Abstract
The tryptophan metabolites indole, indole-3-acetate, and tryptamine were identified in mouse cecal extracts and fecal pellets by mass spectrometry. The aryl hydrocarbon receptor (AHR) agonist and antagonist activities of these microbiota-derived compounds were investigated in CaCo-2 intestinal cells as a model for understanding their interactions with colonic tissue, which is highly aryl hydrocarbon (Ah)–responsive. Activation of Ah-responsive genes demonstrated that tryptamine and indole 3-acetate were AHR agonists, whereas indole was an AHR antagonist that inhibited TCDD (2,3,7,8-tetrachlorodibenzo-p-dioxin)–induced CYP1A1 expression. In contrast, the tryptophan metabolites exhibited minimal anti-inflammatory activities, whereas TCDD decreased phorbol ester-induced CXCR4 [chemokine (C-X-C motif) receptor 4] gene expression, and this response was AHR dependent. These results demonstrate that the tryptophan metabolites indole, tryptamine, and indole-3-acetate modulate AHR-mediated responses in CaCo-2 cells, and concentrations of indole that exhibit AHR antagonist activity (100–250 μM) are detected in the intestinal microbiome.
Introduction
The aryl hydrocarbon receptor (AHR) is a ligand-activated transcription factor that forms a nuclear heterodimer with the AHR nuclear translocator protein to activate gene transcription (Hankinson, 1995; Denison et al., 2011). AHR complex-mediated gene expression involves interaction with dioxin-responsive elements (DREs) in 5′-promoter regions of target genes, and the overall mechanisms of this response have been intensively investigated using the CYP1A1 gene and other drug-metabolizing enzymes as models (Whitlock, 1999). More recent studies demonstrate that this genomic pathway for activation of the AHR is more complex than the classic induction of CYP1A1, and there is also evidence for extranuclear activity of the AHR (Huang and Elferink, 2012; Tanos et al., 2012). TCDD (2,3,7,8-tetrachlordibenzo-p-dioxin) and structurally related halogenated aromatics and polynuclear aromatic hydrocarbons were among the first compounds identified as AHR ligands (Poland et al., 1976; Bigelow and Nebert, 1982). However, subsequent studies have shown that this receptor binds structurally diverse compounds that include industrial compounds, pharmaceuticals, phytochemicals such as flavonoids and indole derivatives, and endogenous biochemicals including indigoids, kynurenine, 7-ketocholesterol, FICZ (6-formylindolo-[3,2-b]-carbazole), and bilirubin (reviewed in Denison and Nagy, 2003; Safe et al., 2012). AHR ligands exhibit both agonist and antagonist activities, and there is evidence that some compounds exhibit tissue/cell-specific agonist or antagonist activities (Jordan, 2007). For example, α-naphthoflavone and 3′-methoxy-4′-nitroflavone were initially characterized as AHR antagonists, but subsequent studies showed that they exhibit both agonist and antagonist activity (Lu et al., 1996; Zhou and Gasiewicz, 2003). The AHR-active pharmaceutical mexiletine induces hepatic CYP1A1 in vivo and in vitro and binds the AHR (Hu et al., 2007); however, in MDA-MB-468 breast cancer cells, mexiletine inhibits induction of CYP1A1 by TCDD and is an AHR antagonist (Jin et al., 2012), and this is typical of a tissue-specific selective AHR modulator (SAhRM).
There is increasing evidence that endogenous AHR plays an important functional role in multiple tissues and organs (Barouki et al., 2007; McMillan and Bradfield, 2007), and this has been amply demonstrated in AHR−/− mice and by AHR silencing in nontransformed and transformed cell lines. For example, in gastrointestinal tissues including lymphoid tissues, the receptor and its ligands modulate inflammatory responses, including those associated with induced colitis in animal models (Kawajiri et al., 2009; Arsenescu et al., 2011; Benson and Shepherd, 2011; Furumatsu et al., 2011; Kiss et al., 2011; Li et al., 2011; Singh et al., 2011). The AHR agonist β-naphthoflavone significantly suppresses dextran sodium sulfate-induced colitis in C57BL/6 mice (Furumatsu et al., 2011) and in the APCmin model of colon cancer, the loss of AHR enhances colon carcinogenesis, and AHR ligands inhibit polyp formation in APCmin/AHR+/+ mice (Kawajiri et al., 2009).
Previous studies in our laboratory have identified the tryptophan metabolite indole as a major extracellular metabolite produced by gut bacteria, such as Escherichia coli (Bansal et al., 2010); in human feces, concentrations of indole can reach millimolar levels (Karlin et al., 1985; Zuccato et al., 1993). Indole exhibits AHR agonist activity in a yeast assay but is inactive in an aryl hydrocarbon (Ah)–responsive liver cancer cell line (Miller, 1997; Heath-Pagliuso et al., 1998). However, other tryptophan-derived compounds, including tryptamine, indole-3-acetate, and 3-indoxyl sulfate, have previously been characterized as AHR agonists (Gillner et al., 1985; Heath-Pagliuso et al., 1998; Miller, 1997; Schroeder et al., 2010; Vikstrom Bergander et al., 2012). It has been well established that the gut microbiome and its metabolites have a direct effect on intestinal homeostasis (Villard et al., 2007; DiNatale et al., 2010; Maynard et al., 2012; Tremaroli and Backhed, 2012), and the AHR plays an important role in maintaining gut homeostasis (Kawajiri et al., 2009; Arsenescu et al., 2011; Benson and Shepherd, 2011; Furumatsu et al., 2011; Kiss et al., 2011; Li et al., 2011; Singh et al., 2011). Therefore, the major objectives of this research were to investigate the AHR agonist or antagonist activity of tryptophan-derived microbiota metabolites in CaCo-2 human epithelial colon cancer cells and other cell lines, and determine their role in modulating inflammation in CaCo-2 cells. The results clearly demonstrate the production of high levels of tryptophan metabolites in the gut microbiome, and the AHR agonist and antagonist activities of these metabolites are response, cell context, and compound dependent, which is typical of a SAhRM.
Materials and Methods
Cell Lines, Antibodies, and Reagents.
CaCo-2 human colon cancer cell line and MDA-MB-468 and MDA-MB-231 human breast cancer cell lines were obtained from the American Type Culture Collection (Manassas, VA). CaCo-2 cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) nutrient mixture supplemented with 20% fetal bovine serum (FBS), 10 ml/l 100× minimum Eagle’s medium (MEM) nonessential amino acid solution (Gibco/Life Technologies, Grand Island, NY), and 10 ml/l 100× antibiotic/antimycotic solution (Sigma-Aldrich, St. Louis, MO). MDA-MB-468 and MDA-MB-231 cells were maintained in DMEM nutrient mixture supplemented with 10% FBS and 10 ml/l 100× antibiotic/antimycotic solution (Sigma-Aldrich). The cells were maintained at 37°C in the presence of 5% CO2, and the solvent (dimethyl sulfoxide [DMSO]) used in the experiments was ≤0.2%. Monoamine oxidase-A small interfering RNA (siRNA) was purchased from Santa Cruz Biotechnology (Santa Cruz, CA). CYP1A1, CYP1B1, AHR, and β-actin antibodies were purchased from Santa Cruz Biotechnology, and β-catenin antibody was purchased from Cell Signaling Technologies (Beverly, MA). The CYP1A1 and CYP1B1 antibodies show minimal cross-reactivity. All compounds used in this study were purchased from Sigma-Aldrich.
Sample Collection.
