Abstract
Exploration of the ubiquitin system in eukaryotes has shown that the chemical modification of proteins by ubiquitin, known as ubiquitylation, is an incredibly important post-translational event that is crucial to numerous cellular processes. Ubiquitylation is carried out by a series of enzymes that specifically target proteins to either change their activity or their location or earmark them for degradation. Using a wide range of genome-wide approaches, the ubiquitin system has been shown to be of particular importance in the survival and propagation of the human malaria parasites. In this review, we highlight our current understanding of the ubiquitin system in Plasmodium, and discuss its possible role in the development of drug resistant malaria strains.
Keywords: malaria, ubiquitin, drug resistance, apicoplast
Introduction
The ubiquitin system is one of the principle pathways used by all eukaryotic cells to regulate protein abundance levels and protein activities. It encompasses a complex network of enzymes that add and remove ubiquitin from a protein substrate. The reversible covalent attachment of ubiquitin is one of the most pervasive post-translational modifications that occur in eukaryotic cells. Similar to phosphorylation, ubiquitylation is involved in a variety of cellular processes, such as protein turnover1, transcriptional regulation2, cell cycle progression3, differentiation4, and signal transduction5. While the ubiquitin system is commonly associated with the proteasome degradation pathway, it is also involved in many processes that are proteasome-independent.
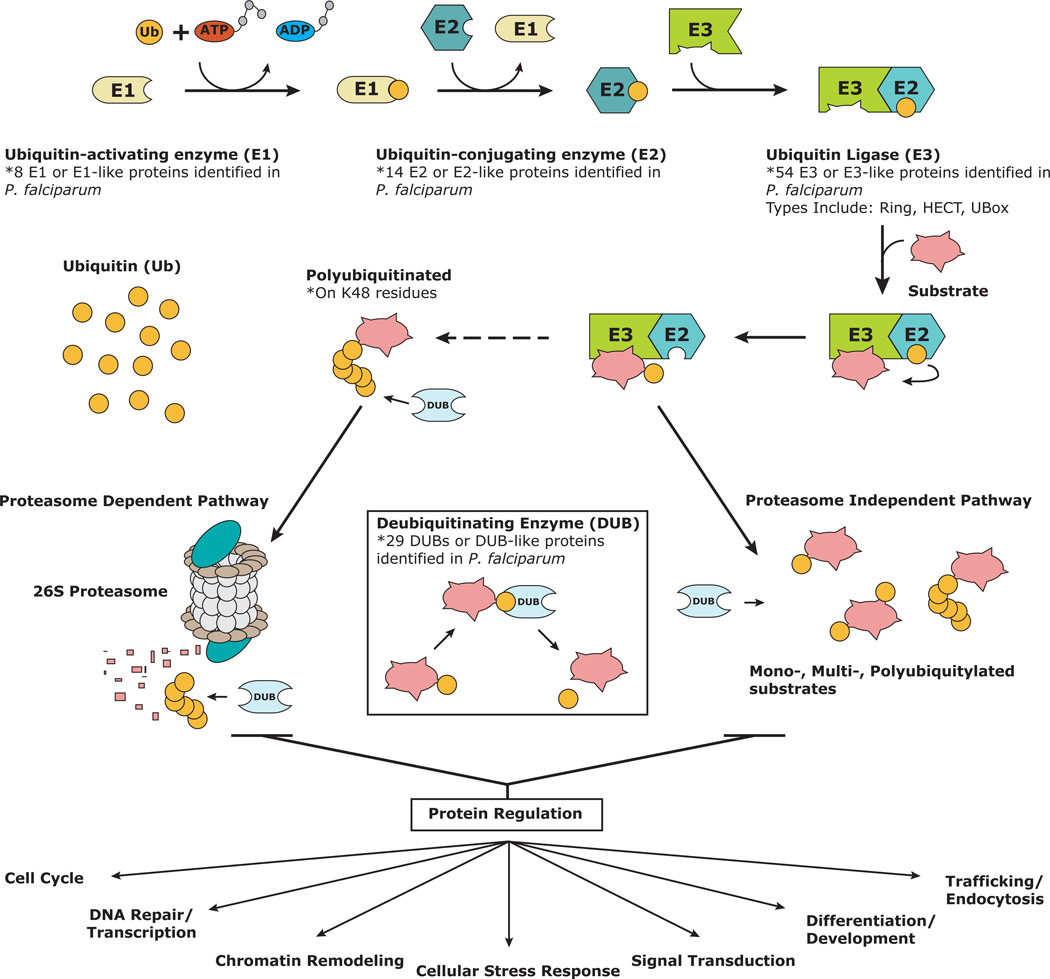
Ubiquitin is a conserved 76-amino-acid polypeptide that is covalently attached to one or more lysine residues on cellular proteins through a complex enzymatic cascade, which includes an E1 ubiquitin-activating enzyme, an E2 ubiquitin-conjugating enzyme and an E3 ubiquitin ligase6 (Figure 1). Ubiquitin itself contains seven lysines that can be further conjugated to the carboxyl terminus of another ubiquitin to form a polyubiquitin chain. Ubiquitylation begins with the activation of ubiquitin via an E1 ubiquitin-activating enzyme. This initial reaction results in a C-terminal thioester linkage between the E1 ubiquitin-activating enzyme and ubiquitin. Subsequently, an E2 ubiquitin-conjugating enzyme catalyzes the transfer of ubiquitin from the E1 enzyme to a conserved cysteine on the E2 enzyme by a trans-esterification reaction. The final step in the ubiquitylation process is the targeted attachment of ubiquitin to a protein via an E3 ubiquitin ligase, creating an isopeptide bond between a lysine on the target protein and the C-terminal glycine on ubiquitin6. E3 proteins are categorized into three major classes of enzymes, Homologous to E6-associated protein C-terminus (HECT) ligases, Really Interesting New Gene (RING) fingers and U-box E3 ligases, which are the main determinants of substrate specificity. The formation of polyubiquitin chains linked through lysine at positions 11 and 48 on ubiquitin (Lys 11 and 48) target protein substrates to the proteasome for degradation; while alternative linkages, such as Lys 6, 9, 27 and 33 have unsubstantiated roles in eukaryotic cells7, 8. Moreover, linkages such as Lys 29 and 63, fulfill other cellular functions, including aiding in lysosomal degradation and many proteolysis-independent mechanisms, respectively8, 9(Figure 1). Like phosphorylation, ubiquitylation can be reversed by the activity of deubiquitylation enzymes (DUBs). It is the cooperative interplay between these enzymes that determines the prevalence of ubiquitylated proteins and their role in the cell.
Figure 1. Depiction of the ubiquitin-mediated pathways in P. falciparum.
