Abstract
AIM: To evaluate the clinicopathological features of colorectal cancer (CRC) with a v-Raf murine sarcoma viral oncogene homolog B1 (BRAF) mutation and its molecular interaction with microsatellite instability (MSI) and v-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog (KRAS) in patients with advanced CRCs.
METHODS: From October 2009 to December 2011, 141 patients with stage III (n = 51) or IV (n = 90) CRCs who were tested for the BRAF mutation at Severance Hospital were included. Among 141 patients, five were excluded due to follow-up loss. Therefore, 136 patients were included in the study. The clinicopathological data, MSI status, and KRAS/BRAF mutation status were reviewed retrospectively. In addition, to evaluating the value of BRAF mutation status, progression-free survival and overall survival in all patients were collected and compared between the BRAF wild-type group and BRAF mutation group.
RESULTS: Of 136 patients, 80 (58.8%) were male and the mean age was 59 years. BRAF and KRAS mutations were detected in 9.6% and 35.3% of patients, respectively. Only 4.3% of patients had MSI-high tumors and there were no MSI-high in tumors with a BRAF mutation. BRAF mutations tended to be more frequent in stage IV than in stage III (11.76% vs 5.88%, P = 0.370). Patients with a BRAF mutation had a lower incidence of KRAS mutation than those without (7.69% vs 38.21%, P = 0.033). Overall survival was significantly shorter in the BRAF mutation group than in the BRAF wild-type group both by univariate analysis (P = 0.041) and multivariate analysis (HR = 2.195; 95%CI: 1.039-4.640; P = 0.039), while progression-free survival was not different according to BRAF mutation status.
CONCLUSION: CRCs with a BRAF mutation have distinct molecular features and resulted in a poor prognosis in Korean patients with advanced CRC.
Keywords: BRAF, Colorectal cancer, Molecular features, Chemotherapy response, Prognosis
Core tip: This study identified the clinicopathological features of colorectal cancer (CRC) with BRAF mutation and its molecular interaction with microsatellite instability and KRAS targeting only to stage III/IV CRCs. These molecular markers enable the classification of CRCs into meaningful subtypes for prognosis. Our data strongly support the prognostic role of BRAF mutation in Korean patients with advanced CRC.
INTRODUCTION
Colorectal cancer (CRC) is the third most common cancer in Western countries. In Korea, CRC is the second most common cancer in males and the third in females, with an estimated 25782 new cases and 7645 deaths each year[1]. TNM staging of CRC has become a promising tool in determining treatment and prognosis, but it is evident that CRC has a significant clinical heterogeneity even within the same pathologic stage[2-4]. Recent advances in molecular genetics enable the classification of CRC using molecular markers, including microsatellite instability (MSI) and v-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog (KRAS) and v-raf murine sarcoma viral oncogene homolog B1 (BRAF) mutations, to predict prognosis and treatment response[5-8].
BRAF is a part of the Ras/Raf/mitogen-activated protein kinase kinase (MAP2K)/mitogen-activated protein kinases (MAPK) signaling pathway. Many of the transcription factors activated by the Ras/Raf/MAP2K/MAPK pathway are involved in cell proliferation and differentiation and many growth factor genes have binding sites for transcription factors activated by the Ras–ERK pathway, located in their promoter regions. Thus, aberrant activation of this pathway may provoke self-sufficiency in proliferative signals and continuous stimulation of cell growth[9]. Mutations of KRAS or BRAF activate this pathway and are an established mechanism that drives colorectal carcinogenesis[10]. The most common mutation of BRAF is the classic GTG-GAG substitution at position 1799 of exon 15, which results in the V600E amino acid change and the subsequent constitutive activation of the EGFR signaling pathway. Generally, 10% to 20% of CRCs have BRAF mutations, and the incidence of BRAF mutation varies according to the status of the MSI. It has been reported that 34% to 70% of CRCs classified as MSI-high have BRAF mutations[11-13]. Compared to KRAS mutations, patients with CRC and BRAF mutations show some different clinical manifestations. It is well known that CRCs with KRAS mutations respond poorly to cetuximab[14]. However, the predictive role of BRAF mutations in response to cetuximab is not yet clear. As for prognostic markers, CRC with BRAF mutations have been shown to have significantly poor prognosis compared to those with BRAF wild-type[15-17], while KRAS mutations have no prognostic role[18].
Although CRCs have rapidly increased in Korea, the incidence of BRAF mutation and its clinical meaning have not yet been explored in Korean patients with CRC. The aim of this study was to evaluate the clinicopathological features of CRC with a BRAF mutation and its molecular interaction with MSI and KRAS mutation status in patients with stage III or IV CRC.
