Abstract
Defects of the oxidative phosphorylation system, in particular of cytochrome-c oxidase (COX, respiratory chain complex IV), are common causes of Leigh syndrome (LS), which is a rare neurodegenerative disorder with severe progressive neurological symptoms that usually present during infancy or early childhood. The COX-deficient form of LS is commonly caused by mutations in genes encoding COX assembly factors, e.g. SURF1, SCO1, SCO2 or COX10. However, other mutations affecting genes that encode proteins not directly involved in COX assembly can also cause LS. The leucine-rich pentatricopeptide repeat containing protein (LRPPRC) regulates mRNA stability, polyadenylation and coordinates mitochondrial translation. In humans, mutations in Lrpprc cause the French Canadian type of LS. Despite the finding that LRPPRC deficiency affects the stability of most mitochondrial mRNAs, its pathophysiological effect has mainly been attributed to COX deficiency. Surprisingly, we show here that the impaired mitochondrial respiration and reduced ATP production observed in Lrpprc conditional knockout mouse hearts is caused by an ATP synthase deficiency. Furthermore, the appearance of inactive subassembled ATP synthase complexes causes hyperpolarization and increases mitochondrial reactive oxygen species production. Our findings shed important new light on the bioenergetic consequences of the loss of LRPPRC in cardiac mitochondria.
INTRODUCTION
Leigh syndrome (LS, MIM 256000) is a genetically and biochemically heterogeneous neurodegenerative entity characterized by subacute necrotizing encephalopathy predominantly affecting the thalamus, brain stem and spinal cord (1). LS patients typically develop sudden onset of severe brain stem dysfunction, psychomotor regression, hypotonia, ataxia, and lactic acidosis during their infancy or early childhood and display various types of biochemical defects, including deficient oxidative phosphorylation (2). The oxidative phosphorylation system (OXPHOS) is located in the mitochondrial inner membrane and is composed of two functional entities, i.e. the respiratory chain (RC) and the phosphorylation system, which includes the ATP synthase and carriers, such as the ATP/ADP carrier (ANT) and the phosphate carrier (PiC). The RC is historically defined as consisting of four complexes, denoted complex I–IV, which perform substrate oxidation to drive proton extrusion from the mitochondrial matrix to the intermembrane space. The proton electrochemical potential across the inner mitochondrial membrane (ΔΨ) is used by the ATP synthase to drive ATP synthesis thus coupling proton transport to ATP production.
Whereas various types of defects in oxidative phosphorylation or the pyruvate dehydrogenase enzyme complex can cause LS (3–7), complex IV (cytochrome-c oxidase, COX) deficiency is one of the most common causes. Mutations in various nuclear genes have been reported in COX-deficient LS, e.g. in Surf1 (8,9), Sco1 (10), Sco2 (11), Cox10 (12) and Cox15 (13), and these genes all encode proteins involved in assembly or biogenesis of COX. Surprisingly, Surf1 knockout (14,15) and Sco2 knock-in (16) mouse models appear healthy and may even have increased lifespan despite a profound COX deficiency (17,18).
An unusual COX-deficient form of LS, the French Canadian type of LS (LSFC, MIM 220111), has been shown to be caused by mutations in the gene for the leucine-rich pentatricopeptide repeat containing protein (Lrpprc) (19), which is a key posttranscriptional regulator of mtDNA expression (20). In comparison with other forms of COX-deficient LS, patients with LSFC seem to have more severe symptoms associated with an acute metabolic crisis leading to high mortality at early ages (21,22). We have previously generated whole body Lrpprc knockout mice and demonstrated that this condition causes embryonic lethality (20), in agreement with results from an independent knockout strain (23). Furthermore, we found that tissue-specific inactivation of Lrpprc in the heart causes a progressive cardiomyopathy resulting in death before the age of 16 weeks (20). Studies of RNAi-knockdown cell lines have shown that LRPPRC is involved in regulating the stability of mtDNA-encoded mRNAs (24,25). However, the in vivo function of LRPPRC is not restricted to regulating mRNA stability because mice with conditional knockout of Lrpprc and fruit flies with knockdown of Bsf, the fly homolog of Lrpprc a.k.a. DmLrpprc1, have additional strong biochemical phenotypes, i.e. impaired polyadenylation of mtDNA-encoded mRNAs and loss of mitochondrial translation coordination (20,26). Intriguingly, unlike the COX-deficient mice mentioned above, the Lrpprc heart-specific knockout mice develop a lethal cardiomyopathy, suggesting that COX deficiency may not provide the sole explanation of the phenotype.
In this study, we decided to carefully characterize the bioenergetic properties of Lrpprc knockout heart mitochondria. Unexpectedly, we show that the COX deficiency per se cannot explain the severe OXPHOS dysfunction observed in Lrpprc heart knockout mice. Instead, we show that LRPPRC deficiency is associated with an ATP synthase assembly defect leading to a drastic loss of function of this enzyme. Our work thus shows that the bioenergetic defect in Lrpprc knockout heart mitochondria is not driven by COX deficiency but rather caused by an ATP synthase deficiency.
RESULTS
Loss of LRPPRC induces defective respiration under phosphorylating conditions
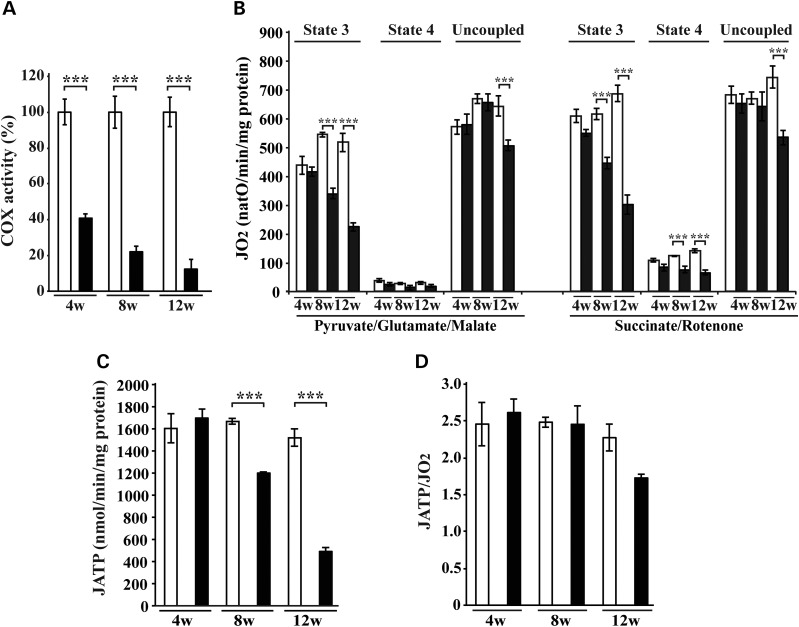
We assessed COX activity in conditional Lrpprc knockout mouse hearts and found a progressive deficiency (Fig. 1A), consistent with our previous results and reports of severe COX deficiency in LSFC patients (20). The COX deficiency in Lrpprc knockout hearts was profound with 40% remaining activity at age 4 weeks and 10% remaining activity at age 12 weeks (Fig. 1A). We proceeded to investigate the bioenergetic consequences of the strong decrease in COX activity by assessing the oxygen consumption rate in freshly isolated cardiac mitochondria (Fig. 1B). Mitochondria were incubated with respiratory substrates whose metabolism result in delivery of electrons at the level of complex I (pyruvate, glutamate, malate) or complex II (succinate + rotenone) and the oxygen consumption rate was recorded in the phosphorylating (state 3: ADP and Pi), non-phosphorylating (state 4: oligomycin to inhibit ATP synthase) and uncoupled state (uncoupling agent CCCP). Interestingly, the respiration in Lrpprc heart knockout mitochondria was profoundly affected in the phosphorylating state, whereas uncoupled respiration was either unaffected or only mildly affected at the latest studied time point (Fig. 1B). The ATP production rate was assessed in the presence of succinate, rotenone and ADP, as previously described (27), and was normal in Lrpprc heart knockout mitochondria at the age of 4 weeks, and strongly impaired at the ages of 8 and 12 weeks (Fig. 1C). We also assessed ATP production rate with pyruvate, glutamate, malate and ADP, and found a strong impairment in Lrpprc heart knockout mitochondria at the age of 12 weeks (Supplementary Material, Fig. S1A). The coupling yield of the oxidative phosphorylation, i.e. the ratio between ATP production and oxygen consumption (JATP/JO2), was normal at all studied ages (Fig. 1D). In summary, these findings show that the massive reduction in COX activity (Fig. 1A) is associated with an impaired respiration in the phosphorylating state (Fig. 1B) and reduced ATP synthesis (Fig. 1C, Supplementary Material, Fig. S1A), which all occur concomitantly with normal coupling of the oxidative phosphorylation system in mitochondria lacking LRPPRC (Fig. 1D). Surprisingly, the overall capacity of the RC was only mildly affected in the uncoupled state in the absence of LRPPRC (Fig. 1B).
Figure 1.
Loss of LRPPRC results in OXPHOS dysfunction. (A) The COX enzyme activity was measured in heart mitochondria from Lrpprc knockout and control at age 4 weeks (4w), 8 weeks (8w) and 12 weeks (12w). Open bars, controls (n = 4); filled bars, knockouts (n = 4). Error bars indicate mean ± SEM (*P < 0.05; **P < 0.01; ***P < 0.001). (B) Oxygen consumption of heart mitochondria from Lrpprc knockout and control at different ages. Isolated mitochondria were incubated with substrates feeding electrons to complex I (pyruvate, glutamate, malate) or complex II (succinate combined with complex I inhibition by rotenone). Each set of substrates was successively combined with ADP (to assess the phosphorylating respiration: state 3), oligomycin (to measure the non-phosphorylating respiration: state 4) and finally uncoupled by adding an increasing concentration of CCCP. Open bars, control [n = 4 (at age 4 and 8 weeks) n = 15 (at age 12 weeks)]; filled bars, knockout [n = 4 (at age 4 and 8 weeks) n = 15 (at age 12 weeks)]. Error bars indicate means ± SEM (*P < 0.05; **P < 0.01; ***P < 0.001). (C) ATP synthesis flux assessed in heart mitochondria from Lrpprc knockout and control at the age of 12 weeks, in the presence of succinate and rotenone. Open bars, control (n = 3–4); filled bars, knockout (n = 3–4). Error bars indicate means ± SEM (*P < 0.05; **P < 0.01; ***P < 0.001). (D) Measurement of the oxidative phosphorylation coupling yield (nmol ATP/natO) in heart mitochondria from Lrpprc knockout and controls at different ages. Open bars, control (n = 4); filled bars, knockout (n = 4). Error bars indicate means ± SEM (*P < 0.05; **P < 0.01; ***P < 0.001).