Female C57BL/6 mice at 5 weeks of age were purchased from The Jackson Laboratory (Bar Harbor, ME) and were allowed to acclimate for 1 week. All mice were maintained in a pathogen-free animal facility located at the Texas A&M University. The animals were handled in accordance with the Institutional Animal Care and Use Committee guidelines under an approved animal use protocol. Mice (n = 7) were sacrificed at 6 weeks of age. The entire cecum (tissue with luminal contents) and fecal pellets were collected from each animal. The samples were weighed, flash frozen, and stored at −80°C before processing for extraction.
Metabolites were extracted from the cecal tissue or fecal pellets using a methanol/chloroform extraction method (Sellick et al., 2010) with minor modifications. Cold methanol/chloroform (2:1, v/v; 1.5 ml) was added to a preweighed cecal or fecal sample and homogenized on ice. The sample tube was centrifuged at 15,000g for 10 minutes at 4°C, and the supernatant was transferred to a new sample tube through a 70-μm cell strainer. Ice-cold water (0.6 ml) was added, and the sample tube was vortexed and centrifuged (15,000g, 5 minutes, 4°C) to obtain phase separation. The upper and lower phases were separately collected in fresh sample tubes with a syringe, taking care not to disturb the interface. The polar (upper) phase (500 μl) was evaporated to dryness in a Savant SpeedVac concentrator (Thermo Scientific, Asheville, NC), and then was reconstituted in 50 μl of methanol/water (1:1, v/v). Extracted metabolites were stored at −80°C until analysis.
Metabolite Analysis.
Before the sample analysis, mass spectrometry (MS) parameters were optimized for each target metabolite to identify the multiple reaction monitoring (MRM) transition (precursor/product fragment ion pair) with the highest intensity under direction injection at 10 μl/min. The following parameters were optimized operating in positive mode: declustering potential, entrance potential, collision energy, and collision cell exit potential. The optimized parameter values for the target metabolites analyzed in this study are shown in Supplemental Table 1. The target metabolites in samples were detected and quantified on a triple quadrupole linear ion trap mass spectrometer (3200 QTRAP; AB Sciex, Foster City, CA) coupled to a binary pump high-performance liquid chromatography (1200 Series; Agilent Technologies, Santa Clara, CA). Peak identification and integration were performed using Analyst software (version 5; Agilent Technologies, Foster City, CA). Samples were maintained at 4°C on an autosampler before injection. Chromatographic separation was achieved on a hydrophilic interaction column (Luna 5 μm NH2 100 Å 250 mm × 2 mm; Phenomenex, Torrance, CA) using a solvent gradient method (Bajad et al., 2006). Solvent A was an ammonium acetate (20 mM) solution in water with 5% acetonitrile (v/v). The pH of solvent A was adjusted to 9.5 immediately before analysis using ammonium hydroxide. Solvent B was pure acetonitrile.
Cell Proliferation Assay.
Cells (5 × 103 per well) were plated in 96-well plates and allowed to attach for 16 hours. The medium was then changed to DMEM containing 2.5% FBS and 1× MEM nonessential amino acid, and either vehicle (DMSO) or different concentrations of the compounds were added. After 24 hours, the treatment medium was replaced with fresh medium containing 0.05 mg of MTT (3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide) per 100 μl and incubated for 4 hours. The medium was then removed, and 100 μl DMSO was added to the wells. The optical density of each sample was read on a microplate reader (FLUOstar Optima; BMG Labtech, Ortenberg, Germany) at 570 nm against a blank prepared from cell-free wells. Cell proliferation was expressed as a percentage of the relative absorbance of untreated controls.
Colonic Crypt Isolation and Three-Dimensional Culture.
Colonic crypts were isolated essentially as described elsewhere (Davidson et al., 2012). Briefly, intact colons were everted on a disposable mouse gavage needle and incubated with 20 mM EDTA in Ca/Mg-free Hanks’ balanced salt solution at 37°C for 30 minutes. Colonic crypts were released by mechanical disruption and purified by a series of phosphate-buffered saline washes and centrifugation steps (Davidson et al., 2012). Purified crypts were kept on ice, and resuspended in cold growth factor reduced, phenol red-free Matrigel (BD Biosciences, San Jose, CA) at a density of 15 crypts/µl. A total of 750 crypts/50 µl Matrigel were plated onto the center of wells in a 24-well plate and were incubated at 37°C for 10 minutes. After polymerization, complete medium containing advanced DMEM/Ham’s F-12 medium (Gibco/Life Technologies), epidermal growth factor (50 ng/ml; Gibco/Life Technologies), Noggin (100 ng/ml; Peprotech, Rocky Hill, NJ), R-Spon (500 ng/ml; Sino Biologic, Beijing, China), N2 supplement (1X; Invitrogen), B27 supplement (1X; Life Technologies), N-acetylcysteine (1 µM; Sigma-Aldrich), and Wnt conditioned medium was added to the wells. After 7 days of culture to allow development of mature organoids, the culture was then incubated for 6 hours at 37°C with 1 nM TCDD, 500 μM indole, or a combination of TCDD and indole.
Chromatin Immunoprecipitation Assay.
The chromatin immunoprecipitation (ChIP) assay was performed using the ChIP-IT Express Magnetic Chromatin Immunoprecipitation kit (Active Motif, Carlsbad, CA) according to the manufacturer’s protocol. CaCo-2 cells (5 × 106 cells) were treated with TCDD and/or compounds for 2 hours. The cells were then fixed with 1% formaldehyde, and the cross-linking reaction was stopped by addition of 0.125 M glycine. After washing twice with phosphate-buffered saline, the cells were scraped and pelleted. The collected cells were hypotonically lysed, and nuclei were collected. Nuclei were then sonicated to the desired chromatin length (∼200–1500 bp). The sonicated chromatin was immunoprecipitated with normal rabbit IgG or AHR antibodies and protein A–conjugated magnetic beads at 4°C for overnight. After the magnetic beads were extensively washed, protein-DNA crosslinks were reversed and eluted. DNA was prepared by proteinase K digestion followed by polymerase chain reaction (PCR) amplification. The CYP1A1 primers were 5′-TCA GGG CTG GGG TCG CAG CGC TTC T-3′ (sense) and 5′-GCT ACA GCC TAC CAG GAC TCG GCA G-3′ (antisense); we then amplified a 122-bp region of human Cyp1A1 promoter, which contained the AHR-binding sequences. The PCR products were resolved on a 2% agarose gel in the presence of ethidium bromide.
Quantitative Real-Time PCR.
We prepared cDNA from the total RNA of cells using amfiRivert cDNA Master Mix Platinum (GenDEPOT, Barker, TX). Each PCR analysis was performed in triplicate in a 20-μl volume using SYBR Green Mastermix (Applied Biosystems, Foster City, CA) for 15 minutes at 95°C for initial denaturing, followed by 40 cycles of 95°C for 15 seconds and 60°C for 1 minute in the Bio-Rad iCycler (MyiQ2) real-time PCR system (Bio-Rad Laboratories, Hercules, CA). The comparative cycle threshold method was used for relative quantitation of samples. Values for each gene were normalized to the expression levels of TATA-binding protein. The sequences of the primers used for real-time PCR were as follows: CYP1A1 sense 5′-GAC CAC AAC CAC CAA GAA C-3′, antisense 5′-AGC GAA GAA TAG GGA TGA AG-3′; chemokine (C-X-C motif) receptor 4 (CXCR4) sense 5′-TTT TCT TCA CGG AAA CAG GG-3′, antisense 5′-GTT ACC ATG GAG GGG ATC AG-3′; intercellular adhesion molecule 1 (ICAM1) sense 5′-TGA TGG GCA GTC AAC AGC TA-3′, antisense 5′-AGG GTA AGG TTC TTG CCC AC-3′; interleukin-1β (IL-1β) sense 5′-GAA GCT GAT GGC CCT AAA CA-3′, antisense 5′-AAG CCC TTG CTG TAG TGG TG-3′; matrix metallopeptidase 9 (MMP-9) sense 5′-TTG GTC CAC CTG GTT CAA CT-3′, antisense 5′-ACG ACG TCT TCC AGT ACC GA-3′; monoamine oxidase-A sense 5′-TGG AGA ATC AAG AGA AGG CG-3′, antisense 5′-CAG TCA AGA GTT TGG CAG CA-3′; and TATA-binding protein sense 5′-TGC ACA GGA GCC AAG AGT GAA-3′, antisense 5′-CAC ATC ACA GCT CCC CAC CA-3′.