MATERIALS AND METHODS
From October 2009 to December 2011, 141 patients in stage III (n = 51) and IV (n = 90) CRC underwent molecular testing, including MSI analysis and determination of KRAS and BRAF mutation status in Severance Hospital. Among the 141 patients, five were excluded due to follow-up loss. In total, 136 patients were included in the present study. After an initial staging work-up including a CT scan, patients without metastasis or with resectable liver or lung metastasis received surgery and adjuvant chemotherapy with FOLFOX. Patients with unresectable metastatic disease received palliative chemotherapy with FOLFOX or FOLFIRI[19,20]. The clinicopathological data, including age, sex, family history of CRC, BMI (body mass index), Eastern Cooperative Oncology Group (ECOG) performance status tumor stage, tumor location, tumor grade, initial CEA and chemotherapeutic regimen were reviewed retrospectively. This study was approved by the Institutional Review Boards of Yonsei University College of Medicine.
DNA extract, MSI analysis, BRAF and KRAS sequencing
Before obtaining tissue samples, written informed consent was obtained from all patients. Tissue samples from the tumor and normal colonic mucosa were obtained from each patient after resection. DNA extracted from each tumor was amplified by a standard polymerase chain reaction using five Bethesda guidelines panel loci (BAT25, BAT26, MFD15, D2S123, and D5S346)[21]. In accordance with the consensus definitions of the National Cancer Institute, tumor samples were classified as displaying high-degree microsatellite instability (MSI-H, instability at 30% or more of the markers tested), low-degree microsatellite instability (MSI-L, instability at less than 30% of the markers tested), or microsatellite stability (MSS, stability at all of the markers tested). Due to the similar biological properties between MSI-L and MSS, these two molecular phenotypes were grouped together in all analyses.
KRAS and BRAF were charged at ISU ABXI (Seoul, South Korea). Genomic DNA was extracted from 10-μm-thick paraffin sections containing a portion of tumor tissue by the QIAamp DNA Mini kit (Qiagen, Hilden, Germany). Fifty nanograms of DNA were amplified in a 20 μL reaction solution containing 10 μL of 2 X concentrated HotStarTaq Master Mix (Qiagen, Hilden, Germany), including polymerase chain reaction (PCR) buffer with 3 mmol/L MgCl2, 400 μmol/L of each dNTP, and 0.3 μmol/L of each primer pair (KRAS F: 5’-ttatgtgtgacatgttctaat, R: 5’-agaatggtcctgcaccagtaa/BRAF, F: 5’-atgcttgctctgataggaaaatga, R: 5’-agcagcatctcagggcca). Amplifications were performed using a 15-min initial denaturation at 95 °C, followed by 35 cycles of 30 s at 94 °C, 30 s at 59 °C, and 30 s at 72 °C, and a 10-min final extension at 72 °C. PCR products separated in 2% gel were purified with a QIAgen gel extraction kit (Qiagen). DNA templates were processed for the DNA sequencing reaction using the ABI-PRISM BigDye Terminator version 3.1 (Applied Biosystems, Foster City, CA, United States) with both forward and reverse sequence-specific primers. Twenty nanograms of purified PCR products were used in a 10 μL sequencing reaction solution containing 1 μL of BigDye Terminator v3.1 and 0.1 μmol/L of the same PCR primer. Sequencing reactions were performed using 25 cycles of 10 s at 96 °C, 5 s at 50 °C, and 4 min at 60 °C. Sequence data were generated with the ABI PRISM 3730 DNA Analyzer (Applied Biosystems) and analyzed by Sequencing Analysis 5.1.1. software (Applied Biosystems) to compare variations.
Statistical analysis
The primary outcome was to compare overall survival (OS) and progression-free survival (PFS) of patients with BRAF mutation to those with BRAF wild-type. The secondary outcome was to evaluate the gene-gene interaction between BRAF mutation and KRAS mutation or MSI. Continuous variables were expressed as mean ± SD. Each patient’s baseline characteristics were analyzed by descriptive statistics. OS was calculated from the time of diagnosis until death or the last follow-up visit, and PFS was calculated from the time of diagnosis until disease recurrence or progression. OS and PFS were analyzed using the Kaplan-Meier method, and survival curves were compared using the log-rank method. Cox proportional hazards modeling was used to control multiple risk factors that have been shown to influence CRC survival by computing 95% confidence intervals (CIs). A P-value < 0.05 was considered statistically significant. All statistical analyses were performed using SPSS version 18.0 (SPSS Inc., Chicago, IL, United States).
