Abstract
We previously demonstrated that the Alzheimer's disease (AD) associated risk allele, rs3865444C, results in a higher surface density of CD33 on monocytes. Here, we find alternative splicing of exon 2 to be the primary mechanism of the genetically driven differential expression of CD33 protein. We report that the risk allele, rs3865444C, is associated with greater cell surface expression of CD33 in both subjects of European and African–American ancestry and that there is a single haplotype influencing CD33 surface expression. A meta-analysis of the two populations narrowed the number of significant SNPs in high linkage disequilibrium (LD) (r2 > 0.8) with rs3865444 to just five putative causal variants associated with increased protein expression. Using gene expression data from flow-sorted CD14+CD16− monocytes from 398 healthy subjects of three populations, we show that the rs3865444C risk allele is strongly associated with greater expression of CD33 exon 2 (pMETA = 2.36 × 10−60). Western blotting confirms increased protein expression of the full-length CD33 isoform containing exon 2 relative to the rs3865444C allele (P < 0.0001). Of the variants in strong LD with rs3865444, rs12459419, which is located in a putative SRSF2 splice site of exon 2, is the most likely candidate to mediate the altered alternative splicing of CD33's Immunoglobulin V-set domain 2 and ultimately influence AD susceptibility.
INTRODUCTION
The CD33 locus has been implicated in Alzheimer's disease (AD) susceptibility (1–9). The best marker for this association is rs3865444, and the ‘C’ allele (rs3865444C) has been reported to be associated with a modest increase in risk of AD (Odds ratio 1.10, P = 2.0 × 10−9) (7). rs3865444C captures the effect of the causal variant(s) and has been used effectively as a surrogate marker in functional studies: we and others have reported higher levels of CD33 protein expression relative to rs3865444C (10–12). More specifically, rs3865444C is associated with a 7-fold increase in the level of CD33 expression on the cell surface of monocytes as well as altered monocyte function that is implicated in the accumulation of amyloid neuropathology in aging individuals (10).
CD33, also known as Siglec-3, is a 67 kDa transmembrane glycoprotein expressed on the surface of myeloid progenitor cells, mature monocytes and macrophages. A lectin, full-length CD33 contains an extracellular immunoglobulin (Ig) V-set sialic-acid binding domain, an extracellular Ig C2-set domain and cytosolic immunoreceptor tyrosine-based inhibitory motifs. Alternative splicing of CD33 generates two isoforms of the protein: full-length CD33M and truncated CD33m which lacks the Ig V-set domain encoded by exon 2 (13). Recently, a quantitative PCR study of brain tissue demonstrated that the ratio of the mRNA isoform lacking exon 2 to total CD33 gene expression was differentially expressed relative to rs3865444, suggesting genotype-induced differential splicing of the CD33 gene (14). Further, these authors propose rs12459419, a single-nucleotide polymorphism (SNP) located in exon 2 that is in linkage disequilibrium (LD) with rs3865444 (r2 = 1 and D′ = 1 in 1000 Genomes European population [CEU]), as the causal variant based on a minigene assay. However, the functional consequence of CD33 splicing has not yet been fully characterized. CD33 has been implicated in modulating multiple cellular functions, including inhibition of cellular proliferation and activation (15). In terms of amyloid biology, the larger isoform, CD33M, modulates A-beta uptake, while the truncated protein isoform, CD33m, has no effect (11).
Here, we expand our understanding of the effect of the CD33 locus by performing an across-population fine-mapping exercise to (1) prioritize candidate causal variants that are in LD with the rs3865444 index SNP and (2) assess the possibility that there are additional variants with independent effects on CD33 protein and mRNA expression. Further, we refine the association of the locus with CD33 protein expression by demonstrating that the risk haplotype's increased inclusion of exon 2 into CD33 mRNA likely mediates the association with increased risk of AD. Finally, we use western blotting and densitometry to confirm that the previously reported difference in CD33 cell surface expression related to the risk allele (10) is primarily due to increased expression of the full-length CD33M protein isoform in monocytes.
