Highlights
-
•
Serine and glycine are essential metabolites for cancer cells.
-
•
Serine and glycine provide precursors for macromolecules and antioxidant defence.
-
•
Metabolic enzymes of serine and glycine biosynthesis are upregulated in cancer.
-
•
Innovative anticancer therapy is aiming to target serine and glycine biosynthesis.
Keywords: cancer metabolism, serine, glycine, one-carbon metabolism, folate
Abstract
Serine and glycine are biosynthetically linked, and together provide the essential precursors for the synthesis of proteins, nucleic acids, and lipids that are crucial to cancer cell growth. Moreover, serine/glycine biosynthesis also affects cellular antioxidative capacity, thus supporting tumour homeostasis. A crucial contribution of serine/glycine to cellular metabolism is through the glycine cleavage system, which refuels one-carbon metabolism; a complex cyclic metabolic network based on chemical reactions of folate compounds. The importance of serine/glycine metabolism is further highlighted by genetic and functional evidence indicating that hyperactivation of the serine/glycine biosynthetic pathway drives oncogenesis. Recent developments in our understanding of these pathways provide novel translational opportunities for drug development, dietary intervention, and biomarker identification of human cancers.
Serine is a central hub of cancer metabolism
Cancer cells undergo specific metabolic reprogramming to sustain cell growth and proliferation [1,2]. In addition to a large energy requirement, cancer cells also accumulate building blocks for the construction of new cellular components, including nucleic acids, proteins, and lipids, as well as important cofactors for the maintenance of cellular redox status [3]. Glucose and glutamine are the main sources used to maintain active essential metabolic pathways, such as glycolysis and anaplerotic flux of the tricarboxylic acid (TCA) cycle. In anabolic pathways, the serine biosynthetic pathway represents a crucial turning point in glucose conversion [4]. Imported serine and serine derived from a branch of glycolysis can be converted to glycine, which in turn provides carbon units for one-carbon metabolism. The synthesis of proteins, lipids, nucleic acids, and other cofactors requires one-carbon metabolism, which is a complex metabolite network that is based on the chemical reactions of folate compounds. The one-carbon unit proceeds in a cyclical pathway from which it is transferred to other metabolic pathways. The importance of this metabolic pathway is underlined by the fact that antifolate chemotherapy is currently widely used in cancer treatment and has been since its discovery more than 50 years ago [5].
In this review, we outline the recent advances in the understanding and implications of serine and glycine metabolism in cancer biology. Genetic and functional evidence suggests that aberrant activation of the serine biosynthetic pathway is an essential process in cancer pathogenesis. Serine and glycine provide precursors for proteins, nucleic acids, and lipids. In addition, regulation of serine biosynthesis directly modulates cellular antioxidant capacity, thus implying a function in the maintenance of tumour homeostasis. All these findings may provide a new translational opportunity for drug development, dietary intervention, and biomarker identification in order to provide targeted antimetabolic therapy.
De novo serine biosynthesis
Glycolysis provides ATP and energy in most cell types, but cancer cells extensively use glycolysis to sustain anabolism, which is necessary for tumour growth. Serine biosynthesis is a component of these glycolysis-diverting pathways. The glycolytic intermediate 3-phosphoglycerate is converted to serine following a three-step enzymatic reaction (Figure 1). Cancer cells use phosphoglycerate dehydrogenase (PHGDH) and NAD to oxidise ∼10% of the 3-phosphoglycerate generated from glycolysis into the serine precursor 3-phosphohydroxypyruvate [6]. Subsequent enzymes in the pathway convert 3-phosphohydroxypyruvate into serine via transamination (PSAT1) and phosphate ester hydrolysis (PSPH) reactions. PHGDH expression is normally upregulated in triple-negative breast cancer and in melanoma [7]. In these tumours, the genomic locus on human chromosome 1p12 that encodes PHGDH is subject to frequent amplification, even though no oncogenes are included in this region [8]. These analyses suggest that tumours containing amplification of PHGDH might exploit serine biosynthesis activity. In support of this, suppression of PHGDH in cell lines with elevated PHGDH expression, but not in those without, causes a strong decrease in cell proliferation and a reduction in serine synthesis. In addition, consistent ectopic expression of PHDGH in the breast epithelial cell line MCF10A disrupts the acinar morphology and induces further phenotypic alterations that predispose to malignant transformation [9,10]. Therefore, PHDGH upregulation and serine biosynthesis can be necessary and/or sufficient to sustain cancer growth and oncogenic transformation.
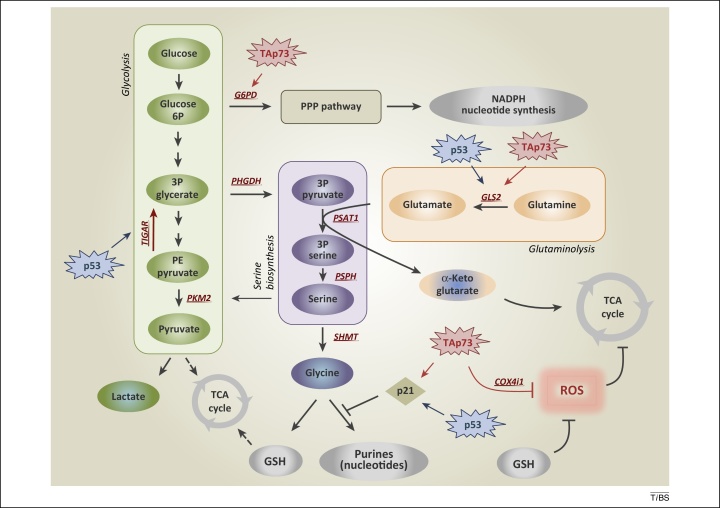
Figure 1.