Ubiquitin is initially activated by an E1 ubiquitin-activating enzyme and then transferred to an E2 ubiquitin-conjugating enzyme. The E2 enzyme may either complex with an E3 ubiquitin-ligase to directly ubiquitylate a target substrate or transfer the ubiquitin to an E3 enzyme, which will subsequently ubiquitylate a target substrate. Substrates polyubiquitinated at lysine K48 are targeted to the proteasome for destruction. Ubiquitylation of substrates at other lysine residues can modulate substrate activity in many cellular processes. Deubiquitylating enzymes (DUBs) remove ubiquitin by cleaving the peptide bonds and replenish the ubiquitin pool.
Over of the last 30 years, our understanding of the components of the ubiquitin system in eukaryotic organisms has grown significantly due to its implications in human disease. Dysregulation of the ubiquitin system has been implicated in numerous human diseases, including neurological and infectious diseases and several different types of cancer10–14. In particular, the ubiquitin system has been demonstrated to be a contributing factor in the etiology of Parkinson’s disease12, in addition to other neurodegenerative disorders13. The ubiquitin ligase (PARK2) that was shown to be dysfunctional in Parkinson’s disease also has a role in ubiquitin-mediated autophagy of intracellular pathogens, specifically M. tuberculosis14. Exploring proteins involved in the spindle assembly checkpoint has also revealed that ubiquitylation enzymes are crucial regulators of the cell cycle15. As a result of unchecked cell cycle progression, many of these mutated ubiquitin-proteasome system (UPS) related proteins are the source of different types of malignancies16, 17. These studies and the possibility of targeting the ubiquitin system as novel therapeutic strategies emphasize the importance of understanding this cellular mechanism in the context of human pathology.
Here we consider the significance of the ubiquitin system in the survival and propagation of the human malaria parasite, Plasmodium falciparum. P. falciparum is the most virulent form of malaria and is responsible for approximately 650,000 deaths every year. It has been projected by the World Health Organization that half of the worlds’ population is still at risk for contracting the parasite18. The human malaria parasite has a complex life cycle consisting of three major stages: the mosquito stage and the human liver and blood stages19. An infected mosquito that bites a human injects parasites called sporozoites into the bloodstream. The sporozoites quickly travel to the liver to invade liver cells. Sporozoites differentiate and divide to produce tens of thousands of merozoites. When the merozoites exit the liver cells and re-enter the blood stream. Subsequently, they invade erythrocytes, and within 48 hours a parasite can differentiate and develop to produce up to 32 new daughter cells that egress and reinvade new red blood cells. This asexual cycle continues until the parasite is stressed in the human host. Parasites then undergo sexual differentiation, which can require up to two weeks for full maturation. When a female mosquito takes a blood meal, sexually mature male and female gametocytes are taken up and undergo sexual reproduction in the mosquito midgut. Sporozoites then migrate to the salivary glands of the mosquito and are injected into a human host during another blood meal, thus completing the cycle19. Throughout its life cycle, the parasite undergoes cell differentiation multiple times, requiring changes in protein activity and degradation of unnecessary stage-specific proteins20, 21. The central role of ubiquitylation in all of these processes as well as the identification of parasite-specific components of the ubiquitin system open up new opportunities for drug discovery. With a continued need for new preventative and curative measures to combat the parasite infection, understanding the mechanisms that control the parasite ubiquitylation pathways at the molecular level may open new doors for the development of novel therapeutic strategies. In the face of increased drug resistance to many of the most efficacious anti-malarials22, the discovery of specific inhibitors targeting different components of the ubiquitin system in the malaria parasite have prompted investigations into this pathway.
In this review, we highlight the current knowledge of the ubiquitin system in Plasmodium spp., focusing on the power of genome-wide approaches to discover the components of the system and its substrates. Additionally, we discuss the recent implications of the ubiquitin pathway in the acquisition of drug resistance.
Identifying components of the ubiquitin system using comparative proteomic analyses
Comparative genomic and proteomic analyses are essential tools for characterizing protein functions in different species. Comparative proteomics allows investigators to search and identify proteins and their potential function based on preexisting knowledge from other species. Functional characterization of proteins can be done computationally and/or experimentally23. Typically, this entails relying on specific protein features, such as functional or structural domains, to determine the function of a given protein in a different species24. This approach has been particularly useful with studying the ubiquitin system in Plasmodium spp. It was not until 2007 that the importance and the functional components of the ubiquitin system were beginning to be understood in the malaria parasite25, 26. While still in its infancy, our understanding of the ubiquitin system in Plasmodium spp. has grown significantly over the last six years, with much of our knowledge attributed to the initial comparative analyses discussed below.
A comprehensive in silico proteomic analysis identified most, if not all components of the ubiquitin system in Plasmodium. A hidden markov model (HMM) analysis was used to discover proteins containing one of the 24 Pfam domains related to the ubiquitin system26. These domains included those found in ubiquitin, ubiquitin-like proteins, and the enzymes involved in activation, conjugation, ligation and deconjugation of ubiquitin to its target substrates. While the study focused on Plasmodium spp., three other apicomplexan species and five non-apicomplexan eukaryotic organisms were included in the analysis to provide a comparative perspective and to evaluate the completeness of the HMM search. Over 100 UPS-related proteins were found in Plasmodium spp., and similar numbers were identified in the other apicomplexan parasites. The number of UPS-related proteins found in the apicomplexans was comparable to the S. cerevisiae proteome suggesting that most of the components of the UPS are well conserved among lower eukaryotes. In contrast, significantly more UPS-related proteins were identified in the multicellular eukaryotic proteomes used in the analysis, C. elegans, A. thaliana, D. melanogaster and H. sapiens.
A total of 114 UPS-related proteins were identified in the P. falciparum genome: 9 were identified as ubiquitin or ubiquitin-like proteins; 8 were E1 or E1-like activating enzymes; 14 were E2 or E2-like conjugating enzymes; 54 were E3 or E3-like ligases and 29 were DUB or DUB-like proteins (DUBLps)26. In addition, three fusion-protein precursors for ubiquitin (PF3D7_1402500, PF3D7_1365900 and PF3D7_1211800) were identified in all of the apicomplexans, and all precursor proteins displayed a high level of conservation among the eukaryotic species examined (Figure 2). Ubiquitin-like modifiers were also identified in the HMM search among most of the Plasmodium spp., which included: neural precursor-cell expressed developmentally and down-regulated 8 (NEDD8), small-ubiquitin-related modifier (SUMO), homologous to ubiquitin 1 (HUB1), ubiquitin-related modifier 1 (URM1) and autophagy 8 (ATG8). These results agree with an earlier study, exploring UBL modifiers in various protozoans species25. Moreover, later studies in Plasmodium have shown sumoylation to be an important regulator in the oxidative stress response27, and ATG8 to be a key UBL modifier in autophagy and possibly in the biogenesis of the apicoplast organelle28, 29. There is no current understanding of the role of NEDD8 in the P. falciparum; however, it likely serves an essential role in cell cycle progression. Similarly, very little is known about the roles of URM1 and HUB1 in eukaryotes (Table 1). Other UBL modifiers, such as ISG15, FAT10, UFM1 and FUB1, were missing in apicomplexan proteomes. These findings are not surprising because some of these UBL modifiers appear to be limited to multicellular eukaryotes and are involved in immunological responses30, 31.