The respiratory defect in the phosphorylating state is not caused by the COX deficiency
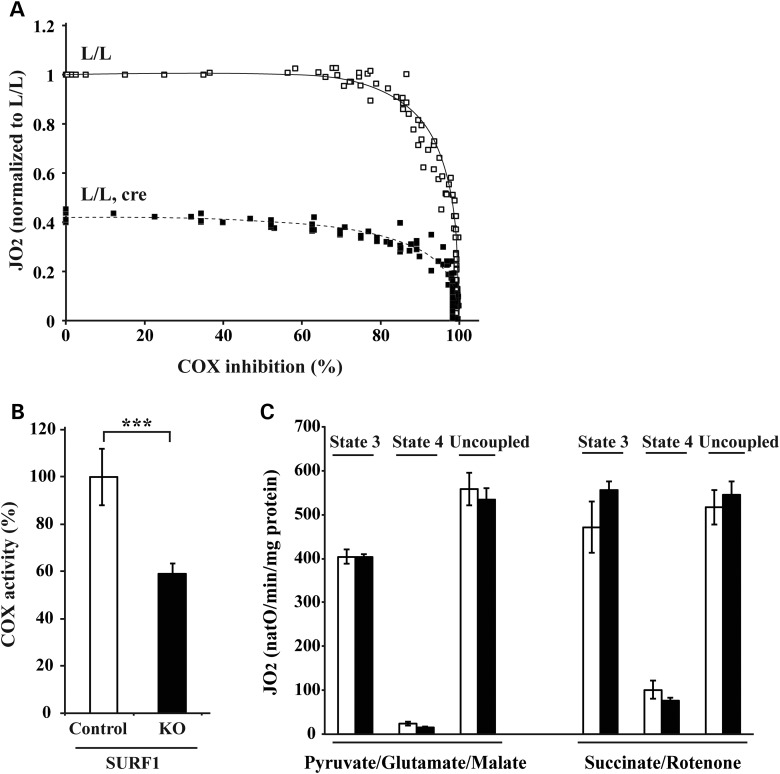
We were intrigued by the finding that loss of ∼90% of the COX enzyme activity only mildly impaired the RC capacity and therefore we proceeded to analyze to what extent inhibition of COX reduces respiration under phosphorylating conditions with the substrates succinate, rotenone and ADP (Fig. 2A) or pyruvate, glutamate, malate and ADP (Supplementary Material, Fig. S1B). We progressively inhibited COX by cyanide treatment of wild-type cardiac mitochondria and found that the respiratory rate (JO2) in the phosphorylating state is only mildly altered until COX is inhibited so that only <10% of the original activity remains (Fig. 2A, Supplementary Material, Fig. S1B). To validate these conclusions by an independent set of experiments, we studied respiration in mitochondria isolated from hearts of Surf1 knockout mice (Fig. 2B and C). The COX activity in heart mitochondria from 40-week-old Surf1 knockout mice is reduced by ∼40% (Fig. 2B), but the respiration is unaffected (Fig. 2C), consistent with the absence of cardiomyopathy in these knockout mice (14). Next, we progressively inhibited the residual COX activity by cyanide treatment of heart mitochondria from conditional Lrpprc knockout mice at age 12 weeks (residual COX activity ∼10%; Fig. 1A) and found no further impairment of respiration until <60% of the residual COX activity remained (corresponding ∼6% of the COX activity in wild-type heart mitochondria) (Fig. 2A, Supplementary Material, Fig. S1B). These results provide additional support for the conclusion that the severe reduction of respiration in the phosphorylating state in heart mitochondria from conditional Lrpprc knockout mice is not explained by the profound COX deficiency.
Figure 2.
COX deficiency does not explain the mitochondrial respiration deficiency in phosphorylating conditions. (A) Threshold curves were performed on heart mitochondria from Lrpprc knockout and control mice at the age of 12 weeks in the presence of succinate, rotenone and ADP. Open square, control (n = 4); filled square, knockout (n = 4). (B) COX enzyme activity was measured in heart mitochondria from Surf1 knockout and control mice at the age of 40 weeks. Open bars, control (n = 4); filled bars, knockout (n = 4). Error bars indicate means ± SEM (*P < 0.05; **P < 0.01; ***P < 0.001). (C) Oxygen consumption was assessed in heart mitochondria from Surf1 knockout mice at age 40 weeks. Open bars, control (n = 4); filled bars, knockout (n = 4). Error bars indicate means ± SEM (*P < 0.05; **P < 0.01; ***P < 0.001).
Loss of LRPPRC impairs assembly and enzyme activity of the ATP synthase
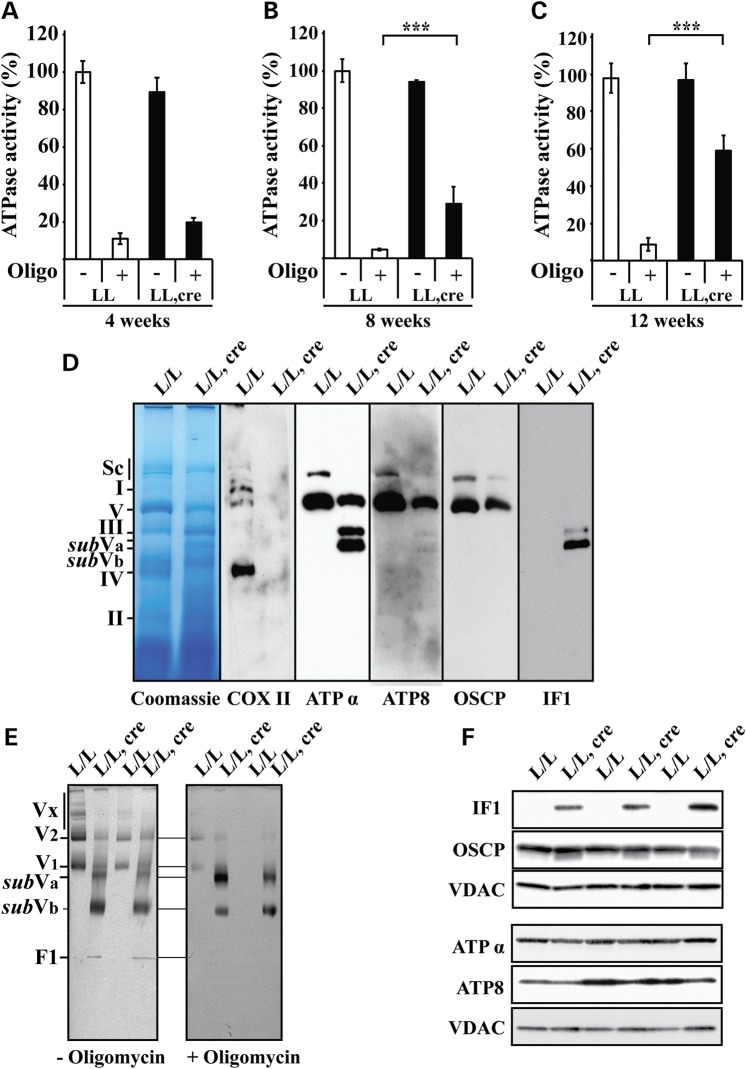
We proceeded to analyze the ATP synthesis system to further investigate the cause of the severe respiratory defect in the phosphorylating state in Lrpprc knockout heart mitochondria. The oligomycin-sensitive ATPase activity was analyzed as previously described (28) and we found an almost complete inhibition by oligomycin treatment of wild-type heart mitochondria (Fig. 3A–C). In contrast, the ATPase activity in Lrpprc knockout heart mitochondria at the age of 8 and 12 weeks was increasingly resistant to oligomycin (Fig. 3A–C). The observation that the total ATPase activity was normal, whereas the oligomycin-resistant activity was increased in Lrpprc knockout heart mitochondria (Fig. 3A–C) prompted us to analyze the assembly status of ATP synthase. To this end, we first determined that a ratio of 1 g/g digitonin to mitochondrial protein provided a good solubilization condition for characterization of the supra-molecular organization of ATP synthase by in-gel enzyme activity assays (Supplementary Material, Fig. S2A). Next, we proceeded to use blue native (BN-PAGE) (Fig. 3D, Supplementary Material, Fig. S2B) and clear native electrophoresis (CN-PAGE) (Fig. 3E, Supplementary Material, Fig. S2A and D) to determine the oligomerization state of the ATP synthase (29,30). As an internal control, we studied COX and found a substantial decrease of both the in-gel enzyme activity (Supplementary Material, Fig. S2C) and the levels of the assembled complex (Fig. 3D, Supplementary Material, Fig. S2B), consistent with the drastic reduction of COX enzyme activity in heart mitochondria of Lrpprc knockout mice (Fig. 1A). ATP synthase oligomers were present in heart mitochondria of Lrpprc conditional knockout mice at the age of 4 weeks (Supplementary Material, Fig. S2D), but these oligomers were almost totally lost in 12 weeks old conditional Lrpprc knockouts and instead sub-assembled complexes (here denoted subVa and subVb) appeared (Fig. 3D and E, Supplementary Material, Fig. S2A and D). The CN-PAGE procedure allowed us to assess the oligomycin sensitivity of the different ATP synthase complexes (Fig. 3E). In control heart mitochondria, the in-gel ATPase activity was almost completely inhibited by oligomycin, whereas the subVa and subVb subcomplexes in Lrpprc knockout heart mitochondria showed an ATPase activity that was resistant to oligomycin (Fig. 3E). To further characterize the basis for the oligomycin resistance in the subassembled ATP-synthase complexes, we investigated their subunit composition. The steady-state levels of the oligomycin sensitivity-conferring protein (OSCP) were not significantly altered on western blot analysis of heart mitochondria from conditional Lrpprc knockout mice at the age of 12 weeks (Fig. 3F). The level of ATPα was unchanged, whereas the level of ATP8 was reduced in total protein extracts form heart mitochondria of conditional Lrpprc knockout mice at the age of 12 weeks (Fig. 3F). Interestingly, despite normal IF1 mRNA levels (data not shown), the IF1 protein levels were dramatically increased in Lrpprc knockout heart mitochondria at the age of 12 weeks (Fig. 3F). Western blot analysis of ATP synthase complexes in heart mitochondria from conditional Lrpprc knockout mice at the age of 12 weeks showed that ATP8 and OSCP were mainly present in the assembled ATP synthase complex (Fig. 3D), ATPα was present in both the assembled complex and the subcomplexes (Fig. 3D), whereas IF1 was only found in the subcomplexes (Fig. 3D). As an independent, non-quantitative way to assess the protein composition of ATP synthase and subcomplexes, we performed mass spectrometry analysis (Table 1). We analyzed five samples of each complex and subcomplex and noted the number of times peptides from a particular protein subunit could be found (Table 1). The OSCP protein conferring oligomycin resistance was detectable in all ATP synthase complexes and subcomplexes (Table 1), however, western blot analysis of complexes separated by BN-PAGE revealed very low levels of the OSCP protein in the ATP synthase subcomplexes (Fig. 3D). These findings argue that most of the subcomplexes lack OSCP, which may explain their insensitivity to oligomycin. The IF1 protein was dramatically increased in Lrpprc knockout heart mitochondria (Fig. 3F) and was associated with the subVa and subVb complexes (Fig. 3D). Western blot (Fig. 3D) and mass spectrometry (Table 1) analyses showed that most membranous domain (Fo) and catalytic domain (F1) subunits were present in the subVa complex, whereas many of the Fo and F1 subunits as well as subunits of the stator domain were absent in the subVb complex. These results show that loss of LRPPRC leads to impaired ATP synthase assembly and the appearance of oligomycin-resistant ATP synthase subcomplexes.
