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 34.Pendergrass SA, Dudek S, Crawford DCC, Ritchie M. Synthesis-View: visualization and interpretation of SNP association results for multi-cohort, multi-phenotype data and meta-analysis. BioData Mining. 2010 doi: 10.1186/1756-0381-3-10. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reiner AP, Carty CL, Carlson CS, Wan JY, Rieder MJ, Smith JD, et al. Association between patterns of nucleotide variation across the three fibrinogen genes and plasma fibrinogen levels: the Coronary Artery Risk Development in Young Adults (CARDIA) study. J Thromb Haemost. 2006 Jun;4(6):1279–1287. doi: 10.1111/j.1538-7836.2006.01907.x. [DOI] [PubMed] [Google Scholar]
- 36.Wassel CL, Lange LA, Keating BJ, Taylor KC, Johnson AD, Palmer CD, et al. Association of genomic loci from a cardiovascular gene SNP array with fibrinogen levels in European Americans and African-Americans from six cohort studies: the Candidate gene Association Resource (CARe) Blood. 2010 Oct 26; doi: 10.1182/blood-2010-06-289546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dehghan A, Yang Q, Peters A, Basu S, Bis JC, Rudnicka AR, et al. Association of novel genetic Loci with circulating fibrinogen levels: a genome-wide association study in 6 population-based cohorts. Circ Cardiovasc Genet. 2009 Apr;2(2):125–133. doi: 10.1161/CIRCGENETICS.108.825224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kottke-Marchant K. Genetic polymorphisms associated with venous and arterial thrombosis: an overview. Archives of Pathology and Laboratory Medcine. 2011;126(3):295–304. doi: 10.5858/2002-126-0295-GPAWVA. [DOI] [PubMed] [Google Scholar]
- 39.Wagner GP, Zhang J. The pleiotropic structure of the genotype-phenotype map: the evolvability of complex organisms. Nat Rev Genet. 2011 Mar;12(3):204–213. doi: 10.1038/nrg2949. [DOI] [PubMed] [Google Scholar]
- 40.Pendergrass SA, Brown-Gentry K, Dudek SM, Torstenson ES, Ambite JL, Avery CL, et al. The use of phenome-wide association studies (PheWAS) for exploration of novel genotype-phenotype relationships and pleiotropy discovery. Genet Epidemiol. 2011 Jul;35(5):410–422. doi: 10.1002/gepi.20589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.David Ginsburg Genetic Risk Factors for Arterial Thrombosis and Inflammation. Hematology. 2005:442–444. doi: 10.1182/asheducation-2005.1.442. [DOI] [PubMed] [Google Scholar]
- 42.Feinbloom D, Bauer KA. Assessment of hemostatic risk factors in predicting arterial thrombotic events. Arterioscler Thromb Vasc Biol. 2005 Oct;25(10):2043–2053. doi: 10.1161/01.ATV.0000181762.31694.da. [DOI] [PubMed] [Google Scholar]
- 43.Lane DA, Grant PJ. Role of hemostatic gene polymorphisms in venous and arterial thrombotic disease. Blood. 2000 Mar 1;95(5):1517–1532. [PubMed] [Google Scholar]
- 44.Lowe GD. Common risk factors for both arterial and venous thrombosis. Br J Haematol. 2008 Mar;140(5):488–495. doi: 10.1111/j.1365-2141.2007.06973.x. [DOI] [PubMed] [Google Scholar]
- 45.Mackman N. Triggers, targets and treatments for thrombosis. Nature. 2008 Feb 21;451(7181):914–918. doi: 10.1038/nature06797. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.