Abstract
Purpose:
The aim of this study was to identify biomarkers that may be predictive for the clinical activity of the redox-active antitumor agent imexon.
Experimental Design:
cDNA microarray and quantitative real-time PCR were used to identify global changes in gene expression in peripheral blood mononuclear cells (PBMC) collected from patients treated with imexon during a phase I trial. Electrophoretic mobility shift assays and Western blot analysis were done using the RPMI8226 myeloma cell line grown in vitro and PBMCs treated ex vivo to investigate the molecular mechanism responsible for these gene changes.
Results:
Both cDNA microarray and quantitative real-time PCR showed the up-regulation of many antioxidant genes, including thioredoxin reductase-1, glutaredoxin-2, and peroxiredoxin-3 in PBMCs collected from patients treated with imexon. Studies in PBMCs treated ex vivo and RPMI8226 myeloma cells showed that imexon increased binding to the activator protein-1 consensus sequence measured by electrophoretic mobility shift assay. Supershift analysis showed that the majority of the activator protein-1DNA binding activity was c-Jun, with minor contribution of Jun-D. Nuclear translocation of the nuclear factor (erythroid-derived 1)-like 2 transcription factor and its binding to the antioxidant response element was also increased after imexon treatment, which correlated with an increase in the message levels for nuclear factor (erythroid-derived 1)-like 2/antioxidant response element – regulated antioxidant genes.
Conclusions:
Together, these results show that a predominant biological effect of imexon is a change in redox state that can be detected in surrogate normal tissues as increased redox-sensitive transcription factor binding and increased antioxidant gene expression.
Imexon is an aziridine-containing iminopyrrolidone that shows promising in vitro and in vivo antitumor activity in myeloma, lymphoma, and pancreatic cancer cell line models (1-3). Imexon was tested for anticancer activity in a limited number of clinical trials in the early 1970s (4). Although it was found to be relatively nontoxic at low, fixed doses in few patients, it was not developed further as an anticancer agent. Recently, imexon has again entered phase 1 clinical trials in patients with advanced solid malignancies.
Preclinical studies have shown that imexon reacts with sulfhydryl groups resulting in cellular thiol depletion, an increase in oxidative stress superoxide anion radical formation, mitochondrial alterations, and apoptosis (5-7). Consistent with this mode of action, an imexon-resistant RPMI8226 myeloma cell line was shown to overexpress thioredoxin-2, a mitochondrial member of the thioredoxin family of redox proteins, which protect against apoptosis (8). In addition, this cell line displayed cross-resistance with other chemotherapeutic agents known to induce oxidative stress, including doxorubicin and arsenic trioxide.
Protection against oxidative stress is provided by reduced glutathione, a major factor responsible for maintaining a reduced cellular environment. Reduced glutathione is reduced by NADPH through glutathione reductase, and reduced glutathione–dependent reactions with electrophilic and oxidizing species are catalyzed by glutaredoxins (9). A second key factor responsible for maintaining a reduced cellular environment is thioredoxin-1, a small (12 kDa) ubiquitously expressed protein that is reduced by electrons from NADPH through thioredoxin reductase (10). Peroxiredoxins (thioredoxin peroxidases) reduce hydrogen peroxide and alkyl hydroperoxides to water and alcohol using reducing equivalents derived from thioredoxin (11).
Oxidative stress results in a shift of the thiol-disulfide equilibrium of cellular proteins (12), activating transcription factors that mediate the induction of a variety of antioxidant enzymes. Activator protein-1 (AP-1) is an immediate early response transcription factor activated in response to oxidative stress. AP-1 is composed of Fos and Jun homodimers and heterodimers (reviewed in ref. 13) and the binding of AP-1 to the AP-1 consensus sequence in the promoter of genes induces antioxidant enzymes, including glutathione S-transferase (GST; ref. 14), diphtheria toxin-diaphorase, and manganese superoxide dismutase (15). The antioxidant response element (ARE), a cis-acting regulator element in promoter region of antioxidant genes, such as GSTs, thioredoxin reductase-1, thioredoxin-1, and ferritin, as well as phase II xenobiotic metabolizing enzymes, is also activated by oxidative stress (reviewed in ref. 16). The transcription factors that bind to the ARE include the NF-E2–related factors 1, 2, and 3 (Nrf1, Nrf2, and Nrf3) that have been shown to positively (Nrf1 and Nrf2; refs. 17, 18) or negatively (Nrf3; ref. 19) regulate ARE-mediated gene expression. Nrf2 is normally sequestered in the cytoplasm by the Keap1-Cul3 complex where it is degraded by the ubiquitin-proteasomal pathway (20). However, under oxidative conditions, Nrf2 binding to Keap1 is inhibited and Nrf2 translocates from the cytoplasm to the nucleus where it heterodimerizes with a variety of basic leucine zipper transcription factors, including Jun (c-Jun, Jun-B, and Jun-D) and Maf (Maf G, Maf K, and Maf F) proteins (18, 21-23). This results in transactivation of AREs and the coordinated induction of antioxidant and phase II detoxification gene expression (18).
In the present study, we used transcription profiling analysis to identify an increase in antioxidant gene expression in peripheral blood mononuclear cells (PBMC) collected from patients treated with imexon during a phase I study. Many of these genes are know to be regulated by AP-1 and the ARE. Mechanistic studies revealed that imexon treatment increases nuclear Nrf2 levels in RPMI8226 myeloma cells and MDA-mb-231 breast cancer cells grown in vitro and increases both AP-1 and the ARE and binding in RPMI8226 cells and in normal PBMCs treated with imexon ex vivo. These findings suggest that imexon increases oxidative stress, which leads to an adaptive response involving up-regulation of several antioxidant defense genes. Part of the mechanism of action of imexon as an antitumor agent may be through an increase in oxidative stress. These antioxidant gene products also represent potential biomarkers for monitoring imexon activity in clinical studies.