RESULTS
Patient characteristics
Of 136 patients, 107 (78.7%) patients were diagnosed with colon cancer and 29 (21.3%) with rectal cancer. Fifty-one (37.5%) patients were stage III and 85 (62.5%) were stage IV. Thirteen patients (9.6%) had BRAF mutations, and 48 (35.3%) patients had KRAS mutations. The most frequent mutation at KRAS was G12D, which accounted for 29.2% of KRAS mutations (14/48). The second most frequent mutation was G13D (12/48), and the remainder occurred in the following order: G12V (9/48), G12C (6/48), G12S (4/48), G13C (2/48) and G12A (1/48). Five (4.3%) patients had MSI-high tumors. Molecular characteristics such as MSI status and KRAS and BRAF mutation status were not significantly different according to the tumor location.
The clinicopathological characteristics of patients according to BRAF mutation are summarized in Table 1. Tumors with BRAF wild-type tended to exhibit more differentiated histology (91.9% vs 76.9%, P = 0.211) and an earlier stage (stage III, 39.0% vs 23.1%; P = 0.370) than tumors with BRAF mutation. There was no MSI-H in tumors with BRAF mutation. Tumors with BRAF wild-type had significantly more KRAS mutations than tumors with BRAF mutation (38.2% vs 7.7%, P = 0.033). Only one (7.7%) tumor with BRAF mutation had KRAS mutation. The other clinicopathological findings, such as sex, age, BMI, family history of CRC, ECOG performance status, tumor location and initial CEA level, were not different between tumors with BRAF wild-type and mutation. Treatment modalities in enrolled patients are summarized in Table 2. Ninety-three (75.6%) patients with BRAF wild-type and nine (69.2%) with BRAF mutation received surgery (P = 0.737). One hundred nineteen (96.7%) patients with BRAF wild-type and 11 (84.6%) with BRAF mutation received chemotherapy (P = 0.182). Chemotherapy regimens and targeted agents were not significantly different between the two groups. The mean ± SD follow-up duration in all patients was 21.5 ± 13.3 mo.
Table 1.
Baseline characteristics of enrolled patients n (%)
| BRAF wild-type (n = 123) | BRAF Mutant (n = 13) | P value | |
| Sex Male/Female | 74 (60.2)/49 (39.8) | 6 (46.2)/7 (53.8) | 0.329 |
| Age (mean ± SD, age) | 58.54 ± 12.91 | 61.08 ± 7.87 | 0.488 |
| Age (< 60 yr) | 60 (48.8) | 5 (38.5) | 0.479 |
| BMI (mean ± SD, kg/m2) | 22.77 ± 3.64 | 22.70 ± 3.31 | 0.950 |
| Family history of colorectal cancer | 25 (20.3) | 1 (7.7) | 0.462 |
| ECOG performance status | 0.421 | ||
| 0-1 | 116 (94.3) | 13 (100.0) | |
| 2-3 | 7 (5.7) | 0 (0.0) | |
| Tumor type | 0.871 | ||
| Colon | 97 (78.9) | 10 (76.9) | |
| Rectum | 26 (21.1) | 3 (23.1) | |
| Tumor location | 0.211 | ||
| Proximal | 35 (28.5) | 6 (46.2) | |
| Distal | 88 (71.5) | 7 (53.8) | |
| Histology | 0.111 | ||
| WD and MD | 113 (91.9) | 10 (76.9) | |
| PD and UD | 10 (8.1) | 3 (23.1) | |
| AJCC tumor stage | 0.370 | ||
| Stage III | 48 (39.0) | 3 (23.1) | |
| IIIA | 5 (4.1) | 1 (7.7) | |
| IIIB | 30 (24.4) | 1 (7.7) | |
| IIIC | 13 (10.6) | 1 (7.7) | |
| Stage IV | 75 (61.0) | 10 (76.9) | |
| IVA | 27 (22.0) | 3 (23.1) | |
| IVB | 48 (39.0) | 7 (53.8) | |
| MSI | 0.604 | ||
| MSS and MS-low | 110 (89.4) | 12 (92.3) | |
| MSI-high | 5 (4.1) | 0 | |
| Unchecked | 8 (6.5) | 1 (7.7) | |
| K-ras | 0.033 | ||
| Wild | 76 (61.8) | 12 (92.3) | |
| Mutant | 47 (38.2) | 1 (7.7) | |
| Initial CEA (mean ± SD, ng/mL) | 251.41 ± 1520.96 | 14.29 ± 23.90 | 0.576 |
SD: Standard deviation; BMI: Body mass index; ECOG: Eastern Cooperative Oncology Group; WD: Well differentiated; MD: Moderately differentiated; PD: Poorly differentiated; UD: Undifferentiated; AJCC: American Joint Committee on Cancer; MSI: Microsatellite instability; MSS: Microsatellite stable; CEA: Carcinoembryonic antigen.