Figure 3.
Loss of LRPPRC is associated with the appearance of an oligomycin-insensitive subassembled ATP synthase complex. (A) The ATPase activity in the presence or absence of oligomycin (Oligo) was normalized to the activity in controls measured in the absence of oligomycin, The ATPase activity was determined in heart mitochondria from Lrpprc knockout and control mice at age 4 weeks. Open bars, control (n = 3); filled bars, knockout (n = 3). Error bars indicate means ± SEM. (B) The ATPase activity in the presence or absence of oligomycin (Oligo) was normalized to the activity in controls measured in the absence of oligomycin. The ATPase activity was determined in heart mitochondria from Lrpprc knockout and control mice at the age of 8 weeks. Open bars, control (n = 3); filled bars, knockout (n = 3). Error bars indicate mean ± SEM (*P < 0.05; **P < 0.01; ***P < 0.001). (C) The ATPase activity in the presence or absence of oligomycin (Oligo) was normalized to the activity in controls measured in the absence of oligomycin. The ATPase activity was determined in heart mitochondria from Lrpprc knockout and control mice at age 12 weeks. Open bars, control (n = 5); filled bars, knockout (n = 5). Error bars indicate means ± SEM (*P < 0.05; **P < 0.01; ***P < 0.001). (D) Blue native polyacrylamide gel electrophoresis analysis of heart mitochondria extracted with a ratio (1 g/g) of digitonin to mitochondrial protein from Lrpprc knockout and control mice at the age of 12 weeks. Immunodetection of COX II, ATPα, ATP8, OSCP and IF1 after transfer of proteins from the BN-PAGE to PVDF membrane. The position of supercomplexes (Sc), complex I (I), the ATP synthase monomer (V), ATP synthase subcomplexes (subVa, subVb), complex IV (IV) and complex II (II) are indicated on the left side. (E) The supra-molecular organization of ATP synthase was determined by clear native polyacrylamide gel electrophoresis (CN-PAGE) stained for in-gel ATPase activity (appear black after color inversion) without (right panel) or with oligomycin (left panel) in heart mitochondria from Lrpprc knockout and control at the age of 12 weeks. Mitochondria were solubilized with a ratio (1.5 g/g) of digitonin to mitochondrial protein (n = 6). The position of ATP synthase oligomers (Vx), dimer (V2), monomer (V1), subassemblies (subVa, subVb) and F1 are indicated on the left side. (F) Steady-state levels of different ATP synthase subunits analyzed by western blots analyses in heart mitochondria from Lrpprc knockout and control mice at age 12 weeks (n = 3).
Table 1.
Subunit composition of dimers, monomers and subassembled ATP synthase complexes determined by mass spectrometry
| Subunit | Mass (da) | PLGS score |
Peptides |
Coverage (%) |
Incidence of detection (/5) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| L/L |
L/L, cre |
|||||||||||||
| L/L | L/L, cre | L/L | L/L, cre | L/L | L/L, cre | V2 | V1 | V2 | V1 | subVa | subVb | F1 | ||
| α | 59 715 | 45 982 | 46 238 | 59 | 51 | 63 | 58 | 5 | 5 | 5 | 5 | 5 | 5 | 5 |
| β | 56 265 | 31 324 | 32 068 | 38 | 32 | 75 | 56 | 5 | 5 | 5 | 5 | 5 | 5 | 5 |
| γ | 16 751 | 8476 | 9171 | 14 | 10 | 36 | 34 | 5 | 5 | 5 | 5 | 5 | 2 | 4 |
| δ | 32 750 | 11 492 | 11 412 | 4 | 5 | 26 | 38 | 4 | 3 | 3 | 5 | 3 | 2 | 0 |
| ɛ | 5834 | 19 006 | 21 785 | 3 | 3 | 37 | 30 | 3 | 5 | 3 | 5 | 4 | 3 | 0 |
| OSCP | 23 348 | 14 405 | 20 730 | 18 | 13 | 68 | 60 | 5 | 5 | 5 | 5 | 5 | 5 | 3 |
| b | 28 930 | 3914 | 2688 | 9 | 7 | 33 | 24 | 5 | 5 | 4 | 5 | 4 | 1 | 0 |
| e | 8230 | 15 448 | 3020 | 5 | 6 | 47 | 53 | 5 | 5 | 5 | 5 | 3 | 2 | 4 |
| f | 8999 | 3516 | 4051 | 3 | 3 | 26 | 21 | 5 | 5 | 4 | 5 | 5 | 2 | 2 |
| g | 11 417 | 12 753 | 10 165 | 7 | 5 | 52 | 41 | 4 | 5 | 4 | 5 | 5 | 0 | 2 |
| ATP8 | 7761 | 6763 | 8802 | 3 | 2 | 28 | 24 | 5 | 5 | 5 | 4 | 4 | 0 | 0 |
The ATP synthase dimer (V2), monomer (V1), subassembled complexes (subVa and subVb) and F1 subunit composition in heart mitochondria from Lrpprc knockout and control mice at the age of 12 weeks (n = 5). The average PLGS score, number of peptides and coverage were calculated from values obtained in ATP synthase dimer (V2) and monomers (V1). The PLGS score was calculated by the Protein Lynx Global Server (PLGS 2.2.5) software and is a statistical measure of accuracy of assignation. A higher score implies greater confidence in protein identity. The incidence of detection in five replicates is presented in the right panel.
Altered mitochondrial cristae morphology
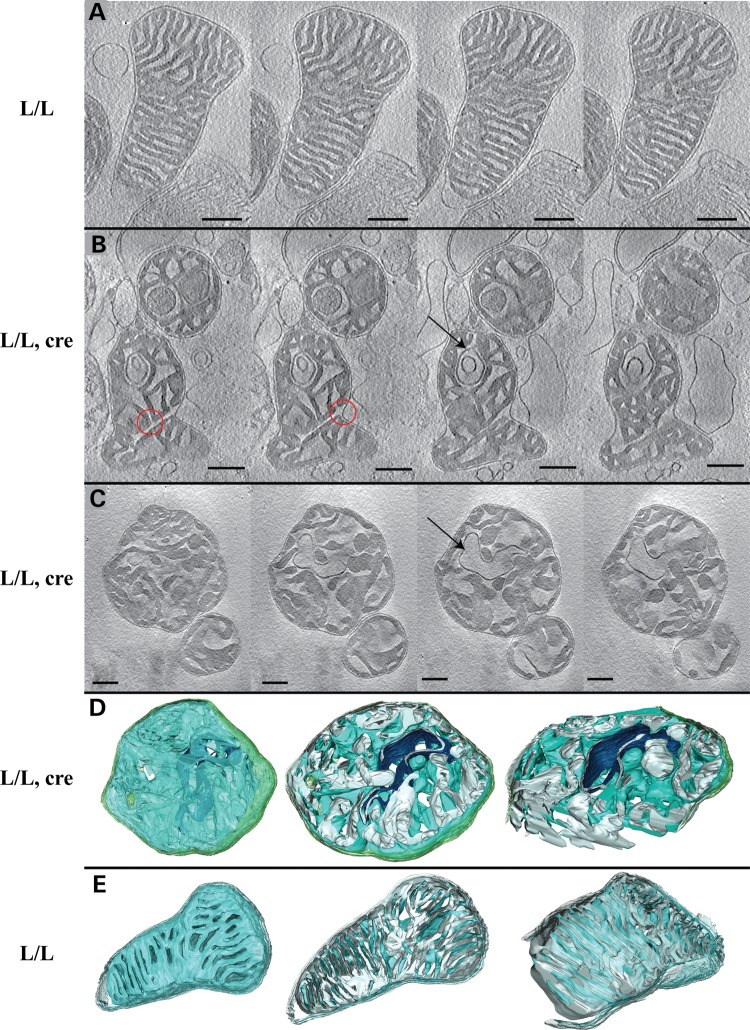
ATP synthase oligomers have recently been shown to cause membrane curvature by forming long dimer rows along the cristae ridges (31–35). The disassembly of ATP synthase dimer rows is accompanied by severe changes of cristae morphology with disappearance of the typical lamellar cristae in yeast mutants lacking dimer-specific subunits (35,36). Our previous electron microscopy studies using thin plastic sections of mouse heart tissue have shown altered cristae morphology in the absence of LRPPRC (20). Here, we further assessed mitochondrial morphology in isolated Lrpprc knockout heart mitochondria by performing electron cryo-tomography to obtain three-dimensional reconstructions of fully hydrated mitochondria at high resolution (Fig. 4 and Supplementary Material, Movies 1 and 2). Wild-type mitochondria from 12-week-old control mice had predominantly thin, stacked, lamellar cristae, with clearly discernible cristae junctions (Fig. 4A and E). In contrast, Lrpprc knockout heart mitochondria showed irregular inner membrane morphology (Fig. 4B–D). In many mitochondria, the shape and distribution of the cristae was irregular and lamellar cristae were absent (Fig. 4B and C). Cristae junctions were wider than in wild-type controls and in some cases not discernible. Furthermore, we observed several mitochondria in which canonical cristae were completely lost and the inner membrane instead formed a network of interconnected vesicles that extended throughout the mitochondrion (Fig. 4B–D). In comparison with regular cristae, the inner-membrane curvature was partially inverted and the characteristic cristae ridges typically occupied by ATP synthase (35) were largely missing. Occasionally, extremely dense and thin matrix compartments, essentially consisting of only two membranes, were observed.
