Abstract
Bladder cancer is a commom malignancy in the urinary tract that is influenced by genetic and environmental factors. The role of functional polymorphisms in the methylenetetrahydrofolate reductase (MTHFR) gene with bladder cancer risk remains to be determined. This meta-analysis was performed to derive a more precise estimation of MTHFR Ala222Val and Glu429Ala polymorphisms and bladder cancer risk. Data were collected with the last report up to September 2013. A total of 3,463 cases and 3,927 controls for Ala222Val, and 3,177 cases and 3,502 controls for Glu429Ala were analyzed. The pooled odds ratios (ORs) and 95% confidence interval (CI) were estimated for the association with bladder cancer risk. Overall, no significant associations of Ala222Val and Glu429Ala polymorphisms with bladder cancer risk were found (for Ala222Val: Val/Val vs. Ala/Ala: OR, 1.02; 95% CI: 0.80–1.29; Val/Ala vs. Ala/Ala: OR, 1.02; 95% CI: 0.92–1.12; dominant model: OR, 1.01; 95% CI: 0.87–1.17; recessive model: OR, 1.00; 95% CI: 0.87–1.15; and for Glu429Ala: Ala/Ala vs. Glu/Glu: OR, 1.11; 95% CI: 0.78–1.58; Ala/Glu vs. Glu/Glu: OR, 1.16; 95% CI: 0.95–1.40; dominant model: OR, 1.15; 95% CI: 0.94–1.41; recessive model: OR, 0.96; 95% CI: 0.79–1.15). In stratified analyses by ethnicity, significant associations were observed for Glu429Ala polymorphism in individuals of Middle Eastern descent (Ala/Glu vs. Glu/Glu: OR, 2.11; 95% CI: 1.26–3.53; dominant model: OR, 2.16; 95% CI: 1.16–4.01; recessive model: OR, 1.82; 95% CI: 1.11–3.01). This meta-analysis demonstrated that overall there was no association of MTHFR Ala222Val and Glu429Ala polymorphisms with bladder cancer risk. However, in the stratified analysis by ethnicity the MTHFR Glu429Ala polymorphism was significantly associated with increased bladder cancer risk in individuals of Middle Eastern descent.
Keywords: methylenetetrahydrofolate reductase, bladder cancer, polymorphism, meta-analysis
Introduction
Bladder cancer is the most common malignant tumor of the urinary tract, with an estimated 386,300 new cases and 150,200 mortalities from bladder cancer annually (1), ranking seventh in men and seventeenth in women worldwide (2). The etiology of bladder cancer involves the interaction between genetic and environmental factors. Common risk factors include cigarette smoking, occupational exposure to aromatic amines and polycyclic aromatic hydrocarbons, inflammation of the urinary tract and consumption of certain drugs (3). Current evidence supports that molecular alterations and DNA polymorphisms potentially alter individual susceptibility to bladder cancer (4–5).
Of all the genetic susceptibility factors of bladder cancer, folate metabolic pathway has received increasing attention (5–7). Since it is the one-carbon groups donor in DNA methylation and synthesis, folate deficiency may induce DNA hypomethylation and potentially induce a proto-oncogene expression leading to cancer.
Methylenetetrahydrofolate reductase (MTHFR) is the key enzyme involved in folate metabolism, which acts as a critical metabolic node in the regulation of methylation reactions. It catalyzes the conversion of 5,10-methylenetetrahydrofolate to 5-methyltetrahydrofolate. The 5,10-methylenetetrahydrofolate is the methyl donor for de novo thymidine synthesis and the 5-methyltetrahydrofolate is used as a cosubstrate to convert homocysteine into methionine, which is the immediate precursor of S-adenosylmethionine and the primary methyl donor of DNA methylation. A less active form of MTHFR may result in hypomethylation, which is a candidate mechanism for the development of cancer (8).
The functional polymorphisms in the MTHFR gene, Ala222Val (C677T) and Glu429Ala (A1298C), have received increasing attention. Ala222Val homozygotes have been associated with reduced enzyme activity of ~30% of the control value (9), while the Glu429Ala homozygotes exhibit ~60% of the control activity. Heterozygotes for the Ala222Val and Glu429Ala mutations had ~50–60% of control activity (10). Since the two variants result in reduced activity of MTHFR, their associations with the susceptibility of a variety of cancers has been evaluated (11–14).
MTHFR Ala222Val and Glu429Ala polymorphisms have also been evaluated in relation to bladder cancer risk. However, the results of previous studies have yielded conflicting results. Given the amount of accumulated data, this meta-analysis was performed to derive a more precise estimation of the association of MTHFR Ala222Val and Glu429Ala and bladder cancer risk.
Materials and methods
Publication search
An exhaustive search of the literature was performed using the electronic databases: PubMed, EBSCO-Medline, Elsevier ScienceDirect and BIOSIS Previews for relevant articles published (up to September 2013). The articles were identified by using the search terms ‘methylenetetrahydrofolate reductase’, ‘MTHFR’, ‘NADPH2’, ‘urinary bladder’ and ‘bladder’. The search was limited to human studies with no language restrictions being applied. Additional studies were obtained through the references cited in retrieved articles on the association between the MTHFR Ala222Val and Glu429Ala polymorphisms and bladder cancer. The searching of the electronic databases and reviewing of the references in retrieved articles were independently achieved by two investigators.