Table 2.
Treatment modality of the patients n (%)
| BRAF wild-type (n = 123) | BRAF Mutant (n = 13) | P value | |
| Surgery | 0.737 | ||
| Yes | 93 (75.6) | 9 (69.2) | |
| No | 30 (24.4) | 4 (30.8) | |
| Chemotherapy | |||
| Yes | 119 (96.7) | 11 (84.6) | |
| No | 4 (3.3) | 2 (15.4) | |
| Chemotherapy regimen | 0.441 | ||
| FOLFOX | 97 (82.2) | 9 (81.8) | |
| FOLFIRI | 5 (4.2) | 0 | |
| FL | 7 (5.9) | 2 (15.4) | |
| Xeloda | 8 (6.8) | 0 | |
| SOX | 1 (0.8) | 0 | |
| Target agent use | 0.128 | ||
| No | 91 (74.0) | 8 (61.5) | |
| Bevacizumab | 19 (15.4) | 5 (38.5) | |
| Cetuximab | 13 (10.6) | 0 |
Tumor response and survival according to BRAF mutation status
To determine the value of BRAF mutation status as a prognostic marker, PFS and OS were compared between patients with BRAF wild-type and BRAF mutation (Figure 1). PFS was not statistically different between the two groups (BRAF wild type vs BRAF mutation, 10.1 ± 7.8 mo vs 7.2 ± 5.0 mo; P = 0.135). The OS was significantly shorter in patients with BRAF mutation than those with BRAF wild-type (BRAF wild type vs BRAF mutation, 22.2 ± 13.2 mo vs 18.8 ± 13.6 mo; P = 0.041). In contrast to BRAF mutation, KRAS mutation status was not associated with PFS or OS in patients with stage III/IV CRCs.
Figure 1.
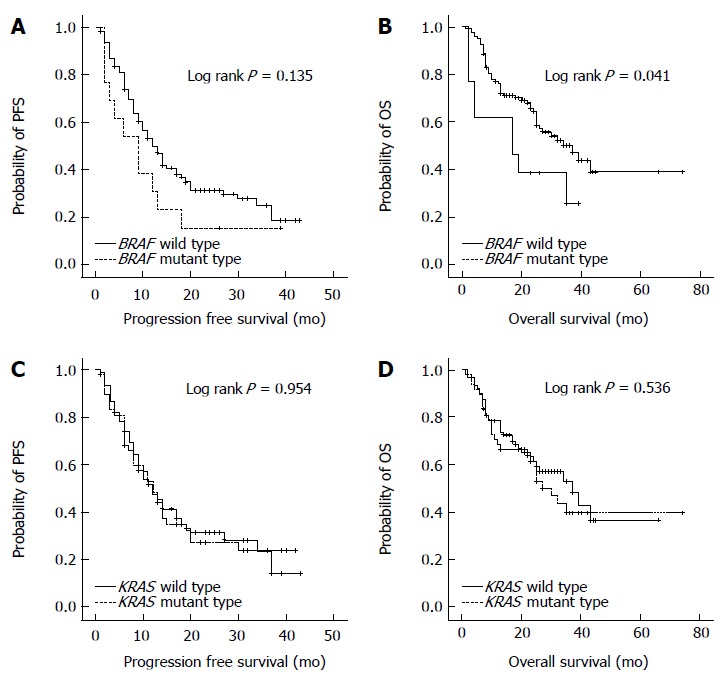
Progression-free survival and overall survival according to BRAF or KRAS mutation status. A: PFS was not significantly different by the BRAF mutation status (P = 0.135); B: OS was significantly poorer in the BRAF mutant group than in the BRAF wild-type group (P = 0.041); PFS (C) and OS (D) were not significantly different by the KRAS mutation status (P = 0.954 and P = 0.536, respectively). PFS: Progression-free survival; OS: Overall survival.
Univariate and multivariate analyses were performed to validate the prognostic factors for survival in patients with stage III/IV CRC. Univariate analysis revealed that histology, tumor stage, surgery and BRAF status were significant prognostic factors for survival (P = 0.001, P < 0.001, P < 0.001 and P = 0.041, respectively). Multivariate analysis showed that TNM stage IV (HR = 3.183; 95%CI: 1.517-6.679; P = 0.002), poor differentiation and lack of differentiation in histology (HR = 2.821; 95%CI: 1.378-5.776; P = 0.005), no surgery (HR = 3.694; 95%CI: 1.972-6.918; P < 0.001) and BRAF mutation (HR = 2.195; 95%CI: 1.039-4.640; P = 0.039) were significant poor prognostic factors for survival in patients with stage III/IV CRC (Table 3).