Figure 4.
Loss of LRPPRC impairs mitochondrial cristae morphology. (A–C) Slices through tomograms of mitochondria analyzed by electron cryo-microscopy. Scale bars correspond to 250 nm. (A) An example of the highly organized cristae observed in heart mitochondria from control at the age of 12 weeks. Cristae are lamellar and form stacks. (B and C) Two examples of abnormal cristae morphology observed in heart mitochondria from Lrpprc knockout at the age of 12 weeks. The organization of the inner membrane with deeply invaginated cristae is lost. Instead, cristae are enlarged and the membrane curvature is partially inverted. The red circles indicate unusually large cristae junctions and connections between cristae. The arrows point towards dense, thin membrane compartments protruding from the matrix. (D and E) Three-dimensional segmentation of tomographic complete volumes (left) and cut-away views through the xy-plane (center) or an oblique slice (right). (D) Heart mitochondria from Lrpprc knockout mice at the age of 12 weeks (compare with C). (E) Heart mitochondria from control mice at the age of 12 weeks (compare with A). The membrane surface facing the cristae is shown in blue, the surface facing the matrix is shown in grey. The outer membrane is shown in transparent green. The dense, thin membrane compartment is shown in dark blue.
Taken together, the abnormal cristae morphology (Fig. 4B–D), the reduced amount of oligomers of ATP synthase and the presence of ATP synthase subcomplexes (Fig. 3D and E) in Lrpprc knockout heart mitochondria support previous reports (36,32) that ATP synthase oligomerization is required for proper cristae organization.
Bioenergetic consequences of ATP synthase subassembly on membrane potential and ROS production
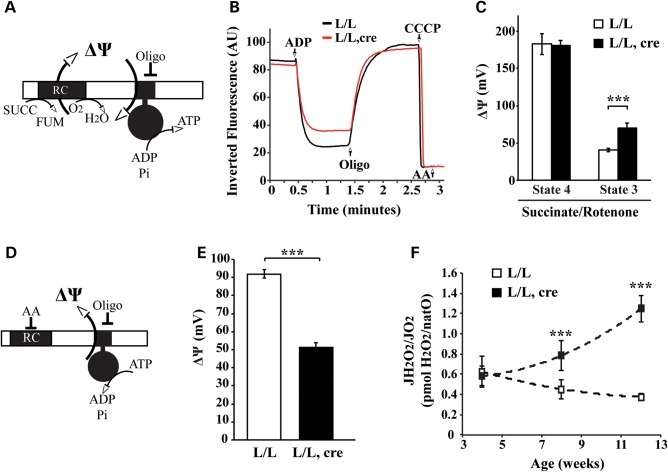
The presence of oligomycin-insensitive, partially assembled ATP synthase complexes in Lrpprc knockout heart mitochondria prompted us to further investigate the bioenergetic properties. Under physiological conditions, ATP synthase couples the membrane potential (ΔΨ) consumption to ATP synthesis. We measured the ΔΨ under phosphorylating and non-phosphorylating conditions (Fig. 5A–C). In agreement with other models harboring a specific ATP synthase deficiency (37–39), mitochondria from Lrpprc knockout hearts can maintain ΔΨ in the non-phosphorylating state, but are hyperpolarized under phosphorylating conditions (Fig. 5B and C, Supplementary Material, Fig. S3A). Consequently, the decrease in ATP production in Lrpprc knockout heart mitochondria (Fig. 1C) is associated with decreased ΔΨ consumption. We proceeded to investigate the ability of ATP synthase to couple proton transfer to ATP usage by assaying the enzyme activity under ATP hydrolyzing conditions, thus uncoupling its enzyme activity from the function of the RC (Fig. 5D). It is well known that specific pH conditions can release inhibitory peptides (IF1) from the ATP synthase (40,41) and thereby allow the generation of an increased ΔΨ by hydrolysis of ATP (Fig. 5D). Our results show that the ATP synthase in Lrpprc knockout heart mitochondria cannot maintain a high ΔΨ when ATP is hydrolyzed (Fig. 5E). Next, we assessed hydrogen peroxide release when the oxidative phosphorylation system is working under phosphorylating conditions (Fig. 5F). The hydrogen peroxide production per oxygen consumed, a.k.a. peroxidic yield (42), was dramatically increased in Lrpprc knockout heart mitochondria at the ages of 8 and 12 weeks (Fig. 5F, Supplementary Material, Fig. S3B). Our data are in accordance with previous observations from studies on patient cell lines that have an ATP synthase deficiency because of a pathogenic mutation in the ATP6 gene of mtDNA (43–45). The ATP synthase defect in these patient cell lines causes a mitochondrial hyperpolarization and an increase in reactive oxygen species (ROS) production. Altogether, the results presented here strongly argue that the partly assembled ATP synthase complexes in Lrpprc knockout heart mitochondria cannot properly couple ATP synthesis or hydrolysis to proton translocation across the inner membrane. Interestingly, the observed hyperpolarization under phosphorylating conditions (Fig. 5C) is associated with an increased ROS production (Fig. 5F). However, this increased ROS production is not associated with increased levels of protein and lipid carbonylation (Supplementary Material, Fig. S3C), or with an increase in the steady-state levels of mitochondrial superoxide dismutase (SOD2) (Supplementary Material, Fig. S3D).
Figure 5.
Subassembled ATP synthase fails to translocate protons and increases ROS production. (A) Scheme of the conditions used to measure the membrane potential (ΔΨ) in isolated mitochondria incubated with succinate and rotenone under phosphorylating conditions. (B) Inverted rhodamine 123 fluorescent signal recording in heart mitochondria from Lrpprc knockout (red line) and control (black line) mice at the age of 12 weeks. Fluorescent signal variation was followed after successive addition of ADP, oligomycin (Oligo), CCCP and Antimycin A (AA). (C) Quantification of the membrane potential (ΔΨ) under non-phosphorylating (State 4) and phosphorylating (State 3) conditions in heart mitochondria from Lrpprc knockout and control mice at the age of 12 weeks. Open bars, control (n = 4); filled bars, knockout (n = 4). Error bars indicate means ± SEM (*P < 0.05; **P < 0.01; ***P < 0.001). (D) Scheme of the conditions used to measure the membrane potential (ΔΨ) in isolated mitochondria incubated with ATP and antimycin A. These conditions allow the ATP synthase to generate membrane potential (ΔΨ) by hydrolyzing ATP. (E) The mitochondrial membrane potential imposed by the ATPase in heart mitochondria from Lrpprc knockout and control mice at age 12 weeks. Open bars, control (n = 4); filled bars, knockout (n = 4). Error bars indicate means ± SEM (*P < 0.05; **P < 0.01; ***P < 0.001). (F) The mitochondrial peroxidic yield, i.e. hydrogen peroxide released per oxygen consumed, was assessed under phosphorylating conditions with succinate and rotenone in heart mitochondria from Lrpprc knockout and control at different ages. Open square, control (n = 4); filled square, knockout (n = 4). Error bars indicate means ± SEM (*P < 0.05; **P < 0.01; ***P < 0.001).
DISCUSSION
The role of LRPPRC in mammalian mitochondria has attracted much interest as mutations in Lrpprc cause LSFC, a severe progressive form of infantile onset neurodegeneration associated with COX deficiency. Recent advances in our understanding of the molecular mode of action of LRPPRC based on studies in knockout mice (20) and knockdown flies (26) have demonstrated that LRPPRC is a key posttranscriptional regulator of mtDNA expression that is necessary for mRNA stability, mRNA polyadenylation and coordination of mitochondrial translation. Despite this progress, the bioenergetic consequences associated with loss of LRPPRC have not been fully elucidated. LSFC patients have a profound COX deficiency and this defect has been considered to be of key importance for explaining the disease pathophysiology. However, the global decrease in mtDNA expression makes it possible that there are other OXPHOS problems, besides the COX deficiency, that are driving the phenotype. To investigate this hypothesis, we carried out an extensive bioenergetic and morphological characterization of mitochondria lacking LRPPRC. Surprisingly, we report here that the main consequence of loss of LRPPRC is deficient ATP production due to direct effects on ATP synthase oligomerization and assembly. In contrast, COX deficiency is not the main cause of the OXPHOS dysfunction.
