Abstract
Histone deacetylases (HDACs) catalyze the deacetylation of lysine residues on histones and non-histone proteins. HDACs have been shown to control the functions of key cell cycle proteins. Consistent with this, the overexpression of HDACs has been observed in multiple cancers, resulting in deregulation of the cell cycle and uncontrolled proliferation. This review focuses on the impact that HDACs have on cell cycle control through the deacetylation of proteins.
Introduction
One of the dynamic natures of chromatin relies on posttranslational modification of its core components, histones. The modification of histones through methylation, phosphorylation, SUMOylation, ubiquitination, acetylation, etc. makes up the histone code that provides the necessary information for gene activation or repression (1–3). Acetylation, which is counterbalanced by deacetylation, is one of the most extensively characterized posttranslational histone modifications (3). The two opposing reactions act in tandem to achieve a closed or open conformation of chromatin. The open conformation results from the addition of acetyl groups to the lysine residues of histone tails by a class of proteins named histone acetyltransferases (HATs) (4). Acetylation relaxes higher order chromatin structure and, in turn, provides access to proteins, such as transcription factors. The removal of the acetyl group, called deacetylation, is catalyzed by a class of highly conserved proteins termed histone deacetylases (HDACs). Histone acetylation/deacetylation paves the way for the recruitment of additional factors and ultimately results in the remodeling of chromatin to activate or repress transcription.
Histone deacetylases were first identified by their ability to deacetylate specific lysine residues on histones. However, HDACs also catalyze the removal of acetyl groups from non-histone proteins. Using a proteomics approach, Choudhary et al. have identified a number of acetylated proteins and potential non-histone targets of HDACs that function in various cellular processes, such as cell cycle regulation, DNA repair, apoptosis, proliferation, and differentiation (5). Thus, HDACs act as a fulcrum to support multiple cellular processes by their ability to regulate protein expression and function. Evidence for this can be garnered from the variety of cell type-specific effects of HDAC inhibitors. Due to their ability to induce a wide range of cytotoxic effects in tumor cells, including apoptosis, cell cycle arrest, and inhibition of proliferation, HDAC inhibitors are actively being developed as potential anti-cancer drugs (6–8).
Using a trapoxin (an inhibitor of histone deacetylase) affinity matrix, Schreiber and colleagues purified and cloned a human 55 kDa protein related to the yeast protein RPD3 (9). Enzymatic assay of this 55 kDa protein, HDAC1, showed that it contains histone deacetylase activity. To date, 18 HDACs have been identified and divided into four classes based on homology to their yeast counterparts (10–13): Classes I, II, III, and IV. Classes I, II, and IV are zinc-dependent HDACs, whereas Class III are nicotinamide adenine dinucleotide-dependent enzymes (reviewed in ref. 13). Class I HDACs, homologous to yeast Rpd3 (14), comprise of HDACs 1–3 and 8. Some Class I HDACs are exclusively nuclear, while others are located both in the nucleus and cytoplasm. Class II HDACs are further subdivided into Class IIa, which consists of HDACs 4, 5, 7, and 9, and Class IIb, which consists of HDACs 6 and 10. Class II HDACs contain an N-terminal NLS and a C-terminal NES and localize to the nucleus and cytoplasm (15). Class II HDACs are homologous to the yeast HDA1 protein and many show tissue specificity (16). Most Class I and some Class II HDACs exist as part of multimeric repressor protein complexes, such as Sin3, NuRD, NCoR (nuclear receptor corepressor), and SMRT (Silencing mediator for retinoid and thyroid receptors) [reviewed in ref. 17, 18]. Class III HDACs comprise the nicotinamide adenine dinucleotide-dependent HDACs, also known as sirtuins due to their homology to the Sir2 proteins of yeast (19). Due to the dependence of Class III HDACs on the metabolic cofactor nicotinamide adenine dinucleotide, their activity is closely linked to the metabolic state of the cell and is known to regulate lifespan (20–23). There are seven members of the sirtuin family, Sirt 1–7. Sirt 1, 6, and 7 are predominantly nuclear in their localization (24). Sirt2 is predominantly cytoplasmic, while Sirt 3, 4, and 5 are localized to the mitochondria (24, 25). Due to their spatial differences, sirtuins display specialized functions in the cell. Some sirtuins possess ADP ribosyl transferase activity, and only Sirtuins 1–3, and 5 have been shown to possess a conserved deacetylase domain (20, 26). HDAC11 is the only member of Class IV HDACs, and it shares a conserved deacetylase domain with Class I and Class II HDAC members (12).