De novo serine biosynthesis diverges from glycolysis. The serine synthesis pathway utilises the glycolytic intermediate 3P-glycerate, which is converted by PHGDH, PSAT-1, and PSPH into serine. Removal of exogenous serine causes activation of its biosynthetic pathway. Serine accumulation accelerates glycolytic flux, although allosteric activation of PKM2 by serine. p53, via TIGAR, and TAp73, via G6PD, facilitate activation of the PPP, promoting NADPH and nucleotide synthesis. p53-dependent activation of p21 induces transient cell cycle arrest, blocking flux to purines, thus maintaining GSH synthesis. TAp73 drives glutamine/glutamate conversion by inducing expression of GLS-2, thus pushing serine biosynthetic pathway, while it represses intracellular ROS controlling COX4i1 subunit expression. Abbreviations: 3P glycerate, glycerate-3-phosphate; PHGDH, phosphoglycerate dehydrogenase; PKM2, pyruvate kinase M2; PPP, pentose phosphate pathway; ROS, reactive oxygen species; COX4i1, cytochrome C oxidase subunit 4 isoform 1; G6PD, glucose-6-phosphate dehydrogenase; GLS-2, glutaminase-2; GSH, glutathione; PSAT-1, phosphoserine aminotransferase 1; PSPH, phosphoserine phosphatase; TIGAR, TP53-inducible glycolysis and apoptosis regulator.
Serine is a central metabolite for biosynthetic reactions, but PHGDH suppression inhibits proliferation, even in cells cultured in media containing exogenous serine. This suggests that, besides the control of the intracellular serine level, additional outputs and outcomes of serine biosynthetic activity push the requirement of PHGDH upregulation in cancer [7]. For instance, PSAT1 uses the PHGDH product 3-phosphohydroxypyruvate to convert glutamate to α-ketoglutarate. α-Ketoglutarate is an anaplerotic intermediate that refuels the TCA cycle and sustains cancer metabolism. A metabolic flux experiment with 13C-glutamine revealed that the conversion of glutamine to glutamate and then α-ketoglutarate, and other TCA intermediates, was significantly reduced in cells in which PHGDH or PSAT-1 was suppressed. Through these pathways the serine synthesis pathway is a major contributor of TCA intermediates; it is responsible for approximately half of the anaplerotic flux to the TCA cycle.
De novo synthesis of serine provides precursors for a variety of biosynthetic pathways, and accordingly, a pivotal aspect of the serine de novo synthesis is the conversion of serine to glycine by serine hydroxymethyltransferase (SHMT). As indicated above, glycine is a major source of methyl groups for the one-carbon pools required for the biosynthesis of glutathione (GSH), protein, purines and DNA/histone methylation. The details of the role of glycine in cancer cell metabolism are discussed in depth later.
Serine and the p53 family
The tumour suppressor p53 is emerging as an important regulator of cellular metabolism. p53 is a key player in the cellular response to stress in the form of numerous challenges, including DNA damage, hypoxia, and oncogene activation [11]. The complexity and diversity of these cellular processes indicate roles for p53 in normal and cancer cell homeostasis, in addition to its traditional role as a tumour suppressor [12]. The ability of p53 to respond to nutrient deficiencies lies in the established function of p53 as a mediator of the cellular stress response. p53 has been associated with the capacity of cancer cells to deal with serine starvation and oxidative stress, and indeed, p53 helps cancer cells to overcome serine starvation, preserving the cellular antioxidant capacity. Cells lacking p53 fail to respond to serine starvation due to oxidative stress, which leads to reduced viability and severely impaired proliferation. During serine starvation, activation of the p53–p21 axis leads to cell cycle arrest, which promotes cell survival by efficiently channelling depleted serine stores to glutathione synthesis [13]. The idea that cancer cells reprogram their metabolism to counteract reactive oxygen species (ROS) production fits with the suggestion that aerobic glycolysis (the Warburg effect) is adopted to avoid the generation of metabolic ROS [14]. These events underscore the relevance of p53 in coordinating metabolic reprogramming in response to metabolic stress. Recently, the p53-family member p73 was reported to play a role in serine biosynthesis. Metabolic profiling of a human cancer cell line revealed that TAp73 activates serine biosynthesis, resulting in increased intracellular levels of serine, glycine, and GSH. TAp73 depletion abrogates cancer cell proliferation during serine/glycine deprivation, supporting the role of p73 in promoting serine biosynthesis to help cancer cells under metabolic stress [15]. Therefore, p73 and p53 together appear to help cancer cells cope with the oxidative stress associated with serine starvation. This concept supports the importance of serine in cancer cell metabolism, indicating that many cancer cells may show sensitivity to serine depletion, in particular when p53 and/or p73 activity is altered. However, some cancer cells may circumvent serine dependence through other genetic alterations, such as PHGDH upregulation, which increases serine biosynthesis. Serine depletion (either by removal from the diet or in vivo enzymatic depletion) may represent an innovative therapeutic strategy that could create additional possibilities for human cancer treatment. The p53 family directly influences various glycolysis-related pathways, enabling cells to respond to metabolic stress. In particular both p53, via TP53-inducible glycolysis and apoptosis regulator (TIGAR) [16], and p73, via glucose-6-phosphate dehydrogenase (G6PD) [17], favour the pentose phosphate shunt and nucleotide biosynthesis. In addition, p53 [18,19], p63 [20], and p73 [21] activate the expression of glutaminase-2 (GLS-2), promoting glutaminolysis, which interferes with serine biosynthesis (Figure 1). Interestingly, likewise, the oncogenic transcription factor c-Myc represses miR-23a and miR-23b, resulting in upregulation of GLS-1 and glutaminolysis [22]. This underlines how a sophisticated integration of oncogene and tumour suppressor signalling might result in alteration of serine/glycine metabolism in cancer cells. The complexity of this scenario highlights the importance of a better understanding of the metabolic functions of oncogenes and tumour suppressors, in particular p53 family, to design novel therapeutic approaches in cancer.
