Abstract
Colorectal cancers (CRCs) with a high level of microsatellite instability (MSI-H) are clinicopathologically distinct tumors characterized by predominance in females, proximal colonic localization, poor differentiation, mucinous histology, tumor-infiltrating lymphocytes, a Crohn’s-like lymphoid reaction and a favorable prognosis. In terms of their molecular features, MSI-H CRCs are heterogeneous tumors associated with various genetic and epigenetic alterations, including DNA mismatch repair deficiency, target microsatellite mutations, BRAF mutations, a CpG island methylator phenotype-high (CIMP-H) status, and a low level of genomic hypomethylation. The molecular heterogeneity of MSI-H CRCs also depends on ethnic differences; for example, in Eastern Asian countries, relatively low frequencies of CIMP-H and BRAF mutations have been observed in MSI-H CRCs compared to Western countries. Although the prognostic features of MSI-H CRCs include a favorable survival of patients and low benefit of adjuvant chemotherapy, there may be prognostic differences based on the molecular heterogeneity of MSI-H CRCs. Here, we have reviewed and discussed the molecular and prognostic features of MSI-H CRCs, as well as several putative prognostic or predictive molecular markers, including HSP110 expression, beta2-microglobulin mutations, myosin 1a expression, CDX2/CK20 expression, SMAD4 expression, CIMP status and LINE-1 methylation levels.
Keywords: Colorectal cancer, Microsatellite instability, DNA mismatch repair, DNA methylation, CpG islands, Prognosis, Adjuvant chemotherapy
Core tip: A high level of microsatellite instability (MSI-H) is a known molecular indicator of a favorable prognosis and low benefit of 5-fluorouracil-based adjuvant chemotherapy in patients with colorectal cancer (CRC). However, MSI-H CRCs are molecularly heterogeneous tumors, which are characterized by DNA mismatch repair deficiency and various genetic and epigenetic alterations. Therefore, we hypothesized that MSI-H CRCs can be divided into prognostic subgroups based on the molecular heterogeneity. This article provides an up-to-date review concerning the underlying molecular features of MSI-H CRCs and potential prognostic or predictive molecular markers for MSI-H CRCs.
INTRODUCTION
Microsatellite instability (MSI) is a unique molecular alteration induced by deficiencies in the DNA mismatch repair (MMR) system and is characterized by unstable (length-changeable) microsatellites, a type of simple DNA sequence repeat. The MSI phenotype has been regarded as one of the main molecular subtypes of colorectal cancers (CRCs) and accounts for 12%-20% and 6%-13% of CRCs in Western and Eastern countries, respectively[1-6]. Hereditary CRCs with a high level of MSI (MSI-H) constitute approximately 3%-5% of all CRCs and arise exclusively in patients with Lynch syndrome. Lynch syndrome was formerly called hereditary non-polyposis colorectal cancer and is caused by a germline mutation in at least one of the MMR genes (MLH1, MSH2, PMS2, and MSH6), frequently resulting in the development of early-onset malignancies, including CRC and endometrial cancer[7,8]. Sporadic MSI-H CRCs account for approximately 3%-15% of all CRCs and develop mainly as a result of inactivation of the MLH1 gene via promoter CpG island hypermethylation[9].
MSI status in CRCs can be determined by DNA testing using microsatellite markers, and five microsatellite markers recommended by the National Cancer Institute (NCI) workshop have been officially used for MSI analysis: BAT25, BAT26, D2S123, D5S346 and D17S250[10]. In DNA analysis using these NCI markers, instability observed in two or more of the five markers corresponds to MSI-H. MSI-H can be interpreted as the presence of MSI. In contrast, a low level of MSI (MSI-L), which is assigned when only one unstable marker is detected, is not regarded as a true MSI-positive status. Microsatellite-stable (MSS) status can be assigned when all of the markers show stability. Immunohistochemistry (IHC) for MMR proteins can be applied as a screening test or a supportive test for MSI analysis.
MSI-H CRC is known to have distinct clinicopathological and molecular features, including preferential localization in the proximal colon, a less advanced cancer stage, extracellular mucin production, medullary carcinoma and poorly differentiated carcinoma, tumor-infiltrating lymphocytes, a Crohn’s-like lymphoid reaction, and a BRAF V600E mutation[4,11-14]. In addition, and more importantly, it has been consistently reported that MSI-H CRC is associated with favorable survival and chemotherapy resistance. In a considerable number of previous studies, patients with MSI-H CRC demonstrated a significantly better survival than MSS/MSI-L CRC patients[15,16], whereas the beneficial effect of 5-fluorouracil (5-FU)-based adjuvant chemotherapy in patients with MSI-H CRC has been controversial[15-19]. These prognostic features of MSI-H CRCs have been increasingly reported, and MSI is currently regarded as a molecular marker indicating favorable prognosis for CRCs[20]. However, because MSI-H CRCs are characterized by various underlying molecular changes, including a defective MMR (dMMR) system and genetic and epigenetic alterations, it is likely that molecular factors for the stratification of patient prognosis and prediction of chemotherapy response in MSI-H CRCs could be identified. On the basis of this hypothesis, we reviewed the literature and provide information about several putative prognostic molecular factors for MSI-H CRCs.
MOLECULAR HETEROGENEITY OF MSI-H CRC
DNA MMR deficiency
Germline mutation or sporadic methylation of MMR genes: As described above, MSI is caused by the inactivation of at least one of the MMR genes. The inactivation of MMR genes can be induced by a germline mutation or by promoter CpG island hypermethylation. Germline mutation of MMR genes, including MLH1, MSH2, PMS2, or MSH6, represents a major cause of hereditary MSI-H CRCs in Lynch syndrome. Among these MMR genes, germline mutations in the MLH1 or MSH2 genes account for the majority of Lynch syndrome CRCs[21]. Promoter methylation of MMR genes is a major cause of sporadic MSI-H CRCs and exclusively involves the MLH1 gene. MLH1 promoter methylation is closely associated with the CpG island methylator phenotype (CIMP) in sporadic CRCs and has been used as one of the major molecular markers for CIMP determination in CRCs[4,22-24]. It is also generally expected that all of the MLH1-methylated MSI-H CRCs are CIMP-high (CIMP-H) tumors, although discordance between MLH1 methylation and CIMP status can be observed in a small subset of MSI-H CRCs[25]. A detailed review and discussion concerning the molecular basis and prognostic implication of CIMP in MSI-H CRCs will be presented in the following sections.
Constitutional (germline) epimutation of MMR genes: Promoter methylation of MSH2 owing to germline deletion of 3’ exons in the EPCAM gene (also known as the TACSTD1 gene), which is located in the region immediately upstream of MSH2, was recently identified as a cause of MSI-H CRC in Lynch syndrome[26,27]. This molecular alteration is associated with a small subset of patients with Lynch syndrome and is also called the MSH2 “epimutation” because of its unique feature of heritable constitutional epigenetic change[28,29]. In addition, constitutional epimutation of the MLH1 gene has also been found in a few patients with Lynch syndrome[30-34]. Although it has been reported that an association between MLH1 epimutation and a family history of CRC is not evident[30], the clinical and pathological significance of epimutations in MMR genes in hereditary MSI-H CRCs remains unclear. According to our data, MLH1 methylation-positive/CIMP-negative tumors account for 7.3% of MSI-H CRCs[25], and these cases were associated with an early age of onset and favorable survival. It cannot be excluded that MLH1 epimutation-associated MSI-H CRCs may be included in these MLH1 methylation-positive/CIMP-negative MSI-H CRCs, and the predominance of young patients can imply the presence of MLH1 epimutation carriers. Thus, additional studies should be performed to investigate the detailed epidemiology and clinical implications of MMR gene epimutations in MSI-H CRC.
Expression profile of MMR proteins: Normal DNA MMR function is executed by MMR protein complexes composed of heterodimers of MutL homologues (the MLH or PMS series) or MutS homologues (the MSH series). The major role of the human DNA MMR system is performed by the MutSα and MutLα complexes. The MutSα complex comprises a MSH2-MSH6 heterodimer, whereas the dimer of MLH1 and PMS2 forms the MutLα complex[21,35]. These complexes are essential components of the human DNA MMR machinery, and defects in any one of these four MMR proteins lead to a dysfunctional MMR system and ultimately result in MSI. Therefore, loss of expression of MMR proteins can serve as a molecular hallmark of a dMMR system and the MSI-H status in tumors. IHC for MMR proteins is a simple and valuable tool for the detection of dMMR CRC and can be helpful for investigating the underlying molecular alteration and hereditary/sporadic status of MSI-H CRCs. The immunohistochemical profile of four MMR proteins in MSI-H CRCs can be summarized as four expression phenotypes: MLH1-negative/PMS2-negative, PMS2-negative only, MSH2-negative/MSH6-negative, and MSH6-negative only[8,36]. These four phenotypes most likely represent inactivation of MLH1, PMS2, MSH2, and MSH6, respectively. The majority of MSI-H CRCs are induced by inactivation of MLH1 or MSH2, whereas inactivation of PMS2 or MSH6 causes only a minor portion of MSI-H CRCs. Interestingly, an inactivating mutation or methylation of MLH1 is accompanied by PMS2 loss, and inactivation of MSH2 is combined with the loss of MSH6 expression due to their heterodimer structures. However, inactivation (mainly through germline mutations) of PMS2 or MSH6 does not accompany a loss of MLH1 or MSH2 expression, respectively[8,36]. Although IHC for MLH1 and MSH2 has generally been used for the screening of MMR status in CRCs, the inclusion of screening for PMS2 and MSH6 would compensate for the equivocal results of MLH1 or MSH2 IHC and aid in the detection of rare MSI-H CRCs with germline mutations of PMS2 or MSH6.
Genetic alterations
Microsatellite mutations: A recent study elucidating the molecular landscape of CRC by The Cancer Genome Atlas Project reported that the hypermutated phenotype mainly overlaps with MSI-H status in CRCs[37]. This finding is not surprising because MSI-H status tumors are highly vulnerable to insertions or deletions in microsatellite sequences, as described above. In CRCs with MSI, many types of genetic mutations can occur, but the majority of mutations that develop under MSI-H status are frameshift mutations because the instability of microsatellite sequences in coding regions can alter entire reading frames adjacent to the insertion or deletion point. Previous investigations have reported frameshift mutations of various genes caused by the instability of microsatellites in MSI-H CRCs. The target genes for microsatellite mutations in MSI-H CRCs include TGFBR2, BAX, ACVR2, IGF2R, BLM, MSH3, MSH6, E2F4, PTEN, AIM2, CASPASE5, MBD4, TCF4, STK11, RAD50, CHK1, AXIN2, WISP3, B2M, MYO1A and CDX2[38-54]. These genes are known to be associated with important biological functions such as signal transduction, apoptosis regulation, cell cycle regulation, cell proliferation, cell differentiation, and DNA MMR; therefore, loss-of-function mutations in these genes can critically contribute to tumorigenesis. However, although the biological and clinical implications of mutations in these genes in CRCs have been explored, their potential use as prognostic markers or therapeutic targets has not been established. For instance, although mutations in TGFBR2 and BAX, which are representative target tumor suppressor genes for microsatellite mutations in MSI-H CRCs, have been previously analyzed to determine their prognostic implications, conflicting results have been reported. A few groups have reported that TGFBR2 and BAX mutations were associated with a favorable prognosis in MSI-H CRCs[55,56], whereas other investigators did not find a prognostic significance of these mutations in MSI-H CRCs[57,58]. Interestingly, microsatellite alterations in intronic regions could induce mutations in genes such as HSP110 and MRE11 in MSI-H CRCs[59,60]. In the recent study by Dorard et al[59], deletions of the T17 mononucleotide repeat located in intron 8 of the HSP110 gene were shown to lead to exon 9 skipping and the production of truncated mutant proteins of HSP110. The HSP110 mutation as a potential prognostic and predictive marker in MSI CRC-H will be discussed briefly in the “Putative prognostic or predictive molecular markers for MSI-H CRC” section.