RESULTS
CD33 surface expression in subjects of European and African–American ancestry
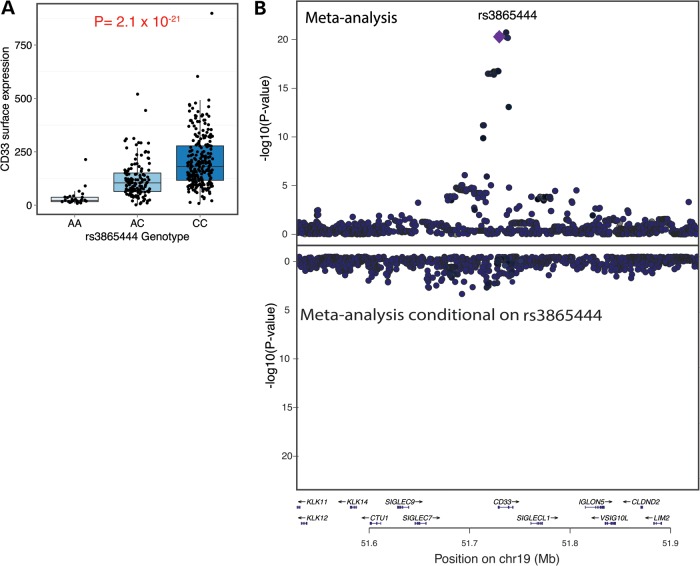
We collected monocyte CD33 cell surface expression data in subjects of European and African–American (AA) ancestry in order to leverage differences in linkage disequilibrium (LD) between European and AA haplotypes and more precisely define the association between the CD33 locus and CD33 surface expression. We measured the level of CD33 surface expression in monocytes of 151 older subjects of European ancestry from two cohorts of aging, the Religious Order Study (ROS) and the Memory and Aging Project (MAP), who have genome-wide genotype data. Additionally, CD33 monocyte surface expression data were generated in 164 subjects of AA ancestry and 75 European American (EA) subjects from the Chicago Health and Aging Project (CHAP). We find that subjects of AA ancestry have a higher mean level of CD33 expression compared with subjects of European ancestry (P = 1.0 × 10−6) and that this difference in CD33 expression is more prominent in men (n = 224, β = 3.0, P = 1.1 × 10−6) than in women (n = 345, β = 1.1, P = 0.04). However, this effect is not related to an AA individual's genome-wide proportion of African ancestry (Supplementary Material, Fig. S1). Instead, adding the rs3865444 variant as a covariate in this analysis abrogates the association of CD33 expression with ancestry (P = 0.57). Therefore, the difference related to ancestry is completely explained by the difference in the frequency of the minor, protective allele, rs3865444A: 0.18 in subjects of European ancestry and 0.01 in subjects of AA ancestry. In both populations, rs3865444C is correlated with greater CD33 surface expression with a similar effect size (EA ROS–MAP ρ = 0.56; AA CHAP ρ = 0.36; EA CHAP ρ = 0.54). In a meta-analysis of the three sets of samples, the association between rs3865444C and CD33 surface expression is highly significant (pMETA = 2.1 × 10−21, ρ = 0.51) (Fig. 1A), consistent with our previous observation in the ROS–MAP subjects (10).
Figure 1.
A single haplotype in the CD33 locus is associated with greater CD33 expression on the cell surface of monocytes. (A) rs3865444C is significantly associated with greater CD33 surface expression in a meta-analysis of monocyte-derived data from ROSMAP/CHAP EA and AA subjects (n = 390, P = 2.1 × 10−21). (B) Upper panel: Within the CD33 locus, meta-analysis of the ROSMAP/CHAP EA and AA CD33 monocyte surface expression data distills the significant SNPs to a group of five variants in high LD, one of which is likely to be the causal variant. rs3865444 is one of those five variants. Lower panel: Using conditional analysis to adjust for the effect of rs3865444, we find no significant residual associations with CD33 surface expression in the CD33 locus. The y-axis scale is inverted relative to the upper panel. This image was produced using LocusZoom (27).
Given the availability of genome-wide genotype data in the EA and AA subjects, we applied a fine-mapping approach to maximize our power and leverage the differences in LD to resolve the role of markers in LD with rs3865444C in the CD33 region. Meta-analysis of the two populations narrowed the number of significant SNPs in high LD (r2 > 0.8 in both AA and EA) with rs3865444C to just five putative causal variants (Fig. 1B, top panel). Of these variants, rs12459419 was recently proposed to alter splicing of CD33 in minigene transfected BV2 microglial-like cells (14).
To identify additional, independent effects within the LD block containing the index SNP, we performed a conditional analysis to adjust for the effect of rs3865444 on CD33 surface expression. We observed no significant signal of association after regressing out the effect of rs3865444, suggesting that a single haplotype mediates the observed effect on CD33 surface expression in both EA and AA subjects within the LD block containing CD33 (Fig. 1B, lower panel; Supplementary Material, Table S1).
In order to assess the likelihood that the association between the risk allele and AD susceptibility is driven by the regulatory effect on CD33 expression, we performed a regulatory trait concordance (RTC) analysis (16). Comparing the distribution of association results for CD33 surface expression in EA subjects to AD susceptibility in subjects of the same ancestry (7), RTC suggests that the two associations are unlikely to be coincidental and thus that the same variant influences both traits (RTC = 0.88) (Fig. 1A). The alteration of CD33 protein expression may therefore be the primary functional consequence of the CD33 locus that influences susceptibility to AD.