Serine and pyruvate kinase (PK)M2
Allosteric positive and negative feedback is widely used to regulate metabolic pathways swiftly, and in this case, serine can allosterically fine-tune the glycolytic rate in cancer cells. PK catalyses the last step of the glycolytic pathway to convert phosphoenolpyruvate (PEP) to pyruvate and produce one molecule of ATP. The M2 isoform of PK (PKM2) is a splice variant of the PKM gene, which is predominantly expressed in proliferating tissue [23,24]. A clear picture of PKM2 function in cancer is still unresolved. Although several pieces of evidence suggest that a switch from PKM1 to PKM2 expression occurs in tumour cells during cancer transformation, the recent generation of PKM2−/− mice has revealed that PKM2 deletion accelerates tumour onset in a Brca1-loss-driven model of breast cancer. In contrast with PKM1, the M2 isoform shows low enzymatic activity. Lower PEP/pyruvate conversion favours accumulation of glycolytic intermediates, refuelling diverging anabolic pathways such as the pentose phosphate pathway (PPP) and serine biosynthesis. Allosteric regulations of PKM2 drive the shift between an active tetrameric form and a less-active dimer [25]. The tetramer has a high affinity for PEP, whereas the low affinity of the dimer results in reduced activity at physiological concentrations of PEP [26]. Serine acts as allosteric activator of PKM2 [27]. Serine can activate recombinant PKM2 with a half-maximal activation concentration (AC50) within the physiological range of intracellular serine concentrations (1.3 mM). Serine binding to PKM2 affects its enzymatic activity, lowering the Michaelis constant (Km) of PKM2 for PEP by 2.3-fold [28]. This regulation implies that when serine is abundant, PKM2 is fully activated, allowing the consumption of glucose through aerobic glycolysis. Upon serine deprivation, PKM2 activity is reduced, and pyruvate is diverted to a fuel-efficient mode in the mitochondria, and glycolysis metabolites are diverted to serine biosynthetic pathways to sustain cell proliferation. Through this mechanism, serine supports aerobic glycolysis and lactate production; events that are critical for cancer cell growth and survival.
Serine fuels glycine biosynthesis
Maintenance of the biosynthetic potential of the cell and of its redox and epigenetic status relies heavily on the shuttling of carbon units by folate derivatives in the one-carbon metabolic network. The complexity of this network and its multifaceted involvement in cell proliferation and cancer biology is now becoming clearer. Central to the aforementioned series of interconnected metabolic pathways is the conversion of serine to glycine that is catalysed by the enzyme serine hydroxymethyltransferase. The reaction catalysed by SHMT represents a major source of methyl groups for the one-carbon pools that are required for de novo nucleotide biosynthesis and DNA methylation. SHMT occupies a critical position at the convergence of two key pathways for chemotherapeutic intervention: serine/glycine metabolism and nucleotide biosynthesis.
Mammalian SHMTs are pyridoxal 5′-phosphate (PLP)-dependent enzymes [29] that catalyse the reversible transfer of the serine Cβ to tetrahydrofolate (tetrahydropteroylglutamate, H4PteGlu or THF) or its polyglutamylated derivatives, yielding glycine and 5,10-CH2-H4PteGlu (N5,N10-MTHF). In the human genome, two SHMT genes are found; SHMT1, encoding the cytoplasmic isozyme (SHMT1), and SHMT2, encoding the mitochondrial one (SHMT2) [30]. SHMT2 encodes a second transcript that lacks the mitochondrial import sequence; this isozyme (i.e., SHMT2α) localises in the cytoplasm with SHMT1 and accounts for the unexpected viability of Shmt1−/− mice [31]. Both SHMT1 and SHMT2 are transcriptional targets of c-Myc [32], and changes in their expression and/or activity have been shown in several tumours [33,34]. Together with c-Myc-dependent upregulation of GLS-1, this indicates that c-Myc plays a direct role in serine/glycine metabolism. In a recent study, the SHMT1 gene was found to be dispensable for Myc-induced lymphomagenesis, but mouse genetic models strongly suggest that the adaptability of tumour cells to alterations in critical metabolic pathways is cell-context dependent. For instance, no major impact of SHMT1 loss was observed on colon adenocarcinoma initiation and development in ApcMin mouse models, whereas acceleration of lymphomagenesis was detected in the λ-Myc mice, indicating a B cell specific event. These data argue against SHMT1 as a target for chemotherapy, although a deeper analysis of the relative role of the two isoforms is still awaited [35]. This evidence is clearly relevant when selecting patients for drug treatments (see below).
The SHMT2 isoform appears to be preferentially involved in the synthesis of mitochondrial dTMP, whereas SHMT1 and, to a lesser extent, SHMT2α participate to the synthesis of nuclear dTMP, undergoing nuclear import during S-phase and supplying N5,N10-MTHF during the thymidylate cycle along with thymidylate synthase (TS) and dihydrofolate reductase (DHFR) [31]. N5,N10-MTHF is also utilised in the de novo biosynthesis of purines. Recently, SHMT2 was also found to be associated with the deubiquitylating complex BRISC (Brcc36-containing isopeptidase complex, which is involved in the interferon response) and to be required for substrate targeting by BRISC, thus suggesting potential new roles for this protein in the cytosol [36].
Glycine can also be generated from threonine by threonine dehydrogenase (TDH) and glycine C-acetyltransferase (GCAT) (Figure 2). In vitro threonine deprivation promotes cell death that is associated with a dramatic reduction of histone methylation. Metabolic flux demonstrates that threonine enters one-carbon metabolism through glycine cleavage [37,38]. Intracellular glycine can also come from many other sources, such as betaine, choline, N-methylglycine and dimethyl-glycine following a series of demethylation reactions. The alternative to de novo synthesis of glycine is extracellular uptake. Direct import from the extracellular environment is enabled by amino acid transporters.