BRAF/KRAS mutations: BRAF is a member of the RAF kinase family of genes and is a downstream effector of the KRAS gene, whereas KRAS is a member of the RAS family of genes and is a downstream effector of the EGFR gene in the Ras-Raf-MEK-ERK signaling pathway. The Ras-Raf-MEK-ERK signaling pathway is commonly involved in cell cycle progression and cell proliferation, and thus, activating mutations of key component genes in this pathway, including mutations in the BRAF or KRAS gene, can bring about uncontrolled cell growth and increased cell survival and may play an important role in tumorigenesis. BRAF and KRAS mutations are mutually exclusive in cancers, and the majority of mutations of BRAF and KRAS in human tumors are hot spot mutations in codon 600 (V600E) and codons 12 or 13, respectively[61,62]. According to previous studies, BRAF mutations were found in 5%-15% of overall CRCs[61,63-66], whereas 32%-40% of CRCs have KRAS mutations[63,64,67-72]. However, it has been revealed that BRAF V600E mutations are highly associated with MSI-H CRCs, although the incidence of KRAS mutations is inversely correlated with MSI-H status in CRCs[36]. The frequencies of BRAF and KRAS mutations in MSI-H CRCs have been reported to be 16%-52% and 12%-20%, respectively, in Western countries[63,73-76]. Notably, because BRAF V600E mutations have been found exclusively in sporadic tumors among MSI-H CRCs, it has been suggested that detection of BRAF mutations in CRCs may be a useful supportive tool for distinguishing sporadic CRCs from Lynch syndrome CRCs[14,73,77]. In fact, it is thought that this observed correlation between BRAF mutations and sporadic MSI-H CRCs is mostly based on the more significant association between BRAF mutations and CIMP-H status in CRCs[13,23]. Regarding the implications for prognosis, several previous studies have reported that BRAF mutations indicate poor survival in patients with CRC[4,78]. However, it has been suggested that the contribution of BRAF mutations to an adverse prognosis is significant for MSI-L/MSS CRCs but not MSI-H CRCs[65,79]. Therefore, the clinical and prognostic significance of BRAF mutations in MSI-H CRCs should be carefully explored in larger samples.
Epigenetic alterations
CIMP: CIMP represents a distinct subset of CRCs that show extensive promoter CpG island methylation and are characterized by transcriptional repression of many tumor suppressor genes as a result of promoter methylation[80]. The CIMP status of CRCs can be classified into three subtypes: CIMP-H, CIMP-low (CIMP-L), and CIMP-zero (CIMP-0)[24,81,82]. Among these subtypes, CIMP-H is generally regarded as the true CIMP-positive status. Previous investigations have revealed that CIMP-H is highly associated with sporadic MSI-H owing to the high frequency of MLH1 promoter methylation in CIMP-H CRCs[4,13,83]. According to data from Western countries, CIMP-H CRCs account for 7%-29% of all CRCs, and 56%-82% of MSI-H CRCs have the CIMP-H subtype (Table 1)[4,13,22,24,63,84-86]. In our previous study investigating differential clinicopathological features of MSI-H CRCs depending on CIMP status, we found that the CIMP-H subtype was significantly associated with older age, frequent BRAF V600E mutations, poor differentiation, medullary carcinoma components, and signet ring cell carcinoma components in MSI-H CRCs[87]. The effect of CIMP status on MSI-H CRC prognosis will be discussed in the “Putative prognostic or predictive molecular markers for MSI CRC” section.
Table 1.
Frequency of CpG island methylator phenotype-high in colorectal cancers: A review of the literature
| Ref. | Country | CIMP-H CRCs/tested CRCs n (%) |
|
| In all CRCs | In MSI-H CRCs | ||
| Western countries | |||
| Samowitz et al[22], 2005 | United States | 250/859 (29.1) | 64/78 (82.1) |
| Weisenberger et al[13], 2006 | United States | 33/187 (17.6) | NA |
| Samowitz et al[84], 2006 | United States | 313/1271 (24.6) | 105/170 (61.8) |
| Barault et al[85], 2008 | France | 95/578 (16.4) | 58/80 (72.5) |
| Ogino et al[4], 2009 | United States | 123/631 (19.5) | 86/118 (72.9) |
| Dahlin et al[86], 2010 | Sweden | 46/411 (11.2) | 34/61 (55.7) |
| Zlobec et al[24], 2011 | Switzerland | 22/314 (7) | NA |
| Lochhead et al[63], 2013 | United States | 205/1173 (17.5) | 140/184 (76.1) |
| Eastern countries | |||
| Koinuma et al[101], 2004 | Japan | NA | 161/28 (57.1) |
| Nagasaka et al[102], 2008 | Japan and Germany | NA | 152/36 (41.7) |
| Kim et al[98], 2009 | South Korea | 29/271 (10.7) | 8/33 (24.2) |
| Min et al[99], 2011 | South Korea | 34/245 (13.9) | NA |
| Bae et al[97], 2013 | South Korea | 47/734 (6.4) | 18/65 (27.7) |
| Kim et al[25], 2013 | South Korea | NA | 64/220 (29.1) |
MLH1-methylated colorectal cancers (CRCs) instead of CpG island methylator phenotype-high (CIMP-H) CRCs;
Sporadic microsatellite instability-high (MSI-H) CRCs instead of CIMP-H CRCs. NA: Not applicable.
Genome-wide DNA methylation: The genomic methylation levels of specific tissues can be estimated by measuring the methylation levels of repetitive DNA elements, such as long interspersed nucleotide element-1 (LINE-1) and Alu, because these repetitive elements are globally distributed and occupy considerable portions of the human genome[88,89]. Of these repetitive elements, the methylation level of LINE-1 has been generally used as a reliable surrogate marker for the global DNA methylation level. In particular, LINE-1 hypomethylation has been regarded as one of the molecular characteristics that distinguishes CRC tumors from normal tissue[90]. Interestingly, several previous investigations have reported that LINE-1 hypomethylation is inversely correlated with MSI-H status in CRCs[91-93]. In a study by Ogino et al[93], a relatively high level of LINE-1 methylation was significantly associated with both MSI-H and CIMP-H statuses in CRCs. Notably, in this study, a correlation between the LINE-1 methylation level and MSI status was significant regardless of CIMP status; furthermore, a low LINE-1 methylation level was associated with 18q loss of heterozygosity (LOH) in CRCs. These findings suggest that genomic hypomethylation may be a characteristic phenomenon of the chromosomal instability (CIN) pathway, rather than the MSI pathway, in colorectal carcinogenesis. However, the mechanistic correlation between MSI status and resistance to genomic hypomethylation in CRCs should be further evaluated. The prognostic value of the LINE-1 methylation level in MSI-H CRCs will be described below, in the “Putative prognostic or predictive molecular markers for MSI-H CRC” section.
Molecular heterogeneity among ethnic groups
As mentioned above, MSI-H CRCs constitute approximately 15% of all CRCs in Western countries[8,9,35]. However, according to previous studies reported by our group and others, a relatively low frequency of MSI-H (5.5%-9.4%) has been consistently observed in Korean patients with CRC, regardless of the institutions at which the study samples were collected[6,25,87,94-97]. Furthermore, the frequencies of CIMP-H and BRAF V600E mutations in CRCs are lower in Koreans (6.4%-13.9% and 4.3%-5.4%, respectively) than in Western populations[97-100]. We hypothesized that the low frequency of MSI-H CRCs in Korea is mainly based on the low prevalence of CIMP-H CRCs and that there are ethnic differences in the major molecular alterations associated with CRCs. Therefore, we performed a literature review to assess the frequencies of CIMP-H and BRAF mutations in CRCs, and the results are summarized in Tables 1 and 2, respectively. CIMP-H CRCs account for 7%-29.1% of all CRCs and 55.7%-82.1% of MSI-H CRCs in Western countries (Table 1)[4,13,22,24,63,84-86], whereas CIMP-H CRCs constitute 6.4%-13.9% of all CRCs and 24.2%-57.1% of MSI-H CRCs in Eastern Asian countries (Table 1)[25,97-99,101,102]. In Western countries, the BRAF V600E mutations were present in 3.7%-20.6% of all CRCs and in 18.4%-70.8% of MSI-H CRCs (Table 2)[4,13,22,24,63,79,84-86,103-112]. These proportions were lower, at 0.7%-11.4% of all CRCs and 6.2%-45.7% of MSI-H CRCs, in Eastern Asian countries (Table 2)[25,97-102,113-121]. These findings suggest that the Eastern Asian ethnicity is associated with a relatively low prevalence of CIMP-H and BRAF mutations in CRCs; consequently, these epidemiologic features may also result in a low frequency of MSI-H status in CRCs because it is thought that the majority of sporadic MSI-H CRCs are derived from CIMP-H CRCs. Thus, the low incidence of CIMP-H and BRAF mutations in Eastern Asian patients with CRC may be due to genetic or environmental differences between Eastern and Western ethnic groups. However, the detailed epidemiology and causal factors of the molecular heterogeneity of CRCs between ethnic groups should be elucidated in further investigations.
Table 2.
Frequency of BRAF V600E mutations in colorectal cancers: A review of the literature
| Ref. | Country | BRAF-mutant CRCs/tested CRCs n (%) |
|
| In all CRCs | In MSI-H CRCs | ||
| Western countries | |||
| Samowitz et al[22], 2005 | United States | 86/859 (10) | 43/78 (55.1) |
| Weisenberger et al[13], 2006 | United States | 26/187 (13.9) | NA |
| Samowitz et al[84], 2006 | United States | 123/1271 (9.7) | 67/170 (39.4) |
| Goel et al[103], 2007 | United States | 26/126 (20.6) | 17/24 (70.8) |
| Maestro et al[104], 2007 | Spain | 12/324 (3.7) | 9/49 (18.4) |
| Barault et al[85], 2008 | France | 76/578 (13.1) | 51/80 (63.8) |
| French et al[105], 2008 | United States | 77/490 (15.7) | 35/58 (60.3) |
| Ogino et al[4], 2009 | United States | 105/631 (16.6) | 53/118 (44.9) |
| Richman et al[106], 2009 | United Kingdom | 56/710 (7.9) | NA |
| Kumar et al[107], 2009 | United States (African) | 7/98 (7.1) | 7/30 (23.3) |
| Vilkin et al[108], 2009 | Israel | 24/128 (18.8) | 6/13 (46.2) |
| Roth et al[79], 2010 | Europe | 103/1307 (7.9) | 45/188 (23.9) |
| Dahlin et al[86], 2010 | Sweden | 55/411 (13.4) | 34/61 (55.7) |
| Zlobec et al[24], 2011 | Switzerland | 42/314 (13.4) | NA |
| Tie et al[109], 2011 | Australia | 52/525 (9.9) | 24/75 (32) |
| Yamauchi et al[110], 2012 | United States | 183/1276 (14.3) | NA |
| Kalady et al[111], 2012 | United States | 56/475 (11.8) | 29/76 (38.2) |
| Tian et al[112], 2013 | The Netherland and Spain | 42/381 (11) | NA |
| Lochhead et al[63], 2013 | United States | 182/1253 (14.5) | 101/193 (52.3) |
| Eastern countries | |||
| Koinuma et al[101], 2004 | Japan | 16/140 (11.4) | 12/28 (42.9) |
| Nagasaka et al[113], 2004 | Japan and Australia | 21/234 (9) | 16/35 (45.7) |
| Chang et al[114], 2006 | Taiwan | 9/213 (4.2) | 7/19 (36.8) |
| Nagasaka et al[102], 2008 | Japan and Germany | 20/243 (8.2) | 10/36 (27.8) |
| Kim et al[98], 2009 | South Korea | 13/271 (4.8) | 3/33 (9.1) |
| Yagi et al[115], 2010 | Japan | 13/149 (8.7) | NA |
| Shen et al[116], 2011 | China | 2/118 (1.7) | NA |
| Liou et al[117], 2011 | Taiwan | 12/314 (3.8) | NA |
| Yokota et al[118], 2011 | Japan | 15/229 (6.6) | NA |
| Aoyagi et al[119], 2011 | Japan | 1/134 (0.7) | NA |
| Kwon et al[100], 2011 | South Korea | 4/92 (4.3) | NA |
| Min et al[99], 2011 | South Korea | 11/245 (4.5) | 6/49 (12.2) |
| Hsieh et al[120], 2012 | Taiwan | 2/182 (1.1) | NA |
| Nakanishi et al[121], 2012 | Japan | 17/254 (6.7) | 11/31 (35.5) |
| Bae et al[97], 2013 | South Korea | 39/728 (5.4) | 4/65 (6.2) |
| Kim et al[25], 2013 | South Korea | NA | 26/219 (11.9) |
CRC: Colorectal cancer; MSI: Microsatellite instability; NA: Not applicable.