Table 3.
Prognostic factors in colorectal cancer patients in multivariate analysis
| HR (95%CI) | P value | |
| Age (older than 60 yr vs younger) | 1.164 (0.682-1.984) | 0.578 |
| Sex (male vs female) | 0.927 (0.541-1.586) | 0.781 |
| Tumor type (rectum vs colon) | 0.931 (0.511-1.697) | 0.816 |
| Initial stage (stage IV vs III) | 3.183 (1.517-6.679) | 0.002 |
| Histology (PD and UD vs WD and MD) | 2.821 (1.378-5.776) | 0.005 |
| Surgical treatment (no vs yes) | 3.694 (1.972-6.918) | < 0.001 |
| BRAF mutation (mutant vs wild-type) | 2.195 (1.039-4.640) | 0.039 |
| KRAS mutation (mutant vs wild-type) | 1.305 (0.766-2.221) | 0.327 |
CI: Confidence interval; PD: Poorly differentiated; UD: Undifferentiated; WD: Well differentiated; MD: Moderately differentiated; CEA: Carcinoembryonic antigen.
DISCUSSION
This retrospective study demonstrated that stage III or IV CRC in Korean patients have distinct molecular characteristics, including exclusive mutations between KRAS and BRAF genes and a low incidence of MSI-H. BRAF mutant tumors showed significantly shorter survival than BRAF wild-type tumors, while the KRAS mutation had no prognostic impact.
It has been reported that 10% to 20% of CRCs have a BRAF mutation[3,22]. The BRAF mutation is associated with MSI-H through hMLH1 promoter hypermethylation, which is known to be associated with a high level of CpG island methylator phenotype (CIMP)[11]. Several reports have revealed a low incidence of BRAF mutations in CRC. Although it is not clear if ethnicity affects the status of the BRAF mutation, previous studies with small sample sizes of Koreans reported only 3.8%-7% of BRAF mutant CRCs in Korean patients[23,24]. A few studies showed that BRAF mutations were extremely uncommon in rectal cancer, with an incidence of 0%-2%[25,26]. However, the present study indicated that the incidence of BRAF mutation was not affected by ethnicity or tumor location. Although it is known that BRAF genes are exclusively mutated with KRAS genes, coincident mutations of KRAS and BRAF rarely occur in CRC with an incidence of 0.001%[27]. Herein, we found that one case had both KRAS (G12V) and BRAF mutations. This patient was a 49-year-old male and had rectosigmoid junction cancer of MSS type in MSI. It is not clear whether these tumors have a different biology and natural history than KRAS or BRAF only mutant tumors or which of the two mutations is the dominant oncogene driving tumor proliferation because coincident KRAS and BRAF mutation were infrequently observed[27].
The present study showed a small number of MSI-H tumors compared to previous studies[5,28] and thus failed to demonstrate the relationship between MSI status and BRAF mutation. This may have been due to the advanced stage of CRCs in the present study. Our previous study demonstrated that MSI-H tumors were strongly associated with early tumor stage, and only 6% of stage III/IV CRC had MSI-H tumors[29]. Compared with colon cancer, rectal cancer has a low incidence of MSI-H tumors and this may have affected the results of the present study[29].
CRC has significant clinical heterogeneity based on several molecular markers such as MSI, KRAS and BRAF[2-4]. It has been well documented that MSI-H tumors have a better prognosis than MSS/MSI-L tumors[5,29]. In contrast, BRAF mutant tumors have a poor prognosis compared to BRAF wild-type tumors[6,7]. The BRAF mutation has been used as a strong prognostic factor for overall survival in patients with CRCs, which was also confirmed in the present study. Thus, CRCs can be classified into four subtypes by these two distinct prognostic markers of MSI status and BRAF mutation[7]. MSS/BRAF mutant tumors are known to exhibit the worst prognosis, while MSI-H/BRAF wild-type tumors have the best prognosis. MSI-H/BRAF mutant or MSS/BRAF wild-type tumors have been suggested as intermediate subtypes[7]. However, several recent studies suggested that the association of BRAF mutation with poor prognosis is limited to MSS tumors[16,30]. Thus, further studies are necessary to adapt this molecular classification for clinical practice.
This study has several limitations. First, a retrospective study design has inherent limitations. Second, because of the small number of MSI-H tumors, the present study failed to evaluate the association of BRAF mutation with MSI status. Finally, even for advanced cancer, the follow-up period was relatively insufficient at a mean of 21 mo.