CD33 mRNA expression and alternative splicing
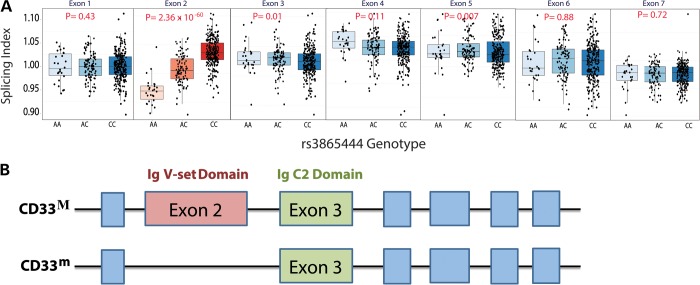
To explore the mechanism of the difference in CD33 surface expression further, we turned to an exon-level analysis of CD33 mRNA expression. Interestingly, two different protein isoforms of CD33 exist (13), full-length CD33M and truncated CD33m which lacks the Ig V-set domain encoded by exon 2 (Fig. 2B). To examine the effect of the CD33 locus on CD33 mRNA expression, we leveraged flow-sorted CD14+CD16− monocyte expression data collected using the Affymetrix GeneChip Human Gene 1.0 ST Array and genotype data imputed to Minor Allele Frequency (MAF) > 0.01 from healthy subjects of EA, AA and East Asian–American (EAA) ancestry as part of the Immunological Variation (ImmVar) project (Raj et al., unpublished data). Using these exon-level mRNA expression data from EA subjects, we find that the rs3865444C risk allele is strongly associated with greater expression of CD33 exon 2 (P = 4.9 × 10−11, ρ = 0.43) and is not significantly associated with the expression of other CD33 exons (Supplementary Material, Table S2). To discriminate exon-specific and gene-level effects, we performed a secondary analysis using each exon's splicing index (SI), a normalized exon expression intensity calculated by dividing each sample's exon expression level by the sample's overall gene expression level (17). Testing the association between genotypes surrounding CD33 and the exon splicing indices, we again find that the rs3865444C risk allele is strongly associated with greater expression of exon 2 (P = 1.6 × 10−23, ρ = 0.62) and is not associated with the expression of other CD33 exons (Fig. 3A, top panel; Supplementary Material, Table S3 and Fig. S2). We confirmed this effect in exon expression data from subjects of AA (P = 2.9 × 10−6, ρ = 0.43) and EAA ancestry (P = 1.5 × 10−6, ρ = 0.51) (Fig. 3A, bottom panels; Supplementary Materials, Tables S2, S3 and Fig. S3, S4). Meta-analysis of the splicing indices of the three populations reveals that rs3865444 has the strongest evidence for association with exon 2 (P = 2.36 × 10−60) (Figs. 2A and 3B, top panel). Consistent with the cell surface analysis, we obtain a high RTC score for rs3865444 (RTC = 0.94) suggesting that altered CD33 splicing may be the primary functional effect of the locus influencing susceptibility to AD.
Figure 2.
The CD33 locus is associated with greater mRNA expression of CD33 exon 2, which encodes an Ig V-set domain. (A) Using a SI to deconvolve gene-level and exon-specific expression, rs3865444C is significantly associated with a greater mRNA expression of CD33 exon 2 (n = 398, P = 2.36 × 10−60) quantified using a microarray platform and RNA from ex vivo, cytometrically sorted monocytes from ImmVar EA, AA and EAA subjects. The CD33 locus is not significantly associated with expression of other CD33 exons in monocytes. (B) CD33 is a type 1 transmembrane glycoprotein expressed on the surface of myeloid cells. Full-length CD33M consists of a signal peptide, an extracellular Ig V-set sialic-acid binding domain, an extracellular Ig C2-set domain, a transmembrane region, and a cytosolic domain containing immunoreceptor tyrosine-based inhibitory motifs. The alternatively spliced CD33m isoform lacks the Ig V-set domain encoded by exon 2 (13). The figure is not drawn to scale.
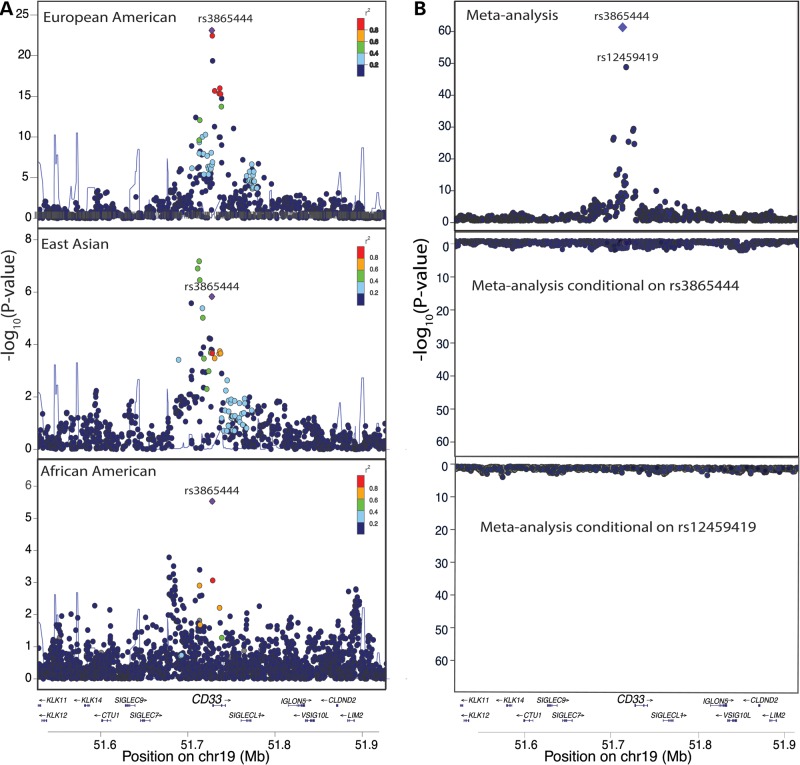
Figure 3.
rs3865444C is the best candidate causal variant for association with the mRNA SI of CD33 exon 2 in monocytes. (A) In the ImmVar EA and AA populations, rs3865444 is the most significant SNP for association with the mRNA SI of CD33 exon 2 (pEA = 1.6 × 10−23; pAA = 2.9 × 10−6). In the ImmVar EAA population, multiple SNPs in high LD with rs3865444 are associated with the SI of exon 2. (B) In a meta-analysis (upper panel) of the EA, AA and EAA populations, rs3865444 is the most significant SNP for association with the SI of CD33 exon 2 (P = 2.36 × 10−60) (top panel), and, after conditioning on rs3865444 and rs12459419, there are no significant genetic associations with the SI of exon 2 (lower panel). These images were produced using LocusZoom (27).