Figure 2. A comparative alignment of the ubiquitin protein sequences in different eukaryotes.
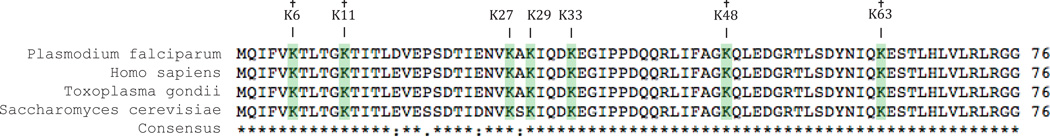
The ubiquitin protein sequences show a high level of conservation among different organisms. Lysine residues that are known sites for ubiquitylation and extension are highlighted. † In P. falciparum K6, K11, K48, and K63 have been detected as sites for ubiquitylation and extension.
Table 1. Validated proteins of the ubiquitin system in Plasmodium.
All discussed proteins of the ubiquitin system are listed in the table above along with reference numbers and what is known about their respective functional role(s) in Plasmodium.
| Plasmodium Gene ID | Enzyme/Protein Classification | Function/Putative Function | References | |
|---|---|---|---|---|
| Ub/Ub-like | ||||
| Ubiquitin | PF3D7_1402500 | Ub | Ubiquitylation of substrates | 26, 38, 42, 44, 65 |
| PF3D7_1365900 | Ub | Ubiquitylation of substrates | 26, 38, 42, 44, 66 | |
| PF3D7_1211800 | Ub | Ubiquitylation of substrates | 26, 38, 42, 44, 67 | |
| ATG8 | Pf3D7_1019900 | Ub-like protein | Autophagy | 28, 29 |
| NEDD8 | Pf3D7_1313000 | Ub-like protein | Unknown | - |
| SUMO | Pf3D7_0505800 | Ub-like protein | Oxidative Stress Response | 27 |
| E1/E1-like | ||||
| UBA1 | Pf3D7_1225800 | E1 | Activation of ubiquitin | 38 |
| UBA2 | Pf3D7_1237000 | E1-like protein | Activation of SUMO | 27 |
| UBA3 | Pf3D7_0817000 | E1-like protein | Activation of NEDD8 | - |
| Unnamed | Pf3D7_1333200 | E1-like protein | Part of the ERAD-like system | 42, 44 |
| Unnamed | Pf3D7_1365400 | E1-like protein | Part of the ERAD-like system | 42 |
| E2/E2-like | ||||
| UBC7 | Pf3D7_1203900 | E2 Conjugating Enzyme | ERAD | 38 |
| Unnamed | Pf3D7_1243700 | E2 Conjugating Enzyme | Implicated in drug resistance | 73 |
| Unnamed | Pf3D7_1345500 | E2-like protein | Part of the ERAD-like system | 42 |
| E3/E3-like | ||||
| HRD1 | Pf3D7_1422500 | E3 Ligase | ERAD | 38 |
| RAD5 | Pf3D7_1343400 | E3 Ligase | Implicated in drug resistance | 72, 74 |
| Unnamed | Pf3D7_0826100 | E3 Ligase | Implicated in drug resistance | 73 |
| Unnamed | Pf3D7_0316900 | E3-like protein | Part of the ERAD-like system | 42 |
| Unnamed | Pf3D7_0312100 | E3-like protein | Part of the ERAD-like system | 42 |
| DUB | ||||
| UCH54 | Pf3D7_1117100 | DUB | DUB with deNeddylating activity | 36 |
| UBP1 | Pf3D7_0104300 | DUB | Implicated in drug resistance | 75, 76 |
| OTU-like cysteine protease | Pf3D7_1031400 | DUB | Targeted to the apicoplast | Unpublished |
To examine the degree of conservation among the ubiquitin system proteins in Plasmodium spp., an all-against-all blast search was conducted for each of the proteins domains between the proteomes of 13 different eukaryotes26. The comparison of the E1 proteins showed relatively high conservation between the species, when examining the ubiquitin-activating (UBA) domain. The UBA domain is characteristic among all E1 enzymes and is responsible for activating ubiquitin or ubiquitin-like proteins and transferring them to their cognate E2 enzymes. Among the E1 proteins, homologs of UBA1–3 were discovered in P. falciparum, as well as in other apicomplexans. UBA1 is the first protein in the ubiquitylation cascade that is responsible for transferring an activated ubiquitin to E2 enzymes32, while UBA2 and UBA3 are ubiquitin-like proteins responsible for activating and transferring SUMO and NEDD8, respectively33, 34 (Table 1). Similar to the E1 proteins, the E2 ubiquitin-conjugating enzymes showed a high degree on conservation between the species26. A total of 14 E2 proteins were identified in P. falciparum, with the other apicomplexans exhibiting a similar number of E2 proteins. While all of the apicomplexans encoded the majority of E2 homologs found in other species, two atypical E2 proteins were discovered within the Plasmodium and Toxoplasma genera that may be unique among the apicomplexan phylum.
In contrast to the E1 and E2 proteins, E3 ubiquitin and E3 ubiquitin-like ligases were found to be very diverse between the species. Approximately 50% of the UPS-related proteins found within P. falciparum were E3 or E3-like proteins. Members from each of the three families of E3 ubiquitin ligases, RING, HECT and U-box, were discovered in all apicomplexans, of which the RING E3 ligases were the most abundant and divergent in P. falciparum26. While also implicated in proteasome-independent mechanisms, RING E3 ligases are key regulators of ubiquitylation and protein turnover in eukaryotes35. Among the E3 ligases identified in the HMM search were homologs of the well-studied Skp1-Cullin-Fbox (SCF)-type ligase and the Anaphase-Promoting Complex (APC). Interestingly, Plasmodium spp. differed from the other eukaryotes in that they lacked one of the homolog subunits of the APC. In other eukaryotes, the APC plays a vital role during mitosis. Plasmodium undergoes a specialized type of cellular division, known as schizogony, which involves four to five rounds of asynchronous multiplication of its genome before the daughter cells separate. Therefore, a divergent composition of the APC is not entirely unexpected. The differences observed in Plasmodium among the E3 and E3-like ligases compared to other eukaryotic organisms suggest that these proteins may represent potential targets for anti-malarials.