Inclusion and exclusion criteria
Inclusion criteria for the studies were: case-control studies on the association between MTHFR Ala222Val and Glu429Ala polymorphisms and bladder cancer risk, and the data of each study was required to be sufficient for statistical analysis of odds ratio (OR) and 95% confidence interval (CI). Studies that had duplicated data and in which genotypes could not be ascertained were excluded.
Data abstraction
Information was extracted from all the eligible publications independently by two investigators as per the inclusion and exclusion criteria. For disagreements, consensus was reached by discussion of the two investigators. The data abstracted from each study were as follows: first author’s name, publication date, country, ethnicity, source of controls, number of cases and controls, and number of cases and controls for MTHFR Ala222Val and Glu429Ala polymorphisms. Individuals of different descents were classified as Chinese, European, American and Middle Eastern.
Statistical analysis
ORs with 95% CIs were applied to evaluate the strength of association between MTHFR Ala222Val and Glu429Ala polymorphisms and bladder cancer risk. The pooled ORs were estimated for the co-dominant model (Val/Val vs. Ala/Ala and Val/Ala vs. Ala/Ala for Ala222Val; Ala/Ala vs. Glu/Glu and Ala/Glu vs. Glu/Glu for Glu429Ala), dominant model (Val/Val + Val/Ala vs. Ala/Ala for Ala222Val; Ala/Ala + Ala/Glu vs. Glu/Glu for Glu429Ala) and recessive model (Val/Val vs. Val/Ala + Ala/Ala for Ala222Val; Ala/Ala vs. Ala/Glu + Glu/Glu for Glu429Ala), respectively. The Chi-square test-based Q-statistic (Q test) was applied to assess heterogeneity among the studies. The fixed-effects model was used to calculate the pooled ORs if no heterogeneity was detected (Ph≥0.05 by Q test) (15). Otherwise, the random-effects model was applied (Ph<0.05 by Q test) (16).
Subgroup analyses were performed by ethnicity and source of controls. The sensitivity analyses were conducted by excluding each study at a time to determine its effect on the overall estimation, since all the studies indicated that they conformed to the Hardy-Weinberg equilibrium (HWE). The publication bias was estimated by the funnel plot, in which the standard error of log (OR) was plotted against its log (OR) for each study. Egger’s linear regression test was applied to assess the asymmetry of the funnel plot, with P<0.05 indicating an asymmetric plot and a possible publication bias (17). In this meta-analysis, the Stata version 11.0 (StataCorp, College Station, TX, USA) was applied for all the statistical tests.
Results
Flow of included studies
A total of 420 articles potentially relevant to the searching terms were screened, including PubMed, 25; EBSCO-Medline, 24; Elsevier ScienceDirect, 338; and BIOSIS Previews, 33. Based on the inclusion criteria, a total of 13 studies (5–7,18–27) with full-text articles on polymorphisms of MTHFR Ala222Val and Glu429Ala and bladder cancer risk were identified as eligible. Of the 13 studies, two studies (22,24) were excluded due to duplicated data with the study by Rouissi et al (6).
Study characteristics
A total of 11 studies (5–7,18–21,23,25–27) were included in this meta-analysis for Ala222Val (including 3,463 cases and 3,.927 controls) and 9 studies (5–7,19–21,23,25,27) were included for Glu429Ala (including 3,177 cases and 3,502 controls) (Table I). Three of the 11 studies were population-based and eight were hospital-based studies. The studies indicated that the distribution of genotypes in controls was consistent with HWE. Detailed data from the included studies were abstracted (Table II).
Table I.
Characteristics of the case-control studies considered in the meta-analysis.
| Authors (refs.) | Year | Ethnicity | Genotyping method | HWE | Control source | Demographic characteristics | |
|---|---|---|---|---|---|---|---|
|
| |||||||
| Cases | Controls | ||||||
| Safarinejad et al (27) | 2011 | Iranian (Asian) | PCR-RFLP | 0.56 | HB | 158 bladder cancer patients, mean age: 62.7±10.6 years | 316 controls, mean age: 61.6±9.4 years |
| Chung et al (26) | 2010 | Chinese-Taiwan (Asian) | PCR-RFLP | 0.26 | HB | 150 bladder cancer patients, mean age: 65.3±1.1 years | 300 controls, mean age: 66.2±0.7 years |
| Cai et al (23) | 2009 | Chinese (Asian) | PCR-RFLP | 0.08 | HB | 312 bladder cancer patients, mean age: 63.1±11.3 years | 325 controls, mean age: 63.7±12.2 years |
| Rouissi et al (6) | 2009 | Tunisian (African) | PCR-RFLP | 0.49 | PB | 185 bladder cancer patients, mean age 67.5±9.7 years | 191 controls, mean age (match the case) |
| Wang et al (25) | 2009 | Chinese (Asian) | PCR-RFLP | 0.07 | HB | 239 bladder cancer patients, pack years <55 (n=42); 55–65 (77); >65 (n=120) | 250 controls, pack years <55 (n=45); 55–65 (n=81); >65 (n=124) |
| Moore et al (7) | 2007 | Spanish (European) | TaqMan and Golden Gate | 0.48 | HB | 1,150 bladder cancer patients, mean age: 66.0±10.0 years | 1,149 controls, mean age: 65.0±10.0 years |
| Karagas et al (21) | 2005 | USA (American) | PCR-RFLP | 0.70 | PB | 352 bladder cancer patients, pack years ≤40 (9); 41–55 (n=60); 56–70 (n=203); >70 (n=80) | 551 controls, pack years ≤40 (31); 41–55 (n=110); 56–70 (n=304); >70 (n=106) |
| Moore et al (19) | 2004 | Argentina (American) | PCR-RFLP | 0.29 | PB | 110 bladder cancer patients, mean age: 68.1 (range, 20–80 years) | 110 controls, mean age: 68.4 years (match the case) |
| Lin et al (5) | 2004 | USA (American) | PCR-RFLP | 0.07 | HB | 457 bladder cancer patients, mean age: 65.0 (range, 18–86 years) | 457 controls, mean age: 64.0 (range, 21–89 years) |
| Sanyal et al (20) | 2004 | Swedish (European) | PCR-RFLP | 0.82 | HB | 327 bladder cancer patients, mean age: 70.0 (range, 33–96 years) | 246 controls, mean age (match the case) |
| Kimura et al (18) | 2001 | Germany (European) | PCR-RFLP | 0.17 | HB | 165 bladder cancer patients, mean age: 67.4±11.5 years | 150 controls, mean age: 62.0±11.4 years |
HWE, Hardy-Weinberg equilibrium; PCR-RFLP, polymerase chain reaction-restriction fragment length polymorphism; HB, hospital-based; PB, population-based.