Materials and Methods
Collection of patient PBMCs
Peripheral blood was obtained from either normal volunteers or patients enrolled in a phase I trial of imexon in patients with metastatic cancer or disseminated malignancy. The study was approved by the local institutional review board and patient informed consent was obtained. Approximately 10 mL of blood were collected immediately before treatment and 3 h after the end of a 1-h infusion of imexon. Samples were collected from a total of seven patients receiving doses of imexon ranging from 80 to 570 mg/m2. The patient tumor types were one melanoma, one colon, two pancreatic, one endometrial, and two ovarian. PBMCs were isolated by Ficoll (Amersham Biosciences, Pittsburgh, PN) centrifugation and stored at −80°C until RNA was isolated as described below.
RNA isolation and preparation
Total RNA was isolated from PBMCs using the GeneElute Mammalian Total RNA Miniprep kit (Sigma-Aldrich, St. Louis, MO) and was DNase treated using the DNA-Free kit (Ambion, Austin, TX). Total RNA was then treated with the RNeasy Clean-up kit (Qiagen, Valencia, CA) to remove any trace buffers from the isolation or DNasing steps. Two micrograms of total RNA were used for amplification. One round of amplification was done using the MessageAmpII aRNA kit (Ambion) according to the manufacturer’s instructions.
Microarray hybridizations
Probes were synthesized by using 5 μg of first round aRNA labeled with Alexa Fluor 647 (experimental) or Alexa Fluor 555 (control; Invitrogen, Carlsbad, CA). Samples were combined for each probe and purified using the QIAquick PCR purification kit (Qiagen). cDNA arrays of 22,000 human genes and expressed sequence tags (EST; Invitrogen) in duplicate were hybridized in triplicate for each sample as described previously (24). After washing, slides were coated with DyeSaver2 (Genisphere, Inc., Hatfield, PA) according to the manufacturer’s instructions. Slides were scanned using a ScanArray Express 5000 (Perkin-Elmer Life Sciences, Boston, MA) and quantitation was done using the Digital Genome AnalyzerDG software package (MolecularWare, Inc., Cambridge, MA). The spot intensity values were normalized according to each grid of the array (n = 48) by multiplying by a correction factor of the average intensity values for the reference channel divided by the test sample. The mean ratio from each duplicate, run in triplicate, was calculated as treated/control corrected values and reported as the average relative expression levels.
Quantitative real-time PCR
Quantitative real-time PCR was done using a PE Biosystems GeneAmp 5700 Sequence Detection System (Perkin-Elmer Life Sciences). Amplification reactions were set up in a reaction volume of 25 μL using the Applied Biosystems Taqman Universal PCR Master Mix (Applera Corp., Applied Biosystems, Foster City, CA). PCR primers and Taqman probes were synthesized and preoptimized by Applied Biosystems (Assays-on-Demand). Each reaction containing PCR Master Mix, 900 nmol of each primer, and 250 nmol of the Fam probe was assayed in triplicate using 50 ng aRNA. Reverse transcription was done at 48°C for 30 min, and samples were incubated for 10 min at 95°C and then amplified over 40 cycles at 15 sec at 95°C followed by 1 min at 60°C. Quantitation was done using the comparative CT method with β2-macroglobulin as the normalization gene.
Identification of ARE consensus sequences
Microarray gene ESTs were referenced to their Genebank accession nos., which were compared with the Genebank data base using BLAST to retrieve human gene sequences, which had an expected value (E) of 1e−30 or less and noncoding regions extrapolated and pattern matched using a PERL script for the consensus ARE (TGAG/CNNNGC) binding site consensus sequences (reviewed in ref. 25). All sequences were searched up to 10,000 bp unless another gene was found to be in that area. Complement sequences were also searched and reported when identified.
Cell culture conditions
The RPMI8226 myeloma cell line and MDA-mb-231 cell lines were obtained from American Type Culture Collection (Manassas, VA). RPMI8226 cells were maintained in RPMI 1640 supplemented with 5% heat-inactivated fetal bovine serum (Omega, Calabasas CA), 1 mmol/L L-glutamine, and 100 units/mL penicillin/streptomycin (Invitrogen), and MDA-mb-231 cells were maintained in DMEM supplemented with 10% fetal bovine serum (Omega). Imexon (4-imino-1,3-diazabicyclo[3.1.0.]-hexan-one; NSC-714597) was obtained from the Pharmaceutical Resources Branch of the National Cancer Institute (Bethesda, MD) under the Rapid Access to Intervention in Development Program. The drug was supplied as a 99.6% pure lyophilized white powder and has water solubility of ∼10 mg/mL. A stock solution (18 mmol/L) was prepared in water, sterile filtered, and stored at −80°C.
Electrophoretic mobility shift assays
Nuclear and cytoplasmic extracts were prepared from RPMI8226 myeloma cells as described previously (26). Double-stranded oligonucleotides (Santa Cruz Biotechnology, Santa Cruz, CA) corresponding to Nrf2 (5′CTACGATTTCTGCTTAGTCATTGTCTTCC3′) or the AP-1 consensus binding sequence (5′-CGCTTGATGACTCAGCCGGAA-3′) were radiolabeled using polynucleotide kinase (Roche, Indianapolis, IN), and unincorporated nucleotide was removed on a G50 spin column. Nuclear extracts, 5 μg per reaction, were incubated with binding buffer [200 mmol/L Tris (pH 8.0), 1 mol/L NaCl, 40 mmol/L EDTA, 1.8 μg salmon sperm, 2% NP40, 40% bovine serum albumin] for 10 min at 4°C before addition of probe and then incubated for 20 min at room temperature. Reaction mixtures were separated on 6% acrylamide/Tris-borate EDTA gels, dried, and analyzed by PhosphorImager analysis using an MD Storm and ImageQuant software (Molecular Dynamics, Sunnyvale, CA). For competition assays, nuclear lysates were incubated with specific rabbit polyclonal antisera (1:40 dilution) for c-Jun or Jun-D (Santa Cruz Biotechnology) for 1 h at room temperature before the addition of the radiolabeled probe.