The bioenergetic characterization of Lrpprc knockout heart mitochondria shows that only a small fraction, <10%, of the COX enzyme activity is required to maintain OXPHOS under phosphorylating conditions. This large excess in COX capacity, also defined as COX reserve capacity, has been described in different tissues and models, but remains poorly understood (46–48). Our results indicate that the substantial COX reserve capacity explains why the Surf1 knockout mouse is largely asymptomatic (14–16). Moreover, the fact that severe myocardial dysfunction cannot be detected in COXVIaH knockout hearts, despite ∼80% reduction in COX activity (17), supports our conclusion that cardiac mitochondria possess a substantial COX capacity reserve.
In contrast to previous models, we show here that loss of Lrpprc in heart does not only impair COX activity but it also affects the ATP synthase complex organization and activity. The electron cryo-tomography analysis of Lrpprc knockout heart mitochondria shows that the loss of ATP synthase oligomers has a severe effect on cristae morphology. Additionally, we show here that the ATP synthase subcomplexes are oligomycin-resistant and fail to properly couple ATP hydrolysis to proton translocation. So far the OSCP protein has been described as key to confer oligomycin sensitivity to the ATP synthase. Recent structural analyses has shown that OSCP links the α subunit of the catalytic part (F1) to the subunit b of the stator (49). This stator-mediated physical connection between the F1 and Fo domains seems essential for inhibiting ATP synthesis or hydrolysis in the presence of oligomycin. The lack of oligomycin-induced inhibition of the ATP synthase subcomplexes can be due to the very low levels of OSCP or lack of the oligomycin-binding sites in subunits c (50) and ATP6 (51–53). The ATP synthase regulatory peptide IF1 has been shown to have a major role on ATP synthase function and structure (54). The IF1 peptide binds the catalytic part of the ATP synthase to prevent futile ATP hydrolysis. Interestingly, the increased level of IF1 and its association with the ATP synthase subcomplexes could be a safeguarding mechanism that explains the absence of ATP hydrolysis under physiological pH conditions in Lrpprc knockout heart mitochondria. The presence of the ATP synthase subcomplexes does not affect the inner membrane proton conductance and does not uncouple the OXPHOS despite impairing the overall ATP production capacity.
Patient cell lines with pathogenic mutations of the ATP synthase subunit 6 (ATP6) (55,56) or subunit 8 (ATP8) genes (57) of mtDNA show defects in the supra-molecular organization of ATP synthase with appearance of ATP synthase subcomplexes (58). A similar type of ATP synthase destabilization has previously been reported in mouse models with impaired mtDNA expression due to mtDNA replication defects (59), impaired mtDNA transcription or impaired mitochondrial translation (60–62). The presence of the ATP synthase subcomplexes, therefore, seems to constitute a sensitive marker for the loss of mtDNA expression.
The severe ATP synthase deficiency reported here sheds important new light on the bioenergetic consequences of LRPPRC loss of function mutants. The ATP synthase activity is rarely measured when investigating OXPHOS defects and has not been investigated in LSFC patient tissue samples (22,63) or fibroblast cultures (25). LSFC patients have a different spectrum of associated symptoms compared with other types of COX-deficient LS and often develop an acute fatal acidosis crisis. Our finding that loss of LRPPRC does not only affect COX activity, but also impairs the ATP synthase may provide an important explanation for the clinical difference between LSFC and forms of LS caused by isolated COX deficiency.
MATERIALS AND METHODS
Mouse breeding
Breeding and genotyping of heart- and skeletal muscle-specific Lrpprc knockout mice were performed as previously described in a C57Bl6/N background (20). Briefly, LrpprcloxP/loxP mice were crossed with transgenic mice expressing cre recombinase under the control of the muscle creatine kinase promoter (Ckmm-cre). The resulting double heterozygous mice (LrpprcloxP/+, +/Ckmm-cre) were mated to LrpprcloxP/loxP mice to generate tissue-specific knockout (LrpprcloxP/loxP, +/Ckmm-cre) and control (LrpprcloxP/loxP) mice. The Surf1 mice were obtained from Surf1+/− intercrosses. Surf1 mice were analyzed at 40 weeks of age.
Mitochondrial isolation from heart
Mice were sacrificed by cervical dislocation, and hearts were quickly collected in ice-cold DPBS (Gibco), minced and homogenized with few strokes of a Potter S homogenizer (Sartorius) in 5 ml of ice-cold mitochondria isolation buffer (MIB; 310 mm sucrose, 20 mm Tris–HCl, 1 mm EGTA, pH 7.2). Mitochondria were purified by differential centrifugation (1200g for 10 min) and supernatants were then centrifuged at 12 000g for 10 min. The crude mitochondrial pellet was resuspended in an appropriate volume of MIB. The mitochondrial protein concentration was determined using the Protein DC Lawry-based assay (Bio-Rad).
Mitochondrial respiratory assay
Mitochondrial oxygen consumption flux was measured as previously described (64) at 37°C using 65–125 μg of crude mitochondria diluted in 2.1 ml of mitochondrial respiration buffer (120 mm sucrose, 50 mm KCl, 20 mm Tris–HCl, 4 mm KH2PO4, 2 mm MgCl2, 1 mm EGTA, pH 7.2) in an Oxygraph-2k (OROBOROS INSTRUMENTS, Innsbruck, Austria). The oxygen consumption rate was measured using either 10 mm pyruvate, 5 mm glutamate and 5 mm malate or 10 mm succinate and 10 nm rotenone. Oxygen consumption was assessed in the phosphorylating state with 1 mm ADP (state 3) or non-phosphorylating state by adding 2.5 μg/ml oligomycin (pseudo state 4). In the control mitochondria, the respiratory control ratio (RCR) values were >10 with pyruvate/glutamate/malate and >5 with succinate/rotenone. Respiration was uncoupled by successive addition of carbonyl cyanide m-chlorophenyl hydrazone (CCCP) up to 3 μM to reach maximal respiration.
Threshold curve of COX control on mitochondrial respiration in phosphorylating condition
As previously described (48), the titration of respiration in the phosphorylating condition with a COX-specific inhibitor (KCN) allows the assessment of the control exerted by this enzyme on the OXPHOS function. In this experiment, we defined the variation of the respiration in phosphorylating condition induced by a decrease in the COX enzyme activity. The mitochondrial respiration was measured with succinate 10 mm and rotenone 10 nm or with pyruvate 10 mm, glutamate 5 mm, malate 5 mm and ADP 1 mm, as described above, in presence of different concentrations of KCN (up to 4 mm). The KCN titration of the COX activity was assessed by measuring the oxygen consumption rate of mitochondria incubated with the same KCN concentration and with TMPD 0.2 mm, ascorbate 1 mm and antimycin A 0.5 µM as described below. The titration curve of the COX activity by KCN in control and Lrpprc knockout heart mitochondria is presented in Supplementary Material, Figure S1C.
Measurement of ATP synthesis flux (JATP)
Isolated mitochondria (65 µg/ml) were suspended in the mitochondrial respiration buffer (see above). After addition of ADP (1 mm), succinate (2 mm) and rotenone (10 nM) or addition of ADP (1 mm), pyruvate (10 mm), glutamate (5 mm) and malate (5 mm), the oxygen consumption and ATP synthesis rates were both measured. Aliquots were collected every 20 s and precipitated in 7% HClO4/25 mm EDTA, centrifuged at 16 000 g for 10 min and then neutralized with KOH 2 M, MOPS 0.3 M. The ATP content in these samples was determined with the ATPlite 1step from PerkinElmer®. In a parallel experiment, oligomycin (2.5 μg/ml protein) was added to the mitochondrial suspension to determine the non-oxidative ATP synthesis rate.
Measurement of isolated enzyme activity (COX and ATPase)
The COX activity was assessed using a classical TMPD/ascorbate assay. Briefly, isolated mitochondria (65 µg/ml) were suspended in mitochondrial respiration buffer (see above). Oxygen consumption was assessed in the presence of TMPD (0.2 mm), ascorbate (1 mm) and antimycin A (0.5 µM). After few minutes of stationary respiration, KCN (2 mm) was injected into the chamber. The COX activity corresponds to the KCN sensitive respiration. ATPase activity was assessed as previously described (65). Shortly, 65 µg/ml freshly isolated mitochondria were incubated at 37°C in the following buffer: triethanolamine 75 mm, MgCl2 2 mm, pH 8.9. Mitochondria were preincubated 2 min with alamethicin 10 µg/ml. The reaction was started by adding 2 mm of ATP. Samples (150 µl) were removed every 2 min and precipitated in 7% HClO4/25 mm EDTA (50 µl). Phosphate was quantified by incubating 150 µl of each aliquot in 1 ml of the following buffer (molybdate 5.34 mm, ferrous sulfate 28.8 mm, and H2SO4 0.75 N). After 2 min incubation, the absorbance was assessed at 600 nm. In a parallel experiment, oligomycin (2.5 μg/ml protein) was added to the mitochondrial suspension to determine the oligomycin-insensitive ATPase activity.
Measurement of reactive oxygen species
The rate of H2O2 production was determined by monitoring the oxidation of the fluorogenic indicator amplex red in the presence of horseradish peroxidase. The concentrations of horseradish peroxidase and amplex red in the incubation medium were 5 U/ml and 1 µM, respectively. Fluorescence was recorded at the following wavelengths: excitation 560 nm and emission 590 nm. A standard curve was obtained by adding known amounts of H2O2 to the assay medium in the presence of the reactants. Mitochondria (65 µg protein/ml) were incubated in the respiratory medium (see above), at 37°C and the H2O2 production rate measurement was initiated by substrate addition. The H2O2 production rate was determined from the slope of a plot of the fluorogenic indicator versus time.
Membrane potential measurement
Trans-membrane potential variations (ΔΨ) in isolated mitochondria were assayed by monitoring the fluorescence quenching of rhodamine 123 with a Hitachi F7000 fluorimeter. The ΔΨ was estimated from the fluorescence quenching of the lipophilic cationic dye rhodamine 123. Isolated mitochondria (65 µg protein/ml) were incubated in the mitochondrial buffer thermostated at 37°C containing succinate (10 mm) or pyruvate (10 mm), glutamate (5 mm), malate (5 mm) and 0.66 µM of rhodamine 123 (Sigma). When added, ADP was 1 mm, ATP 2 mm, oligomycin 2.5 µg/ml.