PROGNOSTIC HETEROGENEITY OF MSI-H CRC
Prognostic features and chemotherapy responses of MSI-H CRC
Although several previous investigations have failed to identify prognostic significance of MSI status in CRCs[18,122], it has been consistently reported that patients with MSI-H CRC show a better survival than MSI-L/MSS CRC patients[15,16]. However, there has been controversy regarding the predictive value of MSI status for the response to adjuvant chemotherapy in patients with CRC[122-124]. Regardless of the controversy, it is generally agreed that there are fewer or no beneficial effects of adjuvant chemotherapy, especially 5-FU-based chemotherapy, for patients with MSI-H CRC compared to patients with MSI-L/MSS CRC[17,19,125-127]. According to previous in vitro experiments, the preservation of MMR function in cancer cells is likely important for inducing the apoptotic effect of 5-FU[128-131], and this finding could explain the molecular basis of resistance to 5-FU-based chemotherapy in MSI-H CRCs. In contrast to the tendencies towards a poor response to 5-FU, and although the findings remain controversial[132], several studies supporting MSI-H as a predictive factor for improved response to irinotecan or irinotecan-based chemotherapy in CRC patients have been reported[133,134]. In previous experiments, it has also been suggested that mutations of MRE11 and/or RAD50 found in MSI-H CRC cells could account for increased sensitivity to irinotecan[135,136]. Concerning the response to the leucovorin/5-FU/oxaliplatin (FOLFOX) regimen in CRC patients, a few investigations have suggested that MSI is associated with improved survival in CRC patients who are treated with adjuvant FOLFOX[137], whereas other studies have reported that MSI status was not significantly associated with a survival benefit of CRC patients after adjuvant FOLFOX treatment[138-141]. Collectively, in patients with CRC, although MSI-H can indicate a better prognosis than MSI-L/MSS status, whether there is an association between MSI status and response to adjuvant chemotherapy remains controversial. Thus, further investigation is needed to confirm the predictive value of MSI status regarding responses to various chemotherapy regimens for CRCs.
Putative prognostic or predictive molecular markers for MSI-H CRC
Although MSI is known to be a molecular factor indicating a favorable prognosis in CRCs, we hypothesized that prognostic heterogeneity based on molecular heterogeneity might be present in MSI-H CRCs. Therefore, we expected that molecular markers stratifying patient prognosis or predicting chemotherapy response among MSI-H CRCs could be identified (Figure 1). By performing a review of the literature, potential prognostic or predictive molecular markers in MSI-H CRC were identified and are summarized in Table 3. These markers are introduced and discussed in following sections.
Figure 1.
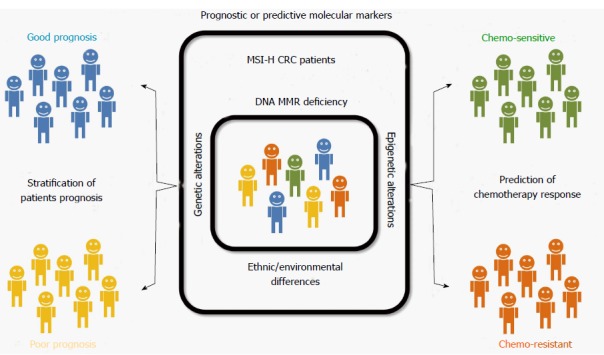
Graphical summary of this review, consisting of a conceptual model for prognostic heterogeneity based on the molecular heterogeneity of microsatellite-unstable colorectal cancers. CRC: Colorectal cancer; MSI-H: Microsatellite instability-high; MMR: Mismatch repair.
Table 3.
Potential prognostic molecular factors for microsatellite instability-high colorectal cancer: A review of the literature
| Molecular factors | Prognostic implication in MSI-H CRC (molecular alteration) | Ref. |
| HSP110 | Favorable (high expression of mutant HSP110/low expression of wild-type HSP110) | Dorard et al[59], 2011 |
| Kim et al[96], 2014 | ||
| Beta2- microglobulin | Favorable (mutation of beta2-microglobulin) | Kloor et al[54], 2007 |
| Tikidzhieva et al[143], 2012 | ||
| Myosin 1a | Unfavorable (low expression of myosin 1a) | Mazzolini et al[53], 2012 |
| CDX2/CK20 | Unfavorable (loss of CDX2/CK20 expression) | Kim et al[149], 2013 |
| SMAD4 | Favorable (high expression of SMAD4) | Isaksson-Mettavainio et al[150], 2012 |
| CIMP | Unfavorable (CIMP-H) | Bae et al[87], 2011 |
| LINE-1 | Unfavorable (low LINE-1 methylation level) | Rhee et al[162], 2012 |
CIMP: CpG island methylator phenotype; CIMP-H: CIMP-high; CRC: Colorectal cancer; LINE-1: Long interspersed nucleotide element-1; MSI-H: Microsatellite instability-high; HSP110: Heat shock protein 110 kDa; CDX2: Caudal-type homeobox 2; CK20: Cytokeratin 20; SMAD4: SMAD family member 4.
HSP110: Dorard et al[59] recently reported that the expression level of mutant HSP110 (heat shock protein 110 kDa) is significantly associated with prognosis and chemotherapy response in MSI-H CRCs. According to this study, the T17 mononucleotide repeat located within intron 8 of HSP110 is vulnerable to deletions under the dMMR condition in CRCs, and these deletions can lead to exon 9 skipping and the generation of a truncated mutant HSP110 (HSP110ΔE9). MSI-H CRC patients with a high mRNA expression level of HSP110ΔE9 survived longer, and this improved survival was maintained in both stage III and adjuvant chemotherapy-treated subgroups[59]. In our recent study, we evaluated the expression status of wild-type HSP110 (HSP110wt) by IHC in MSI-H CRCs[96] and found that reduced expression of HSP110wt was correlated with a large deletion in the HSP110 T17 repeat and favorable prognosis in MSI-H CRCs, which is reasonable because the HSP110wt expression level is expected to be inversely correlated with the HSP110ΔE9 expression level. Mutation of HSP110 and variation in HSP110 expression are representative of the molecular heterogeneity associated with the prognostic heterogeneity of MSI-H CRCs, and it is expected that these molecular alterations could be used as predictive markers and therapeutic targets in MSI-H CRCs[142].
Beta2-microglobulin: According to recent investigations, coding microsatellite mutations in the beta2-microglobulin (B2M) gene occur in approximately 30% of MSI-H CRCs and are significantly associated with a low risk of disease relapse and a low frequency of distant metastasis in MSI-H CRCs[54,143]. Although the molecular mechanism underlying how B2M mutations affect prognosis in MSI-H CRCs is not fully understood, the biological functions of B2M may be associated with the metastatic potential of cancer cells, based on results demonstrating that B2M can induce epithelial-mesenchymal transition in cancer cells and may mediate bone metastasis of cancers[144]. As a putative prognostic marker and potential therapeutic target, the functional and prognostic significance of B2M mutations in MSI-H CRCs should be further evaluated.
Myosin 1a: A recent study by Mazzolini et al[53] reported that the brush border protein myosin 1a (MYO1A) could act as a tumor suppressor in the intestine, and frameshift mutations in the MYO1A gene were detected in 32% of MSI-H CRCs. Interestingly, according to this study, a low expression level of MYO1A was associated with worse survival in patients with MSI-H CRCs, and MYO1A expression was identified as an independent prognostic factor in MSI-H CRCs. However, there is a lack of data elucidating the biological and clinicopathological significance of reduced MYO1A expression in CRCs; additional experimental and clinical studies are therefore needed.
CDX2/CK20: CDX2 (caudal-type homeobox 2) and CK20 (cytokeratin 20) are proteins associated with intestinal differentiation and are also important markers of the normal intestinal epithelium and CRCs. Several previous studies identified that a loss of CDX2 and/or CK20 expression in CRCs was associated with MSI-H or CIMP-H status[24,145-148]. In a recent investigation, we found that a loss of CDX2/CK20 expression was significantly associated with poor differentiation, CIMP-H status, and an unfavorable prognosis in MSI-H CRCs[149]. According to our study, CRC patients with simultaneous loss of CDX2 and CK20 expression in tumor tissue constituted a highly aggressive subgroup of MSI-H CRC patients, with early death or recurrence occurring in this subgroup. Although CDX2/CK20 loss is not a specific molecular alteration associated with MSI-H CRC, these molecular factors can likely be used as markers to classify patients with MSI-H CRCs into prognostic subgroups.
SMAD4: In a recent investigation by Isaksson-Mettävainio et al[150], high SMAD4 (SMAD family member 4) expression was significantly correlated with a favorable prognosis in MSI-H CRCs. Previous studies have also revealed that a loss of SMAD4 expression is associated with advanced stage, metastatic potential and an adverse prognosis in CRCs[151-154]. In fact, loss of SMAD4 has been shown to be associated with 18q LOH[154,155] and may not be correlated with MSI status in CRCs because 18q LOH is a characteristic molecular alteration in the CIN pathway. Although it is thought that the prognostic implication of SMAD4 expression can be applied to all CRCs, a variation in SMAD4 expression and its significance for the prognosis of MSI-H CRCs remains an interesting field of study. Therefore, the underlying mechanism and prognostic value of variations in SMAD4 expression in MSI-H CRCs should be further explored.
CIMP: The prognostic value of CIMP status in CRCs remains unclear. A few previous studies reported that the CIMP-H subtype was associated with poor prognosis in patients with CRC; however, this adverse effect of CIMP-H on CRC prognosis was significant only in the MSS patient subgroup and not in the MSI-H patient subgroup[86,98]. However, Ogino et al[4] provided contrasting data showing that CIMP-H was associated with a low cancer-specific mortality in CRC patients, regardless of both MSI status and BRAF mutations. In addition, although there is a lack of data elucidating the prognostic significance of CIMP-L in CRCs, a study by Dahlin et al[86] found that the CIMP-L subtype was associated with an unfavorable prognosis for CRCs, regardless of MSI status. The dependence of a chemotherapeutic response on CIMP status in CRCs is also controversial. Some investigators have suggested that CIMP-H is associated with a survival benefit in CRC patients receiving 5-FU-based chemotherapy[99,156], whereas the opposite results have also been reported[157,158]. Focusing on the prognostic implication of CIMP for MSI-H CRCs, we previously reported that for MSI-H CRC patients, those with CIMP-H tumors had worse survival than those with CIMP-L/0 tumors[87]. In fact, it is suspected that the differences in survival according to CIMP status in patients with MSI-H CRC may reflect differences in age distribution; in particular, patients with sporadic MSI-H CRC, who are expected to have CIMP-H status, are older and may have various comorbidities. In contrast, those with Lynch syndrome CRC, who have a CIMP-L/0 status, are younger and may be relatively healthy. This age distribution may critically affect patient prognosis, and thus, the prognostic significance of CIMP status in MSI-H CRC may be partly influenced by this age effect. Currently, it is debated whether CIMP status can serve as a true prognostic factor for MSI-H CRCs; therefore, more attention must be paid to analyzing the prognostic and predictive effect of CIMP status in CRCs.