Western Blot Analysis.
Cells (3 × 105) were plated in six-well plates in DMEM media containing 2.5% FBS (and 1× MEM nonessential amino acid for CaCo-2 cells) for 16 hours and then treated with different concentrations of the compounds. Cellular lysates were prepared in a lysis buffer containing 50 mM Tris-HCl (pH 7.5), 2 mM ethylenediaminetetraacetic acid, 150 mM NaCl, 0.5% deoxycholate, 0.1% SDS, 1 mM NaF, 1 mM Na3VO4, 1 mM phenyl methyl sulfonyl fluoride, 10 μl/ml protease inhibitor cocktail (GenDEPOT) and 1% NP-40. The cells were disrupted and extracted at 4°C for 30 minutes. After centrifugation, the supernatant was obtained as the cell lysate. Protein concentrations were measured using the Bio-Rad protein assay. Aliquots of cellular proteins were electrophoresed on 10% SDS-PAGE and transferred to a polyvinylidene fluoride membrane (Bio-Rad Laboratories). The membrane was allowed to react with a specific antibody, and detection of specific proteins was performed by enhanced chemiluminescence. Loading differences were normalized using a polyclonal β-actin antibody.
Transfection of siRNA.
Cells (2 × 105 cells/well) were plated in six-well plates in DMEM medium containing 20% FBS and 1× MEM nonessential amino acid. After 16 hours, the cells were transfected with 20 nM of each siRNA duplex for 24 hours using RiboCellin siRNA Delivery Reagent (BioCellChallenge, Toulon, France) following the manufacturer’s protocol. The medium was then changed and incubated for 24 hours. After incubation, the cells were treated with either vehicle (DMSO) or different concentrations of the compounds, and cells were collected for Western blot analysis and quantitative real-time PCR assay.
Statistical Analysis.
All of the experiments were repeated a minimum of three times. The data are expressed as the mean ± S.E. Statistical significance was analyzed using either Student’s t test or analysis of variance (ANOVA) with Scheffe’s test. The results are expressed as mean with error bars representing 95% confidence intervals for three experiments for each group unless otherwise indicated, and P < 0.05 was considered statistically significant.
Results
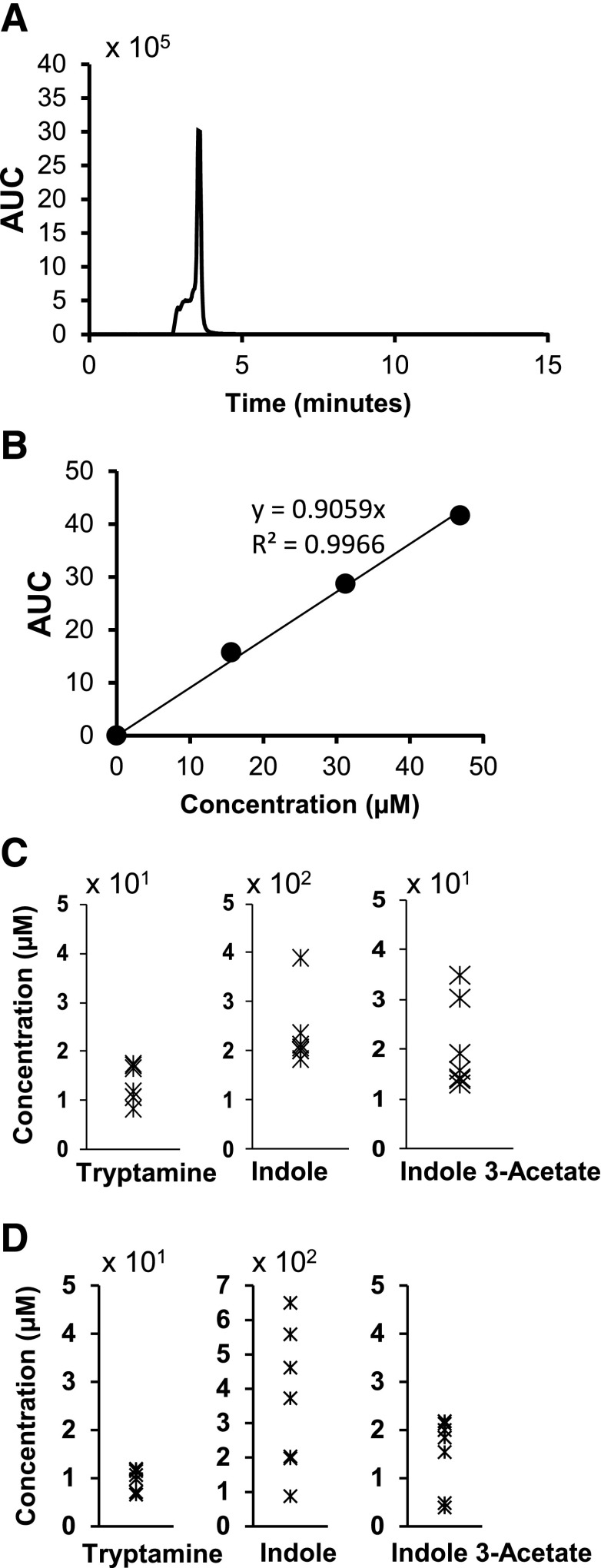
Previous studies have shown that indole is a major bacterial metabolite. In our study, we analyzed the cecal material (luminal contents and tissue) and fecal pellets from 6-week-old C57BL/6 mice for the presence of indole and other tryptophan-derived microbiota metabolites. The major tryptophan metabolites identified included indole, indole-3-acetate, and tryptamine (Fig. 1A). Levels of these metabolites were quantified in both cecal extracts and fecal pellets using MS-MRM, and the concentration of indole in the cecal material ranged between 200 and 400 μM; tryptamine and indole-3-acetate were less abundant, with concentrations ranging from 10–20 and 10–40 μM, respectively. Similar levels of these metabolites were detected in the fecal pellets (Fig. 1D).
Fig. 1.
MRM-MS quantification of tryptophan-derived metabolites in murine cecal contents and feces. (A and B) Detection and quantitation of tryptamine. Pure standards of tryptophan metabolites were used to generate standard curves for determining the concentration of metabolites in the samples using liquid chromatography–MRM-MS, as described in Materials and Methods. (C and D) Various metabolites in cecal extracts (μM) and feces (μM) in 6-week-old mice (n = 5), as quantified by liquid chromatography–MRM-MS. AUC, area under the curve.