In conclusion, CRCs have distinct molecular features, including MSI status and mutations of KRAS and BRAF. These molecular markers enable the classification of CRCs into meaningful subtypes for prognosis. Our data strongly support the prognostic role of BRAF mutation in Korean patients with advanced CRC.
COMMENTS
Background
Approximately 10% of colorectal cancers (CRCs) have BRAF mutations and these CRCs have a worse prognosis than those with BRAF wild-type. In addition, the BRAF gene is thought to be closely associated with several molecular markers such as microsatellite instability (MSI) and KRAS in CRCs. However, although the prevalence of CRCs has rapidly increased in South Korea, the incidence of BRAF mutation and its clinical meaning are unknown in Korean CRC patients.
Research frontiers
Recent advances in molecular genetics enable the classification of CRC by molecular markers, including MSI and KRAS and BRAF mutations, to predict prognosis and treatment response. It is well known that CRCs with KRAS mutations respond poorly to cetuximab. However, the predictive role of BRAF mutations in cetuximab response is not clear. As for prognostic markers, MSI-high tumors have a better prognosis than MSS/MSI-low tumors. In contrast, CRC with BRAF mutations have a worse prognosis than those with BRAF wild-type, while KRAS mutations have no prognostic role. Future research should aim to uncover the role and purpose of molecular markers in CRC and to identify their potential usage clinically.
Innovations and breakthroughs
This study identified the clinicopathological features of CRC with a BRAF mutation and its molecular interaction with MSI and KRAS targeting for stage III/IV CRC.
Applications
Molecular markers such as BRAF, KRAS, MSI may be used to classify cases of colorectal carcinoma into subtypes for prognosis.
Terminology
BRAF: BRAF is a human gene that makes a protein called B-Raf. The gene is also referred to as proto-oncogene B-Raf and v-Raf murine sarcoma viral oncogene homolog B1, while the protein is more formally known as serine/threonine-protein kinase B-Raf. The B-Raf protein is involved in sending signals inside cells, which are involved in directing cell growth. KRAS: KRAS is a human gene that makes a protein called KRAS. Like other members of the Ras family, the KRAS protein is a GTPase and is an early player in many signal transduction pathways. The protein product of the normal KRAS gene performs an essential function in normal tissue signaling, and the mutation of a KRAS gene is an essential step in the development of many cancers.
Peer review
The BRAF mutant tumors had a significantly shorter survival than that of BRAF wild-type tumors, while the KRAS mutation had no prognostic impact. These results are interesting and this finding allows these molecular markers to be used to classify cases of colorectal carcinoma into subtypes for prognosis.
Footnotes
P- Reviewers: Nath G, Vorobjova T S- Editor: Ma YJ L- Editor: Webster JR E- Editor: Liu XM
References
- 1.Jung KW, Won YJ, Kong HJ, Oh CM, Seo HG, Lee JS. Cancer statistics in Korea: incidence, mortality, survival and prevalence in 2010. Cancer Res Treat. 2013;45:1–14. doi: 10.4143/crt.2013.45.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zlobec I, Lugli A. Prognostic and predictive factors in colorectal cancer. J Clin Pathol. 2008;61:561–569. doi: 10.1136/jcp.2007.054858. [DOI] [PubMed] [Google Scholar]
- 3.Jass JR. Molecular heterogeneity of colorectal cancer: Implications for cancer control. Surg Oncol. 2007;16 Suppl 1:S7–S9. doi: 10.1016/j.suronc.2007.10.039. [DOI] [PubMed] [Google Scholar]
- 4.Ribic CM, Sargent DJ, Moore MJ, Thibodeau SN, French AJ, Goldberg RM, Hamilton SR, Laurent-Puig P, Gryfe R, Shepherd LE, et al. Tumor microsatellite-instability status as a predictor of benefit from fluorouracil-based adjuvant chemotherapy for colon cancer. N Engl J Med. 2003;349:247–257. doi: 10.1056/NEJMoa022289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Popat S, Hubner R, Houlston RS. Systematic review of microsatellite instability and colorectal cancer prognosis. J Clin Oncol. 2005;23:609–618. doi: 10.1200/JCO.2005.01.086. [DOI] [PubMed] [Google Scholar]