A vast number of acetylated proteins and, therefore, potential non-histone targets of HDACs have recently been identified using proteomics approaches. Deacetylation of non-histone proteins by HDACs mainly affects the target protein stability, DNA binding, or enzyme activity (27). HDACs are known to deacetylate a wide variety of proteins, including those involved in the cell cycle. This review focuses on the regulation of cell cycle machinery by HDACs through histone modification of promoter regions and/or by direct deacetylation of target non-histone proteins.
The cell cycle consists of G1, S, G2, and M phases. The alternation between S phase and M phase is essential for genomic integrity. Progression from one phase to the other is mediated by cyclins and their associated cyclin dependent kinases (Cdks) (28). The Cdks are in turn regulated by Cdk inhibitors, including p21and p27 (29). To ensure faithful passage through the cell cycle, the cell is equipped with regulatory mechanisms, known as checkpoints, that monitor the transition from one phase to the other and halt progression of the cycle in response to DNA damage or replication stress (30). The activation of the checkpoint pathway results in the phosphorylation and activation of checkpoint proteins, such as ATM, ATR, CHK1, CHK2, and p53 (31, 32). In addition to phosphorylation, the significance of acetylation/deacetylation in the regulation of cell cycle proteins is beginning to be appreciated. HDACs deacetylate and regulate the activity of key cell cycle proteins, such as p53, E2F, and pRb (reviewed in ref. 27). Deregulation of any of these proteins or their pathway results in neoplastic transformation. Here, we discuss the role of various HDACs in the regulation of cell cycle progression.
HDACs and the G1-S transition
The G1 to S transition is controlled by the tumor suppressor, retinoblastoma protein (pRb) (33). pRb interacts with members of the E2F family to drive cell cycle progression (34). There are nine E2F family members of which the activating E2Fs (E2F 1–3) are required for S phase progression and apoptosis (35). E2F4 and E2F5 are known as repressive E2Fs and function to repress the cell cycle at the G0/G1 phase of the cell cycle (36). E2Fs are transcription factors that when activated stimulate entry into the cell cycle. The E2F-responsive genes play a crucial role in cell cycle progression. These genes include cdc6, mcm proteins, PCNA, DNA polymerase α, and thymidine kinase (37, 38). The transcriptional activity of E2Fs (E2F 1–5) is suppressed by its association with the pocket family of proteins, including pRb (39). The phosphorylation of pRb by cyclin-cdk complex dissociates pRb from E2F and relieves the repression of E2F responsive genes (40). pRb represses E2F-mediated transcription by recruiting HDACs to the E2F-responsive promoter. pRb binds to HDAC1 and E2F via two distinct sites. Hence, HDAC1 is simultaneously recruited by pRb to E2F-responsive promoters (41, 42). Lai et al. has shown that the association between HDAC1 and pRb is not direct and that RBP1 forms a bridge to facilitate the interaction (43). In addition to HDAC1, Class I HDACs, HDAC2 and HDAC3, but not Class II HDACs, are recruited by RBP1 via the Sin3 complex to form a multimeric corepressor complex consisting of E2F-pRb-RBP1-Sin3-HDAC at the promoter regions of target genes, which affects the repression of those genes through the deacetylation of histone lysine residues (43).
Sin3-HDAC corepressor complex and the cell cycle
In mammals, two Sin3 isoforms exist, mSin3A and mSin3B, that contain conserved paired amphipathic helix (PAH) motifs that mediate protein-protein interactions (44). The mammalian Sin3 proteins, initially identified as MAD interacting proteins, were required for transcriptional repression, and have distinct functions in the regulation of cell cycle, DNA repair proliferation, and apoptosis (45, 46). The Sin3 complex is recruited by transcription factors/DNA binding proteins to repress the transcription of target genes (46). Class I HDACs form an integral component of the Sin3 repressor complex, and HDAC activity is essential for complete transcriptional repression (46, 47). The involvement of the Sin3 complex in cell cycle regulation and exit has been well documented in Sin3A−/− and Sin3B−/− embryos. Sin3A−/− MEFS show cell cycle defects at all stages, including a block in DNA synthesis, G2/M arrest, and apoptosis. mSin3A−/− cells display increased expression of Myc-, E2F1-, and p53-responsive genes, such as cyclin D2, cyclin E, mcm7, cdc2A, mcm4, mcm5, securin, 53BP1, and Gadd45-γ, indicating an inability of transcription factors to recruit the mSin3/HDAC repressor complex and subsequently upregulate target genes leading to cell cycle defects (46). Aside from this, Sin3A−/− MEFs induce the expression of p21, a cdk inhibitor, through the acetylation of histones at the p21 promoter, indicating the existence of a p53-independent mechanism that regulates p21 expression by the mSin3A-HDAC corepressor complex (46).