Western blotting
Westerns blots were done as described previously (7). A 1:1,000 dilution of rabbit polyclonal Nrf1 antibody, a 1:500 dilution of Nrf2 antibody, a 1:1,000 dilution of lamin A/C antibody, and a 1:2,000 dilution of actin antibody (all were from Santa Cruz Biotechnology) were used as primary antibodies. Antimouse, or antirabbit Ig, or antigoat horseradish peroxidase–linked antibodies (Amersham Biosciences, Piscataway, NJ) were used as secondary antibodies. Detection was by chemiluminescence (NEN Life Science Products, Boston, MA) on Kodak film (Rochester, NY).
Statistical considerations
A one-sample, two-sided t test was used to evaluate gene expression for the five-gene subset analyzed by quantitative real-time PCR testing the hypothesis that the fold elevation in expression of a particular gene was significantly different from 1.0.
The results were adjusted for multiple comparisons using the Bonferroni correction. Thus, a statistically significant difference was 0.05/5 or 0.01.
Results
Antioxidant gene expression is increased in PBMCs after imexon treatment
PBMCs were obtained from seven patients treated with imexon enrolled in a phase I trial of imexon before and 3 h following a 1-h infusion. Samples from two patients, one with ovarian adenocarcinoma who received 140 mg/m2 (patient 1) and one with pancreatic adenocarcinoma who received 430 mg/m2 of imexon (patient 2), were used for cDNA microarray analysis. Patient 1 had a total of 5,856 genes/ESTs expressed, with 1,590 genes/ESTs down-regulated (ratio ≤ 0.5) and 2,008 genes/ESTs up-regulated (ratio ≥ 1.5) following imexon treatment. Patient 2 had 11,730 genes expressed with 1,983 genes/ESTs down-regulated and 2,840 genes/ESTs up-regulated following imexon treatment. Table 1 shows the antioxidant genes up-regulated in these two patients. Treatment with imexon resulted in an increased expression of 27 antioxidant genes, which included 17 known Nrf2/ARE–responsive genes (Table 1). In addition, 8 of the other 10 up-regulated antioxidant genes were found to have a putative ARE consensus sequence in their promoter sequence, suggesting possible Nrf2/ARE regulation.
Table 1.
Antioxidant gene expression in PBMCs elevated following treatment with imexon
| Gene class | Ratios (Pt1, Pt2) | Putative ARE sequence |
|---|---|---|
| Glutathione-based metabolism | ||
| Glutaredoxin-2 * | (1.72, ND) | AGTGAGCTGAGATCGCACCCTTG |
| Glutathione peroxidase-1 (45) | (2.50, 0.32) | TGCAGCCTGAGGAGGCCAGGGAG |
| Glutathione peroxidase-3 * | (1.41, 3.56) | GCGACCCTGAGTGTGCCCCCACC |
| Glutathione synthetase | (1.05, 1.79) | None identified |
| GSTA2 (45) | (0.52, 3.18) | TTGGCTGTGAGACTGCATTTGAT |
| GSTA3 * | (2.81, 0.38) | GTAGAGTTGACACGGCTTGTGTC |
| GSTA4 (41) | (1.33, 2.92) | GGACCGCTGACCTGGCGCTTTGT |
| GSTM1 (41) | (3.17, ND) | GGCGCCCTGACTTCGCTCCCGGA |
| GSTM4 * | (3.03, ND) | GCTTCGGTGACATAGCCTCCATT |
| GSTM5 (45) | (1.63, 1.77) | GGCGCCCTGACCTCGCTCCCGGA |
| GSTπ (31) | (0.79, 2.41) | GGCGCCGTGACTCAGCACTGGGG |
| GSTθ-1 * | (1.96, 20.00) | GCGTGGGTGACAGAGCAAGAATC |
| GSTω-1 (31) | (1.72, 1.43) | GATGCTTTGACACAGCCCCTTAA |
| Microsomal GST1 (31) | (20.00, 4.01) | AGTGGGCTGAGTCTGCAGCCACC |
| Iron handling | ||
| Ferritin light polypeptide (31) | (2.26,7.43) | CTCAGCATGACTCAGCAGTCGCT |
| Heme oxygenase (decycling) 1 (46) | (1.51, 0.15) | GAGGGTGTGAGGAGGCAAGCAGT |
| Transferrin * | (ND, 7.97) | TCCCACCTGAGTCTGCAGTCATG |
| Transferrin receptor (31) | (3.04, 3.13) | GACAAAGTGAGTCCGCCTCAAAA |
| Oxygen-free radical metabolism | ||
| NAD(P)H dehydrogenase, quinone 1 (31) | (0.71, 6.91) | GCCGTGCTGAGACCGCAGCATTT |
| Peroxiredoxin-1 (41) | (0.64, 3.22) | None identified |
| Peroxiredoxin-2 (31) | (1.34, 3.86) | GCCCGGGTGAGAGAGCAGGGCCG |
| Peroxiredoxin-3 * | (1.10, 2.13) | GGGGTGGTGAGCGGGCCCTCTGC |
| Peroxiredoxin-6 * | (0.80, 2.16) | GGCAACGTGACCGAGCCCCGCAT |
| Superoxide dismutase-1 | (0.67, 1.71) | GCCTGGGTGACAGAGCGAGACCC |
| Thioredoxin reductase-1 (47) | (2.90, ND) | AGGAAACTGACCTTGCTGAAACT |
| Thioredoxin reductase-2 * | (ND, 2.20) | GGGAGTGTGACCCTGCAGGCTCC |
| NADPH generation (pentose phosphate pathway) | ||
| NADH dehydrogenase (ubiquinone) * | (2.18, 1.53) | None identified |
| Transketolase (31, 45) | (3.52,1,80) | GCCTTGGTGACAGAGCCAGATCT |
NOTE: cDNA microarray was used to measure message expression pre-imexon infusion and 3 h post-imexon infusion in two patients. Patient 1 received 140 mg/m2 and patient 2 received 430 mg/mg2 imexon. Ratios are of post-imexon/pretreatment values. Genes that have been reported previously to be Nrf2/ARE are referenced. Putative ARE consensus sequences are in italics.