While rs3865444 is the most associated SNP with exon 2, it is located 373 bp upstream of CD33 in the promoter region. It is therefore unlikely to directly affect splicing of exon 2. On the other hand, Malik et al. sequenced the CD33 locus in four rs3865444CC and three rs3865444AA subjects to identify SNPs in LD with rs3865444 which are better located to influence exon 2 splicing. Of the three identified SNPs (rs2459141, rs12459419 and rs2455069), rs12459419, located in a putative SRSF2 splice site of exon 2, was suggested as the most likely candidate to alter CD33 splicing, and this putative mechanism was confirmed using a minigene experiment (14). Unfortunately, these three SNPs are not directly genotyped in our subjects, and the imputed data that we have makes it difficult to distinguish their effects on CD33 splicing. If uncertainty from imputation is not incorporated into our analysis (i.e. allelic dosages are assigned to discrete 0, 1 and 2 genotypes) then the statistical evidence for association with exon 2 splicing and CD33 surface expression is identical for rs12459419 and rs3865444. However, when allelic dosages are used, rs12459419 is the second most associated SNP (P = 7.2 × 10−23, ρ = 0.61) in the EA subjects but is not as significant in the AA (P = 1.2 × 10−3, ρ = 0.30; 18th most associated SNP with exon 2) and EAA (P = 2.1 × 10−4, ρ = 0.41; 17th most associated SNP with exon 2) subjects. To assess the roles of rs12459419 and rs3865444 further, we performed conditional analyses accounting for the effect of each variant on the exon 2 splicing phenotype. We observed no significant effect after regressing out either rs3865444 or rs12459419, suggesting that these two variants have statistically equivalent evidence of association with the exon 2 splicing trait (Fig. 3B, bottom panel). Regardless of the exact identity of the causal variant, the exon-specific association suggests that the difference in global CD33 surface expression is primarily due to the greater expression of the protein isoform that contains exon 2, the full-length CD33M isoform.
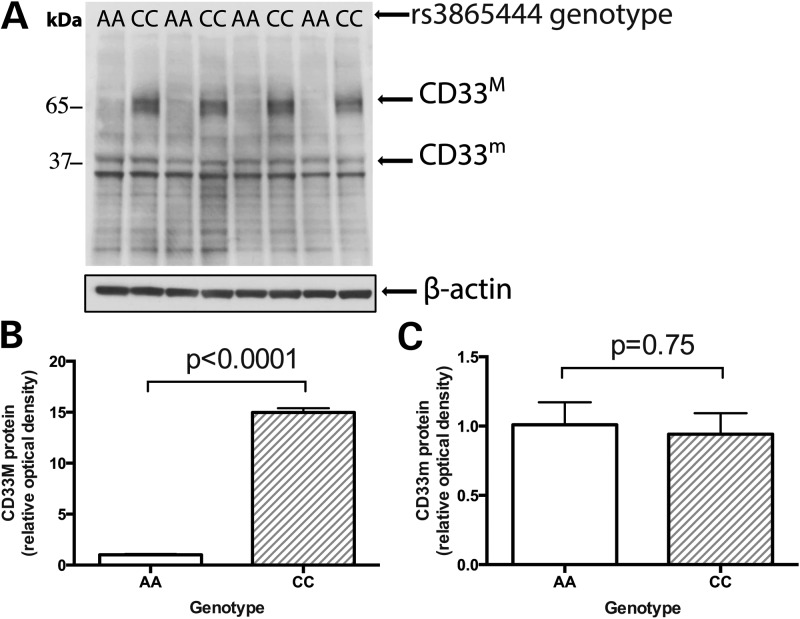
To confirm our hypothesis that the risk haplotype alters CD33 splicing to increase the abundance of the full-length isoform, we used western blotting (Fig. 4A) and densitometry to quantitate the two protein isoforms of CD33 in extracts of purified, ex vivo monocytes. As shown in Figure 4, the CD33M isoform that contains exon 2 is expressed at very low levels in subjects that are homozygous for the protective rs3865444A allele and the rs12459419T allele when they are compared with subjects homozygous for the risk-associated rs3865444C allele and the rs12459419C allele (P < 0.0001) (Fig. 4B). The expression of the truncated CD33m isoform is unaffected by the subjects' genotype (Fig. 4C). Thus, the previously reported difference in monocyte cell surface CD33 expression related to the risk haplotype (10) is due primarily to increased expression of the full-length CD33M isoform which contains exon 2.
Figure 4.