The discovery of DUBs in Plasmodium is of particular importance because, like the E3 ubiqutin ligases, DUBs and DUBLps are less conserved between species. The first DUB to be described in P. falciparum was UCH-54 (PF3D7_1117100, Table 1), which was demonstrated to remove ubiquitin and NEDD8 from conjugated proteins36. This finding was later confirmed in a BLASTP homology search that discovered a total of 11 DUB and DUBLps, representing four of the five subclasses25: JAB1/MPN/Mov34 metalloenzymes, ubiquitin C-terminal hydrolases (UCHs), ubiquitin-specific proteases (USPs) and Machado-Joseph disease (MJD) proteases. Among the most divergent DUBs are the metalloproteases. Eight novel DUB and DUBLps were later identified in the HMM search, including the fifth subclass, known as the otubain proteases26. Interestingly, DUB inhibitors affecting subclasses, such as UCH and USP, have been shown to be effective anti-malarials, with 50% inhibitory concentrations (IC50) in the low micromolar range (Chung, Hamilton, Lee, Prudhomme and Le Roch, unpublished data).
Altogether, these initial findings have shown that Plasmodium contains many of the same ubiquitylation components as other eukaryotes, with some noticeable characteristics. Moreover, differences found in apicomplexan parasites highlight key changes that have occurred over the course of evolution, and support the idea that the ubiquitin system is a prime target that can be exploited for therapeutic purposes.
Novel Targets for Anti-malarials
Among the proteins of the ubiquitin system identified in Plasmodium were components of the classical ER-associated degradation (ERAD) pathway26. ERAD is a highly conserved mechanism that uses the ubiquitin system machinery to selectively target misfolded proteins in the ER, and translocate them to the cytosol where the aberrant proteins are degraded by the proteasome37. With the use of in vitro biochemical assays and cellular localization using immunofluorescence, it was confirmed that PfUBA1, PfUBC7 and PfHRD1 (PF3D7_1225800, PF3D7_1203900 and PF3D7_1422500, respectively, Table 1) were the corresponding homologs of the major proteins of the ERAD pathway38. In two recent studies, the ERAD mechanism was shown to be essential for survival of the human malaria parasite, P. falciparum38, 39. By targeting a crucial protein of the ERAD machinery, known as the signal protein peptidase (SPP), with different inhibitors, it was shown that P. falciparum parasites lost their ability to degrade unstable and misfolded proteins39. While it is suspected that most of the regulation of gene expression in P. falciparum occurs at the transcriptional and post-transcriptional levels40, 41, the malaria parasite may also be particularly sensitive to perturbations in protein abundance levels. Moreover, this suspicion is supported by reported IC50 values that shown inhibitors of the ERAD mechanism are effective antimalarials38, 39. Despite the relatively high conservation among the components in the ERAD pathway in eukaryotes, these proteins remain potential candidates for drug intervention due to their essentiality in parasite survival.
Similarly, proteins encompassing a parasite-specific ERAD-like system were also discovered in the HMM search in apicomplexan parasites26. The ERAD-like pathway contains all of the ubiquitylation enzymes needed for activation (PF3D7_1333200 and PF3D7_1365400), conjugation (PF3D7_1345500), ligation (PF3D7_0316900 and PF3D7_0312100) and deconjugation (PF3D7_1031400)42–44. However, in addition to containing the requisite ubiquitylation domains, ERAD-like proteins contain a signal peptide that is responsible for targeting the proteins to a specialized organelle, known as the apicoplast. The apicoplast is non-photosynthetic plastid that is the result of a secondary endosymbiosis event involving a red alga that created a four membrane-bound plastid. It is an essential organelle that is responsible for fatty acid metabolism, and isoprenoid biosynthesis45–47. While the apicoplast contains a ~35kb-long circular genome encoding ~30 proteins48, most of its function is fulfilled by ~500 nuclear-encoded proteins. The trafficking pathways involved in the import of these proteins through the four apicoplast membranes remains unclear. Apicoplast targeting involves the conventional secretory pathway through the endoplasmic reticulum (ER), and a bipartite leader peptide located at the N-terminus of transported proteins49–51. The bipartite domain consists of a signal peptide (cleaved off after passage into the ER) followed by a transit peptide that avoids secretion of the protein and is sufficient for apicoplast stromal targeting. The discovery of this ERAD-like system in Apicomplexa is similar to the symbiont-specific ERAD-like machinery (SELMA) found in Chromalveolates, a supergroup of plastid-containing unicellular eukaryotes related to red algae44, 52. The SELMA is implicated in pre-protein import to the periplastidal compartment (PPC)53. Similar to the SELMA system, the ERAD-like system found in Plasmodium spp. and Toxoplasma gondii has been showed to be critical for the import of nuclear-encoded proteins into the apicoplast42. While the ERAD-like system in apicomplexans is capable of ubiquitylating proteins in vitro42, the role of ubiquitylation in vivo remains unknown. Despite uncertainties, the functional characterization of parasite-specific ubiquitylation components offers new avenues for antimalarial strategies.
Exploring the Plasmodium ubiquitome
Following the identification of the components of the ubiquitin system in Plasmodium, the next logical step in understanding the ubiquitylation process is identifying the proteins that are ubiquitylated. The addition and removal of ubiquitin and the degradation of ubiquitylated proteins can occur rapidly making identifying the ubiquitome, defined as all proteins that are subject to ubiquitylation, a challenging task. However, several methodologies have been adapted for ubiquitome identification. An in silico prediction model known as UbPred was developed based on experimental evidence in mutant yeast strains to identify similar sequence and structural biases that were common among ubiquitylation sites54. The UbPred model is based on identifying disordered regions within a substrate to predict ubiquitylation sites. While the evidence for this model shows it to be a better global predictor of ubiquitylated substrates, it does not take into account known degradation signals, such as the destruction box (D box) and the KEN-box, and it has been previously suggested to be only a weak signal for proteolysis55. An alternative prediction algorithm, known as UbiPred, that relies on physicochemical properties of the protein sequences has also been developed to predict ubiquitylation sites56. However, UbiPred did not perform as well as UbPred in predicting ubiquitylated substrates54. Moreover, there have been several studies that have identified ubiquitylated proteins experimentally at the proteome-wide level57–64. Many of these studies utilized techniques relying on isolating and identifying ubiquitylated proteins through immunoprecipitation followed by mass spectrometry.