Abbreviation: ND, not detected.
Indicates genes not reported previously to be regulated by Nrf2/ARE.
Five additional patient PBMC samples were examined by quantitative real-time PCR for expression of a subset of the antioxidant genes identified as up-regulated by microarray analysis. Three genes, thioredoxin reductase-2, glutaredoxin-2, and peroxiredoxin-3, were chosen because of their involvement in mitochondrial redox regulation (27-29). Thioredoxin reductase-1 and transferrin receptor were selected based on their previously identified ARE regulatory sequence and because both are Nrf2 regulated (30, 31). Thioredoxin reductase-1 expression was increased in all five patient samples following imexon treatment (P = 0.004; Table 2). However, variable responses were seen in the other gene products examined.
Table 2.
Increased PBMC antioxidant gene expression in patients receiving imexon ratio
| Gene | Patient dose, mg/m2 |
||||
|---|---|---|---|---|---|
| 80 | 105 | 325 | 430 | 570 | |
| Thioredoxin reductase 1 | 5.72 | 3.03 | 9.24 | 3.25 | 7.66 |
| Thioredoxin reductase 2 | 2.00 | 0.61 | 0.25 | 0.95 | 3.28 |
| Glutaredoxin-2 | 1.93 | 1.75 | 0.44 | 1.32 | 2.43 |
| Peroxiredoxin-3 | 3.77 | 0.84 | 1.46 | 1.41 | 1.13 |
| Transferrin receptor | 23.34 | 0.33 | 20.00 | 1.80 | 3.51 |
NOTE: Quantitative real-time PCR was used to measure PBMC message expression in five patients before and 3 h after receiving an imexon infusion at doses shown. Values are the ratio of post-imexon/pretreatment measurements.
Imexon treatment increases AP-1 DNA binding that is primarily mediated by c-Jun
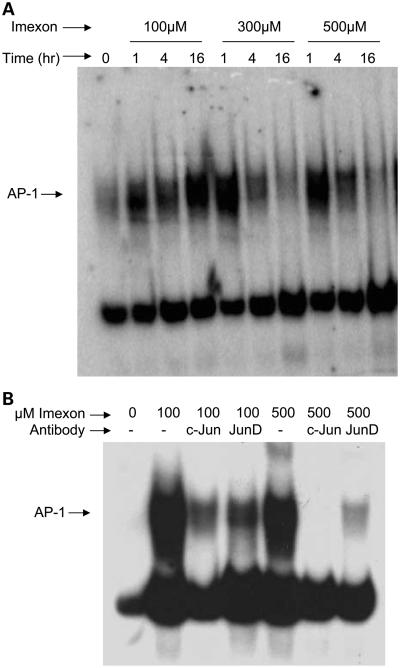
To investigate the molecular pathways responsible for imexon-induced antioxidant gene expression, electrophoretic mobility shift assay (EMSA) analysis was done using nuclear lysates prepared from RPMI8226 myeloma cells treated with 0, 100, 300, or 500 μmol/L imexon for 1, 4, and 16 h in vitro (Fig. 1A). These concentrations reflect a range that is achievable clinically at the MTD of 875 mg/m2, which results in an average Cmax of 479 μmol/L and Css of 34 μmol/L (32). Increased binding was observed to a probe corresponding to the AP-1 consensus sequence as early as 1 h following imexon treatment and remained elevated for up to 16 h. Similar results were obtained when PBMCs obtained from a normal volunteer were treated ex vivo with imexon in the same way (Fig. 2). To identify AP-1 binding proteins responsive to imexon treatment, EMSAs were done in the presence of antibodies to c-Jun and Jun-D (Fig. 1B). The addition of anti-c-Jun antibody resulted in nearly complete clearing of the AP-1 band. A lesser degree of clearing was observed with anti-Jun-D antibody, suggesting that the majority of the AP-1 DNA binding activity is due to c-Jun.
Fig. 1.
AP-1binding activity is increased by imexon treatment. A, EMSA using nuclear extracts prepared from RPMI8226 cells treated with 0, 100, 300, and 500 μmol/L imexon for 1, 4, and 16 h. B, EMSA using nuclear extracts prepared from RPMI8226 cells treated with 0, 100, and 500 μmol/L imexon for 4 h in the presence or absence (−) of antibodies for c-Jun or Jun-D.
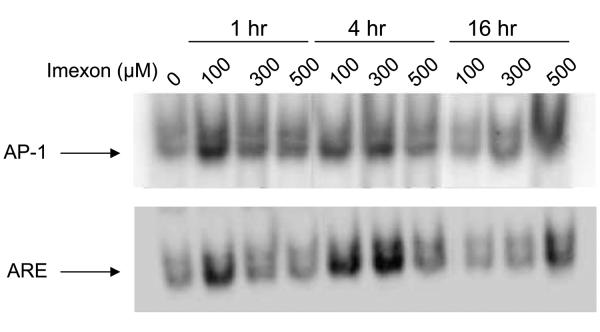
Fig. 2.
AP-1 and Nrf2-ARE binding activity is increased by imexon treatment of normal PBMCs treated ex vivo. EMSAs using nuclear extracts prepared from normal donor PBMCs treated with 0, 100, 300, and 500 μmol/L imexon for 1, 4, and 16 h.