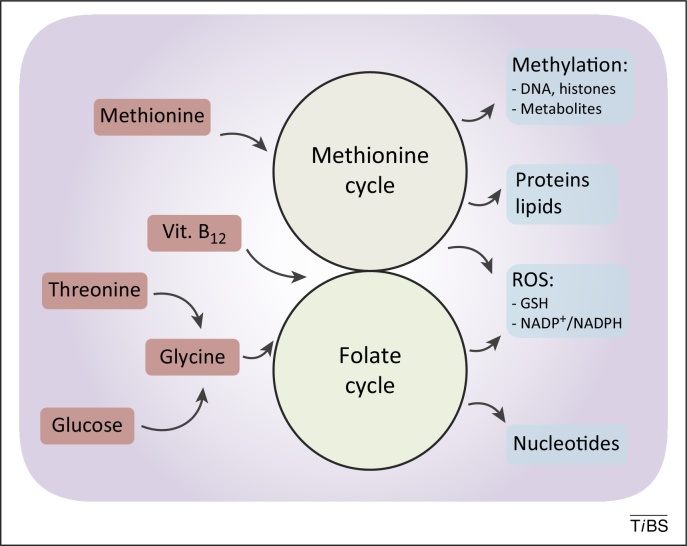
Figure 2.
De novo synthesised or imported serine and glycine refuel one-carbon metabolism. One-carbon metabolism comprises two interconnected metabolic cycles: the folate cycle and the methionine cycle. Different de novo or imported inputs (red boxes), including glycine derived from serine and vitamin B12, converge in this bimodular metabolic pathway. The. complex process produces multiple outputs (blue boxes), including substrates for methylation reactions, proteins, lipids, nucleotides, and reducing power against reactive oxygen species (ROS).
As mentioned above, the folate metabolites provided by SHMT-mediated reactions are also important in maintaining normal methylation patterns and DNA stability, and perturbation of folate levels has been shown to contribute to abnormal methylation patterns and DNA instability in many tumours and in lung cancer patients [33,39]. However, another important contribution of glycine to cell metabolism is associated with its introduction into the glycine cleavage system, which refuels one-carbon metabolism.
One-carbon metabolism
The so-called one-carbon metabolism cycles carbon units from different amino acids, generating several different outputs and integrating several cellular nutrient statuses (Figure 2). A central aspect of one-carbon metabolism is the transformation of folate in all its different states. The coupling of the folate cycle to the methionine cycle constitutes a bicyclic pathway that circulates carbon units and is collectively referred to as one-carbon metabolism. Folate is a B vitamin naturally found in many western foods, whereas the synthetically produced form, defined folic acid, is often included in dietary supplements. In cells, folate is reduced by a series of enzymes, leading to the generation of tetrahydrofolate (THF) (Figure 3). THF contributes to many metabolic reactions that move carbon units from one position to another, generating the folate cycle. The folate cycle is charged by the conversion of serine in glycine (SHMT activity) and by the glycine cleavage system; glycine dehydrogenase (GLDC; also known as GCSP) produces ammonia, carbon dioxide, and a carbon unit for the methylation of THF. The folate cycle is then coupled to the methionine cycle, with methyl-THF (mTHF) donating a carbon to homocysteine, methylating and converting it to methionine.
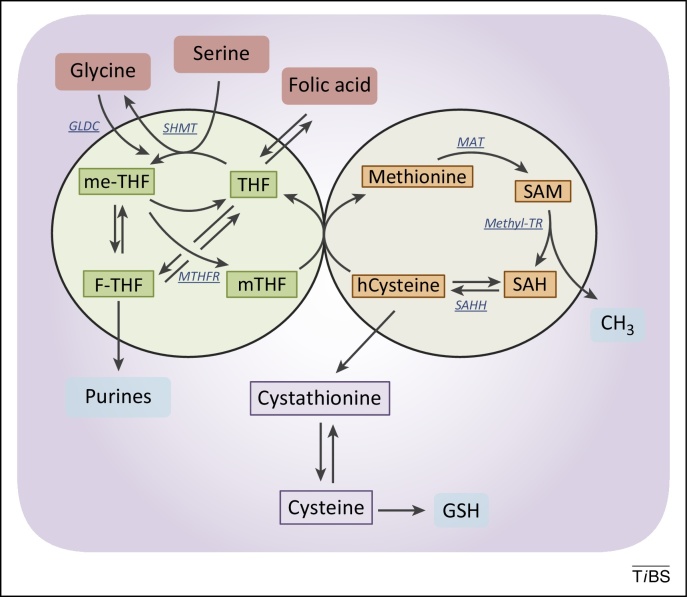
Figure 3.
The folate and the methionine cycles exist and can be modulated independently. Imported folic acid can be converted into reduced to THF and enter the folic cycle. THF is in turn converted to me-THF by SHMT. me-THF is then either converted to F-THF or reduced to mTHF by MTHFR. Demethylation of mTHF completes the folate cycle and begins the methionine cycle, converting hCysteine to methionine. Methionine is used to generate SAM, which is demethylated to form SAH. After deadenylation by SAHH, the cycle performs a full turn with the conversion of SAH in homocysteine. Abbreviations: F-THF, 10-formyltetrahydrofolate; hCysteine, homocysteine; me-THF, 5,10-methylene-THF; mTHF, 5-methyltetrahydrofolate; MTHFR, methylenetetrahydrofolate reductase; SAH, S-adenosylhomocysteine; SAHH, S-adenosyl homocysteine hydrolase; SAM, S-adenosylmethionine; SHMT, serine hydroxymethyl transferase; THF, tetrahydrofolate; MAT, methionine adenosyltransferase; Methyl-TR, methyl-transferase.