Western blot analysis confirms the association between rs3865444C and increased abundance of the full-length CD33M isoform in monocytes. (A) In purified, ex vivo monocytes isolated from healthy subjects in the PhenoGenetic cohort, western blot analysis indicates that full-length CD33M isoform is expressed at low levels in individuals with the protective rs3865444AA genotype compared with individuals with the risk rs3865444CC genotype. (B) As quantified by densitometric analysis, this association is statistically significant (P < 0.0001). (C) The truncated CD33m isoform has no significant association with rs3865444 genotype (P = 0.75).
DISCUSSION
We completed an interrelated set of analyses to characterize the CD33 locus in detail. Fine-mapping of the CD33 cell surface and exon-level mRNA expression traits in monocytes from subjects of multiple ancestries clearly reveals the presence of only one haplotype influencing CD33 expression in this locus. Aggregating all of the assembled evidence, we find that rs3865444, the index SNP that emerged from AD susceptibility GWAS, is the best marker for these associations with CD33 surface expression and exon 2 splicing. However, rs12459419, a SNP in perfect LD (r2 = 1) with rs3865444, was recently proposed as the causal SNP for CD33 splicing of exon 2 using a minigene experiment (14). rs12459419 is located in exon 2 in a predicted binding site for the splicing factor SRSF2. Since we observed no significant signal influencing CD33 surface expression or CD33 exon 2 mRNA expression after regressing out the effect of either rs3865444 or rs12459419, these SNPs have equivalent statistical evidence of association to the splicing and surface expression traits. While the two SNPs are statistically equivalent, the location of rs12459419, in a SRSF2 binding site of exon 2, and the in vitro splicing experiment (14) support rs12459419 as the best candidate causal variant in the CD33 locus.
Our exon-level mRNA and western blot analyses clarify the mechanism by which the CD33 locus influences CD33 expression. We confirm that rs3865444 and rs12459419 are associated with altered splicing of CD33 exon 2, and our protein data are definitive in showing that the risk-associated allele leads to increased protein expression of the full-length CD33M isoform in monocytes and not decreased expression of the CD33m isoform that lacks exon 2, as suggested by Malik et al. (14) who base their interpretation on brain mRNA data normalized to reference genes RPL32 and EIF4H. As CD33M inhibits uptake of amyloid-beta while CD33m has no effect on this function, increased levels of CD33M in subjects at risk for AD suggests that their macrophages and/or microglia are impaired in the removal of amyloid. Our RTC analysis that compares the distribution of this association over the CD33 locus to the reported distribution of the AD susceptibility trait suggests that the effect of the CD33 locus on CD33 splicing may be the susceptibility variant's primary functional consequence in influencing susceptibility to AD.
Our exon-level analysis of CD33 expression highlights an emerging role for an infiltrating macrophage and/or microglial activation network (including the genes TYROBP, CD33 and TREM2) in modulating AD susceptibility and pathology. Both CD33 and TREM2, another transmembrane protein expressed on myeloid and microglial cells and implicated in AD susceptibility (18,19), contain extracellular Ig V-set domains, encoded by exon 2 of CD33 (13) and by exons 2 and 3 of TREM2 (20) (Supplementary Material, Fig. S5). TREM2 binds TYROBP, an adapter protein that was recently proposed to be a key regulator of AD susceptibility networks and is upregulated in late-onset Alzheimer's disease (LOAD). Both TREM2 and CD33 were found in the network regulated by TYROBP (also called DAP12) (21,22). Our study suggests that rs3865444 helps regulate the abundance of the CD33M isoform that contains the Ig V-set domain. Similarly, rs75932628T, a rare missense mutation in the Ig V-set domain of TREM2, confers a significant risk of AD (18). This shared domain alteration may be coincidental but points to one specific functional domain of two different proteins as being potentially important for AD susceptibility. These observations help to formulate new hypotheses with which to investigate the role and the possible interaction of these two proteins in regulating innate immune processes in AD susceptibility, with CD33 generally acting as an inhibitor and TREM2 serving multiple immunomodulatory roles. Overall, our results refine the role of the CD33 locus in altering the innate immune system in ways that ultimately contribute to AD susceptibility and provide a specific structural target for the development of novel therapeutic avenues.
MATERIALS AND METHODS
Study subjects
Informed consent was obtained from all human subjects. All blood draws and data analyses were done in compliance with protocols approved by the Institutional Review Boards of each Institution.
The Brigham and Women's Hospital PhenoGenetic Project
Peripheral venous blood was obtained from healthy control volunteers. The PhenoGenetic Project is a living tissue bank that consists of healthy subjects who are re-contactable and can therefore be recalled based on their genotype. 1741 healthy subjects >18-years-old have been recruited from the general population of Boston. They are free of chronic inflammatory, infectious and metabolic diseases. Their median age is 24, and 62.7% of subjects are women.