Imexon treatment results in increased ARE DNA binding
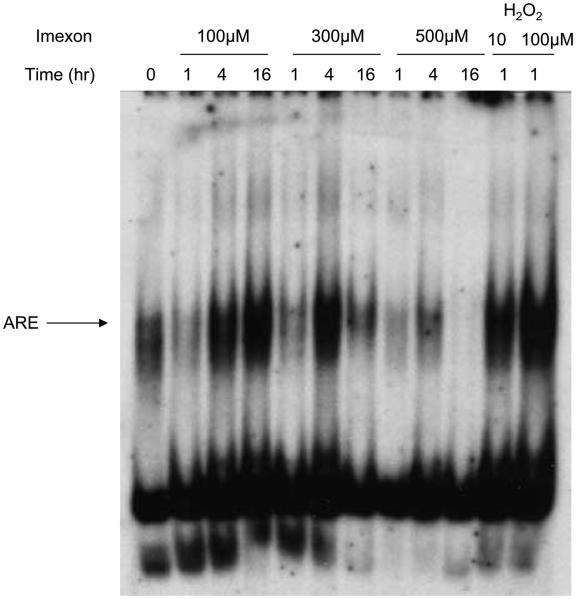
Because the ARE was identified in the promoter region of several genes whose expression was increased in response to imexon treatment, EMSA analysis was done using nuclear lysates prepared from PBMCs isolated from a healthy volunteer (Fig. 2) or RPMI8226 myeloma cells (Fig. 3) treated with 0, 100, 300, and 500 μmol/L imexon for 1, 4, and 16 h in vitro. Increased binding of ARE was seen after 4 h of imexon treatment but was decreased at higher concentration of imexon and with longer incubation times, suggesting an autoinhibition effect under these ex vivo conditions.
Fig. 3.
Nrf2-ARE binding activity is increased by imexon treatment. EMSA gel shift assay using nuclear extracts prepared from RPMI8226 cells treated with 0, 100, 300, and 500 μmol/L imexon for 1, 4, and 16 h. As a positive control, RPMI8226 cells were treated with H2O2 for 1 h.
Imexon treatment increases nuclear expression of Nrf2
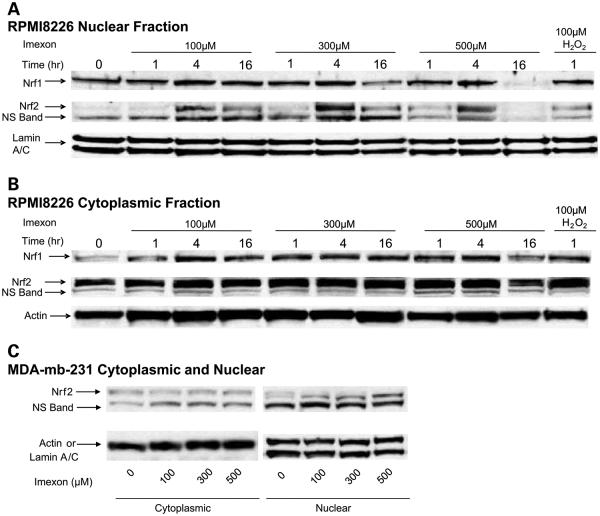
Because the Nrf2 transcription factor has also been shown to be redox-sensitive and to up-regulate antioxidant gene expression through the ARE, we investigated the ability of imexon treatment to translocate Nrf2 protein from the cytoplasm to the nucleus. As shown in Fig. 4A, an increase in Nrf2 in the nucleus was maximal after exposure of RPMI8226 cells to imexon for 4 h but decreased at 16 h and the highest imexon concentrations. Cytoplasmic Nrf2 levels were not changed by imexon treatment (Fig. 4B). Because Nrf1 has also been shown to bind to the ARE and to activate antioxidant gene expression, we measured nuclear and cytoplasmic Nrf1 protein expression following imexon treatment (Fig. 4A and B). There were no significant changes in nuclear or cytoplasmic Nrf1 observed, except for a decrease in expression in both cytoplasmic and nuclear extracts at 16 h of 500 μmol/L imexon. Similarly, Nrf2 protein was increased in the nucleus of the MDA-mb-231 breast cancer cells (Fig. 4C). However, in this cell line, Nrf2 expression was maximal after 16 h compared with 4 h in the RPMI8226 cells.
Fig. 4.
Imexon treatment results in increased nuclear protein expression of Nrf2. A, Western blot of nuclear extracts prepared from RPMI8226 myeloma cells treated with 0, 100, 300, and 500 μmol/L imexon for 1, 4, and 16 h and probed with antibodies for Nrf1 and Nrf2. Lamin A/C was used as loading control. B, Western blot of cytoplasmic extracts prepared as described above. Actin was used as a loading control. NS, nonspecific band. C, Western blot of nuclear and cytoplasmic extracts prepared from MDA-mb-231 breast cancer cells treated with 0, 100, 300, and 500 μmol/L imexon for 16 h and probed with antibodies for Nrf2 and lamin A/C or actin as loading controls.
Discussion
In the present study, we have shown that imexon treatment of patients in a phase 1 clinical trial resulted in activation of an apparent oxidative stress response in PBMCs involving up-regulation of antioxidant gene expression. Studies in RPMI8226 cells suggest that this response may involve both the c-Jun/AP-1 and the Nrf2/ARE response element pathways. It has been reported previously that, in vitro, imexon inhibits mitochondrial function leading to an increase in the formation of reactive oxygen species (5) and it depletes glutathione levels that could be responsible for the oxidative stress response seen (7).