Table II.
Distribution of methylenetetrahydrofolate reductase (MTHFR) gene Ala222Val and Glu429Ala genotypes for bladder cancer patients and controls.
| Study (refs.) | Ethnicity | Ala222Val of MTHFR | Glu429Ala of MTHFR | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
||||||||||||||
| Sample size (case/control) | Case | Control | Sample size (case/control) | Case | Control | ||||||||||
|
|
|
||||||||||||||
| AA | AV | VV | AA | AV | VV | GG | GA | AA | GG | GA | AA | ||||
| Safarinejad et al (27) | Iranian | 158/316 | 67 | 74 | 17 | 144 | 142 | 30 | 158/316 | 48 | 85 | 25 | 178 | 115 | 23 |
| Chung et al (26) | Chinese | 150/300 | 80 | 57 | 13 | 141 | 123 | 36 | N/A | N/A | N/A | N/A | N/A | N/A | N/A |
| Cai et al (23) | Chinese | 312/325 | 82 | 169 | 61 | 113 | 170 | 42 | 312/325 | 215 | 91 | 6 | 226 | 92 | 7 |
| Rouissi et al (6) | Tunisian | 185/191 | 87 | 86 | 12 | 81 | 90 | 20 | 185/191 | 97 | 78 | 10 | 121 | 60 | 10 |
| Wang et al (25) | Chinese | 239/250 | 66 | 128 | 45 | 88 | 132 | 30 | 239/250 | 169 | 67 | 3 | 171 | 75 | 4 |
| Moore et al (7) | Spanish | 1,041/1,049 | 418 | 478 | 145 | 402 | 486 | 161 | 1,068/1,078 | 537 | 457 | 74 | 557 | 429 | 92 |
| Karagas et al (21) | American | 350/543 | 140 | 171 | 39 | 227 | 245 | 71 | 350/542 | 173 | 146 | 31 | 267 | 220 | 55 |
| Moore et al (19) | Argentina | 106/109 | 45 | 42 | 19 | 32 | 59 | 18 | 106/108 | 52 | 45 | 9 | 55 | 45 | 8 |
| Lin et al (5) | American | 448/448 | 199 | 197 | 52 | 218 | 177 | 53 | 448/447 | 219 | 199 | 30 | 213 | 197 | 37 |
| Sanyal et al (20) | Swedish | 309/246 | 173 | 113 | 23 | 121 | 102 | 23 | 311/245 | 145 | 133 | 33 | 110 | 111 | 24 |
| Kimura et al (18) | Germany | 165/150 | 70 | 80 | 15 | 65 | 73 | 12 | N/A | N/A | N/A | N/A | N/A | N/A | N/A |
AA, Ala/Ala; AV, Ala/Val; VV, Val/Val; GG, Glu/Glu; GA, Glu/Ala; N/A, not available.