The rhodamine fluorescence signal at each steady state (F) was recorded using an excitation wavelength of 485 nm. Fluorescence emission was continuously detected at 500 nm. At the end of each experiment, the maximum fluorescence signal (Fmax) was monitored after complete de-energization of the mitochondria following addition of CCCP (6 µm) and antimycin A (0.5 µm). Then, the Fmax-F/Fmax difference at each steady state was calculated.
The membrane potential imposed by the ATPase activity was assessed in the following buffer (155 mm sucrose, 10 mm Tris–HCl, 0.5 mm EGTA, triethanolamine 37.5 mm, MgCl2 1 mm, pH 8.2). After 3 min incubation of the mitochondria in 2 ml of buffer containing pyruvate, glutamate, malate, we added antimycin A (0.5 µg/ml) and ATP 2 mm and the oligomycin-sensitive ΔΨ was recorded.
Clear native PAGE and in gel activity
Clear native PAGE was performed as previously described (66,42). Briefly, 0.1 mg of frozen isolated mitochondria (never thawed) were suspended in 1% digitonin solubilized in the following buffer [potassium acetate 150 mm, HEPES 30 mm, glycerol 12%, 6-aminocaproic acid 2 mm, EDTA 1 mm and 1 pastille of protease inhibitor mini complete (ROCHE), pH 7.4]. Samples were incubated 30 min on ice and centrifuged at 24 000g for 30 min. The supernatant was then supplemented with 6-aminocaproic acid (final concentration: 37.5 mm) and loaded on CN-PAGE (3–13%). Samples migrated overnight at 100–120 V. The gel was incubated at 37°C for 1 h, in the following buffer (triethanolamine 75 mm, MgCl2 5 mm, 0.1% triton X100, lead acetate 0.5 mg/ml, pH 8.9) to obtain the ATPase in gel activity. To detect the oligomycin-insensitive complex, oligomycin (50 µg/ml) was added. The densitometry analysis was performed with the free software FIJI.
Blue native electrophoresis and immunodetection
Blue native electrophoresis (BN-PAGE) was performed in 4–16% gradient gels according to recommendation of the Novex® NativePAGE™ Bis-Tris gel System. Nitrocellulose membranes were used to transfer proteins from SDS gels and PVDF membranes were used to transfer proteins from native gels. Immunodetection of ATP5A1 (Complex V) was performed with specific monoclonal antibodies (Mitoscience). Immunodetection of IF1 was performed using a monoclonal antibody (ab110277; Mitosciences). Immunodetection of OSCP was performed using a polyclonal antibody (sc-74786; Santacruz). VDAC (porin) antibodies were purchased from Calbiochem and polyclonal antisera were used to detect COXII (COX) and ATP8 (67). Protein carbonylation assays were performed according to the Oxyblot protein oxidation kit instructions from Millipore and using the HNE polyclonal antibody from Abcam (ab46544). Immunodetection of SOD2 was performed using a polyclonal antibody from Millipore (AB10346).
Mass spectrometry analysis
The ATP synthase localized by in gel activity bands were excised from the CN-PAGE, chopped into small cubes and transferred to the wells of an OASIS® HLB µElution Plate (Waters Corporation, Milford, CT, USA). The proteins were digested with trypsin (Gold MS grade, Promega, Madison, WI, USA) in an OASIS® plate subsequently (68). Protein identification was performed with a Xevo Q-Tof (Waters Corporation) coupled with a nanoACQUITY UPLC™ (Waters Corporation); 1–4 µl of tryptic digest was directly loaded into an analytical column of 75 µm × 150 mm C18 BEH 1.7 µm (Waters Corporation) with 1% formic acid for 25 min. The loading flow rate was 400 nl/min. The peptides were eluted with a gradient of 3–55% acetonitrile in 0.1% formic acid over 90 min at a flow rate of 400 nl/min. The Xevo Qtof (Waters Corporation) was operated in LC/MSE mode over the m/z range 50–1800 in the Nano-Electrospray mode. The capillary, sample cone, extraction cone and collision energy were 3.6 kV, 25.0, 2.0 and 6.0 V, respectively. During elevated energy scan, the collision energy was ramped from 15 V to 35 V. Glu-fibrinopeptide B of m/z 785.84 was used as Lock Mass for mass correction. Data were collected using MassLynx™ 4.1 and processed using ProteinLynx™ Global Server 2.4. The following are the parameters for database search: minimal fragments ion per peptide matched 3, minimal fragments ion per protein matched 7, missed cleavages 1, fixed modification: carbamidomethyl cysteine; variable modifications: acetyl N-term, oxidation methione.
Mitochondrial electron cryo-tomography
For analysis by electron cryo-microscopy, mitochondria were washed twice with 320 mm trehalose, 20 mm Tris pH 7.3, 1 mm EGTA. Samples were mixed 1:1 with fiducial gold markers (10 nm gold particles conjugated to protein A, Aurion), blotted and immediately plunge-frozen in liquid ethane on Quantifoil holey carbon grids (Quantifoil Micro Tools). Single tilt series (±60°, step size 1.5°) were collected on a FEI Polara (300 kV) using an Ultrascan 4 × 4 k CCD (Gatan) and a post-column Quantum energy filter (Gatan) at −9 µm defocus. The nominal magnification was ×34 000, resulting in a pixel size of 6 Å. A total dose of ∼130 e−/Ų was used. Tilt series alignment using the gold fiducial markers and tomogram reconstruction by back-projection were carried out using the IMOD software package (69). To increase contrast, a final filtering step applying non-linear anisotropic diffusion (70) was performed. Manual segmentation was performed with the program Amira (Mercury Systems).
SUPPLEMENTARY MATERIAL
FUNDING
The study was supported by ERC Advanced Investigator, Cluster of Excellence CECAD and Deutsche Forschungsgemeinschaft, SFB 829, grants to N.G.L. Funding to pay the Open Access publication charges for this article was provided by the Max Planck Society.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Lysann Schmitz for technical assistance with the bioenergetic characterization of heart mitochondria. We also thank Dr Xinping Li and Dr Thomas Franz for technical assistance and advices for the mass spectrometry. We thank Prof. Massimo Zeviani for the gift of the Surf1 knockout mouse model.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Leigh D. Subacute necrotizing encephalomyelopathy in an infant. J. Neurol. Neurosurg. Psychiatr. 1951;14:216–221. doi: 10.1136/jnnp.14.3.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rahman S., Blok R.B., Dahl H.H., Danks D.M., Kirby D.M., Chow C.W., Christodoulou J., Thorburn D.R. Leigh syndrome: clinical features and biochemical and DNA abnormalities. Ann. Neurol. 1996;39:343–351. doi: 10.1002/ana.410390311. [DOI] [PubMed] [Google Scholar]
- 3.López L.C., Schuelke M., Quinzii C.M., Kanki T., Rodenburg R.J.T., Naini A., Dimauro S., Hirano M. Leigh syndrome with nephropathy and CoQ10 deficiency due to decaprenyl diphosphate synthase subunit 2 (PDSS2) mutations. Am. J. Hum. Genet. 2006;79:1125–1129. doi: 10.1086/510023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Taylor R.W., Morris A.A.M., Hutchinson M., Turnbull D.M. Leigh disease associated with a novel mitochondrial DNA ND5 mutation. Eur. J. Hum. Genet. 2002;10:141–144. doi: 10.1038/sj.ejhg.5200773. [DOI] [PubMed] [Google Scholar]
- 5.Horváth R., Abicht A., Holinski-Feder E., Laner A., Gempel K., Prokisch H., Lochmüller H., Klopstock T., Jaksch M. Leigh syndrome caused by mutations in the flavoprotein (Fp) subunit of succinate dehydrogenase (SDHA) J. Neurol. Neurosurg. Psychiatr. 2006;77:74–76. doi: 10.1136/jnnp.2005.067041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Willems J.L., Monnens L.A., Trijbels J.M., Veerkamp J.H., Meyer A.E., van Dam K., van Haelst U. Leigh’s encephalomyelopathy in a patient with cytochrome c oxidase deficiency in muscle tissue. Pediatrics. 1977;60:850–857. [PubMed] [Google Scholar]
- 7.Uziel G., Moroni I., Lamantea E., Fratta G.M., Ciceri E., Carrara F., Zeviani M. Mitochondrial disease associated with the T8993G mutation of the mitochondrial ATPase 6 gene: a clinical, biochemical, and molecular study in six families. J. Neurol. Neurosurg. Psychiatr. 1997;63:16–22. doi: 10.1136/jnnp.63.1.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tiranti V., Hoertnagel K., Carrozzo R., Galimberti C., Munaro M., Granatiero M., Zelante L., Gasparini P., Marzella R., Rocchi M., et al. Mutations of SURF-1 in Leigh disease associated with cytochrome c oxidase deficiency. Am. J. Hum. Genet. 1998;63:1609–1621. doi: 10.1086/302150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu Z., Yao J., Johns T., Fu K., De Bie I., Macmillan C., Cuthbert A.P., Newbold R.F., Wang J., Chevrette M., et al. SURF1, encoding a factor involved in the biogenesis of cytochrome c oxidase, is mutated in Leigh syndrome. Nat. Genet. 1998;20:337–343. doi: 10.1038/3804. [DOI] [PubMed] [Google Scholar]