The output of one-carbon metabolism includes a myriad of components essential to the synthesis of all macromolecules, such as proteins, lipids, and nucleic acids, used in cellular growth and proliferation [40]. DNA synthesis requires nucleotides, a limiting metabolic aspect of cell proliferation. Methylation of dUMP by thymidylate synthase produces dTMP by creating a THF from mTHF. Additionally, purines require a folate pool for their generation through the intermediate 10-formyltetrahydrofolate (F-THF), which is derived from 5,10-methylene-THF. The methionine cycle is required for the synthesis of phosphatidylcholine, which contributes ∼50% of lipid membrane content [41]. Methionine itself is required for protein synthesis, but the adenylation of methionine produces S-adenosylmethionine (SAM). SAM functions as a methyl donor for other metabolic pathways that require methyl moieties, including histone, DNA and RNA methylation, lysine and arginine methylation [42,43], polyamine synthesis [44], and methylation reactions that generate the lipid head group [45]. Glycine/one-carbon metabolism also plays an important role in the cellular redox balance. The reduction of THF by tetrahydrofolate reductase produces one molecule of NADP+ for each turn of the folate cycle. Modulation of the NADP+/NADPH ratio contributes to the maintenance of redox status. Glutathione, a tripeptide comprising cysteine, glycine, and glutamate, is also important for the maintenance of the NADP+/NADPH ratio and is the major contributor to redox balance. Given their contributions to balancing the redox state, cancer cells gain advantages from alterations in these metabolic pathways.
Glycine metabolism is associated with cancer cell proliferation
The second product of the SHMT reaction (i.e., glycine) can be directed to the biosynthesis of purines, where it provides two carbon atoms and a nitrogen atom in the purine ring. Glycine is also an integral component of glutathione, therefore, the main antioxidant molecule of the cell, this amino acid is also required to maintain the cellular redox balance. In the mitochondria, glycine also fuels heme biosynthesis and thus sustains oxidative phosphorylation [46]. It has been recently shown that glycine uptake and catabolism is able to promote tumourigenesis and malignancy, suggesting that glycine metabolism could in principle be a target for therapeutic intervention [47]. By looking at variations in the level of more than 200 metabolites in the NCI-60 cell line, Jain and coworkers showed that both glycine consumption and expression of enzymes in the mitochondrial glycine biosynthetic pathway (but not the corresponding cytosolic enzymes) correlate with the rate of proliferation of cancer cells. Their results indicate a key role for the mitochondria in supporting rapid cancer cell proliferation. A recent meta-analysis, based on mRNA profiles of 1454 metabolic enzymes across 1981 tumours spanning 19 cancer types, further underscored the importance of mitochondrial compartmentalisation of one-carbon metabolism for cancer [48]. Antagonising glycine uptake and its mitochondrial biosynthesis preferentially impaired rapidly proliferating cells; in particular, silencing the mitochondrial SHMT2 gene and deprivation of extracellular glycine slowed down proliferation in HeLa cells and other fast-proliferating cancer cells by prolonging the G1 phase of the cell cycle [47]. Furthermore, upregulation of serine/glycine metabolism correlates with cell proliferation and poor prognosis in several tumours. Additionally, mathematical modelling of the serine/glycine conversion rate in a panel of 60 well-characterised cell lines (NCI60 panel) shows that glycine conversion increases the proliferation rate and contributes significantly to the biosynthetic requirements of purines, ATP, and NADPH in cancer cells [49].
These studies confirm that glycine deprivation (from the diet or by enzymatic depletion) may be a new route for human cancer chemotherapy; thus, targeting SHMT is a high priority in designing innovative therapeutic strategies.
Translational implications of serine and glycine metabolism
A current challenge in cancer biology is the identification of novel tools that may enable the selection of subsets of patients for therapeutic intervention with chemically selective pharmaceutical compounds. A landmark of cancer chemotherapy, and possibly still the most widely used drugs in medical oncology, are antimetabolites (antifolates); drugs that quench the effect of metabolites on cellular processes (Table 1). Among the antifolates, methotrexate and pemetrexed constitute a major class of cancer chemotherapy agents and are currently used as frontline chemotherapy for a diverse range of cancers, including acute lymphoblastic leukaemia, breast cancer, bladder cancer, and lymphomas. Methotrexate and pemetrexed have the ability to bind and inhibit human SHMT in vitro [50]. The action of mitochondrial enzymes in the metabolism of serine and glycine, in addition to those involved in folate metabolism, have been recently shown to be determinants of the response to methotrexate, suggesting that patients whose tumours show upregulation of serine and glycine mitochondrial enzymes are sensitive to folate antagonists targeting thymidylate or purine biosynthesis [51]. Antifolates show selectivity for rapidly proliferating tumours in patients with overexpression of genes coding for mitochondrial folate metabolism enzymes. About 25% of patients with cancers manifesting this phenotype could be selected for treatment with thymidylate and purine biosynthesis inhibitors. The identification of novel strategies, including those specifically targeting SHMT, allow the emergence of new antimetabolites and enlarge the therapeutic window (and efficacy) of current approaches. To design a successful clinical strategy, however, it will be important to select patients and find the right combinations of inhibitors targeting several metabolic enzymes in the serine/glycine pathway.
Table 1.
Antimetabolite drugs in cancer
| Drug | Target enzyme | Status | Cancer |
|---|---|---|---|
| Methotrexate | DHFR | Approved | Multiple cancers |
| Pemetrexed | DHFR, TS | Approved | Multiple cancers |
| Gemcitabine | Ribonucleotide reductase | Approved | Pancreatic cancer |
| 5-FU | Thymidylate reductase | Approved | Colorectal cancer |
| Pralatrexate | DHFR | Approved | Peripheral Tcell lymphoma |
| DMFO | Ornithine decaroboxylase | Clinical trial | Neuroblastoma |
| Azanucleotides | DNA methyltransferases | Approved | Leukaemia |
| Anti-serine biosynthesis | PHGDH PSAT, PSPH | Preclinical studies | |
| Anti-glycine biosynthesis | SHMT1, SHMT2 | Preclinical studies |
Alternative approaches aim to target downstream pathways of serine/glycine/one-carbon metabolism. Potent and clinically approved inhibitors of thymidylate and purine biosynthesis include pralatrexate and 5-fluorouracil (5-FU); methotrexate and pemetrexed, which are wide-selectivity antifolate molecules, also show antagonistic capacity [52]. 5-FU is a standard agent for the treatment of several cancer types. Mimicking uracil, 5-FU inhibits thymidylate synthase, resulting in impairment of the methylation of dUMP to dTMP and folate cycle disruption [53]. 5-FU is also intercellularly converted to 5-fluorouridine (5-FUDR), which incorporates into rRNA molecules and leads to inhibition of rRNA processing. As a result, 5-FUDR induces a p53-dependent cell cycle arrest and/or apoptosis [54].