LINE-1 methylation: Several recent investigations have revealed that a low LINE-1 methylation level is independently associated with an adverse prognosis for CRCs[159-161]. According to one of our studies[162], this prognostic significance of LINE-1 methylation was also maintained in MSI-H CRCs. A low LINE-1 methylation level was an independent factor indicating poor prognosis in MSI-H CRC. These findings indicate that the LINE-1 methylation level can be a useful molecular factor for selecting the poor prognostic subgroup among patients with MSI-H CRC, which is known to be associated with a favorable prognosis, despite the level of LINE-1 hypomethylation being mild in MSI-H CRCs, as described above[92,93].
CONCLUSION
MSI-H CRCs have been characterized as demonstrating a favorable prognosis and low benefit of adjuvant chemotherapy. However, MSI-H CRCs are molecularly heterogeneous tumors; thus, it is strongly suspected that prognostic and predictive molecular factors may be present. To date, several molecular factors, including HSP110, B2M, MYO1A, CDX2/CK20, SMAD4, CIMP, and LINE-1, have been explored as potential prognostic markers for MSI-H CRCs. However, additional investigations are necessary to identify the molecular determinants of patient prognosis and therapeutic response in MSI-H CRCs.
Footnotes
Supported by The National R&D Program for Cancer Control funded by the Ministry of Health and Welfare, South Korea, No. 0720540; the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIP), No. 2011-0030768; Priority Research Centers Program through the NRF grant funded by the Ministry of Education, Science and Technology (MEST), South Korea, No. 2009-0093820; and the Mid-career Researcher Program through the NRF grant funded by MEST, No. 2011-0015646
P- Reviewers: Sipos F, Yu B S- Editor: Gou SX L- Editor: A E- Editor: Wu HL
References
- 1.Aaltonen LA, Salovaara R, Kristo P, Canzian F, Hemminki A, Peltomäki P, Chadwick RB, Kääriäinen H, Eskelinen M, Järvinen H, et al. Incidence of hereditary nonpolyposis colorectal cancer and the feasibility of molecular screening for the disease. N Engl J Med. 1998;338:1481–1487. doi: 10.1056/NEJM199805213382101. [DOI] [PubMed] [Google Scholar]
- 2.Salovaara R, Loukola A, Kristo P, Kääriäinen H, Ahtola H, Eskelinen M, Härkönen N, Julkunen R, Kangas E, Ojala S, et al. Population-based molecular detection of hereditary nonpolyposis colorectal cancer. J Clin Oncol. 2000;18:2193–2200. doi: 10.1200/JCO.2000.18.11.2193. [DOI] [PubMed] [Google Scholar]
- 3.Hampel H, Frankel WL, Martin E, Arnold M, Khanduja K, Kuebler P, Nakagawa H, Sotamaa K, Prior TW, Westman J, et al. Screening for the Lynch syndrome (hereditary nonpolyposis colorectal cancer) N Engl J Med. 2005;352:1851–1860. doi: 10.1056/NEJMoa043146. [DOI] [PubMed] [Google Scholar]
- 4.Ogino S, Nosho K, Kirkner GJ, Kawasaki T, Meyerhardt JA, Loda M, Giovannucci EL, Fuchs CS. CpG island methylator phenotype, microsatellite instability, BRAF mutation and clinical outcome in colon cancer. Gut. 2009;58:90–96. doi: 10.1136/gut.2008.155473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jin HY, Liu X, Li VK, Ding Y, Yang B, Geng J, Lai R, Ding S, Ni M, Zhao R. Detection of mismatch repair gene germline mutation carrier among Chinese population with colorectal cancer. BMC Cancer. 2008;8:44. doi: 10.1186/1471-2407-8-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oh JR, Kim DW, Lee HS, Lee HE, Lee SM, Jang JH, Kang SB, Ku JL, Jeong SY, Park JG. Microsatellite instability testing in Korean patients with colorectal cancer. Fam Cancer. 2012;11:459–466. doi: 10.1007/s10689-012-9536-4. [DOI] [PubMed] [Google Scholar]
- 7.Lynch HT, Lynch PM, Lanspa SJ, Snyder CL, Lynch JF, Boland CR. Review of the Lynch syndrome: history, molecular genetics, screening, differential diagnosis, and medicolegal ramifications. Clin Genet. 2009;76:1–18. doi: 10.1111/j.1399-0004.2009.01230.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pino MS, Chung DC. Microsatellite instability in the management of colorectal cancer. Expert Rev Gastroenterol Hepatol. 2011;5:385–399. doi: 10.1586/egh.11.25. [DOI] [PubMed] [Google Scholar]
- 9.de la Chapelle A, Hampel H. Clinical relevance of microsatellite instability in colorectal cancer. J Clin Oncol. 2010;28:3380–3387. doi: 10.1200/JCO.2009.27.0652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boland CR, Thibodeau SN, Hamilton SR, Sidransky D, Eshleman JR, Burt RW, Meltzer SJ, Rodriguez-Bigas MA, Fodde R, Ranzani GN, et al. A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res. 1998;58:5248–5257. [PubMed] [Google Scholar]
- 11.Jenkins MA, Hayashi S, O’Shea AM, Burgart LJ, Smyrk TC, Shimizu D, Waring PM, Ruszkiewicz AR, Pollett AF, Redston M, et al. Pathology features in Bethesda guidelines predict colorectal cancer microsatellite instability: a population-based study. Gastroenterology. 2007;133:48–56. doi: 10.1053/j.gastro.2007.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gatalica Z, Torlakovic E. Pathology of the hereditary colorectal carcinoma. Fam Cancer. 2008;7:15–26. doi: 10.1007/s10689-007-9146-8. [DOI] [PubMed] [Google Scholar]
- 13.Weisenberger DJ, Siegmund KD, Campan M, Young J, Long TI, Faasse MA, Kang GH, Widschwendter M, Weener D, Buchanan D, et al. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet. 2006;38:787–793. doi: 10.1038/ng1834. [DOI] [PubMed] [Google Scholar]
- 14.Deng G, Bell I, Crawley S, Gum J, Terdiman JP, Allen BA, Truta B, Sleisenger MH, Kim YS. BRAF mutation is frequently present in sporadic colorectal cancer with methylated hMLH1, but not in hereditary nonpolyposis colorectal cancer. Clin Cancer Res. 2004;10:191–195. doi: 10.1158/1078-0432.ccr-1118-3. [DOI] [PubMed] [Google Scholar]
- 15.Popat S, Hubner R, Houlston RS. Systematic review of microsatellite instability and colorectal cancer prognosis. J Clin Oncol. 2005;23:609–618. doi: 10.1200/JCO.2005.01.086. [DOI] [PubMed] [Google Scholar]
- 16.Guastadisegni C, Colafranceschi M, Ottini L, Dogliotti E. Microsatellite instability as a marker of prognosis and response to therapy: a meta-analysis of colorectal cancer survival data. Eur J Cancer. 2010;46:2788–2798. doi: 10.1016/j.ejca.2010.05.009. [DOI] [PubMed] [Google Scholar]
- 17.Benatti P, Gafà R, Barana D, Marino M, Scarselli A, Pedroni M, Maestri I, Guerzoni L, Roncucci L, Menigatti M, et al. Microsatellite instability and colorectal cancer prognosis. Clin Cancer Res. 2005;11:8332–8340. doi: 10.1158/1078-0432.CCR-05-1030. [DOI] [PubMed] [Google Scholar]
- 18.Jover R, Zapater P, Castells A, Llor X, Andreu M, Cubiella J, Balaguer F, Sempere L, Xicola RM, Bujanda L, et al. The efficacy of adjuvant chemotherapy with 5-fluorouracil in colorectal cancer depends on the mismatch repair status. Eur J Cancer. 2009;45:365–373. doi: 10.1016/j.ejca.2008.07.016. [DOI] [PubMed] [Google Scholar]
- 19.Des Guetz G, Schischmanoff O, Nicolas P, Perret GY, Morere JF, Uzzan B. Does microsatellite instability predict the efficacy of adjuvant chemotherapy in colorectal cancer? A systematic review with meta-analysis. Eur J Cancer. 2009;45:1890–1896. doi: 10.1016/j.ejca.2009.04.018. [DOI] [PubMed] [Google Scholar]
- 20.Pritchard CC, Grady WM. Colorectal cancer molecular biology moves into clinical practice. Gut. 2011;60:116–129. doi: 10.1136/gut.2009.206250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grady WM, Carethers JM. Genomic and epigenetic instability in colorectal cancer pathogenesis. Gastroenterology. 2008;135:1079–1099. doi: 10.1053/j.gastro.2008.07.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Samowitz WS, Albertsen H, Herrick J, Levin TR, Sweeney C, Murtaugh MA, Wolff RK, Slattery ML. Evaluation of a large, population-based sample supports a CpG island methylator phenotype in colon cancer. Gastroenterology. 2005;129:837–845. doi: 10.1053/j.gastro.2005.06.020. [DOI] [PubMed] [Google Scholar]
- 23.Ogino S, Cantor M, Kawasaki T, Brahmandam M, Kirkner GJ, Weisenberger DJ, Campan M, Laird PW, Loda M, Fuchs CS. CpG island methylator phenotype (CIMP) of colorectal cancer is best characterised by quantitative DNA methylation analysis and prospective cohort studies. Gut. 2006;55:1000–1006. doi: 10.1136/gut.2005.082933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zlobec I, Bihl M, Foerster A, Rufle A, Lugli A. Comprehensive analysis of CpG island methylator phenotype (CIMP)-high, -low, and -negative colorectal cancers based on protein marker expression and molecular features. J Pathol. 2011;225:336–343. doi: 10.1002/path.2879. [DOI] [PubMed] [Google Scholar]
- 25.Kim JH, Rhee YY, Bae JM, Kwon HJ, Cho NY, Kim MJ, Kang GH. Subsets of microsatellite-unstable colorectal cancers exhibit discordance between the CpG island methylator phenotype and MLH1 methylation status. Mod Pathol. 2013;26:1013–1022. doi: 10.1038/modpathol.2012.241. [DOI] [PubMed] [Google Scholar]
- 26.Ligtenberg MJ, Kuiper RP, Chan TL, Goossens M, Hebeda KM, Voorendt M, Lee TY, Bodmer D, Hoenselaar E, Hendriks-Cornelissen SJ, et al. Heritable somatic methylation and inactivation of MSH2 in families with Lynch syndrome due to deletion of the 3’ exons of TACSTD1. Nat Genet. 2009;41:112–117. doi: 10.1038/ng.283. [DOI] [PubMed] [Google Scholar]
- 27.Kovacs ME, Papp J, Szentirmay Z, Otto S, Olah E. Deletions removing the last exon of TACSTD1 constitute a distinct class of mutations predisposing to Lynch syndrome. Hum Mutat. 2009;30:197–203. doi: 10.1002/humu.20942. [DOI] [PubMed] [Google Scholar]
- 28.Ligtenberg MJ, Kuiper RP, Geurts van Kessel A, Hoogerbrugge N. EPCAM deletion carriers constitute a unique subgroup of Lynch syndrome patients. Fam Cancer. 2013;12:169–174. doi: 10.1007/s10689-012-9591-x. [DOI] [PubMed] [Google Scholar]
- 29.Chan TL, Yuen ST, Kong CK, Chan YW, Chan AS, Ng WF, Tsui WY, Lo MW, Tam WY, Li VS, et al. Heritable germline epimutation of MSH2 in a family with hereditary nonpolyposis colorectal cancer. Nat Genet. 2006;38:1178–1183. doi: 10.1038/ng1866. [DOI] [PubMed] [Google Scholar]