AP-1 activation is an important early cellular response to oxidative stress, leading to the up-regulation of protective genes involved in cell proliferation, differentiation, apoptosis, angiogenesis, tumor invasion, as well as antioxidant defense genes (13, 33). AP-1 activity can be regulated by the type of dimer formation, posttranslational modifications, transcription, and interaction with other proteins, including Nrf2 (34, 35). Recombinant c-Jun DNA binding is increased by oxidant stress leading to AP-1 S-glutathiolation (36). AP-1 activation can also occur through oxidative activation of cytoplasmic mitogen-activated protein kinase family members, which phosphorylate c-Jun (reviewed in ref. 37). Based on the ability of imexon in increased oxidant stress (5, 7), activation of AP-1 by imexon could be due to redox modulation in the cytoplasm or nucleus.
Nrf2 is an activator of ARE-mediated gene expression and mediates the primary antioxidant response to an oxidative challenge in cells (17). Oxidizing stress releases Nrf2 from its cytoplasmic complexation with its cytoplasmic binding partner Keap-1, largely through modification of cysteine257, 273, 299, 297 residues in Keap-1 (38). Some important ARE-regulated genes, which are controlled by Nrf2, include reduced glutathione synthesis enzymes (39), thioredoxin-1 (40), thioredoxin reductase-1 (30), glutathione peroxidase, glutathione reductase (41), and the glutathione transferases (42). We found many of these antioxidant genes to be up-regulated in PBMC of patients treated with imexon (Table 1). We also showed that imexon at the therapeutically achievable concentrations caused an increase in nuclear Nrf2 and binding of Nrf2 to the ARE in both PBMC and RPMI8226 cells. The effect decreased with longer exposure times and higher concentrations of imexon presumable due to the antioxidant defense mounted by the ARE.
Several anticancer drugs have been shown to increase reactive oxygen species, including doxorubicin, mitomycin C, arsenic trioxide, and motexafin-gadolinium, which may contribute to their antitumor activity (43, 44). Apparently, an increase in reactive oxygen species contributes to the antitumor activity of imexon, although this may not be the major mechanism. Thus, the effect of increased reactive oxygen species on antitumor activity may be self-limiting if there is an induced or constitutive increase in antioxidant enzymes. A consequence of an increase in reactive oxygen species is up-regulation of the expression of ARE-regulated antioxidant genes. Some of the genes we found to be up-regulated by imexon were also found to be increased in imexon-resistant RPMI8226 myeloma cells (8). Apparently, an increase in PBMC antioxidant gene expression, particularly of thioredoxin reductase, which showed the most consistent increased expression, could provide an early biomarker for response to imexon or adaptation to oxidative stress and other toxicities, although this possibility will require further studies.
In summary, we have found the up-regulation of many antioxidant genes in PBMCs collected from patients treated with imexon. Thioredoxin reductase-1 expression was the most consistently up-regulated antioxidant gene. Studies in RPMI8226 myeloma cells showed that imexon increased binding to the AP-1 and ARE consensus sequences, which regulate antioxidant gene expression. The results suggest that a prominent biological effect of imexon is a change in redox state that can be detected in surrogate normal tissue as increased antioxidant gene transcription.
Acknowledgments
Grant support: Public Health Service grants U54 CA090824 and CA17094-27 and a “Better Than Ever” Arizona Cancer Center grant to A.F. Baker.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
References
- 1.Salmon SE, Hersh EM. Sensitivity of multiple myeloma to imexon in the human tumor cloning assay. J Natl Cancer Inst. 1994;86:228–30. doi: 10.1093/jnci/86.3.228. [DOI] [PubMed] [Google Scholar]
- 2.Hersh EM, Gschwind CR, Taylor CW, Dorr RT, Taetle R, Salmon SE. Antiproliferative and antitumor activity of the 2-cyanoaziridine compound imexon on tumor cell lines and fresh tumor cells in vitro. J Natl Cancer Inst. 1992;84:1238–44. doi: 10.1093/jnci/84.16.1238. [DOI] [PubMed] [Google Scholar]
- 3.Hersh EM, Grogan TM, Funk CY, Taylor CW. Suppression of human lymphoma development in the severe combined immune-deficient mouse by imexon therapy. J Immunother. 1993;13:77–83. doi: 10.1097/00002371-199302000-00001. [DOI] [PubMed] [Google Scholar]
- 4.Sagaster P, Kokoschka EM, Kokron O, Micksche M. Antitumor activity of imexon. J Natl Cancer Inst. 1995;87:935–6. doi: 10.1093/jnci/87.12.935-a. [DOI] [PubMed] [Google Scholar]
- 5.Dvorakova K, Waltmire CN, Payne CM, Tome ME, Briehl MM, Dorr RT. Induction of mitochondrial changes in myeloma cells by imexon. Blood. 2001;97:3544–51. doi: 10.1182/blood.v97.11.3544. [DOI] [PubMed] [Google Scholar]
- 6.Dorr RT, Raymond MA, Landowski TH, Roman NO, Fukushima S. Induction of apoptosis and cell cycle arrest by imexon in human pancreatic cancer cell lines. Int J Gastrointest Cancer. 2005;36:15–28. doi: 10.1385/IJGC:36:1:015. [DOI] [PubMed] [Google Scholar]