However other approaches targeting mechanisms downstream of serine/glycine/one-carbon metabolism aim to modulate the epigenetic status of tumours. Inhibitors of methyltransferases, which affect post-translational modifications of histones and DNA, belong to this group. Several of these compounds are currently under preclinical evaluation or in early-stage clinical trials [55,56]. Preclinical studies are also ongoing for small molecules targeting the catalytic site of metabolic enzymes such as PHGDH PSAT, PSPH, and GLDC [6]. A different and novel approach to target cancer metabolism is the design of a complementary diet or nutrient modification to add to pharmacological agents. Supporting this idea, low glucose intake can have negative effects on tumour growth and progression [57,58]. Consistent with that effect, high carbohydrate intake is positively correlated with cancer incidence [59,60]. In addition, an in vivo study in mice explored the possibility of restricting serine and glycine metabolism for cancer intervention. In the absence of p53, serine and glycine withdrawal had an even greater effect, suggesting an epistatic interaction between p53 and the availability of serine and glycine [13]. However, the complexity of the relation between diet and one-carbon metabolism is highlighted by the observation that reduced administration of folate is also associated with breast and colorectal cancer [61].
Concluding remarks
A surge of interest in the study of cancer metabolism has occurred in the past few years, resulting in the constant expansion of the compendium of metabolic pathways essential to cancer biology. The recent advances in understanding the relation between cancer and metabolism have highlighted the relevance of serine/glycine biosynthesis and one-carbon metabolism. Genetic and functional evidence support the importance of the activity of these pathways as drivers of oncogenesis. Genetic activation of serine biosynthesis has been proven to drive directly cancer proliferation and predispose normal mammary epithelial cell line to cancer transformation; but how are these metabolic pathways integrated into the mutational landscape of a cancer cell? Little is known in this regard, or of the molecular mechanisms that govern the outcome of serine/glycine biosynthesis and one-carbon metabolism. A biochemical dissection of how these metabolic routes contribute to cancer biology is mandatory to explore its therapeutic potential. How do the inputs and the outputs integrate and balance their contribution to antioxidant response, anabolism, anaplerosis or epigenetic status? The knowledge currently available underlines the relevance of serine biosynthesis to preserve the antioxidant response of cancer cells. That p53−/− tumours cannot mount an antioxidant response in serine/glycine starvation highlights the importance of these metabolites, particularly their integration in the molecular context for the redox homeostasis of cancer cells. In light of their importance for the antioxidant response, it would also be interesting to investigate the contribution of serine/glycine biosynthesis to the early stage of tumourigenesis. Do these pathways affect cancer onset or only contribute to cancer progression and metastasis? Elucidation of these aspects, and others, might provide alternative clinical strategies, potentially enabling the design of personalised therapy for cancer patients (Box 1, outstanding questions). Mathematical models, integrative bioinformatics approaches, and metabolomics should speed development of the field. Building a conceptual framework to understand metabolic regulation in cancer is a challenging but compulsory endeavour that is still in its infancy. To design novel therapeutic approaches with high selectivity and efficacy, we need to understand not only the differences between normal and tumour cells, but also why some cancer cells are more dependent on specific metabolic pathways than others.
Box 1. Outstanding questions.
-
•
How are expression levels of serine/glycine biosynthetic enzymes regulated under basal and stress conditions in cancer cells?
-
•
How is serine/glycine biosynthesis coordinated with the mutational landscape (oncogene signalling) in cancer cells?
-
•
How do serine, glycine, and one-carbon metabolism integrate and balance their contribution to the antioxidant response, anabolism, anaplerosis, or epigenetic status to sustain a cancer cell?
-
•
Do serine and glycine equally contribute to the same phenomenon or do they retain exclusive functions?
-
•
Does serine/glycine biosynthesis pathway affect cancer onset or only contribute to cancer progression?
-
•
Would dietary intervention synergise with classic chemotherapeutic protocols in cancer patient treatments?
Acknowledgements
This work was supported by the Medical Research Council, UK, and grants from AIRC (2011-IG11955), AIRC 5xmille (MCO #9979), Min. Salute (Ric oncol 26/07) and IDI-IRCCS (RF08 c.15, RF07 c.57) to G.M. Funds from Associazione Italiana Ricerca sul Cancro (AIRC-IG2012 n.13150) and Ministero della Università e Ricerca of Italy (20094BJ9R7) to F.C. are gratefully acknowledged.
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial-No Derivative Works License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited.
Contributor Information
Ivano Amelio, Email: ia119@le.ac.uk.
Francesca Cutruzzolá, Email: francesca.cutruzzola@uniroma1.it.
Gerry Melino, Email: gm89@leicester.ac.uk.