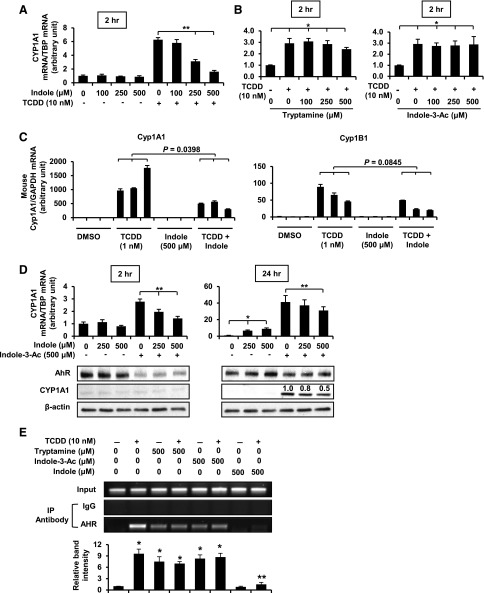
These compounds were investigated as AHR agonists and antagonists in CaCo-2 colon cancer cells. The effects of the tryptophan metabolites alone and in combination with 10 nM TCDD on CYP1A1 mRNA and CYP1A1, CYP1B1, and AHR protein levels (markers of altered Ah-responsive gene expression) were determined. Treatment of CaCo-2 cells with tryptamine (50–1000 μM) induced CYP1A1 mRNA and protein and CYP1B1 protein (Fig. 2A) with significant induction at the lowest concentration (50 μM); AHR levels were unchanged at 50–500 μM concentrations but decreased at the highest concentration (1000 μM) (Fig. 1A). High concentrations of tryptamine also decreased the expression of CYP1A1/CYP1B1 proteins, and this may be due to cytotoxicity (Supplemental Fig. 1). TCDD alone decreased AHR protein levels as has been previously described for other AHR agonists in colon cancer cells (Kawajiri et al., 2009). TCDD also induced CYP1A1 (mRNA and protein) and CYP1B1. In combination with tryptamine, the TCDD-induced responses were slightly attenuated at the two highest concentrations of tryptamine (500 and 1000 μM), indicating that this compound is primarily an AHR agonist in CaCo-2 cells as previously described in other cancer cell lines (Miller, 1997; Heath-Pagliuso et al., 1998; Vikstrom Bergander et al., 2012), with possible partial antagonist activity at the higher concentrations. In contrast to a previous report (Vikstrom Bergander et al., 2012), our data show that the AHR agonist activity of tryptamine (i.e., CYP1A1 induction) was not affected by a monoamine oxidase inhibitor (Supplemental Fig. 2).
Fig. 2.
AHR agonist and antagonist activities of tryptophan metabolites. CaCo-2 cells were treated with tryptamine (A), indole-3-acetate (B), 3-indoxyl sulfate (I3S) (C), or indole (D) alone or in combination with TCDD. After 24 hours, induction of CYP1A1 mRNA and AHR, the CYP1A1 and CYP1B1 proteins were determined by real-time PCR and Western blot analysis, respectively, as outlined in Materials and Methods. Results for mRNA levels are expressed as mean ± S.E. for three separate experiments, and statistically significant (P < 0.05) induction (*) or inhibition of TCDD-induced responses (**) are indicated.
Indole 3-acetate was also identified in the cecal extract and feces (Fig. 1), and the activity of this compound was compared with that of the uremic toxicant 3-indoxyl sulfate, which has previously been described as an AHR agonist in liver cancer cell lines (Schroeder et al., 2010). Both indole-3-acetate and 3-indoxyl sulfate induced CYP1A1 (mRNA and protein) and CYP1B1 (protein); in combination with TCDD, indole-3-acetate did not affect cytochrome P450 induction by TCDD (Fig. 2, B and C). Although 3-indoxyl sulfate was an AHR agonist, induction of CYP1A1 and CYP1B1 protein by TCDD was decreased in CaCo-2 cells cotreated with TCDD plus 3-indoxyl sulfate, suggesting some partial AHR antagonist activity. However, we also observed that 50 μM 3-indoxyl sulfate was cytotoxic (Supplemental Fig. 1), and this may contribute to decreased CYP1A1/CYP1B1 proteins. The two 3-substituted indole derivatives differed with respect to their effects on the AHR; like TCDD, 3-indoxyl sulfate decreased AHR expression, whereas indole-3-acetate did not affect the levels of AHR protein, even though the fold induction of CYP1A1 mRNA by this compound was similar to that observed for TCDD. Treatment of CaCo-2 cells with 50–1000 μM indole significantly induced CYP1A1 mRNA levels only at the two higher concentrations (500–1000 μM); however, this was not accompanied by induction of CYP1A1 and CYP1B1 proteins, and levels of the AHR protein were unchanged (Fig. 2D). In combination experiments, indole (500–1000 μM) inhibited TCDD-mediated induction of CYP1A1 (mRNA and protein) and CYP1B1 protein, and downregulation of AHR demonstrated that indole was an AHR antagonist for these responses (Fig. 2D). The EC50 values for induction of CYP1A1 mRNA by tryptamine indole-3-acetate and indole were 0.048, 0.37, and >1 mM, respectively.
We further investigated the AHR antagonist activities of indole as an inhibitor of TCDD-induced CYP1A1 gene transcription after treatment of CaCo-2 cells for 2 hours (Fig. 3A). The results showed that at this early time point, indole was a full AHR antagonist, whereas tryptamine and indole 3-acetate did not inhibit TCDD-induced CYP1A1 RNA (Fig. 3B). Indole also significantly inhibited TCDD-induced Cyp1a1 and Cyp1b1 mRNA expression in mouse-derived colonic crypts in three-dimensional culture (Fig. 3C), and these data complemented the results in CaCo-2 cells. We also observed that indole inhibited indole 3-acetate–induced CYP1A1 mRNA (2 and 24 hours) and protein (24 hours) expression in CaCo-2 cells (Fig. 3D), demonstrating potential inhibitory interactions among the indole-derived AHR agonists.
Fig. 3.
AHR activity in transactivation and ChIP assays. CaCo-2 cells were treated with (A) indole, (B) indole-3-acetate and tryptamine, alone or in combination with TCDD. After 2 hours, CYP1A1 mRNA and protein levels were determined by real-time PCR. (C) Mouse colonic crypts in culture were treated with indole, TCDD, and their combinations for 6 hours. The Cyp1a1 and Cyp1b1 mRNA levels were determined as outlined in Materials and Methods. The three bars within each treatment group represent three different cultures of colonic crypts. (D) Interactions between indole and indole-3-acetate were determined in CaCo-2 cells treated with these compounds (2 and 24 hours), and CYP1A1 mRNA and protein were determined by real-time PCR and Western blot analysis, as outlined in Materials and Methods. (E) Binding to a CYP1A1 DRE was determined in a ChIP assay, as outlined in Materials and Methods. Studies were performed in triplicate, the results are mean ± S.E., and any statistically significant (P < 0.05) increases (*) (compared with DMSO) or inhibition (**) of TCDD-induced responses are indicated.
The effects of the tryptophan metabolites on TCDD-induced recruitment of the AHR complex to a DRE region on the CYP1A1 promoter were investigated in ChIP assays (Fig. 3E). Treatment of CaCo-2 cells with TCDD for 2 hours resulted in recruitment of the AHR to the CYP1A1 promoter. Similar results were observed for tryptamine and indole-3-acetate. In cells cotreated with TCDD plus tryptamine or indole-3-acetate, the AHR binding to the DRE was essentially unchanged. Indole alone did not induce AHR-DRE binding; in cells cotreated with TCDD plus indole, there was a dramatic decrease in TCDD-induced DRE binding, which was consistent with the AHR antagonist activity of indole observed for transactivation.