To our knowledge, there has only been a single study exploring the ubiquitome in Plasmodium spp. In a recent study by Ponts et al., a combinatorial approach was used to identify all ubiquitylated substrates65. First, both UbPred and UbiPred were used in an in silico approach to determine the ubiquitome in P. falciparum. From the 5,446 protein-coding sequences, UbPred identified 5,036 proteins predicted to contain ubiquitylation sites. In contrast, UbiPred predicted less than half the number of proteins (2,077 proteins) to contain ubiquitylation sites. The authors concluded that the discrepancy observed between the different prediction models was likely attributed to the differences in the ways the models were trained. While ~70% of the P. falciparum proteome was predicted to contain ubiquitylation sites, only 52% and 39% of the S. cerevisiae and A. thaliana proteomes, respectively, were shown to contain ubiquitylation sites65. Collectively, this initial study suggests that P. falciparum contains more proteins that are subject to ubiquitylation relative to these eukaryotic species, A. thaliana and S. cerevisiae; however, additional studies are needed to confirm this finding. One possible explanation for this observation could be the amino acid composition difference between the different species; lysine is the second most abundant amino acid in P. falciparum, and P. falciparum has about twice the amount of lysine relative to the other two eukaryotic species65.
This in silico study was supplemented by a proteome-wide experimental approach that coupled immunoprecipitation to multidimensional protein identification technology (MudPIT). MudPIT is a “shotgun” approach that couples 2-D liquid chromatography to mass spectrometry to identify peptides contained with a complex protein mixture66. Using an anti-ubiquitin antibody, a total of 437 ubiquitylated proteins were detected from the three asexual stages in P. falciparum erythrocytic cycle65. While only 73 of these Plasmodium proteins were detected with significant enrichment, similar percentages in ubiquitin conjugates were observed in human and A. thaliana cells using comparable techniques58, 67. The number of ubiquitylated proteins was highest in the schizont stage with 33 of the proteins being unique to this morphological stage. Among the ubiquitylated proteins discovered was a well-established transcription factor, known as ApiAP2 (PF3D7_0604100, Table 1). This finding strongly suggests that ubiquitylation of transcription machinery may play an important role in the regulation of gene expression in P. falciparum. Chaperone proteins, proteins involved in RNA metabolism and translation had the highest representation among the various detected ubiquitylated proteins. This latest finding also suggests a role for ubiquitylation in the regulation of gene expression at the translational level. Also characterized within this study were the different lysine linkages between ubiquitin and its protein substrates. Ubiquitin contains seven lysine residues at different positions that all can interact to form polyubiquitin chains. Linkages using Lys-6, Lys-9, Lys-11, Lys-27, Lys-33 and Lys-29 were not identified in the in silico analysis; however, Lys-6 and Lys11 were detected experimentally. Lys-48 and Lys-63 linkages were detected using both approaches65. These findings demonstrate that these four lysine linkages, Lys-6, Lys-11, Lys-48 and Lys-63, are likely the most abundant in P. falciparum (Figure 2).
The large discrepancy observed between predicted and experimentally validated ubiquitylated proteins is likely the result of challenges in isolating ubiquitylated substrates. Additional experimental studies will need to be developed to provide a more accurate depiction of the number of ubiquitylated substrates in P. falciparum. Collectively, these studies suggest that a large proportion of the Plasmodium proteome is ubiquitylated and that ubiquitylated proteins are involved in many different cellular processes and are critical contributors of the parasite life cycle progression.
Drug resistance
One of the most exciting findings from recent genome-wide studies in Plasmodium is the recent implication of the ubiquitin system in mechanisms of drug resistance. Drugs like quinine and chloroquine were once the first choice in the treatment of malaria, but since the emergence of resistant strains, novel anti-malarials are continually in demand. Much of the current knowledge surrounding the development of resistant Plasmodium strains to chloroquine, mefloquine, pyrimethamine and artemether-lumefantrine is centered on a small number of genes that mostly encode transporter proteins, which include ATPase6, DHFR, CRT and MDR168–71.
The identification of novel genetic mutations involved in drug resistance employed the use of different mapping techniques, such as linkage analysis and genome-wide association studies (GWAS). Linkage analysis entails crossing organisms with each other, and using genetic markers to calculate recombination rates to pinpoint the location of a causal mutation. Alternatively, GWAS relies on the use of custom genotyping arrays, typically using thousands of SNPs or next generation sequencing technology to sequence hundred of strains, to calculate allele frequencies between resistant and sensitive strains to identify a genetic location. Using these genome-wide methodologies, genes comprising components of ubiquitin system have been implicated in the acquisition of resistance to some of the current anti-malarial drugs, specifically pyrimethamine and artemisinin72–76.
One of the first accounts of the role of the ubiquitin system in drug resistance was the discovery of a high prevalence of mutations in resistant strains in genes encoding components of the ubiquitin system77. In a more recent study, two novel mutations were identified in both an E2 ubiquitin-conjugating enzyme and an HECT E3 ubiquitin ligase73 (Table 1). Using a sequence-based GWAS, 45 Senegalese P. falciparum parasites were sequenced, and the discovered SNPs were interrogated for association with resistance to 12 different anti-malarials. Using both a test for natural selection, known as cross-population extended haplotype homozygosity (XP-EHH), and a mixed model association test, known as EMMA, several of the previous loci conferring resistance were identified, including CRT, MDR1 and DHRF. However, one of the most interesting findings was the identification of an E2 enzyme, encoded by PF3D7_1243700 and an E3 ligase, encoded by PF3D7_0826100, both possibly involved in a mechanism of drug resistance to pyrimethamine.
Currently, artemisinin is the most effective anti-malarial, and over the last few years there has been an aggressive effort to identify the genes(s) responsible for the burgeoning of artemisinin-resistant strains. Using a genome-wide linkage group selection (LGS) analysis, missense mutations were discovered on chromosome 2 in a gene encoding a deubiquitylase (UBP-1) in the P. Chabaudi artemisinin-resistant strain AS-ATN. It was proposed that at least one of these mutations, specifically the V2728F mutation, was conferring artemisinin resistance75, 76. However, the mutation in UPB-1 has not been confirmed in P. falciparum artemisinin-resistant strains78, and additional data will be needed to confirm its role in mechanisms of drug resistance.