Quantitative data synthesis
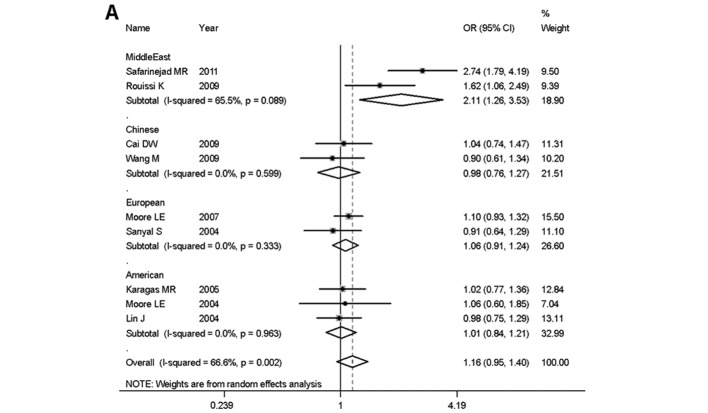
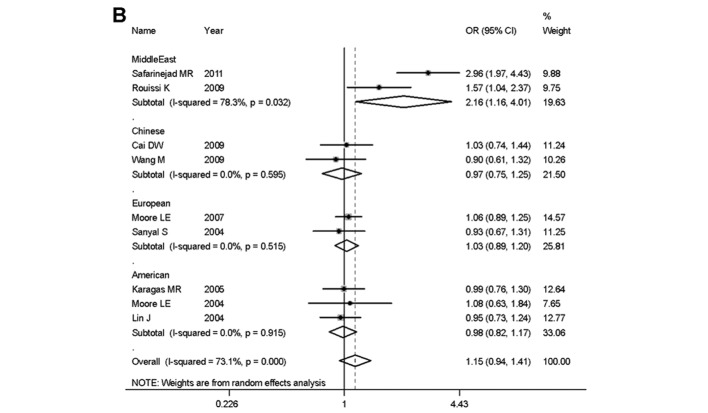
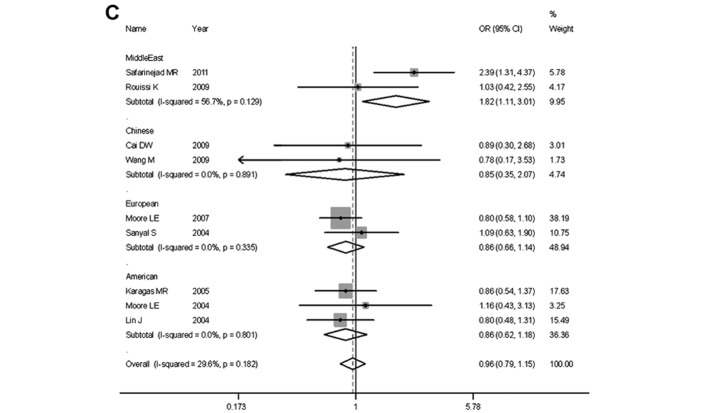
Overall, no significant associations of Ala222Val and Glu429Ala polymorphisms with bladder cancer risk were identified by this meta-analysis (Table III). For MTHFR Ala222Val: Val/Val vs. Ala/Ala: OR, 1.02; 95% CI: 0.80–1.29; Val/Ala vs. Ala/Ala: OR, 1.02; 95% CI: 0.92–1.12; dominant model: OR, 1.01; 95% CI: 0.87–1.17; recessive model: OR, 1.00; 95% CI: 0.87–1.15; and for MTHFR Glu429Ala: Ala/Ala vs. Glu/Glu: OR, 1.11; 95% CI: 0.78–1.58; Ala/Glu vs. Glu/Glu: OR, 1.16; 95% CI: 0.95–1.40; dominant model: OR, 1.15; 95% CI: 0.94–1.41; recessive model: OR, 0.96; 95% CI: 0.79–1.15. In the subgroup analyses by ethnicity, no significant associations were found in any of the genetic models for the MTHFR Ala222Val polymorphisms with bladder cancer risk. By contrast, the Glu429Ala polymorphism was found to be significantly associated with increased bladder cancer risk in individuals of Middle Eastern descent (Ala/Glu vs. Glu/Glu: OR, 2.11; 95% CI: 1.26–3.53; dominant model: OR, 2.16; 95% CI: 1.16–4.01; recessive model: OR, 1.82; 95% CI: 1.11–3.01), whereas no significant associations were identified in individuals of Chinese descent (Ala/Glu vs. Glu/Glu: OR, 0.98; 95% CI: 0.76–1.27; dominant model: OR, 0.97; 95% CI: 0.75–1.25; recessive model: OR, 0.85; 95% CI: 0.35–2.07), European descent (Ala/Glu vs. Glu/Glu: OR, 1.06; 95% CI: 0.91–1.24; dominant model: OR, 1.03; 95% CI: 0.89–1.20; recessive model: OR, 0.86; 95% CI: 0.66–1.14), and American descent (Ala/Glu vs. Glu/Glu: OR, 1.01; 95% CI: 0.84–1.21; dominant model: OR, 0.98; 95% CI: 0.82–1.17; recessive model: OR, 0.86; 95% CI: 0.62–1.18). The results of the subgroup analysis by ethnicity were also shown by forest plots in Fig. 1. In the subgroup analysis by the source of controls, no significant associations were found in any of the genetic models for the two polymorphisms (Table III).
Table III.
Results of meta-analysis for methylenetetrahydrofolate reductase (MTHFR) gene Ala222Val and Glu429Ala polymorphism and bladder cancer risk.