- 30.Hitchins M, Williams R, Cheong K, Halani N, Lin VA, Packham D, Ku S, Buckle A, Hawkins N, Burn J, et al. MLH1 germline epimutations as a factor in hereditary nonpolyposis colorectal cancer. Gastroenterology. 2005;129:1392–1399. doi: 10.1053/j.gastro.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 31.Suter CM, Martin DI, Ward RL. Germline epimutation of MLH1 in individuals with multiple cancers. Nat Genet. 2004;36:497–501. doi: 10.1038/ng1342. [DOI] [PubMed] [Google Scholar]
- 32.Mueller J, Gazzoli I, Bandipalliam P, Garber JE, Syngal S, Kolodner RD. Comprehensive molecular analysis of mismatch repair gene defects in suspected Lynch syndrome (hereditary nonpolyposis colorectal cancer) cases. Cancer Res. 2009;69:7053–7061. doi: 10.1158/0008-5472.CAN-09-0358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goel A, Nguyen TP, Leung HC, Nagasaka T, Rhees J, Hotchkiss E, Arnold M, Banerji P, Koi M, Kwok CT, et al. De novo constitutional MLH1 epimutations confer early-onset colorectal cancer in two new sporadic Lynch syndrome cases, with derivation of the epimutation on the paternal allele in one. Int J Cancer. 2011;128:869–878. doi: 10.1002/ijc.25422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hitchins MP, Owens SE, Kwok CT, Godsmark G, Algar UF, Ramesar RS. Identification of new cases of early-onset colorectal cancer with an MLH1 epimutation in an ethnically diverse South African cohort. Clin Genet. 2011;80:428–434. doi: 10.1111/j.1399-0004.2011.01660.x. [DOI] [PubMed] [Google Scholar]
- 35.Boland CR, Goel A. Microsatellite instability in colorectal cancer. Gastroenterology. 2010;138:2073–2087.e3. doi: 10.1053/j.gastro.2009.12.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Geiersbach KB, Samowitz WS. Microsatellite instability and colorectal cancer. Arch Pathol Lab Med. 2011;135:1269–1277. doi: 10.5858/arpa.2011-0035-RA. [DOI] [PubMed] [Google Scholar]
- 37.Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parsons R, Myeroff LL, Liu B, Willson JK, Markowitz SD, Kinzler KW, Vogelstein B. Microsatellite instability and mutations of the transforming growth factor beta type II receptor gene in colorectal cancer. Cancer Res. 1995;55:5548–5550. [PubMed] [Google Scholar]
- 39.Rampino N, Yamamoto H, Ionov Y, Li Y, Sawai H, Reed JC, Perucho M. Somatic frameshift mutations in the BAX gene in colon cancers of the microsatellite mutator phenotype. Science. 1997;275:967–969. doi: 10.1126/science.275.5302.967. [DOI] [PubMed] [Google Scholar]
- 40.Hempen PM, Zhang L, Bansal RK, Iacobuzio-Donahue CA, Murphy KM, Maitra A, Vogelstein B, Whitehead RH, Markowitz SD, Willson JK, et al. Evidence of selection for clones having genetic inactivation of the activin A type II receptor (ACVR2) gene in gastrointestinal cancers. Cancer Res. 2003;63:994–999. [PubMed] [Google Scholar]
- 41.Calin GA, Gafà R, Tibiletti MG, Herlea V, Becheanu G, Cavazzini L, Barbanti-Brodano G, Nenci I, Negrini M, Lanza G. Genetic progression in microsatellite instability high (MSI-H) colon cancers correlates with clinico-pathological parameters: A study of the TGRbetaRII, BAX, hMSH3, hMSH6, IGFIIR and BLM genes. Int J Cancer. 2000;89:230–235. [PubMed] [Google Scholar]
- 42.Ikeda M, Orimo H, Moriyama H, Nakajima E, Matsubara N, Mibu R, Tanaka N, Shimada T, Kimura A, Shimizu K. Close correlation between mutations of E2F4 and hMSH3 genes in colorectal cancers with microsatellite instability. Cancer Res. 1998;58:594–598. [PubMed] [Google Scholar]
- 43.Shin KH, Park YJ, Park JG. PTEN gene mutations in colorectal cancers displaying microsatellite instability. Cancer Lett. 2001;174:189–194. doi: 10.1016/s0304-3835(01)00691-7. [DOI] [PubMed] [Google Scholar]
- 44.Woerner SM, Kloor M, Schwitalle Y, Youmans H, Doeberitz Mv, Gebert J, Dihlmann S. The putative tumor suppressor AIM2 is frequently affected by different genetic alterations in microsatellite unstable colon cancers. Genes Chromosomes Cancer. 2007;46:1080–1089. doi: 10.1002/gcc.20493. [DOI] [PubMed] [Google Scholar]
- 45.Trojan J, Brieger A, Raedle J, Weber N, Kriener S, Kronenberger B, Caspary WF, Zeuzem S. BAX and caspase-5 frameshift mutations and spontaneous apoptosis in colorectal cancer with microsatellite instability. Int J Colorectal Dis. 2004;19:538–544. doi: 10.1007/s00384-004-0597-1. [DOI] [PubMed] [Google Scholar]
- 46.Bader S, Walker M, Hendrich B, Bird A, Bird C, Hooper M, Wyllie A. Somatic frameshift mutations in the MBD4 gene of sporadic colon cancers with mismatch repair deficiency. Oncogene. 1999;18:8044–8047. doi: 10.1038/sj.onc.1203229. [DOI] [PubMed] [Google Scholar]
- 47.Duval A, Gayet J, Zhou XP, Iacopetta B, Thomas G, Hamelin R. Frequent frameshift mutations of the TCF-4 gene in colorectal cancers with microsatellite instability. Cancer Res. 1999;59:4213–4215. [PubMed] [Google Scholar]
- 48.Nakagawa H, Koyama K, Nakamori S, Kameyama M, Imaoka S, Monden M, Nakamura Y. Frameshift mutation of the STK11 gene in a sporadic gastrointestinal cancer with microsatellite instability. Jpn J Cancer Res. 1999;90:633–637. doi: 10.1111/j.1349-7006.1999.tb00794.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim NG, Choi YR, Baek MJ, Kim YH, Kang H, Kim NK, Min JS, Kim H. Frameshift mutations at coding mononucleotide repeats of the hRAD50 gene in gastrointestinal carcinomas with microsatellite instability. Cancer Res. 2001;61:36–38. doi: 10.1186/bcr362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim CJ, Lee JH, Song JW, Cho YG, Kim SY, Nam SW, Yoo NJ, Park WS, Lee JY. Chk1 frameshift mutation in sporadic and hereditary non-polyposis colorectal cancers with microsatellite instability. Eur J Surg Oncol. 2007;33:580–585. doi: 10.1016/j.ejso.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 51.Thorstensen L, Lind GE, Løvig T, Diep CB, Meling GI, Rognum TO, Lothe RA. Genetic and epigenetic changes of components affecting the WNT pathway in colorectal carcinomas stratified by microsatellite instability. Neoplasia. 2005;7:99–108. doi: 10.1593/neo.04448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wicking C, Simms LA, Evans T, Walsh M, Chawengsaksophak K, Beck F, Chenevix-Trench G, Young J, Jass J, Leggett B, et al. CDX2, a human homologue of Drosophila caudal, is mutated in both alleles in a replication error positive colorectal cancer. Oncogene. 1998;17:657–659. doi: 10.1038/sj.onc.1201971. [DOI] [PubMed] [Google Scholar]
- 53.Mazzolini R, Dopeso H, Mateo-Lozano S, Chang W, Rodrigues P, Bazzocco S, Alazzouzi H, Landolfi S, Hernández-Losa J, Andretta E, et al. Brush border myosin Ia has tumor suppressor activity in the intestine. Proc Natl Acad Sci USA. 2012;109:1530–1535. doi: 10.1073/pnas.1108411109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kloor M, Michel S, Buckowitz B, Rüschoff J, Büttner R, Holinski-Feder E, Dippold W, Wagner R, Tariverdian M, Benner A, et al. Beta2-microglobulin mutations in microsatellite unstable colorectal tumors. Int J Cancer. 2007;121:454–458. doi: 10.1002/ijc.22691. [DOI] [PubMed] [Google Scholar]
- 55.Jung B, Smith EJ, Doctolero RT, Gervaz P, Alonso JC, Miyai K, Keku T, Sandler RS, Carethers JM. Influence of target gene mutations on survival, stage and histology in sporadic microsatellite unstable colon cancers. Int J Cancer. 2006;118:2509–2513. doi: 10.1002/ijc.21710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fernández-Peralta AM, Nejda N, Oliart S, Medina V, Azcoita MM, González-Aguilera JJ. Significance of mutations in TGFBR2 and BAX in neoplastic progression and patient outcome in sporadic colorectal tumors with high-frequency microsatellite instability. Cancer Genet Cytogenet. 2005;157:18–24. doi: 10.1016/j.cancergencyto.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 57.Shima K, Morikawa T, Yamauchi M, Kuchiba A, Imamura Y, Liao X, Meyerhardt JA, Fuchs CS, Ogino S. TGFBR2 and BAX mononucleotide tract mutations, microsatellite instability, and prognosis in 1072 colorectal cancers. PLoS One. 2011;6:e25062. doi: 10.1371/journal.pone.0025062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Samowitz WS, Curtin K, Neuhausen S, Schaffer D, Slattery ML. Prognostic implications of BAX and TGFBRII mutations in colon cancers with microsatellite instability. Genes Chromosomes Cancer. 2002;35:368–371. doi: 10.1002/gcc.10125. [DOI] [PubMed] [Google Scholar]
- 59.Dorard C, de Thonel A, Collura A, Marisa L, Svrcek M, Lagrange A, Jego G, Wanherdrick K, Joly AL, Buhard O, et al. Expression of a mutant HSP110 sensitizes colorectal cancer cells to chemotherapy and improves disease prognosis. Nat Med. 2011;17:1283–1289. doi: 10.1038/nm.2457. [DOI] [PubMed] [Google Scholar]
- 60.Giannini G, Rinaldi C, Ristori E, Ambrosini MI, Cerignoli F, Viel A, Bidoli E, Berni S, D’Amati G, Scambia G, et al. Mutations of an intronic repeat induce impaired MRE11 expression in primary human cancer with microsatellite instability. Oncogene. 2004;23:2640–2647. doi: 10.1038/sj.onc.1207409. [DOI] [PubMed] [Google Scholar]
- 61.Rajagopalan H, Bardelli A, Lengauer C, Kinzler KW, Vogelstein B, Velculescu VE. Tumorigenesis: RAF/RAS oncogenes and mismatch-repair status. Nature. 2002;418:934. doi: 10.1038/418934a. [DOI] [PubMed] [Google Scholar]
- 62.Siena S, Sartore-Bianchi A, Di Nicolantonio F, Balfour J, Bardelli A. Biomarkers predicting clinical outcome of epidermal growth factor receptor-targeted therapy in metastatic colorectal cancer. J Natl Cancer Inst. 2009;101:1308–1324. doi: 10.1093/jnci/djp280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lochhead P, Kuchiba A, Imamura Y, Liao X, Yamauchi M, Nishihara R, Qian ZR, Morikawa T, Shen J, Meyerhardt JA, et al. Microsatellite instability and BRAF mutation testing in colorectal cancer prognostication. J Natl Cancer Inst. 2013;105:1151–1156. doi: 10.1093/jnci/djt173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.De Roock W, Claes B, Bernasconi D, De Schutter J, Biesmans B, Fountzilas G, Kalogeras KT, Kotoula V, Papamichael D, Laurent-Puig P, et al. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. Lancet Oncol. 2010;11:753–762. doi: 10.1016/S1470-2045(10)70130-3. [DOI] [PubMed] [Google Scholar]