Confirmation of the AHR agonist/antagonist activity of the tryptophan metabolites was determined in MDA-MB-468 and MDA-MB-231 cells, which we have previously used to investigate the activities of several AHR-active pharmaceuticals (Jin et al., 2012). Figure 4A shows that the pattern of CYP1A1 induction by tryptamine was similar in both breast cancer cell lines. Tryptamine alone was an AHR agonist, but in combination with TCDD, it decreased expression of CYP1A1 mRNA and protein and partially blocked TCDD-mediated AHR downregulation at the higher concentrations. Indole-3-acetate was a partial AHR agonist in MDA-MB-468 and MDA-MB-231 cells, and in cells cotreated with this compound plus TCDD, there was no evidence of AHR antagonist activity (Fig. 4B). Indole exhibited weak AHR agonist activity in both breast cancer cell lines; in combination studies, indole inhibited TCDD-induced CYP1A1 protein and mRNA levels and blocked TCDD-mediated downregulation of the AHR. In MDA-MB-468 cells, a highly Ah-responsive cell line, indole exhibited almost complete AHR antagonist activity. Thus, the results for the three tryptophan metabolites as AHR agonists/antagonists were comparable in the colon and breast cancer cell lines.
Fig. 4.
AHR agonist and antagonist activities of tryptophan metabolites in MDA-MB-468 and MDA-MB-231 breast cancer cells. Cells were treated with (A) tryptamine, (B) indole-3-acetate, or (C) indole, alone or in combination with TCDD for 24 hours. The effects on CYP1A1 mRNA and AHR and CYP1A1 proteins were determined by real-time PCR or Western blot analysis, respectively, as outlined in Materials and Methods. The mRNA data are mean ± S.E. for three replicate determinations, and statistically significant (P < 0.05) induction (*) or inhibition (**) of TCDD-induced responses are indicated.
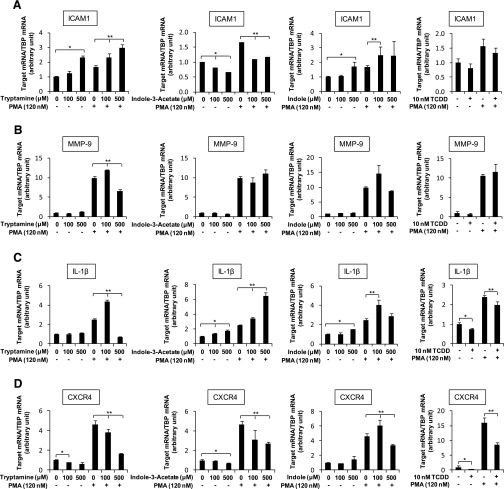
Previous studies in colon cancer cell lines show that AHR agonists have variable effects on endogenous and induced inflammatory responses, including ICAM-1, IL-1β, MMP-9, and β-catenin (Villard et al., 2007; Kawajiri et al., 2009; Furumatsu et al., 2011), and CXCR4 is suppressed by AHR ligands in breast cancer cells (Hsu et al., 2007). Treatment of CaCo-2 cells with tryptamine or indole slightly increased ICAM-1 mRNA; indole-3-acetate and TCDD decreased (Fig. 5A) and 3-indoxyl sulfate had no effect on expression of this gene (Supplemental Fig. 3). PMA (phorbol-12-myristate 13-acetate) only slightly induced ICAM1 in CaCo-2 cells; in combination with the tryptophan metabolites, expression was increased (tryptamine, indole), decreased (indole-3-acetate), or unchanged (3-indoxyl sulfate), and similar results were observed for TCDD. Expression of MMP-9 in CaCo-2 cells was low and unaffected by the indole derivatives or TCDD (Fig. 5B); MMP-9 was induced 10-fold by PMA, and in the cotreatment experiment, only tryptamine (slight decrease) affected the PMA-induced response. IL-β expression was slightly increased (indole and indole-3-acetate), decreased (TCDD), or unaffected (indole and 3-indoxyl sulfate) by the AHR ligands, and PMA only slightly induced (1.5–2.5-fold) IL-1β mRNA levels (Fig. 5C). Cotreatment with PMA plus the indole derivatives resulted in dose-dependent effects for some compounds; 100 and 500 μM tryptamine enhanced and inhibited IL-1β mRNA levels, respectively; indole and indole-3-acetate enhanced and 3-indoxyl sulfate decreased IL-1β mRNA, and TCDD slightly decreased the PMA-induced response. Tryptamine, indole-3-acetate, TCDD, and 3-indoxyl sulfate decreased and indole had no effect on endogenous CXCR4 gene expression. PMA significantly induced CXCR4 mRNA levels, and cotreatment with the highest concentration of all four tryptophan metabolites and TCDD decreased PMA-induced CXCR4 mRNA levels (note: 100 μM indole increased this response). The most consistent effect of TCDD and the tryptophan metabolites was the decreased expression of CXCR4, and this response has previously been observed for other AHR agonists in breast cancer cells (Hsu et al., 2007).
Fig. 5.
Anti-inflammatory activities of tryptophan metabolites and TCDD in CaCo-2 cells. Cells were treated with the tryptophan metabolites or TCDD alone or in combination with PMA for 24 hours, and expression of ICAM1 (A), MMP-9 (B), IL-1β (C), and CXCR4 (D) mRNA was determined by real-time PCR, as outlined in Materials and Methods. Results are expressed as mean ± S.E. for three replicate determinations, and statistically significant (P < 0.05) modulation of basal (DMSO) (*) or PMA-induced (**) responses are indicated.
Results in Fig. 6A show that the AHR antagonist CH223191 [2-methyl-2H-pyrazole-3-carboxylic acid (2-methyl-4-o-tolylazo-phenyl)-amide] alone did not affect basal or PMA-induced CXCR4 mRNA levels but significantly blocked the effects (downregulation) of TCDD, and CH223191 also inhibited TCDD (±PMA)-induced CYP1A1 mRNA levels in CaCo-2 cells. CH223191 partially reversed tryptamine-mediated downregulation of (induced) CXCR4 but did not significantly alter the effects of indole-3-acetate or 3-indoxyl sulfate (Fig. 6B). It was also evident that CH223191 was less effective as an inhibitor of tryptamine-/indole-3-acetate-/3-indoxyl sulfate–induced CYP1A1 mRNA (Fig. 6C) than observed for TCDD (Fig. 6A), and these results are consistent with the reported activity of CH223191 as a specific inhibitor of TCDD and related halogenated aryl hydrocarbons (Zhao et al., 2010). However, AHR knockdown by RNA interference shows CYP1A1 inducibility is decreased for both TCDD and the tryptophan metabolites (Fig. 6D). It should also be noted that CH223191 inhibits induction of Ah-responsive luciferase activity by the natural ligand 6-formylindo[3,2b]carbazole and methyl 2-(1-H′-indolo-3′-carbonyl)-thiazole-4-carboxylate in HepG2 liver cells, suggesting a broader spectrum of AHR antagonist activity for this compound (Choi et al., 2012).
Fig. 6.
Role of the AHR in inhibition of CXCR4 expression. (A) Effect of TCDD on CXCR4 and CYP1A1 mRNA expression. CaCo-2 cells were treated with TCDD, PMA, or CH223191 alone or in combination, and the expression of CXCR4 and CYP1A1 mRNA levels were determined by real-time PCR, as outlined in Materials and Methods. CaCo-2 cells were treated with DMSO, PMA, CH223191, or tryptamine (100 μM), indole-3-acetate (100 μM), and indole (1000 μM), alone or in combination. The CXCR4 (B) and CYP1A1 (C) mRNA levels were determined by real-time PCR. (D) RNA interference. CaCo-2 cells were transfected with an oligonucleotide that targets the AHR. After 48 hours, the cells were treated with TCDD or the tryptophan metabolites for 24 hours, and the CYP1A1, AHR mRNA levels were determined by real-time PCR. Results are expressed as mean ± S.E. for at least three separate experiments, and statistically significant (P < 0.05) induced CYP1A1 or decreased CXCR4 in the cells treated with PMA plus the AHR ligand (*) and reversal of these responses by CH223191 (**) are indicated. A statistically significant (P < 0.05) decrease in CYP1A1 and AHR mRNA (D) is also indicated (*).