While the role of UBP-1 in drug resistance has not yet been validated in P. falciparum, other genes involved in the ubiquitin system have been identified as possibly involved in artemisinin resistance. An E3 ubiquitin-ligase, known as RAD5 (Table 1), has been implicated in two independent genome-wide association studies (GWAS)72, 74. Both studies showed a significant association with resistance to artemisinin in a similar location on chromosome 13. No causal mutations were confirmed in the genes interrogated in the region in either study; however, artemisinin resistance may be multifactorial and identifying perfectly correlative SNPs could be arduous. Moreover, another gene discovered in these GWAS was DNA polymerase delta (Table 1). Both of these enzymes, DNA polymerase delta and RAD5 are part of a pathway known as postreplication repair. Interestingly, UBP1 is also part of the postreplication repair pathway, and it is conceivable that genes involved in this particular pathway could drive artemisinin resistance.
Conclusion
Plasmodium spp. are incredibly pervasive and some of them are lethal human pathogens. Understanding the cellular components critical to parasite development is essential in the search for innovative therapeutic strategies. Similar to other eukaryotes, the ubiquitin system in Plasmodium is essential to parasite survival. The system is critical in numerous cellular processes, as such degradation of misfolded proteins, oxidative stress response, trafficking of proteins to the apicoplast, as well as mechanisms of drug resistance. Exploration of the ubiquitin system using genome-wide approaches has greatly advanced our understanding of this pathway in apicomplexan parasites in general and in particular Plasmodium. Moreover, these large-scale studies have demonstrated that ubiquitylation is a common post-translational modification in P. falciparum. Furthermore, the recent implications of the ubiquitin system in mechanisms of drug resistance highlight the importance of this particular pathway in parasite biology. Malaria research would greatly benefit from studies exploring the ubiquitin system that focused on components of the ubiquitin system that are parasite-specific. Successful identification of these targets will likely yield new and effective therapies to combat malaria infections. Additional in depth studies exploring the role of the ubiquitin system in the acquisition of drug resistance is another area within malaria research that strongly warrants further investigation. In the near future, we suspect more researchers will explore ubiquitylation events in Plasmodium, as we are just now scratching the surface of this overlooked pathway.
Acknowledgments
This work was supported by the National Institutes of Health (grant R01 AI85077-01A1 to KLR.
References
- 1.Eytan E, Ganoth D, Armon T, Hershko A. Proceedings of the National Academy of Sciences of the United States of America. 1989;86:7751–7755. doi: 10.1073/pnas.86.20.7751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kallio PJ, Wilson WJ, O'Brien S, Makino Y, Poellinger L. The Journal of biological chemistry. 1999;274:6519–6525. doi: 10.1074/jbc.274.10.6519. [DOI] [PubMed] [Google Scholar]
- 3.Koepp DM, Harper JW, Elledge SJ. Cell. 1999;97:431–434. doi: 10.1016/s0092-8674(00)80753-9. [DOI] [PubMed] [Google Scholar]
- 4.Guo W, Shang F, Liu Q, Urim L, West-Mays J, Taylor A. Investigative ophthalmology & visual science. 2004;45:1194–1201. doi: 10.1167/iovs.03-0830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dupre DJ, Chen Z, Le Gouill C, Theriault C, Parent JL, Rola-Pleszczynski M, Stankova J. The Journal of biological chemistry. 2003;278:48228–48235. doi: 10.1074/jbc.M304082200. [DOI] [PubMed] [Google Scholar]
- 6.Hershko A, Ciechanover A. Annual review of biochemistry. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 7.Kerscher O, Felberbaum R, Hochstrasser M. Annual review of cell and developmental biology. 2006;22:159–180. doi: 10.1146/annurev.cellbio.22.010605.093503. [DOI] [PubMed] [Google Scholar]
- 8.Komander D. Biochemical Society transactions. 2009;37:937–953. doi: 10.1042/BST0370937. [DOI] [PubMed] [Google Scholar]
- 9.Mukhopadhyay D, Riezman H. Science. 2007;315:201–205. doi: 10.1126/science.1127085. [DOI] [PubMed] [Google Scholar]
- 10.Hede SM, Savov V, Weishaupt H, Sangfelt O, Swartling FJ. Oncogene. 2013 doi: 10.1038/onc.2013.445. [DOI] [PubMed] [Google Scholar]
- 11.Crawford LJ, Irvine AE. Blood reviews. 2013 doi: 10.1016/j.blre.2013.10.002. [DOI] [PubMed] [Google Scholar]
- 12.Lopez G, Sidransky E. Biomarkers in medicine. 2010;4:713–721. doi: 10.2217/bmm.10.96. [DOI] [PubMed] [Google Scholar]
- 13.Lin Z, Zhao D, Yang L. Acta biochimica et biophysica Sinica. 2013;45:477–484. doi: 10.1093/abbs/gmt020. [DOI] [PubMed] [Google Scholar]
- 14.Manzanillo PS, Ayres JS, Watson RO, Collins AC, Souza G, Rae CS, Schneider DS, Nakamura K, Shiloh MU, Cox JS. Nature. 2013;501:512–516. doi: 10.1038/nature12566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sanchez M, Calzada A, Bueno A. The Journal of biological chemistry. 1999;274:9092–9097. doi: 10.1074/jbc.274.13.9092. [DOI] [PubMed] [Google Scholar]
- 16.Roos FC, Evans AJ, Brenner W, Wondergem B, Klomp J, Heir P, Roche O, Thomas C, Schimmel H, Furge KA, Teh BT, Thuroff JW, Hampel C, Ohh M. The American journal of pathology. 2011;178:853–860. doi: 10.1016/j.ajpath.2010.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Ree JH, Jeganathan KB, Malureanu L, van Deursen JM. The Journal of cell biology. 2010;188:83–100. doi: 10.1083/jcb.200906147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.WHO. World Malaria Report 2013. Switzerland: 2013. [Google Scholar]
- 19.Wirth CC, Pradel G. International journal of medical microbiology : IJMM. 2012;302:172–178. doi: 10.1016/j.ijmm.2012.07.003. [DOI] [PubMed] [Google Scholar]