- 6.French AJ, Sargent DJ, Burgart LJ, Foster NR, Kabat BF, Goldberg R, Shepherd L, Windschitl HE, Thibodeau SN. Prognostic significance of defective mismatch repair and BRAF V600E in patients with colon cancer. Clin Cancer Res. 2008;14:3408–3415. doi: 10.1158/1078-0432.CCR-07-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lochhead P, Kuchiba A, Imamura Y, Liao X, Yamauchi M, Nishihara R, Qian ZR, Morikawa T, Shen J, Meyerhardt JA, et al. Microsatellite instability and BRAF mutation testing in colorectal cancer prognostication. J Natl Cancer Inst. 2013;105:1151–1156. doi: 10.1093/jnci/djt173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Laurent-Puig P, Cayre A, Manceau G, Buc E, Bachet JB, Lecomte T, Rougier P, Lievre A, Landi B, Boige V, et al. Analysis of PTEN, BRAF, and EGFR status in determining benefit from cetuximab therapy in wild-type KRAS metastatic colon cancer. J Clin Oncol. 2009;27:5924–5930. doi: 10.1200/JCO.2008.21.6796. [DOI] [PubMed] [Google Scholar]
- 9.McCubrey JA, Steelman LS, Chappell WH, Abrams SL, Wong EW, Chang F, Lehmann B, Terrian DM, Milella M, Tafuri A, et al. Roles of the Raf/MEK/ERK pathway in cell growth, malignant transformation and drug resistance. Biochim Biophys Acta. 2007;1773:1263–1284. doi: 10.1016/j.bbamcr.2006.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Siena S, Sartore-Bianchi A, Di Nicolantonio F, Balfour J, Bardelli A. Biomarkers predicting clinical outcome of epidermal growth factor receptor-targeted therapy in metastatic colorectal cancer. J Natl Cancer Inst. 2009;101:1308–1324. doi: 10.1093/jnci/djp280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Samowitz WS, Albertsen H, Herrick J, Levin TR, Sweeney C, Murtaugh MA, Wolff RK, Slattery ML. Evaluation of a large, population-based sample supports a CpG island methylator phenotype in colon cancer. Gastroenterology. 2005;129:837–845. doi: 10.1053/j.gastro.2005.06.020. [DOI] [PubMed] [Google Scholar]
- 12.Saridaki Z, Papadatos-Pastos D, Tzardi M, Mavroudis D, Bairaktari E, Arvanity H, Stathopoulos E, Georgoulias V, Souglakos J. BRAF mutations, microsatellite instability status and cyclin D1 expression predict metastatic colorectal patients’ outcome. Br J Cancer. 2010;102:1762–1768. doi: 10.1038/sj.bjc.6605694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nosho K, Irahara N, Shima K, Kure S, Kirkner GJ, Schernhammer ES, Hazra A, Hunter DJ, Quackenbush J, Spiegelman D, et al. Comprehensive biostatistical analysis of CpG island methylator phenotype in colorectal cancer using a large population-based sample. PLoS One. 2008;3:e3698. doi: 10.1371/journal.pone.0003698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karapetis CS, Khambata-Ford S, Jonker DJ, O’Callaghan CJ, Tu D, Tebbutt NC, Simes RJ, Chalchal H, Shapiro JD, Robitaille S, et al. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med. 2008;359:1757–1765. doi: 10.1056/NEJMoa0804385. [DOI] [PubMed] [Google Scholar]
- 15.Tol J, Nagtegaal ID, Punt CJ. BRAF mutation in metastatic colorectal cancer. N Engl J Med. 2009;361:98–99. doi: 10.1056/NEJMc0904160. [DOI] [PubMed] [Google Scholar]
- 16.Roth AD, Tejpar S, Delorenzi M, Yan P, Fiocca R, Klingbiel D, Dietrich D, Biesmans B, Bodoky G, Barone C, et al. Prognostic role of KRAS and BRAF in stage II and III resected colon cancer: results of the translational study on the PETACC-3, EORTC 40993, SAKK 60-00 trial. J Clin Oncol. 2010;28:466–474. doi: 10.1200/JCO.2009.23.3452. [DOI] [PubMed] [Google Scholar]