| Genetic model | Recessive model | Dominant model | Homozygote | Heterozygote | |||||
|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
|
|||||
| Ala222Val | No. of study (sample size case/control) | Val/Val vs. Val/Ala + Ala/Ala | Val/Val + Val/Ala vs. Ala/Ala | Val/Val vs. Ala/Ala | Val/Ala vs. Ala/Ala | ||||
|
|
|
|
|
||||||
| OR (95% CI) | Ph | OR (95% CI) | Ph | OR (95% CI) | Ph | OR (95% CI) | Ph | ||
| Overall | 11 (3,463/3,927) | 1.00 (0.87–1.15) | 0.130 | 1.01 (0.87–1.17) | 0.024 | 1.02 (0.80–1.29) | 0.019 | 1.02 (0.92–1.12) | 0.103 |
| Ethnicity | |||||||||
| Chinese | 3 (701/875) | 1.32 (0.80–2.15) | 0.070 | 1.19 (0.79–1.78) | 0.029 | 1.42 (0.72–2.78) | 0.016 | 1.16 (0.93–1.45) | 0.146 |
| European | 3 (1,515/1,445) | 0.89 (0.72–1.11) | 0.747 | 0.90 (0.78–1.05) | 0.485 | 0.86 (0.68–1.09) | 0.629 | 0.92 (0.79–1.07) | 0.558 |
| American | 3 (904/1,100) | 0.93 (0.71–1.22) | 0.759 | 0.98 (0.71–1.36) | 0.063 | 0.95 (0.71–1.26) | 0.690 | 0.97 (0.66–1.43) | 0.031 |
| Middle East | 2 (343/507) | 0.87 (0.54–1.40) | 0.184 | 0.98 (0.74–1.30) | 0.270 | 0.87 (0.53–1.44) | 0.134 | 1.00 (0.75–1.34) | 0.440 |
| Source of control | |||||||||
| PB | 3 (641/843) | 0.83 (0.60–1.14) | 0.496 | 0.92 (0.74–1.13) | 0.109 | 0.78 (0.56–1.11) | 0.589 | 0.86 (0.57–1.29) | 0.057 |
| HB | 8 (2,822/3,084) | 1.08 (0.86–1.35) | 0.099 | 1.06 (0.89–1.25) | 0.033 | 1.12 (0.83–1.50) | 0.013 | 1.03 (0.92–1.15) | 0.203 |
|
| |||||||||
| Glu429Ala | No. of study (sample size case/control) | Ala/Ala vs. Ala/Glu + Glu/Glu | Ala/Ala + Ala/Glu vs. Glu/Glu | Ala/Ala vs. Glu/Glu | Ala/Glu vs. Glu/Glu | ||||
|
|
|
|
|
||||||
| OR (95% CI) | Ph | OR (95% CI) | Ph | OR (95% CI) | Ph | OR (95% CI) | Ph | ||
|
| |||||||||
| Overall | 9 (3,177/3,257) | 0.96 (0.79–1.15) | 0.182 | 1.15 (0.94–1.41) | <0.001 | 1.11 (0.78–1.58) | 0.009 | 1.16 (0.95–1.40) | 0.002 |
| Ethnicity | |||||||||
| Chinese | 2 (551/575) | 0.85 (0.35–2.07) | 0.891 | 0.97 (0.75–1.25) | 0.595 | 0.85 (0.35–2.07) | 0.857 | 0.98 (0.76–1.27) | 0.599 |
| European | 2 (1,379/1,078) | 0.86 (0.66–1.14) | 0.335 | 1.03 (0.89–1.20) | 0.515 | 0.88 (0.66–1.17) | 0.512 | 1.06 (0.91–1.24) | 0.333 |
| American | 3 (904/1,097) | 0.86 (0.62–1.18) | 0.801 | 0.98 (0.82–1.17) | 0.915 | 0.86 (0.62–1.20) | 0.781 | 1.01 (0.84–1.21) | 0.963 |
| Middle East | 2 (343/507) | 1.82 (1.11–3.01) | 0.129 | 2.16 (1.16–4.01) | 0.032 | 2.35 (0.75–7.40) | 0.040 | 2.11 (1.26–3.53) | 0.089 |
| Source of control | |||||||||
| PB | 3 (641/841) | 0.93 (0.64–1.36) | 0.839 | 1.13 (0.92–1.39) | 0.188 | 0.97 (0.66–1.44) | 0.726 | 1.16 (0.93–1.44) | 0.204 |
| HB | 6 (2,536/2,416) | 1.04 (0.72–1.51) | 0.052 | 1.15 (0.87–1.52) | <0.001 | 1.15 (0.68–1.93) | 0.001 | 1.14 (0.89–1.48) | 0.001 |
OR, odds ratio; CI, confidence interval; Ph, P-values for heterogeneity from Q test; PB, population-based; HB, hospital-based. Random-effects model was used when Ph<0.05; otherwise, fixed-model was used.
Figure 1.
Forest plots of methylenetetrahydrofolate reductase (MTHFR) Glu429Ala polymorphism and bladder cancer risk when stratified by ethnicity. (A) Ala/Glu vs. Glu/Glu, (B) dominant model and (C) recessive model, respectively.
Heterogeneity and sensitivity analysis
In the heterogeneity analysis, the Val/Val vs. Ala/Ala model and dominant genetic model for the Ala222Val polymorphism, as well as the Ala/Ala vs. Glu/Glu, Ala/Glu vs. Glu/Glu and dominant genetic models for the Glu429Ala polymorphism were found to be significant (Ph<0.05 by Q test, Table III). The Ph value of the subgroup analysis showed that the heterogeneity was effectively decreased in some of the comparisons and the major source of heterogeneity may stem from the hospital-based controls and ethnicity, such as the Chinese and American subgroups. In the sensitivity analyses, with each study been excluded one at a time during the analysis, the overall results were not altered and no different conclusions were obtained, although the heterogeneity of the analysis was obviously decreased during the exclusion.