- 20.Carruthers VB, Blackman MJ. Molecular microbiology. 2005;55:1617–1630. doi: 10.1111/j.1365-2958.2005.04483.x. [DOI] [PubMed] [Google Scholar]
- 21.Atul R Prasad, Kolla VK, Legac J, Singhal N, Navale R, Rosenthal PJ, Sijwali PS. PloS one. 2013;8:e73530. doi: 10.1371/journal.pone.0073530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klein EY. International journal of antimicrobial agents. 2013;41:311–317. doi: 10.1016/j.ijantimicag.2012.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Coppel RL, Black CG. International journal for parasitology. 2005;35:465–479. doi: 10.1016/j.ijpara.2005.01.010. [DOI] [PubMed] [Google Scholar]
- 24.Doyle S. FEMS microbiology letters. 2011;321:1–9. doi: 10.1111/j.1574-6968.2011.02292.x. [DOI] [PubMed] [Google Scholar]
- 25.Ponder EL, Bogyo M. Eukaryotic cell. 2007;6:1943–1952. doi: 10.1128/EC.00282-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ponts N, Yang J, Chung DW, Prudhomme J, Girke T, Horrocks P, Le Roch KG. PloS one. 2008;3:e2386. doi: 10.1371/journal.pone.0002386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reiter K, Mukhopadhyay D, Zhang H, Boucher LE, Kumar N, Bosch J, Matunis MJ. The Journal of biological chemistry. 2013;288:27724–27736. doi: 10.1074/jbc.M113.498410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tomlins AM, Ben-Rached F, Williams RA, Proto WR, Coppens I, Ruch U, Gilberger TW, Coombs GH, Mottram JC, Muller S, Langsley G. Autophagy. 2013;9 doi: 10.4161/auto.25832. [DOI] [PubMed] [Google Scholar]
- 29.SB Cervantes E, Saraf A, Conner C, Escalante A, Sardiu M, Ponts N, Prudhomme J, Florens L, Le Roch K. Autophagy. 2014;10 doi: 10.4161/auto.26743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hipp MS, Kalveram B, Raasi S, Groettrup M, Schmidtke G. Molecular and cellular biology. 2005;25:3483–3491. doi: 10.1128/MCB.25.9.3483-3491.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Durfee LA, Huibregtse JM. Methods in molecular biology. 2012;832:141–149. doi: 10.1007/978-1-61779-474-2_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McGrath JP, Jentsch S, Varshavsky A. The EMBO journal. 1991;10:227–236. doi: 10.1002/j.1460-2075.1991.tb07940.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen Y, Neve RL, Liu H. Journal of cellular and molecular medicine. 2012;16:2583–2591. doi: 10.1111/j.1582-4934.2012.01604.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schwienhorst I, Johnson ES, Dohmen RJ. Molecular & general genetics : MGG. 2000;263:771–786. doi: 10.1007/s004380000254. [DOI] [PubMed] [Google Scholar]
- 35.Zhao Y, Sun Y. Current pharmaceutical design. 2013;19:3215–3225. doi: 10.2174/13816128113199990300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Artavanis-Tsakonas K, Misaghi S, Comeaux CA, Catic A, Spooner E, Duraisingh MT, Ploegh HL. Molecular microbiology. 2006;61:1187–1195. doi: 10.1111/j.1365-2958.2006.05307.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sommer T, Jentsch S. Nature. 1993;365:176–179. doi: 10.1038/365176a0. [DOI] [PubMed] [Google Scholar]
- 38.Chung DW, Ponts N, Prudhomme J, Rodrigues EM, Le Roch KG. PloS one. 2012;7:e43477. doi: 10.1371/journal.pone.0043477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Harbut MB, Patel BA, Yeung BK, McNamara CW, Bright AT, Ballard J, Supek F, Golde TE, Winzeler EA, Diagana TT, Greenbaum DC. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:21486–21491. doi: 10.1073/pnas.1216016110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fennell C, Babbitt S, Russo I, Wilkes J, Ranford-Cartwright L, Goldberg DE, Doerig C. Malaria journal. 2009;8:99. doi: 10.1186/1475-2875-8-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang M, Mishra S, Sakthivel R, Rojas M, Ranjan R, Sullivan WJ, Jr, Fontoura BM, Menard R, Dever TE, Nussenzweig V. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:3956–3961. doi: 10.1073/pnas.1121567109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Agrawal S, Chung DW, Ponts N, van Dooren GG, Prudhomme J, Brooks CF, Rodrigues EM, Tan JC, Ferdig MT, Striepen B, Le Roch KG. PLoS pathogens. 2013;9:e1003426. doi: 10.1371/journal.ppat.1003426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Agrawal S, van Dooren GG, Beatty WL, Striepen B. The Journal of biological chemistry. 2009;284:33683–33691. doi: 10.1074/jbc.M109.044024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Spork S, Hiss JA, Mandel K, Sommer M, Kooij TW, Chu T, Schneider G, Maier UG, Przyborski JM. Eukaryotic cell. 2009;8:1134–1145. doi: 10.1128/EC.00083-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Foth BJ, Stimmler LM, Handman E, Crabb BS, Hodder AN, McFadden GI. Molecular microbiology. 2005;55:39–53. doi: 10.1111/j.1365-2958.2004.04407.x. [DOI] [PubMed] [Google Scholar]
- 46.Jomaa H, Wiesner J, Sanderbrand S, Altincicek B, Weidemeyer C, Hintz M, Turbachova I, Eberl M, Zeidler J, Lichtenthaler HK, Soldati D, Beck E. Science. 1999;285:1573–1576. doi: 10.1126/science.285.5433.1573. [DOI] [PubMed] [Google Scholar]
- 47.Sato S, Wilson RJ. Current genetics. 2002;40:391–398. doi: 10.1007/s00294-002-0273-3. [DOI] [PubMed] [Google Scholar]
- 48.Wilson RJ, Denny PW, Preiser PR, Rangachari K, Roberts K, Roy A, Whyte A, Strath M, Moore DJ, Moore PW, Williamson DH. Journal of molecular biology. 1996;261:155–172. doi: 10.1006/jmbi.1996.0449. [DOI] [PubMed] [Google Scholar]
- 49.Roos DS, Crawford MJ, Donald RG, Kissinger JC, Klimczak LJ, Striepen B. Current opinion in microbiology. 1999;2:426–432. doi: 10.1016/S1369-5274(99)80075-7. [DOI] [PubMed] [Google Scholar]
- 50.Waller RF, Reed MB, Cowman AF, McFadden GI. The EMBO journal. 2000;19:1794–1802. doi: 10.1093/emboj/19.8.1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van Dooren GG, Schwartzbach SD, Osafune T, McFadden GI. Biochimica et biophysica acta. 2001;1541:34–53. doi: 10.1016/s0167-4889(01)00154-9. [DOI] [PubMed] [Google Scholar]