ROS, MAP, and CHAP
Study participants were free of known dementia at enrollment and agreed to annual clinical evaluations. ROS, started in 1994, enrolls Catholic priests, nuns and brothers, aged 53 or older from about 40 groups in 12 states. Since January 1994, >1150 participants completed their baseline evaluation, of whom 87% are non-Hispanic white, and the follow-up rate of survivors and autopsy rate among the deceased both exceed 90%. MAP, started in 1997, enrolls men and women aged 55 or older and without known dementia at baseline from retirement communities in Chicago. Since October 1997, >1,650 participants completed their baseline evaluation, of which 87% were non-Hispanic white. The follow-up rate of survivors exceeds 90% and the autopsy rate exceeds 80%. CHAP, begun in 1993, is a biracial population study enrolling AA and EA residents of a geographical defined area of the city of Chicago. More detailed descriptions of ROS, MAP and CHAP can be found in prior publications (23,24). The median age of subjects used in the CD33 expression experiments at sampling was 79.9, (range = 65.8–94.8).
Protocols for each study have been approved by the Institutional Review Board of RUSH University.
Flow cytometry/cell surface expression
Aliquots of frozen Peripheral blood mononuclear cells (PBMCs) from the ROS, MAP and CHAP cohorts were thawed and washed in 10 ml PBS. PBMCs were stained with anti-human CD33 (clone AC104.3E3; Miltenyi, Auburn, CA) or mouse IgG1 isotype (Miltenyi) in PBS plus 1% fetal calf serum (FCS). The monocyte gate was defined based on their distinct forward and side-scatter profile. The MFI was acquired on a FACSCalibur (BD Immunocytometry Systems, San Jose, CA) and analyzed with FlowJo software (Tree Star, Ashland, OR). An additive model was used in the analysis, adjusting for age and sex.
Exon expression association analysis
Gene expression levels were quantified on Affymetrix GeneChip Human Gene 1.0 ST Arrays using mRNA derived from CD14+CD16− monocytes obtained from 211 individuals of EA ancestry, 109 individuals of AA ancestry and 78 individuals of EAA ancestry as part of the Immological Variation (ImmVar) project (Raj et al. unpublished data). The Affymetrix arrays have 764,885 distinct 25-mer oligonucleotide probes with annotation at the exon and transcript level including 7 exon probesets for CD33. The raw expression intensity values were normalized using RMA normalization.
Genotyping of the ImmVar samples was performed on the Illumina Human OmniExpress + Exome Chip, a whole-genome genotyping DNA microarray with allele-specific oligonucleotides for 951 117 markers. The genotype success rate was ≥97%. We applied rigorous quality control (QC) that includes (1) gender misidentification (2) subject relatedness (3) Hardy–Weinberg Equilibrium testing (4) use concordance to infer SNP quality (5) genotype call rate (6) heterozygosity outlier and (7) subject mismatches.
We used the BEAGLE software (version: 3.3.2 (25)) to imputed the post-QC genotyped markers using reference Haplotype panels from the 1000 Genomes Project (The 1000 Genomes Project Consortium Phase I Integrated Release Version 3), which contain a total of 37.9 Million SNPs in 1092 individuals with ancestry from West Africa, East Asia and Europe. For subjects of European and East Asian ancestry, we used haplotypes from Utah residents (CEPH) with Northern and Western European ancestry (CEU), and combined panels from Han Chinese in Beijing (CHB) and Japanese in Tokyo (JPT), respectively. For imputing genotypes from AA subjects, we used a combined haplotype reference panels consisting of CEU and Yoruba in Ibadan, Nigeria (YRI). Only SNPs with MAF > 0.01 and imputation quality r2 > 0.4 were kept for subsequent analysis.
For our association analysis, we extracted all SNPs within 200 kb of the rs3865444 index SNP. Associations between SNP genotypes and exon expression values were conducted by Spearman rank correlation (SRC). In a secondary analysis controlling for gene-level expression effects, we calculated a SI for each exon, normalizing exon expression intensity by dividing each sample's exon expression levels by the sample's overall gene expression level. For example, the SI for exon i in individual j is SIij = Eij/Gj where Eij is the expression level for exon i in individual j and Gj is the overall CD33 gene expression level in individual j (17). Again, association tests between SNP genotypes and exon SIs were conducted using SRC.
Meta-analysis
We used the METASOFT software (26) to perform multi-ethnic meta-analysis using a random effects (RE) model. The effect size (estimated using Spearman's rho for eQTL analysis) and standard error of the effect size were used as an input to METASOFT.
Relative trait concordance
We used the RTC method to integrate QTL and AD GWAS data to detect disease-causing cis-regulatory effects as previously described in Nica et al. 2010 (16).
Western blotting and densitometry
PBMCs from the PhenoGenetic cohort were separated by Ficoll-Paque PLUS (GE Healthcare) gradient centrifugation. PBMCs were frozen in 10% DMSO (Sigma-Aldrich)/90% fetal calf serum (vol/vol, Atlanta Biologicals). Monocytes were isolated from frozen PBMCs using CD14 positive microbeads (Miltenyi Biotech). Cells were lysed in IP buffer (Thermo Scientific) with a protease inhibitor mixture (Roche Diagnostics) and a phosphatase inhibitor mixture (Sigma-Aldrich). After 20 min on ice, cells were centrifuged at 12 000 rpm for 10 min and diluted in electrophoresis sample buffer. Samples were heated at 80°C for 5 min and 20 µg total protein was loaded into each well of an SDS-PAGE gel for separation by electrophoresis. Proteins were transferred on to a PVDF membrane and probed with anti-CD33 rabbit polyclonal IgG antibody (H-110) and goat anti-rabbit HRP-conjugated antibody (Santa Cruz Biotechnologies). Membranes were developed with Immobilon Western Chemiluminescent HRP substrate (Millipore). Bands were quantified by densitometric analysis using ImageJ software (Wayne Rasband, NIH, USA).