Publication bias test
Begg’s funnel plot and Egger’s test were applied to assess the publication bias of the studies. No obvious asymmetry was identified by the Begg’s plots. The funnel plot for ORs of the recessive model for Glu429Ala (Ala/Ala vs. Ala/Glu + Glu/Glu) was shown in Fig. 2 as an example. Furthermore, the results of Egger’s test did not show any evidence of publication bias (for the Ala222Val polymorphism: P=0.961 for Val/Val vs. Ala/Ala, 0.558 for Val/Ala vs. Ala/Ala, 0.884 for the recessive genetic model and 0.810 for the dominant genetic model; for the Glu429Ala polymorphism: P=0.468 for Ala/Ala vs. Glu/Glu, 0.457 for Ala/Glu vs. Glu/Glu, 0.440 for the recessive genetic model and 0.362 for the dominant genetic model, respectively).
Figure 2.
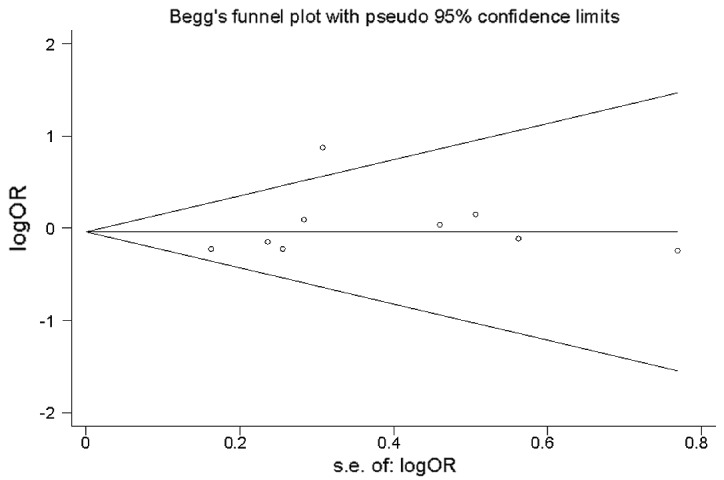
Funnel plot analysis for odds ratios (ORs). Recessive genetic model of methylenetetrahydrofolate reductase (MTHFR) Glu429Ala polymorphism in overall studies is shown as an example.
Discussion
The MTHFR Ala222Val and Glu429Ala polymorphisms have been found to be a risk factor for a variety of cancers including colon cancer (11), acute lymphoblastic leukemia (12), gastric cancer (13) and head and neck squamous cell carcinoma (14). As for bladder cancer, several epidemiological studies (5–7, 18–27) have been investigated for their association with cancer risk. However, the results are not conclusive due to small sample-sized association studies that lack statistical power. In the current meta-analysis, a more precise estimation of MTHFR Ala222Val and Glu429Ala and bladder cancer was derived by including a pooled total of 3,463 cases and 3,927 controls for Ala222Val in 11 studies and 3,177 cases and 3,502 controls for Glu429Ala in 9 studies.
The data showed that the Ala222Val and Glu429Ala polymorphisms were not significantly associated with bladder cancer susceptibility in the entire population. Furthermore, the role of Ala222Val and Glu429Ala polymorphism was evaluated in different subgroups (Chinese, European, American and individuals of Middle Eastern descent), and the results indicated that the Ala222Val polymorphism was not significantly associated with bladder cancer risk in any of the genetic models. Notably, Glu429Ala polymorphism was significantly associated with bladder cancer risk in individuals of Middle Eastern descent. Individuals of Middle Eastern descent who had the Ala/Ala allele were ~82% more likely to have bladder cancer than those who had Ala/Glu or Glu/Glu genotype. Our results suggest that the Glu429Ala polymorphism was not significantly associated with bladder cancer risk in individuals of Chinese, European or American descent. The reason may be attributed to genetic and environmental factors. Since cancer is a complicated multi-genetic disease, different genetic backgrounds may contribute to the discrepancy that the same polymorphisms play different roles among different ethnic populations (28).
As a potential problem when interpreting the results of all meta-analyses (29), heterogeneity was reduced in this study. Significant between-study heterogeneity existed in the Val/Val vs. Ala/Ala and dominant genetic models of the Ala222Val polymorphism, as well as Ala/Ala vs. Glu/Glu, Ala/Glu vs. Glu/Glu and the dominant genetic model for the Glu429Ala polymorphism when all studies were pooled. Following the subgroup analyses by source of control, the heterogeneity among the Val/Val vs. Ala/Ala and dominant genetic models of the Ala222Val polymorphism arose from the hospital-based subgroup. By contrast, heterogeneity of the Ala/Ala vs. Glu/Glu, Ala/Glu vs. Glu/Glu and dominant genetic models of the Glu429Ala polymorphism arose from hospital-based subgroups, as well as from ethnicity, which was effectively decreased in the Chinese, European and American subgroup analyses. There are two reasons for the heterogeneity: i) hospital-based controls did not represent the entire population and ii) differences of genetic backgrounds and environmental factors potentially exist among different ethnical subgroups.
The associations between the Ala222Val and Glu429Ala polymorphisms and bladder cancer risk have been previously studies. For the variant genotype of Ala222Val polymorphism, Lin et al (5), Cai et al (23) and Wang et al (25) reported that it was associated with a higher risk of bladder cancer. Safarinejad et al (27) reported that the polymorphism was not associated with bladder cancer but was associated with increased risk of muscle-invasive bladder cancer. By contrast, Moore et al (19) reported conflicting results, suggesting a lower risk of bladder cancer was observed in individuals carrying either the Ala/Val or Val/Val genotype compared to those carrying the Ala/Ala genotype. The remaining 6 studies reported that there were no statistically significant associations between the Ala222Val polymorphism and bladder cancer risk (6–7,18,20–21,26). For the variant genotypes of Glu429Ala polymorphism, Rouissi et al (6) reported that it was associated with a higher risk of bladder cancer. However, Safarinejad et al (27) reported that it was not associated with bladder cancer but associated with increased risk of muscle-invasive bladder cancer. The remaining 7 studies reported that there were no statistically significant associations between the Glu429Ala polymorphism and bladder cancer risk (5,7,19–21,23,25).