- 65.Samowitz WS, Sweeney C, Herrick J, Albertsen H, Levin TR, Murtaugh MA, Wolff RK, Slattery ML. Poor survival associated with the BRAF V600E mutation in microsatellite-stable colon cancers. Cancer Res. 2005;65:6063–6069. doi: 10.1158/0008-5472.CAN-05-0404. [DOI] [PubMed] [Google Scholar]
- 66.Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 67.Lièvre A, Bachet JB, Boige V, Cayre A, Le Corre D, Buc E, Ychou M, Bouché O, Landi B, Louvet C, et al. KRAS mutations as an independent prognostic factor in patients with advanced colorectal cancer treated with cetuximab. J Clin Oncol. 2008;26:374–379. doi: 10.1200/JCO.2007.12.5906. [DOI] [PubMed] [Google Scholar]
- 68.Lièvre A, Bachet JB, Le Corre D, Boige V, Landi B, Emile JF, Côté JF, Tomasic G, Penna C, Ducreux M, et al. KRAS mutation status is predictive of response to cetuximab therapy in colorectal cancer. Cancer Res. 2006;66:3992–3995. doi: 10.1158/0008-5472.CAN-06-0191. [DOI] [PubMed] [Google Scholar]
- 69.Edkins S, O’Meara S, Parker A, Stevens C, Reis M, Jones S, Greenman C, Davies H, Dalgliesh G, Forbes S, et al. Recurrent KRAS codon 146 mutations in human colorectal cancer. Cancer Biol Ther. 2006;5:928–932. doi: 10.4161/cbt.5.8.3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vaughn CP, Zobell SD, Furtado LV, Baker CL, Samowitz WS. Frequency of KRAS, BRAF, and NRAS mutations in colorectal cancer. Genes Chromosomes Cancer. 2011;50:307–312. doi: 10.1002/gcc.20854. [DOI] [PubMed] [Google Scholar]
- 71.Janakiraman M, Vakiani E, Zeng Z, Pratilas CA, Taylor BS, Chitale D, Halilovic E, Wilson M, Huberman K, Ricarte Filho JC, et al. Genomic and biological characterization of exon 4 KRAS mutations in human cancer. Cancer Res. 2010;70:5901–5911. doi: 10.1158/0008-5472.CAN-10-0192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Grossmann AH, Samowitz WS. Epidermal growth factor receptor pathway mutations and colorectal cancer therapy. Arch Pathol Lab Med. 2011;135:1278–1282. doi: 10.5858/arpa.2011-0047-RA. [DOI] [PubMed] [Google Scholar]
- 73.Lubomierski N, Plotz G, Wormek M, Engels K, Kriener S, Trojan J, Jungling B, Zeuzem S, Raedle J. BRAF mutations in colorectal carcinoma suggest two entities of microsatellite-unstable tumors. Cancer. 2005;104:952–961. doi: 10.1002/cncr.21266. [DOI] [PubMed] [Google Scholar]
- 74.Domingo E, Laiho P, Ollikainen M, Pinto M, Wang L, French AJ, Westra J, Frebourg T, Espín E, Armengol M, et al. BRAF screening as a low-cost effective strategy for simplifying HNPCC genetic testing. J Med Genet. 2004;41:664–668. doi: 10.1136/jmg.2004.020651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Loughrey MB, Waring PM, Tan A, Trivett M, Kovalenko S, Beshay V, Young MA, McArthur G, Boussioutas A, Dobrovic A. Incorporation of somatic BRAF mutation testing into an algorithm for the investigation of hereditary non-polyposis colorectal cancer. Fam Cancer. 2007;6:301–310. doi: 10.1007/s10689-007-9124-1. [DOI] [PubMed] [Google Scholar]
- 76.Samowitz WS, Holden JA, Curtin K, Edwards SL, Walker AR, Lin HA, Robertson MA, Nichols MF, Gruenthal KM, Lynch BJ, et al. Inverse relationship between microsatellite instability and K-ras and p53 gene alterations in colon cancer. Am J Pathol. 2001;158:1517–1524. doi: 10.1016/S0002-9440(10)64102-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.McGivern A, Wynter CV, Whitehall VL, Kambara T, Spring KJ, Walsh MD, Barker MA, Arnold S, Simms LA, Leggett BA, et al. Promoter hypermethylation frequency and BRAF mutations distinguish hereditary non-polyposis colon cancer from sporadic MSI-H colon cancer. Fam Cancer. 2004;3:101–107. doi: 10.1023/B:FAME.0000039861.30651.c8. [DOI] [PubMed] [Google Scholar]
- 78.Fariña-Sarasqueta A, van Lijnschoten G, Moerland E, Creemers GJ, Lemmens VE, Rutten HJ, van den Brule AJ. The BRAF V600E mutation is an independent prognostic factor for survival in stage II and stage III colon cancer patients. Ann Oncol. 2010;21:2396–2402. doi: 10.1093/annonc/mdq258. [DOI] [PubMed] [Google Scholar]
- 79.Roth AD, Tejpar S, Delorenzi M, Yan P, Fiocca R, Klingbiel D, Dietrich D, Biesmans B, Bodoky G, Barone C, et al. Prognostic role of KRAS and BRAF in stage II and III resected colon cancer: results of the translational study on the PETACC-3, EORTC 40993, SAKK 60-00 trial. J Clin Oncol. 2010;28:466–474. doi: 10.1200/JCO.2009.23.3452. [DOI] [PubMed] [Google Scholar]
- 80.Toyota M, Ahuja N, Ohe-Toyota M, Herman JG, Baylin SB, Issa JP. CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci USA. 1999;96:8681–8686. doi: 10.1073/pnas.96.15.8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kambara T, Simms LA, Whitehall VL, Spring KJ, Wynter CV, Walsh MD, Barker MA, Arnold S, McGivern A, Matsubara N, et al. BRAF mutation is associated with DNA methylation in serrated polyps and cancers of the colorectum. Gut. 2004;53:1137–1144. doi: 10.1136/gut.2003.037671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ogino S, Kawasaki T, Kirkner GJ, Loda M, Fuchs CS. CpG island methylator phenotype-low (CIMP-low) in colorectal cancer: possible associations with male sex and KRAS mutations. J Mol Diagn. 2006;8:582–588. doi: 10.2353/jmoldx.2006.060082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hawkins N, Norrie M, Cheong K, Mokany E, Ku SL, Meagher A, O’Connor T, Ward R. CpG island methylation in sporadic colorectal cancers and its relationship to microsatellite instability. Gastroenterology. 2002;122:1376–1387. doi: 10.1053/gast.2002.32997. [DOI] [PubMed] [Google Scholar]
- 84.Samowitz WS, Albertsen H, Sweeney C, Herrick J, Caan BJ, Anderson KE, Wolff RK, Slattery ML. Association of smoking, CpG island methylator phenotype, and V600E BRAF mutations in colon cancer. J Natl Cancer Inst. 2006;98:1731–1738. doi: 10.1093/jnci/djj468. [DOI] [PubMed] [Google Scholar]
- 85.Barault L, Charon-Barra C, Jooste V, de la Vega MF, Martin L, Roignot P, Rat P, Bouvier AM, Laurent-Puig P, Faivre J, et al. Hypermethylator phenotype in sporadic colon cancer: study on a population-based series of 582 cases. Cancer Res. 2008;68:8541–8546. doi: 10.1158/0008-5472.CAN-08-1171. [DOI] [PubMed] [Google Scholar]
- 86.Dahlin AM, Palmqvist R, Henriksson ML, Jacobsson M, Eklöf V, Rutegård J, Oberg A, Van Guelpen BR. The role of the CpG island methylator phenotype in colorectal cancer prognosis depends on microsatellite instability screening status. Clin Cancer Res. 2010;16:1845–1855. doi: 10.1158/1078-0432.CCR-09-2594. [DOI] [PubMed] [Google Scholar]
- 87.Bae JM, Kim MJ, Kim JH, Koh JM, Cho NY, Kim TY, Kang GH. Differential clinicopathological features in microsatellite instability-positive colorectal cancers depending on CIMP status. Virchows Arch. 2011;459:55–63. doi: 10.1007/s00428-011-1080-3. [DOI] [PubMed] [Google Scholar]
- 88.Weisenberger DJ, Campan M, Long TI, Kim M, Woods C, Fiala E, Ehrlich M, Laird PW. Analysis of repetitive element DNA methylation by MethyLight. Nucleic Acids Res. 2005;33:6823–6836. doi: 10.1093/nar/gki987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yang AS, Estécio MR, Doshi K, Kondo Y, Tajara EH, Issa JP. A simple method for estimating global DNA methylation using bisulfite PCR of repetitive DNA elements. Nucleic Acids Res. 2004;32:e38. doi: 10.1093/nar/gnh032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Suter CM, Martin DI, Ward RL. Hypomethylation of L1 retrotransposons in colorectal cancer and adjacent normal tissue. Int J Colorectal Dis. 2004;19:95–101. doi: 10.1007/s00384-003-0539-3. [DOI] [PubMed] [Google Scholar]
- 91.Matsuzaki K, Deng G, Tanaka H, Kakar S, Miura S, Kim YS. The relationship between global methylation level, loss of heterozygosity, and microsatellite instability in sporadic colorectal cancer. Clin Cancer Res. 2005;11:8564–8569. doi: 10.1158/1078-0432.CCR-05-0859. [DOI] [PubMed] [Google Scholar]
- 92.Estécio MR, Gharibyan V, Shen L, Ibrahim AE, Doshi K, He R, Jelinek J, Yang AS, Yan PS, Huang TH, et al. LINE-1 hypomethylation in cancer is highly variable and inversely correlated with microsatellite instability. PLoS One. 2007;2:e399. doi: 10.1371/journal.pone.0000399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ogino S, Kawasaki T, Nosho K, Ohnishi M, Suemoto Y, Kirkner GJ, Fuchs CS. LINE-1 hypomethylation is inversely associated with microsatellite instability and CpG island methylator phenotype in colorectal cancer. Int J Cancer. 2008;122:2767–2773. doi: 10.1002/ijc.23470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kim HR, Kim HC, Yun HR, Kim SH, Park CK, Cho YB, Yun SH, Lee WY, Chun HK. An alternative pathway in colorectal carcinogenesis based on the mismatch repair system and p53 expression in Korean patients with sporadic colorectal cancer. Ann Surg Oncol. 2014;20:4031–4040. doi: 10.1245/s10434-012-2455-7. [DOI] [PubMed] [Google Scholar]
- 95.Hong SP, Min BS, Kim TI, Cheon JH, Kim NK, Kim H, Kim WH. The differential impact of microsatellite instability as a marker of prognosis and tumour response between colon cancer and rectal cancer. Eur J Cancer. 2012;48:1235–1243. doi: 10.1016/j.ejca.2011.10.005. [DOI] [PubMed] [Google Scholar]
- 96.Kim JH, Kim KJ, Rhee YY, Oh S, Cho NY, Lee HS, Kang GH. Expression status of wild-type HSP110 correlates with HSP110 T17 deletion size and patient prognosis in microsatellite-unstable colorectal cancer. Mod Pathol. 2014;27:443–453. doi: 10.1038/modpathol.2013.160. [DOI] [PubMed] [Google Scholar]
- 97.Bae JM, Kim JH, Cho NY, Kim TY, Kang GH. Prognostic implication of the CpG island methylator phenotype in colorectal cancers depends on tumour location. Br J Cancer. 2013;109:1004–1012. doi: 10.1038/bjc.2013.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kim JH, Shin SH, Kwon HJ, Cho NY, Kang GH. Prognostic implications of CpG island hypermethylator phenotype in colorectal cancers. Virchows Arch. 2009;455:485–494. doi: 10.1007/s00428-009-0857-0. [DOI] [PubMed] [Google Scholar]