A previous study reported that several AHR agonists decreased β-catenin expression in DLD-1, SW480, and HCT116 colon cancer cells (Kawajiri et al., 2009); however, TCDD did not affect β-catenin levels in CaCo-2 cells, and among the tryptophan metabolites, only the higher concentrations of tryptamine decreased β-catenin protein levels (Supplemental Fig. 4A), which may be due to cytotoxicity (Supplemental Fig. 1). However, the AHR antagonist CH223191 did not modulate downregulation of β-catenin by tryptamine but clearly inhibited induction of CYP1A1 protein by tryptamine (500 μM) (Supplemental Fig. 4B). The effects of 100 μM tryptamine on CYP1A1 downregulation were not affected by CH223191. These results demonstrate that the AHR-dependent effects of TCDD and the indole derivatives depend on response and cell context, which is consistent with their activity as SAhRMs.
Discussion
AHR and its ligands play an increasingly important role in gut homeostasis and in various intestinal diseases, including colitis and colon cancer (Kawajiri et al., 2009; Arsenescu et al., 2011; Benson and Shepherd, 2011; Furumatsu et al., 2011; Kiss et al., 2011; Li et al., 2011; Singh et al., 2011). For example, AHR is required for the postnatal expansion of CD4-RORγT+ innate lymphoid cells, and dietary AHR ligands, such as indole-3-carbinol (I3C), promote the postnatal expansion of these cells which protect against intestinal infections (Bansal et al., 2010). β-Naphthoflavone, a relatively nontoxic AHR agonist, protects against dextran sodium sulfate–induced colitis in a mouse model and decreased lipopolysaccharide-induced inflammatory responses in SW480 colon cancer cells (Furumatsu et al., 2011). APCmin/+ mice are extensively used as a model for intestinal carcinogenesis, and loss of AHR enhances cecal tumor incidence; in wild-type mice, dietary administration of I3C or diindolylmethane (AHR agonists) protected against intestinal carcinogenesis (Kawajiri et al., 2009). These results clearly demonstrate an important role for the AHR and AHR agonists in maintaining gut health and protecting against intestinal diseases.
Tryptophan metabolites such as the indole derivatives have previously been investigated as AHR ligands, and a recent study showed that the AHR agonist kynurenine (Davarinos and Pollenz, 1999) plays an important pro-oncogenic role in glioma (Opitz et al., 2011). Therefore, the expression of tryptophan metabolites by gut microflora and their AHR agonist or antagonist activities could significantly impact Ah-responsive intestinal functions. Previous studies in our laboratory have identified indole as a major microbial metabolite (Bansal et al., 2010), and indole concentrations in human feces can be in the low millimole range (Karlin et al., 1985; Zuccato et al., 1993). In this study, we used liquid chromatography–MRM-MS and identified the tryptophan metabolites indole, tryptamine, and indole-3-acetate in the cecum and feces of 6-week old C57BL/6 mice, at concentrations comparable to those reported in human feces (Karlin et al., 1985; Zuccato et al., 1993). The concentration of indole was almost an order of magnitude greater than tryptamine or indole-3-acetate in both cecal extracts and fecal samples, and concentrations of tryptamine and indole in the cecum and fecal pellets were comparable to those needed for AHR agonist and antagonist activity, respectively. Concentrations of these tryptophan metabolites (Fig. 1) are probably underestimated due to the overall efficiencies of extraction which are <100%.
We used the CaCo-2 intestinal colon cell line as a model to investigate the AHR agonist and antagonist activities of indole, indole-3-acetate, and tryptamine. Their effects on CYP1A1 mRNA and protein and CYP1B1 protein expression were determined as a measure of their Ah-responsiveness. Moreover, in CaCo-2 cells we also examined the effects of 3-indoxyl sulfate, which has previously been characterized as an AHR agonist (Schroeder et al., 2010). TCDD served as a prototypical agonist, and we also compared the effects of TCDD versus the tryptophan metabolites on proteasome-dependent downregulation of the AHR protein, which is observed for some (e.g., TCDD) but not all AHR ligands (Davarinos and Pollenz, 1999; Kawajiri et al., 2009; Jin et al., 2012). All the tryptophan metabolites exhibit full or partial (indole) AHR agonist activity in CaCo-2 cells and induced CYP1A1 mRNA and protein, whereas induction of CYP1B1 was highly variable. However, after treatment of CaCo-2 cells with tryptamine for 24 hours, there was some indication that this compound may be a partial AHR antagonist (i.e., inhibition of TCDD-induced CYP1A1 and AHR downregulation). In short-term experiments, neither tryptamine or indole-3-acetate inhibited TCDD-induced CYP1A1 gene expression (Fig. 3B). In contrast, indole exhibited AHR antagonist activity at both the 24- and 2-hour treatment times in CaCo-2 cells using TCDD (Figs. 2D and 3A) and indole-3-acetate (Fig. 3D) as agonists, confirming the AHR antagonist activity of indole and indicating potential interactions among the tryptophan/microbiome AHR ligands.
Moreover, we also observed that indole inhibited TCDD-induced Cyp1a1 and Cyp1b1 expression in mouse colonic crypts in three-dimensional culture. Thus, at least for the classic AHR-mediated induction of CYP1A1, 100–250 μM indole exhibited weak agonist/partial antagonist activity in colon cancer cells, suggesting that the relative levels of the tryptophan-derived microbiota metabolites could affect the overall intestinal balance of AHR ligands from dietary or contaminant sources. Previous studies have shown that dietary AHR agonists mitigate induced colitis in mouse models (Li et al., 2011), and we are currently further investigating their interactive effects with microbiota tryptophan metabolites.
Previous reports suggested that AHR agonists modulate inflammatory response genes in colon epithelial and in colon cancer cells (Villard et al., 2007; Kawajiri et al., 2009; Furumatsu et al., 2011) and this included AHR-dependent inhibition of IL-1β in SW480 cells by β-naphthoflavone (Furumatsu et al., 2011). In contrast IL-1β and MMP-9 were induced by 3-methylcholanthrene in CaCo-2 cells (Villard et al., 2007); however, the role of the AHR in mediating these responses was not confirmed. Our results for the tryptophan metabolites in CaCo-2 cells showed highly variable effects of these compounds on endogenous and PMA-induced stress response genes (Fig. 5). Moreover, TCDD did not affect endogenous or PMA-induced ICAM1, MMP-9, or IL-1β gene expression, and the only significant response observed was inhibition of CXCR4 (Fig. 6A). TCDD-induced downregulation of CXCR4 was blocked by the AHR antagonist CH223191. Interestingly, tryptamine, indole-3-acetate, and 3-indoxyl sulfate also decreased endogenous and PMA-induced CXCR4 mRNA levels and exhibited the expected TCDD-like AHR agonist activity; however, cotreatment with the AHR antagonist did not reverse CXCR4 downregulation (Fig. 6, B and C), suggesting that the effects of these compounds were AHR independent.
An AHR-dependent response previously observed in SW480, DLD-1, and HCT116 colon cancer cells was the downregulation of β-catenin by several AHR agonists (not TCDD) (Kawajiri et al., 2009), whereas in CaCo-2 cells, TCDD, indole, indole-3-acetate, and 3-indoxyl sulfate did not affect β-catenin levels and the tryptamine response was AHR independent (Supplemental Fig. 4). Tissue- and response-specific agonist or antagonist activity of receptor ligands is due to several factors, including different ligand-induced conformational changes in the receptor and differential expression of cofactors in various tissues/cells. We previously observed the SAhRM-like activity for a series of AHR-active pharmaceuticals in breast cancer cells (Jin et al., 2012).