- 7.Dvorakova K, Payne CM, Tome ME, et al. Induction of oxidative stress and apoptosis in myeloma cells by the aziridine-containing agent imexon. Biochem Pharmacol. 2000;60:749–58. doi: 10.1016/s0006-2952(00)00380-4. [DOI] [PubMed] [Google Scholar]
- 8.Dvorakova K, Payne CM, Tome ME, et al. Molecular and cellular characterization of imexon-resistant RPMI8226/I myeloma cells. Mol Cancer Ther. 2002;1:185–95. [PubMed] [Google Scholar]
- 9.Meyer AJ, Hell R. Glutathione homeostasis and redox-regulation by sulfhydryl groups. Photosynth Res. 2005;86:435–57. doi: 10.1007/s11120-005-8425-1. [DOI] [PubMed] [Google Scholar]
- 10.Powis G, Mustacich D, Coon A. The role of the redox protein thioredoxin in cell growth and cancer. Free Radic Biol Med. 2000;29:312–22. doi: 10.1016/s0891-5849(00)00313-0. [DOI] [PubMed] [Google Scholar]
- 11.Wood ZA, Schroder E, Robin HJ, Poole LB. Structure, mechanism, and regulation of peroxiredoxins. Trends Biochem Sci. 2003;28:32–40. doi: 10.1016/s0968-0004(02)00003-8. [DOI] [PubMed] [Google Scholar]
- 12.Boonstra J, Post JA. Molecular events associated with reactive oxygen species and cell cycle progression in mammalian cells. Gene. 2004;337:1–13. doi: 10.1016/j.gene.2004.04.032. [DOI] [PubMed] [Google Scholar]
- 13.Eferl R, Wagner EF. AP-1: a double-edged sword in tumorigenesis. Nat Rev Cancer. 2003;3:859–68. doi: 10.1038/nrc1209. [DOI] [PubMed] [Google Scholar]
- 14.Ainbinder E, Bergelson S, Pinkus R, Daniel V. Regulatory mechanisms involved in activator-protein-1 (AP-1)-mediated activation of glutathione-S-transferase gene expression by chemical agents. Eur J Biochem. 1997;243:49–57. doi: 10.1111/j.1432-1033.1997.0049a.x. [DOI] [PubMed] [Google Scholar]
- 15.Bossy-Wetzel E, Bakiri L, Yaniv M. Induction of apoptosis by the transcription factor c-Jun. EMBO J. 1997;16:1695–709. doi: 10.1093/emboj/16.7.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dhakshinamoorthy S, Long DJ, Jaiswal AK. Antioxidant regulation of genes encoding enzymes that detoxify xenobiotics and carcinogens. Curr Top Cell Regul. 2000;36:201–16. doi: 10.1016/s0070-2137(01)80009-1. [DOI] [PubMed] [Google Scholar]
- 17.Jaiswal AK. Nrf2 signaling in coordinated activation of antioxidant gene expression. Free Radic Biol Med. 2004;36:1199–207. doi: 10.1016/j.freeradbiomed.2004.02.074. [DOI] [PubMed] [Google Scholar]
- 18.Venugopal R, Jaiswal AK. Nrf2 and Nrf1in association with Jun proteins regulate antioxidant response element-mediated expression and coordinated induction of genes encoding detoxifying enzymes. Oncogene. 1998;17:3145–56. doi: 10.1038/sj.onc.1202237. [DOI] [PubMed] [Google Scholar]
- 19.Sankaranarayanan K, Jaiswal AK. Nrf3 negatively regulates antioxidant-response element-mediated expression and antioxidant induction of NAD(P)H:quinone oxidoreductase1 gene. J Biol Chem. 2004;279:50810–7. doi: 10.1074/jbc.M404984200. [DOI] [PubMed] [Google Scholar]
- 20.Kobayashi A, Kang MI, Watai Y, et al. Oxidative and electrophilic stresses activate Nrf2 through inhibition of ubiquitination activity of Keap1. Mol Cell Biol. 2006;26:221–9. doi: 10.1128/MCB.26.1.221-229.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Venugopal R, Jaiswal AK. Nrf1 and Nrf2 positively and c-Fos and Fra1 negatively regulate the human antioxidant response element-mediated expression of NAD(P)H:quinone oxidoreductase1 gene. Proc Natl Acad Sci U S A. 1996;93:14960–5. doi: 10.1073/pnas.93.25.14960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nguyen T, Huang HC, Pickett CB. Transcriptional regulation of the antioxidant response element. Activation by Nrf2 and repression by MafK. J Biol Chem. 2000;275:15466–73. doi: 10.1074/jbc.M000361200. [DOI] [PubMed] [Google Scholar]
- 23.Itoh K, Chiba T, Takahashi S, et al. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem Biophys Res Commun. 1997;236:313–22. doi: 10.1006/bbrc.1997.6943. [DOI] [PubMed] [Google Scholar]
- 24.Meuillet EJ, Ihle N, Baker AF, et al. In vivo molecular pharmacology and antitumor activity of the targeted Akt inhibitor PX-316. Oncol Res. 2004;14:513–27. doi: 10.3727/0965040042380487. [DOI] [PubMed] [Google Scholar]
- 25.Motohashi H, Yamamoto M. Nrf2-1 defines a physiologically important stress response mechanism. Trends Mol Med. 2004;10:549–57. doi: 10.1016/j.molmed.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 26.Landowski TH, Olashaw NE, Agrawal D, Dalton WS. Cell adhesion-mediated drug resistance (CAM-DR) is associated with activation of NF-κB (RelB/p50) in myeloma cells. Oncogene. 2003;22:2417–21. doi: 10.1038/sj.onc.1206315. [DOI] [PubMed] [Google Scholar]
- 27.Patenaude A, Ven Murthy MR, Mirault ME. Mitochondrial thioredoxin system: effects of TrxR2 overexpression on redox balance, cell growth, and apoptosis. J Biol Chem. 2004;279:27302–14. doi: 10.1074/jbc.M402496200. [DOI] [PubMed] [Google Scholar]