- 17.Yokota T, Ura T, Shibata N, Takahari D, Shitara K, Nomura M, Kondo C, Mizota A, Utsunomiya S, Muro K, et al. BRAF mutation is a powerful prognostic factor in advanced and recurrent colorectal cancer. Br J Cancer. 2011;104:856–862. doi: 10.1038/bjc.2011.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bengala C, Bettelli S, Bertolini F, Sartori G, Fontana A, Malavasi N, Depenni R, Zironi S, Del Giovane C, Luppi G, et al. Prognostic role of EGFR gene copy number and KRAS mutation in patients with locally advanced rectal cancer treated with preoperative chemoradiotherapy. Br J Cancer. 2010;103:1019–1024. doi: 10.1038/sj.bjc.6605853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Douillard JY, Cunningham D, Roth AD, Navarro M, James RD, Karasek P, Jandik P, Iveson T, Carmichael J, Alakl M, et al. Irinotecan combined with fluorouracil compared with fluorouracil alone as first-line treatment for metastatic colorectal cancer: a multicentre randomised trial. Lancet. 2000;355:1041–1047. doi: 10.1016/s0140-6736(00)02034-1. [DOI] [PubMed] [Google Scholar]
- 20.Giacchetti S, Perpoint B, Zidani R, Le Bail N, Faggiuolo R, Focan C, Chollet P, Llory JF, Letourneau Y, Coudert B, et al. Phase III multicenter randomized trial of oxaliplatin added to chronomodulated fluorouracil-leucovorin as first-line treatment of metastatic colorectal cancer. J Clin Oncol. 2000;18:136–147. doi: 10.1200/JCO.2000.18.1.136. [DOI] [PubMed] [Google Scholar]
- 21.Sargent DJ, Marsoni S, Monges G, Thibodeau SN, Labianca R, Hamilton SR, French AJ, Kabat B, Foster NR, Torri V, et al. Defective mismatch repair as a predictive marker for lack of efficacy of fluorouracil-based adjuvant therapy in colon cancer. J Clin Oncol. 2010;28:3219–3226. doi: 10.1200/JCO.2009.27.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vaughn CP, Zobell SD, Furtado LV, Baker CL, Samowitz WS. Frequency of KRAS, BRAF, and NRAS mutations in colorectal cancer. Genes Chromosomes Cancer. 2011;50:307–312. doi: 10.1002/gcc.20854. [DOI] [PubMed] [Google Scholar]
- 23.Kang GH. Four molecular subtypes of colorectal cancer and their precursor lesions. Arch Pathol Lab Med. 2011;135:698–703. doi: 10.5858/2010-0523-RA.1. [DOI] [PubMed] [Google Scholar]
- 24.Rhee YY, Kim MJ, Bae JM, Koh JM, Cho NY, Juhnn YS, Kim D, Kang GH. Clinical outcomes of patients with microsatellite-unstable colorectal carcinomas depend on L1 methylation level. Ann Surg Oncol. 2012;19:3441–3448. doi: 10.1245/s10434-012-2410-7. [DOI] [PubMed] [Google Scholar]
- 25.Slattery ML, Curtin K, Wolff RK, Boucher KM, Sweeney C, Edwards S, Caan BJ, Samowitz W. A comparison of colon and rectal somatic DNA alterations. Dis Colon Rectum. 2009;52:1304–1311. doi: 10.1007/DCR.0b013e3181a0e5df. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yamauchi M, Morikawa T, Kuchiba A, Imamura Y, Qian ZR, Nishihara R, Liao X, Waldron L, Hoshida Y, Huttenhower C, et al. Assessment of colorectal cancer molecular features along bowel subsites challenges the conception of distinct dichotomy of proximal versus distal colorectum. Gut. 2012;61:847–854. doi: 10.1136/gutjnl-2011-300865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sahin IH, Kazmi SM, Yorio JT, Bhadkamkar NA, Kee BK, Garrett CR. Rare Though Not Mutually Exclusive: A Report of Three Cases of Concomitant KRAS and BRAF Mutation and a Review of the Literature. J Cancer. 2013;4:320–322. doi: 10.7150/jca.3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brim H, Mokarram P, Naghibalhossaini F, Saberi-Firoozi M, Al-Mandhari M, Al-Mawaly K, Al-Mjeni R, Al-Sayegh A, Raeburn S, Lee E, et al. Impact of BRAF, MLH1 on the incidence of microsatellite instability high colorectal cancer in populations based study. Mol Cancer. 2008;7:68. doi: 10.1186/1476-4598-7-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hong SP, Min BS, Kim TI, Cheon JH, Kim NK, Kim H, Kim WH. The differential impact of microsatellite instability as a marker of prognosis and tumour response between colon cancer and rectal cancer. Eur J Cancer. 2012;48:1235–1243. doi: 10.1016/j.ejca.2011.10.005. [DOI] [PubMed] [Google Scholar]
- 30.Samowitz WS, Sweeney C, Herrick J, Albertsen H, Levin TR, Murtaugh MA, Wolff RK, Slattery ML. Poor survival associated with the BRAF V600E mutation in microsatellite-stable colon cancers. Cancer Res. 2005;65:6063–6069. doi: 10.1158/0008-5472.CAN-05-0404. [DOI] [PubMed] [Google Scholar]