- 10.Valnot I., Osmond S., Gigarel N., Mehaye B., Amiel J., Cormier-Daire V., Munnich A., Bonnefont J.P., Rustin P., Rötig A. Mutations of the SCO1 gene in mitochondrial cytochrome c oxidase deficiency with neonatal-onset hepatic failure and encephalopathy. Am. J. Hum. Genet. 2000;67:1104–1109. doi: 10.1016/s0002-9297(07)62940-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Papadopoulou L.C., Sue C.M., Davidson M.M., Tanji K., Nishino I., Sadlock J.E., Krishna S., Walker W., Selby J., Glerum D.M., et al. Fatal infantile cardioencephalomyopathy with COX deficiency and mutations in SCO2, a COX assembly gene. Nat. Genet. 1999;23:333–337. doi: 10.1038/15513. [DOI] [PubMed] [Google Scholar]
- 12.Valnot I., von Kleist-Retzow J.C., Barrientos A., Gorbatyuk M., Taanman J.W., Mehaye B., Rustin P., Tzagoloff A., Munnich A., Rötig A. A mutation in the human heme A: farnesyltransferase gene (COX10) causes cytochrome c oxidase deficiency. Hum. Mol. Genet. 2000;9:1245–1249. doi: 10.1093/hmg/9.8.1245. [DOI] [PubMed] [Google Scholar]
- 13.Oquendo C.E., Antonicka H., Shoubridge E.A., Reardon W., Brown G.K. Functional and genetic studies demonstrate that mutation in the COX15 gene can cause Leigh syndrome. J. Med. Genet. 2004;41:540–544. doi: 10.1136/jmg.2003.017426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Agostino A., Invernizzi F., Tiveron C., Fagiolari G., Prelle A., Lamantea E., Giavazzi A., Battaglia G., Tatangelo L., Tiranti V., et al. Constitutive knockout of Surf1 is associated with high embryonic lethality, mitochondrial disease and cytochrome c oxidase deficiency in mice. Hum. Mol. Genet. 2003;12:399–413. doi: 10.1093/hmg/ddg038. [DOI] [PubMed] [Google Scholar]
- 15.Dell'agnello C., Leo S., Agostino A., Szabadkai G., Tiveron C., Zulian A., Prelle A., Roubertoux P., Rizzuto R., Zeviani M. Increased longevity and refractoriness to Ca(2+)-dependent neurodegeneration in Surf1 knockout mice. Hum. Mol. Genet. 2007;16:431–444. doi: 10.1093/hmg/ddl477. [DOI] [PubMed] [Google Scholar]
- 16.Yang H., Brosel S., Acin-Perez R., Slavkovich V., Nishino I., Khan R., Goldberg I.J., Graziano J., Manfredi G., Schon E.A. Analysis of mouse models of cytochrome c oxidase deficiency owing to mutations in Sco2. Hum. Mol. Genet. 2010;19:170–180. doi: 10.1093/hmg/ddp477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Radford N.B., Wan B., Richman A., Szczepaniak L.S., Li J.-L., Li K., Pfeiffer K., Schägger H., Garry D.J., Moreadith R.W. Cardiac dysfunction in mice lacking cytochrome-c oxidase subunit VIaH. Am. J. Physiol. Heart. Circ. Physiol. 2002;282:H726–H733. doi: 10.1152/ajpheart.00308.2001. [DOI] [PubMed] [Google Scholar]
- 18.Hüttemann M., Klewer S., Lee I., Pecinova A., Pecina P., Liu J., Lee M., Doan J.W., Larson D., Slack E., et al. Mice deleted for heart-type cytochrome c oxidase subunit 7a1 develop dilated cardiomyopathy. Mitochondrion. 2012;12:294–304. doi: 10.1016/j.mito.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mootha V.K., Lepage P., Miller K., Bunkenborg J., Reich M., Hjerrild M., Delmonte T., Villeneuve A., Sladek R., Xu F., et al. Identification of a gene causing human cytochrome c oxidase deficiency by integrative genomics. Proc. Natl. Acad. Sci., USA. 2003;100:605–610. doi: 10.1073/pnas.242716699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ruzzenente B., Metodiev M.D., Wredenberg A., Bratic A., Park C.B., Cámara Y., Milenkovic D., Zickermann V., Wibom R., Hultenby K., et al. LRPPRC is necessary for polyadenylation and coordination of translation of mitochondrial mRNAs. EMBO J. 2012;31:443–456. doi: 10.1038/emboj.2011.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Finsterer J. Leigh and Leigh-like syndrome in children and adults. Pediatr. Neurol. 2008;39:223–235. doi: 10.1016/j.pediatrneurol.2008.07.013. [DOI] [PubMed] [Google Scholar]
- 22.Debray F.-G., Morin C., Janvier A., Villeneuve J., Maranda B., Laframboise R., Lacroix J., Decarie J.-C., Robitaille Y., Lambert M., et al. LRPPRC mutations cause a phenotypically distinct form of Leigh syndrome with cytochrome c oxidase deficiency. J. Med. Genet. 2011;48:183–189. doi: 10.1136/jmg.2010.081976. [DOI] [PubMed] [Google Scholar]
- 23.Xu F., Addis J.B.L., Cameron J.M., Robinson B.H. LRPPRC mutation suppresses cytochrome oxidase activity by altering mitochondrial RNA transcript stability in a mouse model. Biochem. J. 2012;441:275–283. doi: 10.1042/BJ20110985. [DOI] [PubMed] [Google Scholar]
- 24.Gohil V.M., Nilsson R., Belcher-Timme C.A., Luo B., Root D.E., Mootha V.K. Mitochondrial and nuclear genomic responses to loss of LRPPRC expression. J. Biol. Chem. 2010;285:13742–13747. doi: 10.1074/jbc.M109.098400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sasarman F., Brunel-Guitton C., Antonicka H., Wai T., Shoubridge E.A. LSFC Consortium. LRPPRC and SLIRP interact in a ribonucleoprotein complex that regulates posttranscriptional gene expression in mitochondria. Mol. Biol. Cell. 2010;21:1315–1323. doi: 10.1091/mbc.E10-01-0047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bratic A., Wredenberg A., Grönke S., Stewart J.B., Mourier A., Ruzzenente B., Kukat C., Wibom R., Habermann B., Partridge L., et al. The bicoid stability factor controls polyadenylation and expression of specific mitochondrial mRNAs in Drosophila melanogaster. PLoS Genet. 2011;7:e1002324. doi: 10.1371/journal.pgen.1002324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mourier A., Devin A., Rigoulet M. Active proton leak in mitochondria: a new way to regulate substrate oxidation. Biochim. Biophys. Acta. 2010;1797:255–261. doi: 10.1016/j.bbabio.2009.10.011. [DOI] [PubMed] [Google Scholar]
- 28.Tzagoloff A., Byington K.H., MacLennan D.H. Studies on the mitochondrial adenosine triphosphatase system. II. The isolation and characterization of an oligomycin-sensitive adenosine triphosphatase from bovine heart mitochondria. J. Biol. Chem. 1968;243:2405–2412. [PubMed] [Google Scholar]
- 29.Wittig I., Schägger H. Advantages and limitations of clear-native PAGE. Proteomics. 2005;5:4338–4346. doi: 10.1002/pmic.200500081. [DOI] [PubMed] [Google Scholar]
- 30.Grandier-Vazeille X., Guérin M. Separation by blue native and colorless native polyacrylamide gel electrophoresis of the oxidative phosphorylation complexes of yeast mitochondria solubilized by different detergents: specific staining of the different complexes. Anal. Biochem. 1996;242:248–254. doi: 10.1006/abio.1996.0460. [DOI] [PubMed] [Google Scholar]
- 31.Allen R.D., Schroeder C.C., Fok A.K. An investigation of mitochondrial inner membranes by rapid-freeze deep-etch techniques. J. Cell. Biol. 1989;108:2233–2240. doi: 10.1083/jcb.108.6.2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Davies K.M., Strauss M., Daum B., Kief J.H., Osiewacz H.D., Rycovska A., Zickermann V., Kühlbrandt W. Macromolecular organization of ATP synthase and complex I in whole mitochondria. Proc. Natl Acad. Sci. USA. 2011;108:14121–14126. doi: 10.1073/pnas.1103621108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Strauss M., Hofhaus G., Schröder R.R., Kühlbrandt W. Dimer ribbons of ATP synthase shape the inner mitochondrial membrane. EMBO J. 2008;27:1154–1160. doi: 10.1038/emboj.2008.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Habersetzer J., Ziani W., Larrieu I., Stines-Chaumeil C., Giraud M.-F., Brèthes D., Dautant A., Paumard P. ATP synthase oligomerization: from the enzyme models to the mitochondrial morphology. Int. J. Biochem. Cell Biol. 2013;45:99–105. doi: 10.1016/j.biocel.2012.05.017. [DOI] [PubMed] [Google Scholar]
- 35.Davies K.M., Anselmi C., Wittig I., Faraldo-Gómez J.D., Kühlbrandt W. Structure of the yeast F1Fo-ATP synthase dimer and its role in shaping the mitochondrial cristae. Proc. Natl Acad. Sci. USA. 2012;109:13602–13607. doi: 10.1073/pnas.1204593109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paumard P., Vaillier J., Coulary B., Schaeffer J., Soubannier V., Mueller D.M., Brèthes D., di Rago J.-P., Velours J. The ATP synthase is involved in generating mitochondrial cristae morphology. EMBO J. 2002;21:221–230. doi: 10.1093/emboj/21.3.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goyon V., Fronzes R., Salin B., di Rago J.-P., Velours J., Brèthes D. Yeast cells depleted in Atp14p fail to assemble Atp6p within the ATP synthase and exhibit altered mitochondrial cristae morphology. J. Biol. Chem. 2008;283:9749–9758. doi: 10.1074/jbc.M800204200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jesina P., Tesarová M., Fornůsková D., Vojtísková A., Pecina P., Kaplanová V., Hansíková H., Zeman J., Houstek J. Diminished synthesis of subunit a (ATP6) and altered function of ATP synthase and cytochrome c oxidase due to the mtDNA 2 bp microdeletion of TA at positions 9205 and 9206. Biochem. J. 2004;383:561–571. doi: 10.1042/BJ20040407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mayr J.A., Havlícková V., Zimmermann F., Magler I., Kaplanová V., Jesina P., Pecinova A., Nůsková H., Koch J., Sperl W., et al. Mitochondrial ATP synthase deficiency due to a mutation in the ATP5E gene for the F1 epsilon subunit. Hum. Mol. Genet. 2010;19:3430–3439. doi: 10.1093/hmg/ddq254. [DOI] [PubMed] [Google Scholar]