- 28.Nonn L, Berggren M, Powis G. Increased expression of mitochondrial peroxiredoxin-3 (thioredoxin peroxidase-2) protects cancer cells against hypoxia and drug-induced hydrogen peroxide-dependent apoptosis. Mol Cancer Res. 2003;1:682–9. [PubMed] [Google Scholar]
- 29.Lundberg M, Johansson C, Chandra J, et al. Cloning and expression of a novel human glutaredoxin (Grx2) with mitochondrial and nuclear isoforms. J Biol Chem. 2001;276:26269–75. doi: 10.1074/jbc.M011605200. [DOI] [PubMed] [Google Scholar]
- 30.Sakurai A, Nishimoto M, Himeno S, et al. Transcriptional regulation of thioredoxin reductase 1 expression by cadmium in vascular endothelial cells: role of NF-E2-related factor-2. J Cell Physiol. 2005;203:529–37. doi: 10.1002/jcp.20246. [DOI] [PubMed] [Google Scholar]
- 31.Lee JM, Calkins MJ, Chan K, Kan YW, Johnson JA. Identification of the NF-E2-related factor-2-dependent genes conferring protection against oxidative stress in primary cortical astrocytes using oligonucleotide micro-array analysis. J Biol Chem. 2003;278:12029–38. doi: 10.1074/jbc.M211558200. [DOI] [PubMed] [Google Scholar]
- 32.Dragovich T, Dragovich D, Mendelson M, et al. Phase I trial of imexon in patients with advanced solid tumors. J Clin Oncol. 2005;23:3153. doi: 10.1200/JCO.2006.08.9672. [DOI] [PubMed] [Google Scholar]
- 33.Lapperre TS, Jimenez LA, Antonicelli F, et al. Apocynin increases glutathione synthesis and activates AP-1 in alveolar epithelial cells. FEBS Lett. 1999;443:235–9. doi: 10.1016/s0014-5793(98)01723-2. [DOI] [PubMed] [Google Scholar]
- 34.Macian F, Lopez-Rodriguez C, Rao A. Partners in transcription: NFAT and AP-1. Oncogene. 2001;20:2476–89. doi: 10.1038/sj.onc.1204386. [DOI] [PubMed] [Google Scholar]
- 35.Nishinaka T, Yabe-Nishimura C. Transcription factor Nrf2 regulates promoter activity of mouse aldose reductase (AKR1B3) gene. J Pharmacol Sci. 2005;97:43–51. doi: 10.1254/jphs.fp0040404. [DOI] [PubMed] [Google Scholar]
- 36.Klatt P, Molina EP, De Lacoba MG, et al. Redox regulation of c-Jun DNA binding by reversible S-glutathiolation. FASEB J. 1999;13:1481–90. doi: 10.1096/fasebj.13.12.1481. [DOI] [PubMed] [Google Scholar]
- 37.Filosto M, Tonin P, Vattemi G, Savio C, Rizzuto N, Tomelleri G. Transcription factors c-Jun/activator protein-1 and nuclear factor-κB in oxidative stress response in mitochondrial diseases. Neuropathol Appl Neurobiol. 2003;29:52–9. doi: 10.1046/j.1365-2990.2003.00411.x. [DOI] [PubMed] [Google Scholar]
- 38.Nguyen T, Sherratt PJ, Pickett CB. Regulatory mechanisms controlling gene expression mediated by the antioxidant response element. Annu Rev Pharmacol Toxicol. 2003;43:233–60. doi: 10.1146/annurev.pharmtox.43.100901.140229. [DOI] [PubMed] [Google Scholar]
- 39.Moinova HR, Mulcahy RT. Up-regulation of the human γ-glutamylcysteine synthetase regulatory subunit gene involves binding of Nrf-2 to an electrophile responsive element. Biochem Biophys Res Commun. 1999;261:661–8. doi: 10.1006/bbrc.1999.1109. [DOI] [PubMed] [Google Scholar]
- 40.Kim YC, Yamaguchi Y, Kondo N, Masutani H, Yodoi J. Thioredoxin-dependent redox regulation of the anti-oxidant responsive element (ARE) in electrophile response. Oncogene. 2003;22:1860–5. doi: 10.1038/sj.onc.1206369. [DOI] [PubMed] [Google Scholar]
- 41.Kwak MK, Wakabayashi N, Itoh K, Motohashi H, Yamamoto M, Kensler TW. Modulation of gene expression by cancer chemopreventive dithiolethiones through the Keap1-2 pathway. Identification of novel gene clusters for cell survival. J Biol Chem. 2003;278:8135–45. doi: 10.1074/jbc.M211898200. [DOI] [PubMed] [Google Scholar]
- 42.Hayes JD, Chanas SA, Henderson CJ, et al. The Nrf2 transcription factor contributes both to the basal expression of glutathione S-transferases in mouse liver and to their induction by the chemopreventive synthetic antioxidants, butylated hydroxyanisole, and ethoxyquin. Biochem Soc Trans. 2000;28:33–41. doi: 10.1042/bst0280033. [DOI] [PubMed] [Google Scholar]
- 43.Kovacic P, Osuna JA., Jr. Mechanisms of anti-cancer agents: emphasis on oxidative stress and electron transfer. Curr Pharm Des. 2000;6:277–309. doi: 10.2174/1381612003401046. [DOI] [PubMed] [Google Scholar]
- 44.Lecane PS, Karaman MW, Sirisawad M, et al. Motexafin gadolinium and zinc induce oxidative stress responses and apoptosis in B-cell lymphoma lines. Cancer Res. 2005;65:11676–88. doi: 10.1158/0008-5472.CAN-05-2754. [DOI] [PubMed] [Google Scholar]
- 45.Thimmulappa RK, Mai KH, Srisuma S, Kensler TW, Yamamoto M, Biswal S. Identification of Nrf2-regulatedgenes inducedby the chemopreventive agentsulforaphane by oligonucleotide microarray. Cancer Res. 2002;62:5196–203. [PubMed] [Google Scholar]
- 46.Cheng GC, Schulze PC, Lee RT, Sylvan J, Zetter BR, Huang H. Oxidative stress and thioredoxin-interacting protein promote intravasation of melanoma cells. Exp Cell Res. 2004;300:297–307. doi: 10.1016/j.yexcr.2004.07.014. [DOI] [PubMed] [Google Scholar]
- 47.Kwak MK, Wakabayashi N, Greenlaw JL, Yamamoto M, Kensler TW. Antioxidants enhance mammalian proteasome expression through the Keap1-2 signaling pathway. Mol Cell Biol. 2003;23:8786–94. doi: 10.1128/MCB.23.23.8786-8794.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]