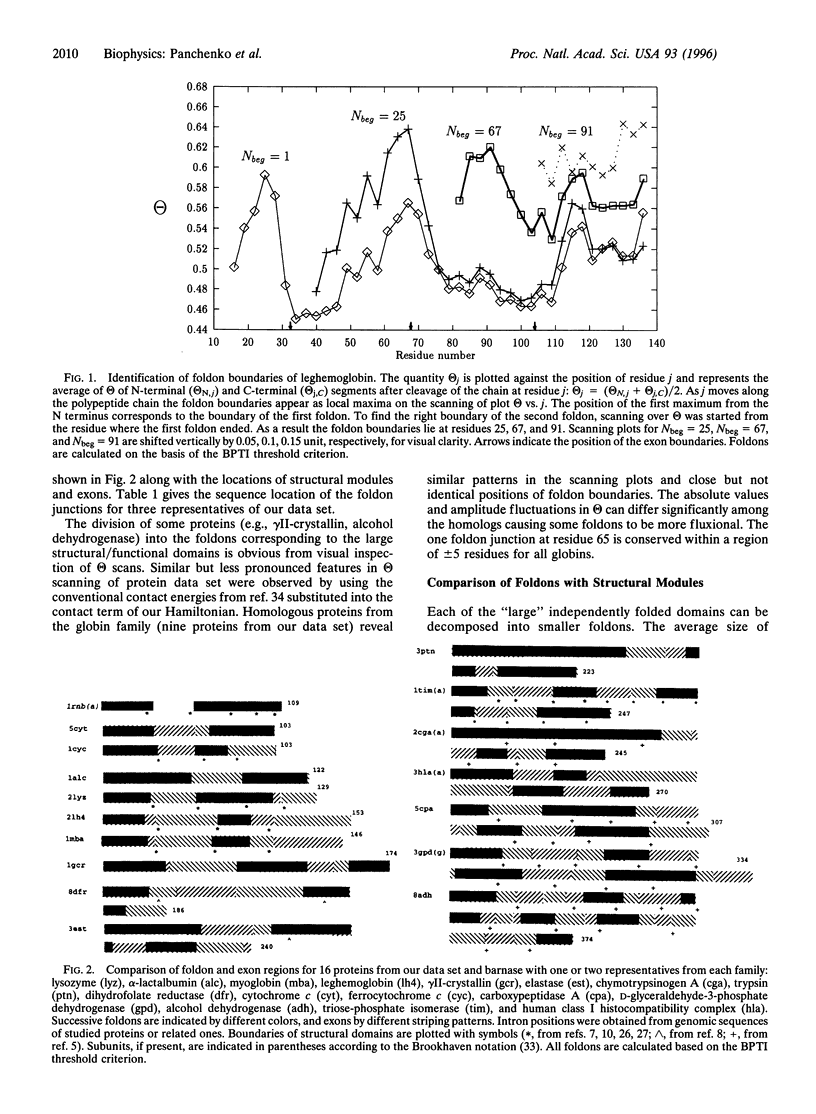
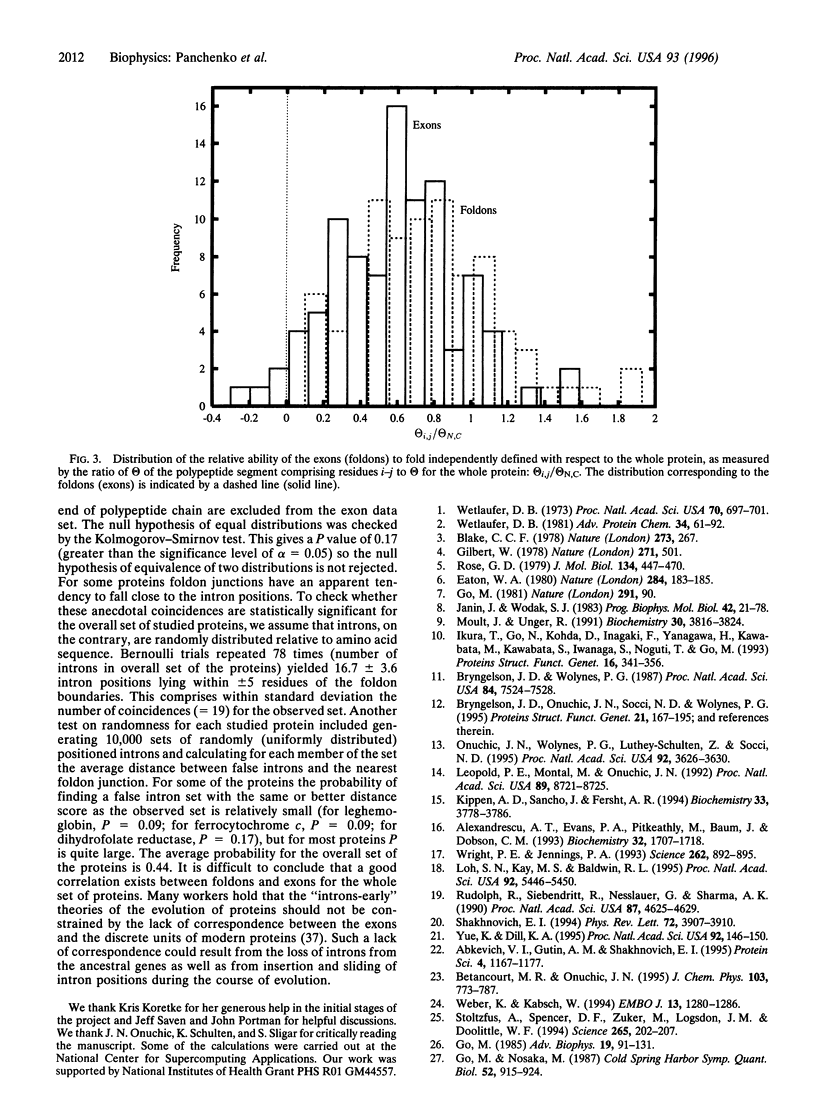
Abstract
Foldons, which are kinetically competent, quasi-independently folding units of a protein, may be defined using energy landscape analysis. Foldons can be identified by maxima in a scan of the ratio of a contiguous segment's energetic stability gap to the energy variance of that segment's molten globule states, reflecting the requirement of minimal frustration. The predicted foldons are compared with the exons and structural modules for 16 of the 30 proteins studied. Statistical analysis indicates a strong correlation between the energetically determined foldons and Go's geometrically defined structural modules, but there are marked sequence-dependent effects. There is only a weak correlation of foldons to exons. For gammaII-crystallin, myoglobin, barnase, alpha-lactalbumin, and cytochrome c the foldons and some noncontiguous clusters of foldons compare well with intermediates observed in experiment.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abkevich V. I., Gutin A. M., Shakhnovich E. I. Domains in folding of model proteins. Protein Sci. 1995 Jun;4(6):1167–1177. doi: 10.1002/pro.5560040615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrescu A. T., Evans P. A., Pitkeathly M., Baum J., Dobson C. M. Structure and dynamics of the acid-denatured molten globule state of alpha-lactalbumin: a two-dimensional NMR study. Biochemistry. 1993 Feb 23;32(7):1707–1718. doi: 10.1021/bi00058a003. [DOI] [PubMed] [Google Scholar]
- Bai Y., Sosnick T. R., Mayne L., Englander S. W. Protein folding intermediates: native-state hydrogen exchange. Science. 1995 Jul 14;269(5221):192–197. doi: 10.1126/science.7618079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein F. C., Koetzle T. F., Williams G. J., Meyer E. F., Jr, Brice M. D., Rodgers J. R., Kennard O., Shimanouchi T., Tasumi M. The Protein Data Bank: a computer-based archival file for macromolecular structures. J Mol Biol. 1977 May 25;112(3):535–542. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- Bryngelson J. D., Onuchic J. N., Socci N. D., Wolynes P. G. Funnels, pathways, and the energy landscape of protein folding: a synthesis. Proteins. 1995 Mar;21(3):167–195. doi: 10.1002/prot.340210302. [DOI] [PubMed] [Google Scholar]