- 40.Pullman M.E., Monroy G.C. A naturally occuring inhibitor of mitochondrial adenosine triphosphatase. J. Biol. Chem. 1963;238:3762–3769. [PubMed] [Google Scholar]
- 41.Venard R., Brèthes D., Giraud M.-F., Vaillier J., Velours J., Haraux F. Investigation of the role and mechanism of IF1 and STF1 proteins, twin inhibitory peptides which interact with the yeast mitochondrial ATP synthase. Biochemistry. 2003;42:7626–7636. doi: 10.1021/bi034394t. [DOI] [PubMed] [Google Scholar]
- 42.Rigoulet M., Mourier A., Galinier A., Casteilla L., Devin A. Electron competition process in respiratory chain: regulatory mechanisms and physiological functions. Biochim. Biophys. Acta. 2010;1797:671–677. doi: 10.1016/j.bbabio.2010.01.030. [DOI] [PubMed] [Google Scholar]
- 43.Mrácek T., Pecina P., Vojtísková A., Kalous M., Sebesta O., Houstek J. Two components in pathogenic mechanism of mitochondrial ATPase deficiency: energy deprivation and ROS production. Exp. Gerontol. 2006;41:683–687. doi: 10.1016/j.exger.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 44.Mattiazzi M., Vijayvergiya C., Gajewski C.D., DeVivo D.C., Lenaz G., Wiedmann M., Manfredi G. The mtDNA T8993G (NARP) mutation results in an impairment of oxidative phosphorylation that can be improved by antioxidants. Hum. Mol. Genet. 2004;13:869–879. doi: 10.1093/hmg/ddh103. [DOI] [PubMed] [Google Scholar]
- 45.Houstek J., Pícková A., Vojtísková A., Mrácek T., Pecina P., Jesina P. Mitochondrial diseases and genetic defects of ATP synthase. Biochim. Biophys. Acta. 2006;1757:1400–1405. doi: 10.1016/j.bbabio.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 46.Villani G., Greco M., Papa S., Attardi G. Low reserve of cytochrome c oxidase capacity in vivo in the respiratory chain of a variety of human cell types. J. Biol. Chem. 1998;273:31829–31836. doi: 10.1074/jbc.273.48.31829. [DOI] [PubMed] [Google Scholar]
- 47.Kunz W.S., Kudin A., Vielhaber S., Elger C.E., Attardi G., Villani G. Flux control of cytochrome c oxidase in human skeletal muscle. J. Biol. Chem. 2000;275:27741–27745. doi: 10.1074/jbc.M004833200. [DOI] [PubMed] [Google Scholar]
- 48.Rossignol R., Malgat M., Mazat J.P., Letellier T. Threshold effect and tissue specificity. Implication for mitochondrial cytopathies. J. Biol. Chem. 1999;274:33426–33432. doi: 10.1074/jbc.274.47.33426. [DOI] [PubMed] [Google Scholar]
- 49.Rees D.M., Leslie A.G.W., Walker J.E. The structure of the membrane extrinsic region of bovine ATP synthase. Proc. Natl Acad. Sci. USA. 2009;106:21597–21601. doi: 10.1073/pnas.0910365106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Symersky J., Osowski D., Walters D.E., Mueller D.M. Oligomycin frames a common drug-binding site in the ATP synthase. Proc. Natl Acad. Sci. USA. 2012;109:13961–13965. doi: 10.1073/pnas.1207912109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Macino G., Tzagoloff A. Assembly of the mitochondrial membrane system: sequence analysis of a yeast mitochondrial ATPase gene containing the oli-2 and oli-4 loci. Cell. 1980;20:507–517. doi: 10.1016/0092-8674(80)90637-6. [DOI] [PubMed] [Google Scholar]
- 52.Ooi B.G., Novitski C.E., Nagley P. DNA sequence analysis of the oli1 gene reveals amino acid changes in mitochondrial ATPase subunit 9 from oligomycin-resistant mutants of Saccharomyces cerevisiae. Eur. J. Biochem. 1985;152:709–714. doi: 10.1111/j.1432-1033.1985.tb09251.x. [DOI] [PubMed] [Google Scholar]
- 53.John U.P., Nagley P. Amino acid substitutions in mitochondrial ATPase subunit 6 of Saccharomyces cerevisiae leading to oligomycin resistance. FEBS Lett. 1986;207:79–83. doi: 10.1016/0014-5793(86)80016-3. [DOI] [PubMed] [Google Scholar]
- 54.Campanella M., Casswell E., Chong S., Farah Z., Wieckowski M.R., Abramov A.Y., Tinker A., Duchen M.R. Regulation of mitochondrial structure and function by the F1Fo-ATPase inhibitor protein, IF1. Cell Metab. 2008;8:13–25. doi: 10.1016/j.cmet.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 55.Nijtmans L.G., Henderson N.S., Attardi G., Holt I.J. Impaired ATP synthase assembly associated with a mutation in the human ATP synthase subunit 6 gene. J. Biol. Chem. 2001;276:6755–6762. doi: 10.1074/jbc.M008114200. [DOI] [PubMed] [Google Scholar]
- 56.Houstek J., Klement P., Hermanská J., Houstková H., Hansíková H., Van den Bogert C., Zeman J. Altered properties of mitochondrial ATP-synthase in patients with a T-->G mutation in the ATPase 6 (subunit a) gene at position 8993 of mtDNA. Biochim. Biophys. Acta. 1995;1271:349–357. doi: 10.1016/0925-4439(95)00063-a. [DOI] [PubMed] [Google Scholar]
- 57.Jonckheere A.I., Hogeveen M., Nijtmans L.G.J., van den Brand M.A.M., Janssen A.J.M., Diepstra J.H.S., van den Brandt F.C.A., van den Heuvel L.P., Hol F.A., Hofste T.G.J., et al. A novel mitochondrial ATP8 gene mutation in a patient with apical hypertrophic cardiomyopathy and neuropathy. J. Med. Genet. 2008;45:129–133. doi: 10.1136/jmg.2007.052084. [DOI] [PubMed] [Google Scholar]
- 58.Wittig I., Meyer B., Heide H., Steger M., Bleier L., Wumaier Z., Karas M., Schägger H. Assembly and oligomerization of human ATP synthase lacking mitochondrial subunits a and A6L. Biochim. Biophys. Acta. 2010;1797:1004–1011. doi: 10.1016/j.bbabio.2010.02.021. [DOI] [PubMed] [Google Scholar]
- 59.Milenkovic D., Matic S., Kühl I., Ruzzenente B., Freyer C., Jemt E., Park C.B., Falkenberg M., Larsson N.-G. TWINKLE is an essential mitochondrial helicase required for synthesis of nascent D-loop strands and complete mtDNA replication. Hum. Mol. Genet. 2013;22:1983–1993. doi: 10.1093/hmg/ddt051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Metodiev M.D., Lesko N., Park C.B., Cámara Y., Shi Y., Wibom R., Hultenby K., Gustafsson C.M., Larsson N.-G. Methylation of 12S rRNA is necessary for in vivo stability of the small subunit of the mammalian mitochondrial ribosome. Cell Metab. 2009;9:386–397. doi: 10.1016/j.cmet.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 61.Cámara Y., Asin-Cayuela J., Park C.B., Metodiev M.D., Shi Y., Ruzzenente B., Kukat C., Habermann B., Wibom R., Hultenby K., et al. MTERF4 regulates translation by targeting the methyltransferase NSUN4 to the mammalian mitochondrial ribosome. Cell Metab. 2011;13:527–539. doi: 10.1016/j.cmet.2011.04.002. [DOI] [PubMed] [Google Scholar]
- 62.Park C.B., Asin-Cayuela J., Cámara Y., Shi Y., Pellegrini M., Gaspari M., Wibom R., Hultenby K., Erdjument-Bromage H., Tempst P., et al. MTERF3 is a negative regulator of mammalian mtDNA transcription. Cell. 2007;130:273–285. doi: 10.1016/j.cell.2007.05.046. [DOI] [PubMed] [Google Scholar]
- 63.Merante F., Petrova-Benedict R., MacKay N., Mitchell G., Lambert M., Morin C., De Braekeleer M., Laframboise R., Gagné R., Robinson B.H. A biochemically distinct form of cytochrome oxidase (COX) deficiency in the Saguenay-Lac-Saint-Jean region of Quebec. Am. J. Hum. Genet. 1993;53:481–487. [PMC free article] [PubMed] [Google Scholar]
- 64.Freyer C., Cree L.M., Mourier A., Stewart J.B., Koolmeister C., Milenkovic D., Wai T., Floros V.I., Hagström E., Chatzidaki E.E., et al. Variation in germline mtDNA heteroplasmy is determined prenatally but modified during subsequent transmission. Nat. Genet. 2012;44:1282–1285. doi: 10.1038/ng.2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Somlo M. Induction and repression of mitochondrial ATPase in yeast. Eur. J. Biochem. 1968;5:276–284. doi: 10.1111/j.1432-1033.1968.tb00368.x. [DOI] [PubMed] [Google Scholar]
- 66.Habersetzer J., Larrieu I., Priault M., Salin B., Rossignol R., Brèthes D., Paumard P. Human F1F0 ATP synthase, mitochondrial ultrastructure and OXPHOS impairment: A (Super-)complex matter? PLoS One. 2013;8:e75429. doi: 10.1371/journal.pone.0075429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Larsson N.G., Wang J., Wilhelmsson H., Oldfors A., Rustin P., Lewandoski M., Barsh G.S., Clayton D.A. Mitochondrial transcription factor A is necessary for mtDNA maintenance and embryogenesis in mice. Nat. Genet. 1998;18:231–236. doi: 10.1038/ng0398-231. [DOI] [PubMed] [Google Scholar]
- 68.Franz T., Li X. The OASIS® HLB μElution plate as a one-step platform for manual high-throughput in-gel digestion of proteins and peptide desalting. Proteomics. 2012;12:2487–2492. doi: 10.1002/pmic.201100354. [DOI] [PubMed] [Google Scholar]
- 69.Kremer J.R., Mastronarde D.N., McIntosh J.R. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- 70.Frangakis A.S., Hegerl R. Noise reduction in electron tomographic reconstructions using nonlinear anisotropic diffusion. J. Struct. Biol. 2001;135:239–250. doi: 10.1006/jsbi.2001.4406. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.