In summary, our results demonstrate that the mouse microbiome produces relatively high concentrations of tryptophan metabolites tryptamine, indole-3-acetate, and indole, and these compounds differentially activate markers of Ah-responsiveness including induction of CYP1A1 and CYP1B1. Previous studies had demonstrated that AHR plays a critical role in intestinal homeostasis and disease, and dietary AHR agonists, such as I3C and diindolylmethane can protect against induced colitis and colon cancer in mouse models (Kawajiri et al., 2009; Arsenescu et al., 2011; Benson and Shepherd, 2011; Furumatsu et al., 2011; Kiss et al., 2011; Li et al., 2011; Singh et al., 2011). Our results demonstrate that microbiota-generated tryptophan metabolites exhibit both AHR agonist and antagonist activities, and these may also be response- and cell context–specific as observed for other SAhRMS (Jin et al., 2012). Thus, the AHR-active tryptophan metabolites along with other dietary and potential exogenous (i.e., contaminants) AHR ligands can potentially influence host intestinal responses and changes in the levels and ratios of microbiota-derived tryptophan metabolites could affect the overall AHR ligand-induced impact on intestinal functions.
Supplementary Material
Abbreviations
- Ah
aryl hydrocarbon
- AHR
aryl hydrocarbon receptor
- CH223191
2-methyl-2H-pyrazole-3-carboxylic acid (2-methyl-4-o-tolylazo-phenyl)-amide
- ChIP
chromatin immunoprecipitation
- CXCR4
chemokine (C-X-C motif) receptor 4
- DMEM
Dulbecco’s modified Eagle’s medium
- DMSO
dimethyl sulfoxide
- DREs
dioxin-response elements
- FBS
fetal bovine serum
- FICZ
6-formylindolo-[3,2-b]-carbazole
- I3C
indole-3-carbinol
- ICAM
intercellular adhesion molecule 1
- IL-1β
interleukin-1β
- MEM
minimum Eagle’s medium
- MMP-9
matrix metallopeptidase 9
- MRM
multiple reaction monitoring
- MS
mass spectrometry
- MTT
3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide
- PCR
polymerase chain reaction
- PMA
phorbol-12-myristate 13-acetate
- SAhRM
selective aryl hydrocarbon receptor modulator
- siRNA
small interfering RNA
- TCDD
2,3,7,8-tetrachlorodibenzo-p-dioxin
Authorship Contributions
Participated in research design: Jin, Chapkin, Alaniz, Jayaraman, Safe.
Conducted experiments: Jin, S.-O. Lee, Sridharan, K. Lee, Davidson, Jayaraman, Alaniz, Safe.
Contributed new reagents or analytic tools: Jayaraman, Chapkin, Safe.
Performed data analysis: Jin, S.-O. Lee, Sridharan, K. Lee, Davidson, Jayaraman, Alaniz, Safe.
Wrote or contributed to the writing of the manuscript: Jin, Sridharan, K. Lee, Jayaraman, Chapkin, Alaniz, Safe.
Footnotes
The work was supported by the National Institutes of Health National Cancer Institute [Grants R01-CA142697 (to S.S.), R21-A1095788 (to R.A. and A.J.), and NSF 084653 (to A.J.)]; and Texas AgriLife Research (to S.S.).
A.J., R.A., and S.S. are senior authors.
 This article has supplemental material available at molpharm.aspetjournals.org.
This article has supplemental material available at molpharm.aspetjournals.org.
References
- Arsenescu R, Arsenescu V, Zhong J, Nasser M, Melinte R, Dingle RW, Swanson H, de Villiers WJ. (2011) Role of the xenobiotic receptor in inflammatory bowel disease. Inflamm Bowel Dis 17:1149–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajad SU, Lu W, Kimball EH, Yuan J, Peterson C, Rabinowitz JD. (2006) Separation and quantitation of water soluble cellular metabolites by hydrophilic interaction chromatography-tandem mass spectrometry. J Chromatogr A 1125:76–88 [DOI] [PubMed] [Google Scholar]
- Bansal T, Alaniz RC, Wood TK, Jayaraman A. (2010) The bacterial signal indole increases epithelial-cell tight-junction resistance and attenuates indicators of inflammation. Proc Natl Acad Sci USA 107:228–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barouki R, Coumoul X, Fernandez-Salguero PM. (2007) The aryl hydrocarbon receptor, more than a xenobiotic-interacting protein. FEBS Lett 581:3608–3615 [DOI] [PubMed] [Google Scholar]
- Benson JM, Shepherd DM. (2011) Aryl hydrocarbon receptor activation by TCDD reduces inflammation associated with Crohn’s disease. Toxicol Sci 120:68–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigelow SW, Nebert DW. (1982) The Ah regulatory gene product. Survey of nineteen polycyclic aromatic compounds’ and fifteen benzo[a]pyrene metabolites’ capacity to bind to the cytosolic receptor. Toxicol Lett 10:109–118 [DOI] [PubMed] [Google Scholar]
- Choi EY, Lee H, Dingle RW, Kim KB, Swanson HI. (2012) Implications and development of AHR-based therapeutic agents. Mol Cell Pharmacol 4:53–60 DOI: 10.4255/mcpharmacol.12.05. [Google Scholar]
- Davarinos NA, Pollenz RS. (1999) Aryl hydrocarbon receptor imported into the nucleus following ligand binding is rapidly degraded via the cytosplasmic [sic] proteasome following nuclear export. J Biol Chem 274:28708–28715 [DOI] [PubMed] [Google Scholar]
- Davidson LA, Goldsby JS, Callaway ES, Shah MS, Barker N, Chapkin RS. (2012) Alteration of colonic stem cell gene signatures during the regenerative response to injury. Biochim Biophys Acta 1822:1600–1607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denison MS, Nagy SR. (2003) Activation of the aryl hydrocarbon receptor by structurally diverse exogenous and endogenous chemicals. Annu Rev Pharmacol Toxicol 43:309–334 [DOI] [PubMed] [Google Scholar]
- Denison MS, Soshilov AA, He G, DeGroot DE, Zhao B. (2011) Exactly the same but different: promiscuity and diversity in the molecular mechanisms of action of the aryl hydrocarbon (dioxin) receptor. Toxicol Sci 124:1–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiNatale BC, Murray IA, Schroeder JC, Flaveny CA, Lahoti TS, Laurenzana EM, Omiecinski CJ, Perdew GH. (2010) Kynurenic acid is a potent endogenous aryl hydrocarbon receptor ligand that synergistically induces interleukin-6 in the presence of inflammatory signaling. Toxicol Sci 115:89–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furumatsu K, Nishiumi S, Kawano Y, Ooi M, Yoshie T, Shiomi Y, Kutsumi H, Ashida H, Fujii-Kuriyama Y, Azuma T, et al. (2011) A role of the aryl hydrocarbon receptor in attenuation of colitis. Dig Dis Sci 56:2532–2544 [DOI] [PubMed] [Google Scholar]
- Gillner M, Bergman J, Cambillau C, Fernström B, Gustafsson JA. (1985) Interactions of indoles with specific binding sites for 2,3,7,8-tetrachlorodibenzo-p-dioxin in rat liver. Mol Pharmacol 28:357–363 [PubMed] [Google Scholar]
- Hankinson O. (1995) The aryl hydrocarbon receptor complex. Annu Rev Pharmacol Toxicol 35:307–340 [DOI] [PubMed] [Google Scholar]