References
- 1.Vander Heiden M.G. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cairns R.A. Regulation of cancer cell metabolism. Nat. Rev. Cancer. 2011;11:85–95. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- 3.Schulze A., Harris A.L. How cancer metabolism is tuned for proliferation and vulnerable to disruption. Nature. 2012;491:364–373. doi: 10.1038/nature11706. [DOI] [PubMed] [Google Scholar]
- 4.Kalhan S.C., Hanson R.W. Resurgence of serine: an often neglected but indispensable amino Acid. J. Biol. Chem. 2012;287:19786–19791. doi: 10.1074/jbc.R112.357194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Farber S., Diamond L.K. Temporary remissions in acute leukemia in children produced by folic acid antagonist, 4-aminopteroyl-glutamic acid. N. Engl. J. Med. 1948;238:787–793. doi: 10.1056/NEJM194806032382301. [DOI] [PubMed] [Google Scholar]
- 6.DeBerardinis R.J. Serine metabolism: some tumors take the road less traveled. Cell Metab. 2011;14:285–286. doi: 10.1016/j.cmet.2011.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Possemato R. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature. 2011;476:346–350. doi: 10.1038/nature10350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beroukhim R. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Locasale J.W. Phosphoglycerate dehydrogenase diverts glycolytic flux and contributes to oncogenesis. Nat. Genet. 2011;43:869–874. doi: 10.1038/ng.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Locasale J.W., Cantley L.C. Genetic selection for enhanced serine metabolism in cancer development. Cell Cycle. 2011;10:3812–3813. doi: 10.4161/cc.10.22.18224. [DOI] [PubMed] [Google Scholar]
- 11.Vousden K.H., Prives C. Blinded by the light: the growing complexity of p53. Cell. 2009;137:413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- 12.Vousden K.H., Ryan K.M. p53 and metabolism. Nat. Rev. Cancer. 2009;9:691–700. doi: 10.1038/nrc2715. [DOI] [PubMed] [Google Scholar]
- 13.Maddocks O.D. Serine starvation induces stress and p53-dependent metabolic remodelling in cancer cells. Nature. 2013;493:542–546. doi: 10.1038/nature11743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lunt S.Y., Vander Heiden M.G. Aerobic glycolysis: meeting the metabolic requirements of cell proliferation. Annu. Rev. Cell Dev. Biol. 2011;27:441–464. doi: 10.1146/annurev-cellbio-092910-154237. [DOI] [PubMed] [Google Scholar]
- 15.Amelio I. p73 regulates serine biosynthesis in cancer. Oncogene. 2013 doi: 10.1038/onc.2013.456. [DOI] [PubMed] [Google Scholar]
- 16.Bensaad K. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell. 2006;126:107–120. doi: 10.1016/j.cell.2006.05.036. [DOI] [PubMed] [Google Scholar]
- 17.Du W. TAp73 enhances the pentose phosphate pathway and supports cell proliferation. Nat. Cell Biol. 2013;15:991–1000. doi: 10.1038/ncb2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu W. Glutaminase 2, a novel p53 target gene regulating energy metabolism and antioxidant function. Proc. Natl. Acad. Sci. U.S.A. 2010;107:7455–7460. doi: 10.1073/pnas.1001006107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Suzuki S. Phosphate-activated glutaminase (GLS2), a p53-inducible regulator of glutamine metabolism and reactive oxygen species. Proc. Natl. Acad. Sci. U.S.A. 2010;107:7461–7466. doi: 10.1073/pnas.1002459107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Giacobbe A. p63 regulates glutaminase 2 expression. Cell Cycle. 2013;12:1395–1405. doi: 10.4161/cc.24478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Velletri T. GLS2 is transcriptionally regulated by p73 and contributes to neuronal differentiation. Cell Cycle. 2013;12:3564–3573. doi: 10.4161/cc.26771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao P. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature. 2009;458:762–765. doi: 10.1038/nature07823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Noguchi T. The L- and R-type isozymes of rat pyruvate kinase are produced from a single gene by use of different promoters. J. Biol. Chem. 1987;262:14366–14371. [PubMed] [Google Scholar]
- 24.Mazurek S. Pyruvate kinase type M2: a key regulator of the metabolic budget system in tumor cells. Int. J. Biochem. Cell Biol. 2011;43:969–980. doi: 10.1016/j.biocel.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 25.Mazurek S. Metabolic cooperation between different oncogenes during cell transformation: interaction between activated ras and HPV-16 E7. Oncogene. 2001;20:6891–6898. doi: 10.1038/sj.onc.1204792. [DOI] [PubMed] [Google Scholar]
- 26.Dombrauckas J.D. Structural basis for tumor pyruvate kinase M2 allosteric regulation and catalysis. Biochemistry. 2005;44:9417–9429. doi: 10.1021/bi0474923. [DOI] [PubMed] [Google Scholar]
- 27.Ye J. Pyruvate kinase M2 promotes de novo serine synthesis to sustain mTORC1 activity and cell proliferation. Proc. Natl. Acad. Sci. U.S.A. 2012;109:6904–6909. doi: 10.1073/pnas.1204176109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chaneton B. Serine is a natural ligand and allosteric activator of pyruvate kinase M2. Nature. 2012;491:458–462. doi: 10.1038/nature11540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Renwick S.B. The crystal structure of human cytosolic serine hydroxymethyltransferase: a target for cancer chemotherapy. Structure. 1998;6:1105–1116. doi: 10.1016/s0969-2126(98)00112-9. [DOI] [PubMed] [Google Scholar]
- 30.Garrow T.A. Cloning of human cDNAs encoding mitochondrial and cytosolic serine hydroxymethyltransferases and chromosomal localization. J. Biol. Chem. 1993;268:11910–11916. [PubMed] [Google Scholar]
- 31.Anderson D.D., Stover P.J. SHMT1 and SHMT2 are functionally redundant in nuclear de novo thymidylate biosynthesis. PLoS ONE. 2009;4:e5839. doi: 10.1371/journal.pone.0005839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nikiforov M.A. A functional screen for Myc-responsive genes reveals serine hydroxymethyltransferase, a major source of the one-carbon unit for cell metabolism. Mol. Cell. Biol. 2002;22:5793–5800. doi: 10.1128/MCB.22.16.5793-5800.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Piskac-Collier A.L. Variants in folate pathway genes as modulators of genetic instability and lung cancer risk. Genes Chromosomes Cancer. 2011;50:1–12. doi: 10.1002/gcc.20826. [DOI] [PubMed] [Google Scholar]