- 99.Min BH, Bae JM, Lee EJ, Yu HS, Kim YH, Chang DK, Kim HC, Park CK, Lee SH, Kim KM, et al. The CpG island methylator phenotype may confer a survival benefit in patients with stage II or III colorectal carcinomas receiving fluoropyrimidine-based adjuvant chemotherapy. BMC Cancer. 2011;11:344. doi: 10.1186/1471-2407-11-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kwon MJ, Lee SE, Kang SY, Choi YL. Frequency of KRAS, BRAF, and PIK3CA mutations in advanced colorectal cancers: Comparison of peptide nucleic acid-mediated PCR clamping and direct sequencing in formalin-fixed, paraffin-embedded tissue. Pathol Res Pract. 2011;207:762–768. doi: 10.1016/j.prp.2011.10.002. [DOI] [PubMed] [Google Scholar]
- 101.Koinuma K, Shitoh K, Miyakura Y, Furukawa T, Yamashita Y, Ota J, Ohki R, Choi YL, Wada T, Konishi F, et al. Mutations of BRAF are associated with extensive hMLH1 promoter methylation in sporadic colorectal carcinomas. Int J Cancer. 2004;108:237–242. doi: 10.1002/ijc.11523. [DOI] [PubMed] [Google Scholar]
- 102.Nagasaka T, Koi M, Kloor M, Gebert J, Vilkin A, Nishida N, Shin SK, Sasamoto H, Tanaka N, Matsubara N, et al. Mutations in both KRAS and BRAF may contribute to the methylator phenotype in colon cancer. Gastroenterology. 2008;134:1950–1960, 1960.e1. doi: 10.1053/j.gastro.2008.02.094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Goel A, Nagasaka T, Arnold CN, Inoue T, Hamilton C, Niedzwiecki D, Compton C, Mayer RJ, Goldberg R, Bertagnolli MM, et al. The CpG island methylator phenotype and chromosomal instability are inversely correlated in sporadic colorectal cancer. Gastroenterology. 2007;132:127–138. doi: 10.1053/j.gastro.2006.09.018. [DOI] [PubMed] [Google Scholar]
- 104.Maestro ML, Vidaurreta M, Sanz-Casla MT, Rafael S, Veganzones S, Martínez A, Aguilera C, Herranz MD, Cerdán J, Arroyo M. Role of the BRAF mutations in the microsatellite instability genetic pathway in sporadic colorectal cancer. Ann Surg Oncol. 2007;14:1229–1236. doi: 10.1245/s10434-006-9111-z. [DOI] [PubMed] [Google Scholar]
- 105.French AJ, Sargent DJ, Burgart LJ, Foster NR, Kabat BF, Goldberg R, Shepherd L, Windschitl HE, Thibodeau SN. Prognostic significance of defective mismatch repair and BRAF V600E in patients with colon cancer. Clin Cancer Res. 2008;14:3408–3415. doi: 10.1158/1078-0432.CCR-07-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Richman SD, Seymour MT, Chambers P, Elliott F, Daly CL, Meade AM, Taylor G, Barrett JH, Quirke P. KRAS and BRAF mutations in advanced colorectal cancer are associated with poor prognosis but do not preclude benefit from oxaliplatin or irinotecan: results from the MRC FOCUS trial. J Clin Oncol. 2009;27:5931–5937. doi: 10.1200/JCO.2009.22.4295. [DOI] [PubMed] [Google Scholar]
- 107.Kumar K, Brim H, Giardiello F, Smoot DT, Nouraie M, Lee EL, Ashktorab H. Distinct BRAF (V600E) and KRAS mutations in high microsatellite instability sporadic colorectal cancer in African Americans. Clin Cancer Res. 2009;15:1155–1161. doi: 10.1158/1078-0432.CCR-08-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Vilkin A, Niv Y, Nagasaka T, Morgenstern S, Levi Z, Fireman Z, Fuerst F, Goel A, Boland CR. Microsatellite instability, MLH1 promoter methylation, and BRAF mutation analysis in sporadic colorectal cancers of different ethnic groups in Israel. Cancer. 2009;115:760–769. doi: 10.1002/cncr.24019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Tie J, Gibbs P, Lipton L, Christie M, Jorissen RN, Burgess AW, Croxford M, Jones I, Langland R, Kosmider S, et al. Optimizing targeted therapeutic development: analysis of a colorectal cancer patient population with the BRAF(V600E) mutation. Int J Cancer. 2011;128:2075–2084. doi: 10.1002/ijc.25555. [DOI] [PubMed] [Google Scholar]
- 110.Yamauchi M, Morikawa T, Kuchiba A, Imamura Y, Qian ZR, Nishihara R, Liao X, Waldron L, Hoshida Y, Huttenhower C, et al. Assessment of colorectal cancer molecular features along bowel subsites challenges the conception of distinct dichotomy of proximal versus distal colorectum. Gut. 2012;61:847–854. doi: 10.1136/gutjnl-2011-300865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kalady MF, Dejulius KL, Sanchez JA, Jarrar A, Liu X, Manilich E, Skacel M, Church JM. BRAF mutations in colorectal cancer are associated with distinct clinical characteristics and worse prognosis. Dis Colon Rectum. 2012;55:128–133. doi: 10.1097/DCR.0b013e31823c08b3. [DOI] [PubMed] [Google Scholar]
- 112.Tian S, Simon I, Moreno V, Roepman P, Tabernero J, Snel M, van’t Veer L, Salazar R, Bernards R, Capella G. A combined oncogenic pathway signature of BRAF, KRAS and PI3KCA mutation improves colorectal cancer classification and cetuximab treatment prediction. Gut. 2013;62:540–549. doi: 10.1136/gutjnl-2012-302423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Nagasaka T, Sasamoto H, Notohara K, Cullings HM, Takeda M, Kimura K, Kambara T, MacPhee DG, Young J, Leggett BA, et al. Colorectal cancer with mutation in BRAF, KRAS, and wild-type with respect to both oncogenes showing different patterns of DNA methylation. J Clin Oncol. 2004;22:4584–4594. doi: 10.1200/JCO.2004.02.154. [DOI] [PubMed] [Google Scholar]
- 114.Chang SC, Lin JK, Yang SH, Wang HS, Li AF, Chi CW. Relationship between genetic alterations and prognosis in sporadic colorectal cancer. Int J Cancer. 2006;118:1721–1727. doi: 10.1002/ijc.21563. [DOI] [PubMed] [Google Scholar]
- 115.Yagi K, Akagi K, Hayashi H, Nagae G, Tsuji S, Isagawa T, Midorikawa Y, Nishimura Y, Sakamoto H, Seto Y, et al. Three DNA methylation epigenotypes in human colorectal cancer. Clin Cancer Res. 2010;16:21–33. doi: 10.1158/1078-0432.CCR-09-2006. [DOI] [PubMed] [Google Scholar]
- 116.Shen H, Yuan Y, Hu HG, Zhong X, Ye XX, Li MD, Fang WJ, Zheng S. Clinical significance of K-ras and BRAF mutations in Chinese colorectal cancer patients. World J Gastroenterol. 2011;17:809–816. doi: 10.3748/wjg.v17.i6.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Liou JM, Wu MS, Shun CT, Chiu HM, Chen MJ, Chen CC, Wang HP, Lin JT, Liang JT. Mutations in BRAF correlate with poor survival of colorectal cancers in Chinese population. Int J Colorectal Dis. 2011;26:1387–1395. doi: 10.1007/s00384-011-1229-1. [DOI] [PubMed] [Google Scholar]
- 118.Yokota T, Ura T, Shibata N, Takahari D, Shitara K, Nomura M, Kondo C, Mizota A, Utsunomiya S, Muro K, et al. BRAF mutation is a powerful prognostic factor in advanced and recurrent colorectal cancer. Br J Cancer. 2011;104:856–862. doi: 10.1038/bjc.2011.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Aoyagi H, Iida S, Uetake H, Ishikawa T, Takagi Y, Kobayashi H, Higuchi T, Yasuno M, Enomoto M, Sugihara K. Effect of classification based on combination of mutation and methylation in colorectal cancer prognosis. Oncol Rep. 2011;25:789–794. doi: 10.3892/or.2010.1118. [DOI] [PubMed] [Google Scholar]
- 120.Hsieh LL, Er TK, Chen CC, Hsieh JS, Chang JG, Liu TC. Characteristics and prevalence of KRAS, BRAF, and PIK3CA mutations in colorectal cancer by high-resolution melting analysis in Taiwanese population. Clin Chim Acta. 2012;413:1605–1611. doi: 10.1016/j.cca.2012.04.029. [DOI] [PubMed] [Google Scholar]
- 121.Nakanishi R, Harada J, Tuul M, Zhao Y, Ando K, Saeki H, Oki E, Ohga T, Kitao H, Kakeji Y, et al. Prognostic relevance of KRAS and BRAF mutations in Japanese patients with colorectal cancer. Int J Clin Oncol. 2013;18:1042–1048. doi: 10.1007/s10147-012-0501-x. [DOI] [PubMed] [Google Scholar]
- 122.Kim GP, Colangelo LH, Wieand HS, Paik S, Kirsch IR, Wolmark N, Allegra CJ. Prognostic and predictive roles of high-degree microsatellite instability in colon cancer: a National Cancer Institute-National Surgical Adjuvant Breast and Bowel Project Collaborative Study. J Clin Oncol. 2007;25:767–772. doi: 10.1200/JCO.2006.05.8172. [DOI] [PubMed] [Google Scholar]
- 123.Kim GP, Colangelo LH, Paik S, O’Connell MJ, Kirsch IR, Allegra C, Wolmark N. Predictive value of microsatellite instability-high remains controversial. J Clin Oncol. 2007;25:4857; author reply 4857–4858. doi: 10.1200/JCO.2007.13.2019. [DOI] [PubMed] [Google Scholar]
- 124.Des Guetz G, Uzzan B, Nicolas P, Schischmanoff O, Perret GY, Morere JF. Microsatellite instability does not predict the efficacy of chemotherapy in metastatic colorectal cancer. A systematic review and meta-analysis. Anticancer Res. 2009;29:1615–1620. [PubMed] [Google Scholar]
- 125.Sargent DJ, Marsoni S, Monges G, Thibodeau SN, Labianca R, Hamilton SR, French AJ, Kabat B, Foster NR, Torri V, et al. Defective mismatch repair as a predictive marker for lack of efficacy of fluorouracil-based adjuvant therapy in colon cancer. J Clin Oncol. 2010;28:3219–3226. doi: 10.1200/JCO.2009.27.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Carethers JM, Smith EJ, Behling CA, Nguyen L, Tajima A, Doctolero RT, Cabrera BL, Goel A, Arnold CA, Miyai K, et al. Use of 5-fluorouracil and survival in patients with microsatellite-unstable colorectal cancer. Gastroenterology. 2004;126:394–401. doi: 10.1053/j.gastro.2003.12.023. [DOI] [PubMed] [Google Scholar]
- 127.Ribic CM, Sargent DJ, Moore MJ, Thibodeau SN, French AJ, Goldberg RM, Hamilton SR, Laurent-Puig P, Gryfe R, Shepherd LE, et al. Tumor microsatellite-instability status as a predictor of benefit from fluorouracil-based adjuvant chemotherapy for colon cancer. N Engl J Med. 2003;349:247–257. doi: 10.1056/NEJMoa022289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Tajima A, Hess MT, Cabrera BL, Kolodner RD, Carethers JM. The mismatch repair complex hMutS alpha recognizes 5-fluorouracil-modified DNA: implications for chemosensitivity and resistance. Gastroenterology. 2004;127:1678–1684. doi: 10.1053/j.gastro.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 129.Arnold CN, Goel A, Boland CR. Role of hMLH1 promoter hypermethylation in drug resistance to 5-fluorouracil in colorectal cancer cell lines. Int J Cancer. 2003;106:66–73. doi: 10.1002/ijc.11176. [DOI] [PubMed] [Google Scholar]
- 130.Meyers M, Wagner MW, Hwang HS, Kinsella TJ, Boothman DA. Role of the hMLH1 DNA mismatch repair protein in fluoropyrimidine-mediated cell death and cell cycle responses. Cancer Res. 2001;61:5193–5201. [PubMed] [Google Scholar]