- Heath-Pagliuso S, Rogers WJ, Tullis K, Seidel SD, Cenijn PH, Brouwer A, Denison MS. (1998) Activation of the Ah receptor by tryptophan and tryptophan metabolites. Biochemistry 37:11508–11515 [DOI] [PubMed] [Google Scholar]
- Hsu EL, Yoon D, Choi HH, Wang F, Taylor RT, Chen N, Zhang R, Hankinson O. (2007) A proposed mechanism for the protective effect of dioxin against breast cancer. Toxicol Sci 98:436–444 [DOI] [PubMed] [Google Scholar]
- Hu W, Sorrentino C, Denison MS, Kolaja K, Fielden MR. (2007) Induction of cyp1a1 is a nonspecific biomarker of aryl hydrocarbon receptor activation: results of large scale screening of pharmaceuticals and toxicants in vivo and in vitro. Mol Pharmacol 71:1475–1486 [DOI] [PubMed] [Google Scholar]
- Huang G, Elferink CJ. (2012) A novel nonconsensus xenobiotic response element capable of mediating aryl hydrocarbon receptor-dependent gene expression. Mol Pharmacol 81:338–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin UH, Lee SO, Safe S. (2012) Aryl hydrocarbon receptor (AHR)-active pharmaceuticals are selective AHR modulators in MDA-MB-468 and BT474 breast cancer cells. J Pharmacol Exp Ther 343:333–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan VC. (2007) SERMs: meeting the promise of multifunctional medicines. J Natl Cancer Inst 99:350–356 [DOI] [PubMed] [Google Scholar]
- Karlin DA, Mastromarino AJ, Jones RD, Stroehlein JR, Lorentz O. (1985) Fecal skatole and indole and breath methane and hydrogen in patients with large bowel polyps or cancer. J Cancer Res Clin Oncol 109:135–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawajiri K, Kobayashi Y, Ohtake F, Ikuta T, Matsushima Y, Mimura J, Pettersson S, Pollenz RS, Sakaki T, Hirokawa T, et al. (2009) Aryl hydrocarbon receptor suppresses intestinal carcinogenesis in ApcMin/+ mice with natural ligands. Proc Natl Acad Sci USA 106:13481–13486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss EA, Vonarbourg C, Kopfmann S, Hobeika E, Finke D, Esser C, Diefenbach A. (2011) Natural aryl hydrocarbon receptor ligands control organogenesis of intestinal lymphoid follicles. Science 334:1561–1565 [DOI] [PubMed] [Google Scholar]
- Li Y, Innocentin S, Withers DR, Roberts NA, Gallagher AR, Grigorieva EF, Wilhelm C, Veldhoen M. (2011) Exogenous stimuli maintain intraepithelial lymphocytes via aryl hydrocarbon receptor activation. Cell 147:629–640 [DOI] [PubMed] [Google Scholar]
- Lu YF, Santostefano M, Cunningham BD, Threadgill MD, Safe S. (1996) Substituted flavones as aryl hydrocarbon (Ah) receptor agonists and antagonists. Biochem Pharmacol 51:1077–1087 [DOI] [PubMed] [Google Scholar]
- Maynard CL, Elson CO, Hatton RD, Weaver CT. (2012) Reciprocal interactions of the intestinal microbiota and immune system. Nature 489:231–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillan BJ, Bradfield CA. (2007) The aryl hydrocarbon receptor sans xenobiotics: endogenous function in genetic model systems. Mol Pharmacol 72:487–498 [DOI] [PubMed] [Google Scholar]
- Miller CA., 3rd (1997) Expression of the human aryl hydrocarbon receptor complex in yeast. Activation of transcription by indole compounds. J Biol Chem 272:32824–32829 [DOI] [PubMed] [Google Scholar]
- Opitz CA, Litzenburger UM, Sahm F, Ott M, Tritschler I, Trump S, Schumacher T, Jestaedt L, Schrenk D, Weller M, et al. (2011) An endogenous tumour-promoting ligand of the human aryl hydrocarbon receptor. Nature 478:197–203 [DOI] [PubMed] [Google Scholar]
- Poland A, Glover E, Kende AS. (1976) Stereospecific, high affinity binding of 2,3,7,8-tetrachlorodibenzo-p-dioxin by hepatic cytosol. Evidence that the binding species is receptor for induction of aryl hydrocarbon hydroxylase. J Biol Chem 251:4936–4946 [PubMed] [Google Scholar]
- Safe S, Chadalapaka G, Jutooru I. (2012) AHR-reactive compounds in the human diet, in The Ah Receptor in Biology and Toxicology (Pohjanvirta R, ed) pp 331–342, John Wiley, Hoboken, NJ [Google Scholar]
- Schroeder JC, Dinatale BC, Murray IA, Flaveny CA, Liu Q, Laurenzana EM, Lin JM, Strom SC, Omiecinski CJ, Amin S, et al. (2010) The uremic toxin 3-indoxyl sulfate is a potent endogenous agonist for the human aryl hydrocarbon receptor. Biochemistry 49:393–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellick CA, Knight D, Croxford AS, Maqsood AR, Stephens GM, Goodacre R, Dickson AJ. (2010) Evaluation of extraction processes for intracellular metabolite profiling of mammalian cells: matching extraction approaches to cell type and metabolite targets. Metabolomics 6:427–438 DOI: 10.1007/s11306-010-0216-9. [Google Scholar]
- Singh NP, Singh UP, Singh B, Price RL, Nagarkatti M, Nagarkatti PS. (2011) Activation of aryl hydrocarbon receptor (AhR) leads to reciprocal epigenetic regulation of FoxP3 and IL-17 expression and amelioration of experimental colitis. PLoS ONE 6:e23522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanos R, Patel RD, Murray IA, Smith PB, Patterson AD, Perdew GH. (2012) Aryl hydrocarbon receptor regulates the cholesterol biosynthetic pathway in a dioxin response element-independent manner. Hepatology 55:1994–2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremaroli V, Bäckhed F. (2012) Functional interactions between the gut microbiota and host metabolism. Nature 489:242–249 [DOI] [PubMed] [Google Scholar]
- Vikström Bergander L, Cai W, Klocke B, Seifert M, Pongratz I. (2012) Tryptamine serves as a proligand of the AhR transcriptional pathway whose activation is dependent of monoamine oxidases. Mol Endocrinol 26:1542–1551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villard PH, Caverni S, Baanannou A, Khalil A, Martin PG, Penel C, Pineau T, Seree E, Barra Y. (2007) PPARalpha transcriptionally induces AhR expression in Caco-2, but represses AhR pro-inflammatory effects. Biochem Biophys Res Commun 364:896–901 [DOI] [PubMed] [Google Scholar]
- Whitlock JP., Jr (1999) Induction of cytochrome P4501A1. Annu Rev Pharmacol Toxicol 39:103–125 [DOI] [PubMed] [Google Scholar]
- Zhao B, Degroot DE, Hayashi A, He G, Denison MS. (2010) CH223191 is a ligand-selective antagonist of the Ah (Dioxin) receptor. Toxicol Sci 117:393–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Gasiewicz TA. (2003) 3′-Methoxy-4′-nitroflavone, a reported aryl hydrocarbon receptor antagonist, enhances Cyp1a1 transcription by a dioxin responsive element-dependent mechanism. Arch Biochem Biophys 416:68–80 [DOI] [PubMed] [Google Scholar]
- Zuccato E, Venturi M, Di Leo G, Colombo L, Bertolo C, Doldi SB, Mussini E. (1993) Role of bile acids and metabolic activity of colonic bacteria in increased risk of colon cancer after cholecystectomy. Dig Dis Sci 38:514–519 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.