SUPPLEMENTARY MATERIAL
FUNDING
This work is supported by the National Institutes of Health (F32 AG043267, R01 AG031553, R01 AG30146, R01 AG17917, R01 AG15819, P30 AG10161, R01 AG11101, R01 NS067305, RC2 GM093080 and R01 AG043617).
Supplementary Material
ACKNOWLEDGEMENTS
The authors are grateful to the participants in the ROS, MAP and CHAP. We also thank the participants of the Brigham and Women's PhenoGenetic Project.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Logue M.W., Schu M., Vardarajan B.N., Buros J., Green R.C., Go R.C., Griffith P., Obisesan T.O., Shatz R., Borenstein A., et al. A comprehensive genetic association study of Alzheimer disease in African Americans. Arch. Neurol. 2011;68:1569–1579. doi: 10.1001/archneurol.2011.646. doi:10.1001/archneurol.2011.646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Deng Y.L., Liu L.H., Wang Y., Tang H.D., Ren R.J., Xu W., Ma J.F., Wang L.L., Zhuang J.P., Wang G., et al. The prevalence of CD33 and MS4A6A variant in Chinese Han population with Alzheimer’s disease. Hum. Genet. 2012;131:1245–1249. doi: 10.1007/s00439-012-1154-6. doi:10.1007/s00439-012-1154-6. [DOI] [PubMed] [Google Scholar]
- 3.Kamboh M.I., Demirci F.Y., Wang X., Minster R.L., Carrasquillo M.M., Pankratz V.S., Younkin S.G., Saykin A.J., Jun G., Baldwin C., et al. Genome-wide association study of Alzheimer’s disease. Transl. Psychiatry. 2012;2:e117. doi: 10.1038/tp.2012.45. doi:10.1038/tp.2012.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chung S.J., Lee J.H., Kim S.Y., You S., Kim M.J., Lee J.Y., Koh J. Association of GWAS top hits with late-onset Alzheimer disease in Korean population. Alzheimer Dis. Assoc. Disord. 2012;27:250–257. doi: 10.1097/WAD.0b013e31826d7281. [DOI] [PubMed] [Google Scholar]
- 5.Reitz C., Jun G., Naj A., Rajbhandary R., Vardarajan B.N., Wang L.S., Valladares O., Lin C.F., Larson E.B., Graff-Radford N.R., et al. Variants in the ATP-binding cassette transporter (ABCA7), apolipoprotein E 4, and the risk of late-onset Alzheimer disease in African Americans. JAMA. 2013;309:1483–1492. doi: 10.1001/jama.2013.2973. doi:10.1001/jama.2013.2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hollingworth P., Harold D., Sims R., Gerrish A., Lambert J.C., Carrasquillo M.M., Abraham R., Hamshere M.L., Pahwa J.S., Moskvina V., et al. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer’s disease. Nat. Genet. 2011;43:429–435. doi: 10.1038/ng.803. doi:10.1038/ng.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Naj A.C., Jun G., Beecham G.W., Wang L.S., Vardarajan B.N., Buros J., Gallins P.J., Buxbaum J.D., Jarvik G.P., Crane P.K., et al. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer's disease. Nat. Genet. 2011;43:436–441. doi: 10.1038/ng.801. doi:10.1038/ng.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carrasquillo M.M., Belbin O., Hunter T.A., Ma L., Bisceglio G.D., Zou F., Crook J.E., Pankratz V.S., Sando S.B., Aasly J.O., et al. Replication of EPHA1 and CD33 associations with late-onset Alzheimer’s disease: a multi-centre case–control study. Mol. Neurodegener. 2011;6:54. doi: 10.1186/1750-1326-6-54. doi:10.1186/1750-1326-6-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bertram L., Lange C., Mullin K., Parkinson M., Hsiao M., Hogan M.F., Schjeide B.M., Hooli B., Divito J., Ionita I., et al. Genome-wide association analysis reveals putative Alzheimer’s disease susceptibility loci in addition to APOE. Am. J. Hum. Genet. 2008;83:623–632. doi: 10.1016/j.ajhg.2008.10.008. doi:10.1016/j.ajhg.2008.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bradshaw E.M., Chibnik L.B., Keenan B.T., Ottoboni L., Raj T., Tang A., Rosenkrantz L.L., Imboywa S., Lee M., Von Korff A., et al. CD33 Alzheimer’s disease locus: altered monocyte function and amyloid biology. Nat. Neurosci. 2013;16:848–850. doi: 10.1038/nn.3435. doi:10.1038/nn.3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Griciuc A., Serrano-Pozo A., Parrado A.R., Lesinski A.N., Asselin C.N., Mullin K., Hooli B., Choi S.H., Hyman B.T., Tanzi R.E. Alzheimer’s disease risk gene CD33 inhibits microglial uptake of amyloid beta. Neuron. 2013;78:631–643. doi: 10.1016/j.neuron.2013.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lourdusamy A., Newhouse S., Lunnon K., Proitsi P., Powell J., Hodges A., Nelson S.K., Stewart A., Williams S., Kloszewska I., et al. Identification of cis-regulatory variation influencing protein abundance levels in human plasma. Hum. Mol. Genet. 2012;21:3719–3726. doi: 10.1093/hmg/dds186. doi:10.1093/hmg/dds186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hernandez-Caselles T., Martinez-Esparza M., Perez-Oliva A.B., Quintanilla-Cecconi A.M., Garcia-Alonso A., Alvarez-Lopez D.M., Garcia-Penarrubia P. A study of CD33 (SIGLEC-3) antigen expression and function on activated human T and NK cells: two isoforms of CD33 are generated by alternative splicing. J. Leukoc. Biol. 2006;79:46–58. doi: 10.1189/jlb.0205096. doi:10.1189/jlb.0205096. [DOI] [PubMed] [Google Scholar]