- 131.Carethers JM, Chauhan DP, Fink D, Nebel S, Bresalier RS, Howell SB, Boland CR. Mismatch repair proficiency and in vitro response to 5-fluorouracil. Gastroenterology. 1999;117:123–131. doi: 10.1016/s0016-5085(99)70558-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Kim JE, Hong YS, Ryu MH, Lee JL, Chang HM, Lim SB, Kim JH, Jang SJ, Kim MJ, Yu CS, et al. Association between deficient mismatch repair system and efficacy to irinotecan-containing chemotherapy in metastatic colon cancer. Cancer Sci. 2011;102:1706–1711. doi: 10.1111/j.1349-7006.2011.02009.x. [DOI] [PubMed] [Google Scholar]
- 133.Bertagnolli MM, Niedzwiecki D, Compton CC, Hahn HP, Hall M, Damas B, Jewell SD, Mayer RJ, Goldberg RM, Saltz LB, et al. Microsatellite instability predicts improved response to adjuvant therapy with irinotecan, fluorouracil, and leucovorin in stage III colon cancer: Cancer and Leukemia Group B Protocol 89803. J Clin Oncol. 2009;27:1814–1821. doi: 10.1200/JCO.2008.18.2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Fallik D, Borrini F, Boige V, Viguier J, Jacob S, Miquel C, Sabourin JC, Ducreux M, Praz F. Microsatellite instability is a predictive factor of the tumor response to irinotecan in patients with advanced colorectal cancer. Cancer Res. 2003;63:5738–5744. [PubMed] [Google Scholar]
- 135.Vilar E, Scaltriti M, Balmaña J, Saura C, Guzman M, Arribas J, Baselga J, Tabernero J. Microsatellite instability due to hMLH1 deficiency is associated with increased cytotoxicity to irinotecan in human colorectal cancer cell lines. Br J Cancer. 2008;99:1607–1612. doi: 10.1038/sj.bjc.6604691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Rodriguez R, Hansen LT, Phear G, Scorah J, Spang-Thomsen M, Cox A, Helleday T, Meuth M. Thymidine selectively enhances growth suppressive effects of camptothecin/irinotecan in MSI+ cells and tumors containing a mutation of MRE11. Clin Cancer Res. 2008;14:5476–5483. doi: 10.1158/1078-0432.CCR-08-0274. [DOI] [PubMed] [Google Scholar]
- 137.Zaanan A, Fléjou JF, Emile JF, Des GG, Cuilliere-Dartigues P, Malka D, Lecaille C, Validire P, Louvet C, Rougier P, et al. Defective mismatch repair status as a prognostic biomarker of disease-free survival in stage III colon cancer patients treated with adjuvant FOLFOX chemotherapy. Clin Cancer Res. 2011;17:7470–7478. doi: 10.1158/1078-0432.CCR-11-1048. [DOI] [PubMed] [Google Scholar]
- 138.Han SW, Lee HJ, Bae JM, Cho NY, Lee KH, Kim TY, Oh DY, Im SA, Bang YJ, Jeong SY, et al. Methylation and microsatellite status and recurrence following adjuvant FOLFOX in colorectal cancer. Int J Cancer. 2013;132:2209–2216. doi: 10.1002/ijc.27888. [DOI] [PubMed] [Google Scholar]
- 139.Kim ST, Lee J, Park SH, Park JO, Lim HY, Kang WK, Kim JY, Kim YH, Chang DK, Rhee PL, et al. Clinical impact of microsatellite instability in colon cancer following adjuvant FOLFOX therapy. Cancer Chemother Pharmacol. 2010;66:659–667. doi: 10.1007/s00280-009-1206-3. [DOI] [PubMed] [Google Scholar]
- 140.Li P, Fang YJ, Li F, Ou QJ, Chen G, Ma G. ERCC1, defective mismatch repair status as predictive biomarkers of survival for stage III colon cancer patients receiving oxaliplatin-based adjuvant chemotherapy. Br J Cancer. 2013;108:1238–1244. doi: 10.1038/bjc.2013.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Des Guetz G, Lecaille C, Mariani P, Bennamoun M, Uzzan B, Nicolas P, Boisseau A, Sastre X, Cucherousset J, Lagorce C, et al. Prognostic impact of microsatellite instability in colorectal cancer patients treated with adjuvant FOLFOX. Anticancer Res. 2010;30:4297–4301. [PubMed] [Google Scholar]
- 142.Chan AT. Turning up the heat on colorectal cancer. Nat Med. 2011;17:1186–1188. doi: 10.1038/nm.2500. [DOI] [PubMed] [Google Scholar]
- 143.Tikidzhieva A, Benner A, Michel S, Formentini A, Link KH, Dippold W, von Knebel Doeberitz M, Kornmann M, Kloor M. Microsatellite instability and Beta2-Microglobulin mutations as prognostic markers in colon cancer: results of the FOGT-4 trial. Br J Cancer. 2012;106:1239–1245. doi: 10.1038/bjc.2012.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Josson S, Nomura T, Lin JT, Huang WC, Wu D, Zhau HE, Zayzafoon M, Weizmann MN, Gururajan M, Chung LW. β2-microglobulin induces epithelial to mesenchymal transition and confers cancer lethality and bone metastasis in human cancer cells. Cancer Res. 2011;71:2600–2610. doi: 10.1158/0008-5472.CAN-10-3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Hinoi T, Tani M, Lucas PC, Caca K, Dunn RL, Macri E, Loda M, Appelman HD, Cho KR, Fearon ER. Loss of CDX2 expression and microsatellite instability are prominent features of large cell minimally differentiated carcinomas of the colon. Am J Pathol. 2001;159:2239–2248. doi: 10.1016/S0002-9440(10)63074-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.McGregor DK, Wu TT, Rashid A, Luthra R, Hamilton SR. Reduced expression of cytokeratin 20 in colorectal carcinomas with high levels of microsatellite instability. Am J Surg Pathol. 2004;28:712–718. doi: 10.1097/01.pas.0000126757.58474.12. [DOI] [PubMed] [Google Scholar]
- 147.Lugli A, Tzankov A, Zlobec I, Terracciano LM. Differential diagnostic and functional role of the multi-marker phenotype CDX2/CK20/CK7 in colorectal cancer stratified by mismatch repair status. Mod Pathol. 2008;21:1403–1412. doi: 10.1038/modpathol.2008.117. [DOI] [PubMed] [Google Scholar]
- 148.Baba Y, Nosho K, Shima K, Freed E, Irahara N, Philips J, Meyerhardt JA, Hornick JL, Shivdasani RA, Fuchs CS, et al. Relationship of CDX2 loss with molecular features and prognosis in colorectal cancer. Clin Cancer Res. 2009;15:4665–4673. doi: 10.1158/1078-0432.CCR-09-0401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Kim JH, Rhee YY, Bae JM, Cho NY, Kang GH. Loss of CDX2/CK20 expression is associated with poorly differentiated carcinoma, the CpG island methylator phenotype, and adverse prognosis in microsatellite-unstable colorectal cancer. Am J Surg Pathol. 2013;37:1532–1541. doi: 10.1097/PAS.0b013e31829ab1c1. [DOI] [PubMed] [Google Scholar]
- 150.Isaksson-Mettävainio M, Palmqvist R, Dahlin AM, Van Guelpen B, Rutegård J, Oberg A, Henriksson ML. High SMAD4 levels appear in microsatellite instability and hypermethylated colon cancers, and indicate a better prognosis. Int J Cancer. 2012;131:779–788. doi: 10.1002/ijc.26473. [DOI] [PubMed] [Google Scholar]
- 151.Alazzouzi H, Alhopuro P, Salovaara R, Sammalkorpi H, Järvinen H, Mecklin JP, Hemminki A, Schwartz S, Aaltonen LA, Arango D. SMAD4 as a prognostic marker in colorectal cancer. Clin Cancer Res. 2005;11:2606–2611. doi: 10.1158/1078-0432.CCR-04-1458. [DOI] [PubMed] [Google Scholar]
- 152.Tanaka T, Watanabe T, Kazama Y, Tanaka J, Kanazawa T, Kazama S, Nagawa H. Loss of Smad4 protein expression and 18qLOH as molecular markers indicating lymph node metastasis in colorectal cancer--a study matched for tumor depth and pathology. J Surg Oncol. 2008;97:69–73. doi: 10.1002/jso.20896. [DOI] [PubMed] [Google Scholar]
- 153.Tanaka T, Watanabe T, Kazama Y, Tanaka J, Kanazawa T, Kazama S, Nagawa H. Chromosome 18q deletion and Smad4 protein inactivation correlate with liver metastasis: A study matched for T- and N- classification. Br J Cancer. 2006;95:1562–1567. doi: 10.1038/sj.bjc.6603460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Miyaki M, Iijima T, Konishi M, Sakai K, Ishii A, Yasuno M, Hishima T, Koike M, Shitara N, Iwama T, et al. Higher frequency of Smad4 gene mutation in human colorectal cancer with distant metastasis. Oncogene. 1999;18:3098–3103. doi: 10.1038/sj.onc.1202642. [DOI] [PubMed] [Google Scholar]
- 155.Thiagalingam S, Lengauer C, Leach FS, Schutte M, Hahn SA, Overhauser J, Willson JK, Markowitz S, Hamilton SR, Kern SE, et al. Evaluation of candidate tumour suppressor genes on chromosome 18 in colorectal cancers. Nat Genet. 1996;13:343–346. doi: 10.1038/ng0796-343. [DOI] [PubMed] [Google Scholar]
- 156.Van Rijnsoever M, Elsaleh H, Joseph D, McCaul K, Iacopetta B. CpG island methylator phenotype is an independent predictor of survival benefit from 5-fluorouracil in stage III colorectal cancer. Clin Cancer Res. 2003;9:2898–2903. [PubMed] [Google Scholar]
- 157.Jover R, Nguyen TP, Pérez-Carbonell L, Zapater P, Payá A, Alenda C, Rojas E, Cubiella J, Balaguer F, Morillas JD, et al. 5-Fluorouracil adjuvant chemotherapy does not increase survival in patients with CpG island methylator phenotype colorectal cancer. Gastroenterology. 2011;140:1174–1181. doi: 10.1053/j.gastro.2010.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Shen L, Catalano PJ, Benson AB, O’Dwyer P, Hamilton SR, Issa JP. Association between DNA methylation and shortened survival in patients with advanced colorectal cancer treated with 5-fluorouracil based chemotherapy. Clin Cancer Res. 2007;13:6093–6098. doi: 10.1158/1078-0432.CCR-07-1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Ahn JB, Chung WB, Maeda O, Shin SJ, Kim HS, Chung HC, Kim NK, Issa JP. DNA methylation predicts recurrence from resected stage III proximal colon cancer. Cancer. 2011;117:1847–1854. doi: 10.1002/cncr.25737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Baba Y, Huttenhower C, Nosho K, Tanaka N, Shima K, Hazra A, Schernhammer ES, Hunter DJ, Giovannucci EL, Fuchs CS, et al. Epigenomic diversity of colorectal cancer indicated by LINE-1 methylation in a database of 869 tumors. Mol Cancer. 2010;9:125. doi: 10.1186/1476-4598-9-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Ogino S, Nosho K, Kirkner GJ, Kawasaki T, Chan AT, Schernhammer ES, Giovannucci EL, Fuchs CS. A cohort study of tumoral LINE-1 hypomethylation and prognosis in colon cancer. J Natl Cancer Inst. 2008;100:1734–1738. doi: 10.1093/jnci/djn359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Rhee YY, Kim MJ, Bae JM, Koh JM, Cho NY, Juhnn YS, Kim D, Kang GH. Clinical outcomes of patients with microsatellite-unstable colorectal carcinomas depend on L1 methylation level. Ann Surg Oncol. 2012;19:3441–3448. doi: 10.1245/s10434-012-2410-7. [DOI] [PubMed] [Google Scholar]