- Bryngelson J. D., Wolynes P. G. Spin glasses and the statistical mechanics of protein folding. Proc Natl Acad Sci U S A. 1987 Nov;84(21):7524–7528. doi: 10.1073/pnas.84.21.7524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton W. A. The relationship between coding sequences and function in haemoglobin. Nature. 1980 Mar 13;284(5752):183–185. doi: 10.1038/284183a0. [DOI] [PubMed] [Google Scholar]
- Gilbert W., Glynias M. On the ancient nature of introns. Gene. 1993 Dec 15;135(1-2):137–144. doi: 10.1016/0378-1119(93)90058-b. [DOI] [PubMed] [Google Scholar]
- Gilbert W. Why genes in pieces? Nature. 1978 Feb 9;271(5645):501–501. doi: 10.1038/271501a0. [DOI] [PubMed] [Google Scholar]
- Go M. Correlation of DNA exonic regions with protein structural units in haemoglobin. Nature. 1981 May 7;291(5810):90–92. doi: 10.1038/291090a0. [DOI] [PubMed] [Google Scholar]
- Go M., Nosaka M. Protein architecture and the origin of introns. Cold Spring Harb Symp Quant Biol. 1987;52:915–924. doi: 10.1101/sqb.1987.052.01.100. [DOI] [PubMed] [Google Scholar]
- Go M. Protein structures and split genes. Adv Biophys. 1985;19:91–131. doi: 10.1016/0065-227x(85)90052-8. [DOI] [PubMed] [Google Scholar]
- Goldstein R. A., Luthey-Schulten Z. A., Wolynes P. G. Protein tertiary structure recognition using optimized Hamiltonians with local interactions. Proc Natl Acad Sci U S A. 1992 Oct 1;89(19):9029–9033. doi: 10.1073/pnas.89.19.9029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikura T., Go N., Kohda D., Inagaki F., Yanagawa H., Kawabata M., Kawabata S., Iwanaga S., Noguti T., Go M. Secondary structural features of modules M2 and M3 of barnase in solution by NMR experiment and distance geometry calculation. Proteins. 1993 Aug;16(4):341–356. doi: 10.1002/prot.340160404. [DOI] [PubMed] [Google Scholar]
- Jackson S. E., Fersht A. R. Folding of chymotrypsin inhibitor 2. 1. Evidence for a two-state transition. Biochemistry. 1991 Oct 29;30(43):10428–10435. doi: 10.1021/bi00107a010. [DOI] [PubMed] [Google Scholar]