Although Wang et al (25) previously published a case-control study with meta-analysis, in which the data included were up to 2007 and 7 case-control studies were analyzed with conclusions suggesting no significant associations between Ala222Val and Glu429Ala polymorphisms and bladder cancer risk, which was in accordance with results of this meta-analysis. However, in the demographic characteristics of that study (25), all the cases and controls of different studies, including individuals from the USA, Argentina and Tunis (mistaken for Turkey in that study) were considered Caucasian, rendering the subgroup analyses by ethnicity impossible, thereby discounting the statistical power of analysis for the estimation of genetic effects. In the present study, the Glu429Ala polymorphism was found to be significantly associated with increased bladder cancer risk in individuals of Middle Eastern descent.
Given the data was abstracted from these publications, there are some limitations that should be addressed with regard to this meta-analysis. Firstly, the ethnicity of the cases and controls were not uniformly defined. The Glu429Ala polymorphism for the Middle Eastern individuals, which was found to be significant, had only 2 publications with 343 cases and 507 controls in total. Secondly, the controls may not truly represent the populations, since most of the controls included in this study were hospital-based controls and the Ala222Val and Glu429Ala polymorphism is known to be potentially associated with other diseases. Thirdly, some of the environmental risk factors of bladder cancer, such as cigarette smoking habits and occupational exposure were not uniformly provided by the original studies. Furthermore, some inevitable publication bias might exist in the publications. Therefore, the results of this study should be interpreted with caution and additional studies with a large amount of data are needed for evaluation.
In conclusion, the results of the present meta-analysis suggest that heterozygous and homozygous variant carriers of the MTHFR Glu429Ala polymorphism are significantly associated with increased bladder cancer risk in individuals of Middle Eastern descent, with no evidence of an association between this polymorphism and bladder cancer risk in individuals of Chinese, European and American descent. Since the Ala/Ala genotype is relatively infrequent in the population, more well-designed studies with larger sample sizes are required to confirm the findings of the present study.
Acknowledgements
The study was supported by the National Natural Science Foundation of China (grant nos. 81101536 and 81201565), the Program of the Pearl River Young Talents of Science and Technology in Guangzhou, China (no. 2011J2200070) and the Specialized Research Fund for the Doctoral Program of Higher Education (nos. 20104433120001 and 20124433120001).
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Ploeg M, Aben KK, Kiemeney LA. The present and future burden of urinary bladder cancer in the world. World J Urol. 2009;27:289–293. doi: 10.1007/s00345-009-0383-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chu H, Wang M, Zhang Z. Bladder cancer epidemiology and genetic susceptibility. J Biomed Res. 2013;27:170–178. doi: 10.7555/JBR.27.20130026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cordon-Cardo C. Molecular alterations associated with bladder cancer initiation and progression. Scand J Urol Nephrol Suppl. 2008:154–165. doi: 10.1080/03008880802291915. [DOI] [PubMed] [Google Scholar]
- 5.Lin J, Spitz MR, Wang Y, et al. Polymorphisms of folate metabolic genes and susceptibility to bladder cancer: a case-control study. Carcinogenesis. 2004;25:1639–1647. doi: 10.1093/carcin/bgh175. [DOI] [PubMed] [Google Scholar]
- 6.Rouissi K, Ouerhani S, Oliveira E, et al. Polymorphisms in one-carbon metabolism pathway genes and risk for bladder cancer in a Tunisian population. Cancer Genet Cytogenet. 2009;195:43–53. doi: 10.1016/j.cancergencyto.2009.06.007. [DOI] [PubMed] [Google Scholar]
- 7.Moore LE, Malats N, Rothman N, et al. Polymorphisms in one-carbon metabolism and trans-sulfuration pathway genes and susceptibility to bladder cancer. Int J Cancer. 2007;120:2452–2458. doi: 10.1002/ijc.22565. [DOI] [PubMed] [Google Scholar]
- 8.Bailey LB. Folate, methyl-related nutrients, alcohol, and the MTHFR 677C→T polymorphism affect cancer risk: intake recommendations. J Nutr. 2003;133(Suppl 1):S3748–S3753. doi: 10.1093/jn/133.11.3748S. [DOI] [PubMed] [Google Scholar]
- 9.Frosst P, Blom HJ, Milos R, et al. A candidate genetic risk factor for vascular disease: a common mutation in methylenetetrahydrofolate reductase. Nat Genet. 1995;10:111–113. doi: 10.1038/ng0595-111. [DOI] [PubMed] [Google Scholar]