- 14.Malik M., Simpson J.F., Parikh I., Wilfred B.R., Fardo D.W., Nelson P.T., Estus S. CD33 Alzheimer’s risk-altering polymorphism, CD33 expression, and exon 2 splicing. J. Neurosci. 2013;33:13320–13325. doi: 10.1523/JNEUROSCI.1224-13.2013. doi:10.1523/JNEUROSCI.1224-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crocker P.R., Paulson J.C., Varki A. Siglecs and their roles in the immune system. Nat. Rev. Immunol. 2007;7:255–266. doi: 10.1038/nri2056. doi:10.1038/nri2056. [DOI] [PubMed] [Google Scholar]
- 16.Nica A.C., Montgomery S.B., Dimas A.S., Stranger B.E., Beazley C., Barroso I., Dermitzakis E.T. Candidate causal regulatory effects by integration of expression QTLs with complex trait genetic associations. PLoS Genet. 2010;6:e1000895. doi: 10.1371/journal.pgen.1000895. doi:10.1371/journal.pgen.1000895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clark T.A., Sugnet C.W., Ares M., Jr Genomewide analysis of mRNA processing in yeast using splicing-specific microarrays. Science. 2002;296:907–910. doi: 10.1126/science.1069415. doi:10.1126/science.1069415. [DOI] [PubMed] [Google Scholar]
- 18.Guerreiro R., Wojtas A., Bras J., Carrasquillo M., Rogaeva E., Majounie E., Cruchaga C., Sassi C., Kauwe J.S., Younkin S., et al. TREM2 variants in Alzheimer’s disease. N. Engl. J. Med. 2013;368:117–127. doi: 10.1056/NEJMoa1211851. doi:10.1056/NEJMoa1211851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jonsson T., Stefansson H., Steinberg S., Jonsdottir I., Jonsson P.V., Snaedal J., Bjornsson S., Huttenlocher J., Levey A.I., Lah J.J., et al. Variant of TREM2 associated with the risk of Alzheimer’s disease. N. Engl. J. Med. 2013;368:107–116. doi: 10.1056/NEJMoa1211103. doi:10.1056/NEJMoa1211103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bouchon A., Dietrich J., Colonna M. Cutting edge: inflammatory responses can be triggered by TREM-1, a novel receptor expressed on neutrophils and monocytes. J. Immunol. 2000;164:4991–4995. doi: 10.4049/jimmunol.164.10.4991. [DOI] [PubMed] [Google Scholar]
- 21.Zhang B., Gaiteri C., Bodea L.G., Wang Z., McElwee J., Podtelezhnikov A.A., Zhang C., Xie T., Tran L., Dobrin R., et al. Integrated systems approach identifies genetic nodes and networks in late-onset Alzheimer’s disease. Cell. 2013;153:707–720. doi: 10.1016/j.cell.2013.03.030. doi:10.1016/j.cell.2013.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gandy S., Heppner F.L. Microglia as dynamic and essential components of the amyloid hypothesis. Neuron. 2013;78:575–577. doi: 10.1016/j.neuron.2013.05.007. doi:10.1016/j.neuron.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bennett D.A., Schneider J.A., Arvanitakis Z., Wilson R.S. Overview and findings from the religious orders study. Curr. Alzheimer Res. 2012;9:628–645. doi: 10.2174/156720512801322573. doi:10.2174/156720512801322573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bennett D.A., Schneider J.A., Buchman A.S., Barnes L.L., Boyle P.A., Wilson R.S. Overview and findings from the rush memory and aging project. Curr. Alzheimer Res. 2012;9:646–663. doi: 10.2174/156720512801322663. doi:10.2174/156720512801322663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Browning S.R., Browning B.L. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am. J. Hum. Genet. 2007;81:1084–1097. doi: 10.1086/521987. doi:10.1086/521987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Han B., Eskin E. Random-effects model aimed at discovering associations in meta-analysis of genome-wide association studies. Am. J. Hum. Genet. 2011;88:586–598. doi: 10.1016/j.ajhg.2011.04.014. doi:10.1016/j.ajhg.2011.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pruim R.J., Welch R.P., Sanna S., Teslovich T.M., Chines P.S., Gliedt T.P., Boehnke M., Abecasis G.R., Willer C.J. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–2337. doi: 10.1093/bioinformatics/btq419. doi:10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.