- Janin J., Wodak S. J. Structural domains in proteins and their role in the dynamics of protein function. Prog Biophys Mol Biol. 1983;42(1):21–78. doi: 10.1016/0079-6107(83)90003-2. [DOI] [PubMed] [Google Scholar]
- Jennings P. A., Wright P. E. Formation of a molten globule intermediate early in the kinetic folding pathway of apomyoglobin. Science. 1993 Nov 5;262(5135):892–896. doi: 10.1126/science.8235610. [DOI] [PubMed] [Google Scholar]
- Kikuchi T., Némethy G., Scheraga H. A. Prediction of the location of structural domains in globular proteins. J Protein Chem. 1988 Aug;7(4):427–471. doi: 10.1007/BF01024890. [DOI] [PubMed] [Google Scholar]
- Kippen A. D., Sancho J., Fersht A. R. Folding of barnase in parts. Biochemistry. 1994 Mar 29;33(12):3778–3786. doi: 10.1021/bi00178a039. [DOI] [PubMed] [Google Scholar]
- Leopold P. E., Montal M., Onuchic J. N. Protein folding funnels: a kinetic approach to the sequence-structure relationship. Proc Natl Acad Sci U S A. 1992 Sep 15;89(18):8721–8725. doi: 10.1073/pnas.89.18.8721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh S. N., Kay M. S., Baldwin R. L. Structure and stability of a second molten globule intermediate in the apomyoglobin folding pathway. Proc Natl Acad Sci U S A. 1995 Jun 6;92(12):5446–5450. doi: 10.1073/pnas.92.12.5446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moult J., Unger R. An analysis of protein folding pathways. Biochemistry. 1991 Apr 23;30(16):3816–3824. doi: 10.1021/bi00230a003. [DOI] [PubMed] [Google Scholar]
- Onuchic J. N., Wolynes P. G., Luthey-Schulten Z., Socci N. D. Toward an outline of the topography of a realistic protein-folding funnel. Proc Natl Acad Sci U S A. 1995 Apr 11;92(8):3626–3630. doi: 10.1073/pnas.92.8.3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose G. D. Hierarchic organization of domains in globular proteins. J Mol Biol. 1979 Nov 5;134(3):447–470. doi: 10.1016/0022-2836(79)90363-2. [DOI] [PubMed] [Google Scholar]