- 10.Weisberg I, Tran P, Christensen B, Sibani S, Rozen R. A second genetic polymorphism in methylenetetrahydrofolate reductase (MTHFR) associated with decreased enzyme activity. Mol Genet Metab. 1998;64:169–172. doi: 10.1006/mgme.1998.2714. [DOI] [PubMed] [Google Scholar]
- 11.Yin G, Kono S, Toyomura K, et al. Methylenetetrahydrofolate reductase C677T and A1298C polymorphisms and colorectal cancer: the Fukuoka Colorectal Cancer Study. Cancer Sci. 2004;95:908–913. doi: 10.1111/j.1349-7006.2004.tb02201.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tong N, Fang Y, Li J, et al. Methylenetetrahydrofolate reductase polymorphisms, serum methylenetetrahydrofolate reductase levels, and risk of childhood acute lymphoblastic leukemia in a Chinese population. Cancer Sci. 2010;101:782–786. doi: 10.1111/j.1349-7006.2009.01429.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boccia S, Gianfagna F, Persiani R, et al. Methylenetetrahydrofolate reductase C677T and A1298C polymorphisms and susceptibility to gastric adenocarcinoma in an Italian population. Biomarkers. 2007;12:635–644. doi: 10.1080/13547500701546766. [DOI] [PubMed] [Google Scholar]
- 14.Galbiatti AL, Ruiz MT, Rodrigues JO, et al. Polymorphisms and haplotypes in methylenetetrahydrofolate reductase gene and head and neck squamous cell carcinoma risk. Mol Biol Rep. 2012;39:635–643. doi: 10.1007/s11033-011-0781-7. [DOI] [PubMed] [Google Scholar]
- 15.Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22:719–748. [PubMed] [Google Scholar]
- 16.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 17.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kimura F, Florl AR, Steinhoff C, et al. Polymorphic methyl group metabolism genes in patients with transitional cell carcinoma of the urinary bladder. Mutat Res. 2001;458:49–54. doi: 10.1016/s1383-5726(01)00010-3. [DOI] [PubMed] [Google Scholar]
- 19.Moore LE, Wiencke JK, Bates MN, Zheng S, Rey OA, Smith AH. Investigation of genetic polymorphisms and smoking in a bladder cancer case-control study in Argentina. Cancer Lett. 2004;211:199–207. doi: 10.1016/j.canlet.2004.04.011. [DOI] [PubMed] [Google Scholar]
- 20.Sanyal S, Festa F, Sakano S, et al. Polymorphisms in DNA repair and metabolic genes in bladder cancer. Carcinogenesis. 2004;25:729–734. doi: 10.1093/carcin/bgh058. [DOI] [PubMed] [Google Scholar]
- 21.Karagas MR, Park S, Nelson HH, et al. Methylenetetrahydrofolate reductase (MTHFR) variants and bladder cancer: a population-based case-control study. Int J Hyg Environ Health. 2005;208:321–327. doi: 10.1016/j.ijheh.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 22.Ouerhani S, Oliveira E, Marrakchi R, et al. Methylenetetrahydrofolate reductase and methionine synthase polymorphisms and risk of bladder cancer in a Tunisian population. Cancer Genet Cytogenet. 2007;176:48–53. doi: 10.1016/j.cancergencyto.2007.03.007. [DOI] [PubMed] [Google Scholar]
- 23.Cai DW, Liu XF, Bu RG, et al. Genetic polymorphisms of MTHFR and aberrant promoter hypermethylation of the RASSF1A gene in bladder cancer risk in a Chinese population. J Int Med Res. 2009;37:1882–1889. doi: 10.1177/147323000903700625. [DOI] [PubMed] [Google Scholar]
- 24.Ouerhani S, Rouissi K, Marrakchi R, et al. Combined effect of NAT2, MTR and MTHFR genotypes and tobacco on bladder cancer susceptibility in Tunisian population. Cancer Detect Prev. 2009;32:395–402. doi: 10.1016/j.canep.2009.04.005. [DOI] [PubMed] [Google Scholar]
- 25.Wang M, Zhu H, Fu G, Wang M, Zhang Z, Lu Q, Wang S, Zhang Z. Polymorphisms of methylenetetrahydrofolate reductase and methionine synthase genes and bladder cancer risk: a case-control study with meta-analysis. Clin Exp Med. 2009;9:9–19. doi: 10.1007/s10238-008-0013-1. [DOI] [PubMed] [Google Scholar]
- 26.Chung CJ, Pu YS, Su CT, et al. Polymorphisms in one-carbon metabolism pathway genes, urinary arsenic profile, and urothelial carcinoma. Cancer Causes Control. 2010;21:1605–1613. doi: 10.1007/s10552-010-9589-3. [DOI] [PubMed] [Google Scholar]
- 27.Safarinejad MR, Shafiei N, Safarinejad S. Genetic susceptibility of methylenetetrahydrofolate reductase (MTHFR) gene C677T, A1298C, and G1793A polymorphisms with risk for bladder transitional cell carcinoma in men. Med Oncol. 2011;28(Suppl 1):S398–S412. doi: 10.1007/s12032-010-9723-9. [DOI] [PubMed] [Google Scholar]
- 28.Hirschhorn JN, Lohmueller K, Byrne E, Hirschhorn K. A comprehensive review of genetic association studies. Genet Med. 2002;4:45–61. doi: 10.1097/00125817-200203000-00002. [DOI] [PubMed] [Google Scholar]
- 29.Munafo MR, Flint J. Meta-analysis of genetic association studies. Trends Genet. 2004;20:439–444. doi: 10.1016/j.tig.2004.06.014. [DOI] [PubMed] [Google Scholar]