- 52.Stoebe B, Maier UG. Protoplasma. 2002;219:123–130. doi: 10.1007/s007090200013. [DOI] [PubMed] [Google Scholar]
- 53.Sommer MS, Gould SB, Lehmann P, Gruber A, Przyborski JM, Maier UG. Molecular biology and evolution. 2007;24:918–928. doi: 10.1093/molbev/msm008. [DOI] [PubMed] [Google Scholar]
- 54.Radivojac P, Vacic V, Haynes C, Cocklin RR, Mohan A, Heyen JW, Goebl MG, Iakoucheva LM. Proteins. 2010;78:365–380. doi: 10.1002/prot.22555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tompa P, Prilusky J, Silman I, Sussman JL. Proteins. 2008;71:903–909. doi: 10.1002/prot.21773. [DOI] [PubMed] [Google Scholar]
- 56.Tung CW, Ho SY. BMC bioinformatics. 2008;9:310. doi: 10.1186/1471-2105-9-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tomlinson E, Palaniyappan N, Tooth D, Layfield R. Proteomics. 2007;7:1016–1022. doi: 10.1002/pmic.200601008. [DOI] [PubMed] [Google Scholar]
- 58.Manzano C, Abraham Z, Lopez-Torrejon G, Del Pozo JC. Plant molecular biology. 2008;68:145–158. doi: 10.1007/s11103-008-9358-9. [DOI] [PubMed] [Google Scholar]
- 59.Tan F, Lu L, Cai Y, Wang J, Xie Y, Wang L, Gong Y, Xu BE, Wu J, Luo Y, Qiang B, Yuan J, Sun X, Peng X. Proteomics. 2008;8:2885–2896. doi: 10.1002/pmic.200700887. [DOI] [PubMed] [Google Scholar]
- 60.Hjerpe R, Aillet F, Lopitz-Otsoa F, Lang V, England P, Rodriguez MS. EMBO reports. 2009;10:1250–1258. doi: 10.1038/embor.2009.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Golebiowski F, Tatham MH, Nakamura A, Hay RT. Nature protocols. 2010;5:873–882. doi: 10.1038/nprot.2010.40. [DOI] [PubMed] [Google Scholar]
- 62.Kim W, Bennett EJ, Huttlin EL, Guo A, Li J, Possemato A, Sowa ME, Rad R, Rush J, Comb MJ, Harper JW, Gygi SP. Molecular cell. 2011;44:325–340. doi: 10.1016/j.molcel.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wagner SA, Beli P, Weinert BT, Nielsen ML, Cox J, Mann M, Choudhary C. Molecular & cellular proteomics : MCP. 2011;10 doi: 10.1074/mcp.M111.013284. M111 013284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xu G, Paige JS, Jaffrey SR. Nature biotechnology. 2010;28:868–873. doi: 10.1038/nbt.1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ponts N, Saraf A, Chung DW, Harris A, Prudhomme J, Washburn MP, Florens L, Le Roch KG. The Journal of biological chemistry. 2011;286:40320–40330. doi: 10.1074/jbc.M111.238790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kislinger T, Emili A. Expert review of proteomics. 2005;2:27–39. doi: 10.1586/14789450.2.1.27. [DOI] [PubMed] [Google Scholar]
- 67.Matsumoto M, Hatakeyama S, Oyamada K, Oda Y, Nishimura T, Nakayama KI. Proteomics. 2005;5:4145–4151. doi: 10.1002/pmic.200401280. [DOI] [PubMed] [Google Scholar]
- 68.Legrand E, Volney B, Meynard JB, Mercereau-Puijalon O, Esterre P. Antimicrobial agents and chemotherapy. 2008;52:288–298. doi: 10.1128/AAC.00263-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Andriantsoanirina V, Durand R, Pradines B, Baret E, Bouchier C, Ratsimbasoa A, Menard D. Malaria journal. 2011;10:283. doi: 10.1186/1475-2875-10-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Djimde A, Doumbo OK, Cortese JF, Kayentao K, Doumbo S, Diourte Y, Coulibaly D, Dicko A, Su XZ, Nomura T, Fidock DA, Wellems TE, Plowe CV. The New England journal of medicine. 2001;344:257–263. doi: 10.1056/NEJM200101253440403. [DOI] [PubMed] [Google Scholar]
- 71.Borges S, Cravo P, Creasey A, Fawcett R, Modrzynska K, Rodrigues L, Martinelli A, Hunt P. Antimicrobial agents and chemotherapy. 2011;55:4858–4865. doi: 10.1128/AAC.01748-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cheeseman IH, Miller BA, Nair S, Nkhoma S, Tan A, Tan JC, Al Saai S, Phyo AP, Moo CL, Lwin KM, McGready R, Ashley E, Imwong M, Stepniewska K, Yi P, Dondorp AM, Mayxay M, Newton PN, White NJ, Nosten F, Ferdig MT, Anderson TJ. Science. 2012;336:79–82. doi: 10.1126/science.1215966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Park DJ, Lukens AK, Neafsey DE, Schaffner SF, Chang HH, Valim C, Ribacke U, Van Tyne D, Galinsky K, Galligan M, Becker JS, Ndiaye D, Mboup S, Wiegand RC, Hartl DL, Sabeti PC, Wirth DF, Volkman SK. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:13052–13057. doi: 10.1073/pnas.1210585109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Takala-Harrison S, Clark TG, Jacob CG, Cummings MP, Miotto O, Dondorp AM, Fukuda MM, Nosten F, Noedl H, Imwong M, Bethell D, Se Y, Lon C, Tyner SD, Saunders DL, Socheat D, Ariey F, Phyo AP, Starzengruber P, Fuehrer HP, Swoboda P, Stepniewska K, Flegg J, Arze C, Cerqueira GC, Silva JC, Ricklefs SM, Porcella SF, Stephens RM, Adams M, Kenefic LJ, Campino S, Auburn S, MacInnis B, Kwiatkowski DP, Su XZ, White NJ, Ringwald P, Plowe CV. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:240–245. doi: 10.1073/pnas.1211205110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hunt P, Afonso A, Creasey A, Culleton R, Sidhu AB, Logan J, Valderramos SG, McNae I, Cheesman S, do Rosario V, Carter R, Fidock DA, Cravo P. Molecular microbiology. 2007;65:27–40. doi: 10.1111/j.1365-2958.2007.05753.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hunt P, Martinelli A, Modrzynska K, Borges S, Creasey A, Rodrigues L, Beraldi D, Loewe L, Fawcett R, Kumar S, Thomson M, Trivedi U, Otto TD, Pain A, Blaxter M, Cravo P. BMC genomics. 2010;11:499. doi: 10.1186/1471-2164-11-499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Volkman SK, Neafsey DE, Schaffner SF, Park DJ, Wirth DF. Nature reviews. Genetics. 2012;13:315–328. doi: 10.1038/nrg3187. [DOI] [PubMed] [Google Scholar]
- 78.Imwong M, Dondorp AM, Nosten F, Yi P, Mungthin M, Hanchana S, Das D, Phyo AP, Lwin KM, Pukrittayakamee S, Lee SJ, Saisung S, Koecharoen K, Nguon C, Day NP, Socheat D, White NJ. Antimicrobial agents and chemotherapy. 2010;54:2886–2892. doi: 10.1128/AAC.00032-10. [DOI] [PMC free article] [PubMed] [Google Scholar]