- 34.Zhang W.C. Glycine decarboxylase activity drives non-small cell lung cancer tumor-initiating cells and tumorigenesis. Cell. 2012;148:259–272. doi: 10.1016/j.cell.2011.11.050. [DOI] [PubMed] [Google Scholar]
- 35.Nilsson L.M. Mouse genetics suggests cell-context dependency for Myc-regulated metabolic enzymes during tumorigenesis. PLoS Genet. 2012;8:e1002573. doi: 10.1371/journal.pgen.1002573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zheng H. A BRISC-SHMT complex deubiquitinates IFNAR1 and regulates interferon responses. Cell Rep. 2013;5:180–193. doi: 10.1016/j.celrep.2013.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shyh-Chang N. Influence of threonine metabolism on S-adenosylmethionine and histone methylation. Science. 2013;339:222–226. doi: 10.1126/science.1226603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sassone-Corsi P. Physiology. When metabolism and epigenetics converge. Science. 2013;339:148–150. doi: 10.1126/science.1233423. [DOI] [PubMed] [Google Scholar]
- 39.Duthie S.J. Folate and cancer: how DNA damage, repair and methylation impact on colon carcinogenesis. J. Inherit. Metab. Dis. 2011;34:101–109. doi: 10.1007/s10545-010-9128-0. [DOI] [PubMed] [Google Scholar]
- 40.Deberardinis R.J. Brick by brick: metabolism and tumor cell growth. Curr. Opin. Genet. Dev. 2008;18:54–61. doi: 10.1016/j.gde.2008.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Spector A.A., Yorek M.A. Membrane lipid composition and cellular function. J. Lipid Res. 1985;26:1015–1035. [PubMed] [Google Scholar]
- 42.Teperino R. Histone methyl transferases and demethylases; can they link metabolism and transcription? Cell Metab. 2010;12:321–327. doi: 10.1016/j.cmet.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang Y., Bedford M.T. Protein arginine methyltransferases and cancer. Nat. Rev. Cancer. 2013;13:37–50. doi: 10.1038/nrc3409. [DOI] [PubMed] [Google Scholar]
- 44.Heby O., Persson L. Molecular genetics of polyamine synthesis in eukaryotic cells. Trends Biochem. Sci. 1990;15:153–158. doi: 10.1016/0968-0004(90)90216-x. [DOI] [PubMed] [Google Scholar]
- 45.Hickman M.J. Coordinated regulation of sulfur and phospholipid metabolism reflects the importance of methylation in the growth of yeast. Mol. Biol. Cell. 2011;22:4192–4204. doi: 10.1091/mbc.E11-05-0467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.di Salvo M.L. Glycine consumption and mitochondrial serine hydroxymethyltransferase in cancer cells: the heme connection. Med. Hypotheses. 2013;80:633–636. doi: 10.1016/j.mehy.2013.02.008. [DOI] [PubMed] [Google Scholar]
- 47.Jain M. Metabolite profiling identifies a key role for glycine in rapid cancer cell proliferation. Science. 2012;336:1040–1044. doi: 10.1126/science.1218595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nilsson R. Metabolic enzyme expression highlights a key role for MTHFD2 and the mitochondrial folate pathway in cancer. Nat. Commun. 2014;5:3128. doi: 10.1038/ncomms4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tedeschi P.M. Contribution of serine, folate and glycine metabolism to the ATP, NADPH and purine requirements of cancer cells. Cell Death Dis. 2013;4:e877. doi: 10.1038/cddis.2013.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Daidone F. In silico and in vitro validation of serine hydroxymethyltransferase as a chemotherapeutic target of the antifolate drug pemetrexed. Eur. J. Med. Chem. 2011;46:1616–1621. doi: 10.1016/j.ejmech.2011.02.009. [DOI] [PubMed] [Google Scholar]
- 51.Vazquez A. Overexpression of the mitochondrial folate and glycine-serine pathway: a new determinant of methotrexate selectivity in tumors. Cancer Res. 2013;73:478–482. doi: 10.1158/0008-5472.CAN-12-3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Goldman I.D. The antifolates: evolution, new agents in the clinic, and how targeting delivery via specific membrane transporters is driving the development of a next generation of folate analogs. Curr. Opin. Investig. Drugs. 2010;11:1409–1423. [PubMed] [Google Scholar]
- 53.Spears C.P. In vivo kinetics of thymidylate synthetase inhibition of 5-fluorouracil-sensitive and -resistant murine colon adenocarcinomas. Cancer Res. 1982;42:450–456. [PubMed] [Google Scholar]
- 54.Ghoshal K., Jacob S.T. Specific inhibition of pre-ribosomal RNA processing in extracts from the lymphosarcoma cells treated with 5-fluorouracil. Cancer Res. 1994;54:632–636. [PubMed] [Google Scholar]
- 55.Stresemann C. Functional diversity of DNA methyltransferase inhibitors in human cancer cell lines. Cancer Res. 2006;66:2794–2800. doi: 10.1158/0008-5472.CAN-05-2821. [DOI] [PubMed] [Google Scholar]
- 56.Spannhoff A. The emerging therapeutic potential of histone methyltransferase and demethylase inhibitors. ChemMedChem. 2009;4:1568–1582. doi: 10.1002/cmdc.200900301. [DOI] [PubMed] [Google Scholar]
- 57.Myers A.P., Cantley L.C. Sugar free, cancer free? Nutrition. 2012;28:1036. doi: 10.1016/j.nut.2012.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fine E.J. Targeting insulin inhibition as a metabolic therapy in advanced cancer: a pilot safety and feasibility dietary trial in 10 patients. Nutrition. 2012;28:1028–1035. doi: 10.1016/j.nut.2012.05.001. [DOI] [PubMed] [Google Scholar]
- 59.Fedirko V. Glycemic index, glycemic load, dietary carbohydrate, and dietary fiber intake and risk of liver and biliary tract cancers in Western Europeans. Ann. Oncol. 2013;24:543–553. doi: 10.1093/annonc/mds434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nagle C.M. Carbohydrate intake, glycemic load, glycemic index, and risk of ovarian cancer. Ann. Oncol. 2011;22:1332–1338. doi: 10.1093/annonc/mdq595. [DOI] [PubMed] [Google Scholar]
- 61.Christensen B.C. Breast cancer DNA methylation profiles are associated with tumor size and alcohol and folate intake. PLoS Genet. 2010;6:e1001043. doi: 10.1371/journal.pgen.1001043. [DOI] [PMC free article] [PubMed] [Google Scholar]