- Rudolph R., Siebendritt R., Nesslaŭer G., Sharma A. K., Jaenicke R. Folding of an all-beta protein: independent domain folding in gamma II-crystallin from calf eye lens. Proc Natl Acad Sci U S A. 1990 Jun;87(12):4625–4629. doi: 10.1073/pnas.87.12.4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidel H. M., Pompliano D. L., Knowles J. R. Exons as microgenes? Science. 1992 Sep 11;257(5076):1489–1490. doi: 10.1126/science.1523407. [DOI] [PubMed] [Google Scholar]
- Shakhnovich EI. Proteins with selected sequences fold into unique native conformation. Phys Rev Lett. 1994 Jun 13;72(24):3907–3910. doi: 10.1103/PhysRevLett.72.3907. [DOI] [PubMed] [Google Scholar]
- Staley J. P., Kim P. S. Complete folding of bovine pancreatic trypsin inhibitor with only a single disulfide bond. Proc Natl Acad Sci U S A. 1992 Mar 1;89(5):1519–1523. doi: 10.1073/pnas.89.5.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoltzfus A., Spencer D. F., Zuker M., Logsdon J. M., Jr, Doolittle W. F. Testing the exon theory of genes: the evidence from protein structure. Science. 1994 Jul 8;265(5169):202–207. doi: 10.1126/science.8023140. [DOI] [PubMed] [Google Scholar]
- Weber K., Kabsch W. Intron positions in actin genes seem unrelated to the secondary structure of the protein. EMBO J. 1994 Mar 15;13(6):1280–1286. doi: 10.1002/j.1460-2075.1994.tb06380.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetlaufer D. B. Folding of protein fragments. Adv Protein Chem. 1981;34:61–92. doi: 10.1016/s0065-3233(08)60518-5. [DOI] [PubMed] [Google Scholar]
- Wetlaufer D. B. Nucleation, rapid folding, and globular intrachain regions in proteins. Proc Natl Acad Sci U S A. 1973 Mar;70(3):697–701. doi: 10.1073/pnas.70.3.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagawa H., Yoshida K., Torigoe C., Park J. S., Sato K., Shirai T., Go M. Protein anatomy: functional roles of barnase module. J Biol Chem. 1993 Mar 15;268(8):5861–5865. [PubMed] [Google Scholar]
- Yue K., Dill K. A. Forces of tertiary structural organization in globular proteins. Proc Natl Acad Sci U S A. 1995 Jan 3;92(1):146–150. doi: 10.1073/pnas.92.1.146. [DOI] [PMC free article] [PubMed] [Google Scholar]