In contrast to the cell cycle arrest associated with Sin3A loss, loss of Sin3B has no profound effect on the cell cycle. However, Sin3B is required for cell cycle exit, as Sin3B−/− MEFs fail to arrest in G0 upon serum deprivation (48). Sin3B is involved in the repression of E2F4 target genes that control the G0/G1 stage of the cell cycle (48). Thus, HDACs regulate cell cycle progression by associating with multicomponent corepressor complexes to alter the expression of cell cycle regulatory genes.
E2F and HDAC
In addition to the repressive effect of HDACs caused by the deacetylation of histone residues and the resulting change in chromatin conformation, HDACs are also capable of directly inhibiting protein function by deacetylating target non-histone proteins. E2Fs are non-histone targets of HATs and HDACs. The acetylation of E2F1 by p300 and CBP results in increased DNA binding and transcriptional activation of E2F1, which is reversed by the deacetylation of the same residues by HDAC1 (49, 50). Of the E2F family, only E2F1–3 have been reported to be acetylated by p300 (50). Thus, HDACs modulate cell cycle progression by regulating E2F family-mediated transcription via two mechanisms: first, by modification of chromatin at the promoter of E2F-responsive gene and second, by directly deacetylating E2F and suppressing its activity.
Loss of HDACs and cell cycle progression
The absence of HDAC1 in mouse embryonic stem cells (ES) results in reduced cell proliferation. Further investigation of HDAC1−/− cells have attributed the proliferation defect to increased levels of p21, since loss of p21 rescues this phenotype of HDAC1 null ES cells. HDAC1 mediates repression of p21 by localizing to and deacetylating histones in the p21 promoter region, resulting in inhibition of p21 gene expression (51). Increased expression of p27 was also observed in HDAC1 null cells, which further contributes to the proliferation defect displayed by HDAC1 null ES cells (51, 52.
Double knockout (DKO) of HDAC2 and HDAC1 MEFS revealed the functional redundancy between HDAC1 and HDAC2 in controlling the G1-S transition, as expression of either HDAC1 or HDAC2 is sufficient to rescue the growth arrest phenotype observed in DKO MEFS. Also, knockout of HDAC2 results in increased levels of HDAC1 protein and vice versa (53). Thus, it can be inferred that both HDAC2 and HDAC1 cooperatively regulate cell cycle progression by repressing p21 transcription.
The role of HDAC3 during DNA replication has been demonstrated by decreased BrdU uptake and impaired cycling of HDAC3 null cells. HDAC3 null cells display defects in DNA double strand break repair elicited during replication stress and S phase-associated DNA damage. Defective DNA repair results in the accumulation of DNA damage and triggers the S phase checkpoint leading to S phase arrest. HDAC3 null cells also display increased acetylation of histones H3 and H4 (54). Newly synthesized H3 and H4 are acetylated prior to their deposition on newly synthesized DNA. These histones must be deacetylated during the process of chromatin maturation, failure of which results in a defect in chromatin assembly that could lead to DNA damage. Similar to HDAC3, a role for HDAC1 in nucleosome assembly is indicated by its association with PCNA. PCNA is a protein that mediates protein-protein interactions and recruit proteins to DNA to coordinate events in replication, repair, epigenetic inheritance, and cell cycle control (55). PCNA associates with HDAC1 to link histone deacetylation of newly assembled nucleosomes with DNA synthesis (56). It could be speculated that PCNA interacts with other HDACs, such as HDAC3, to achieve proper chromatin assembly following DNA replication.
HDACs and cancer cell division
The overexpression of Class I HDACs, HDAC1, HDAC2, and HDAC3 has been implicated in multiple human cancers. Consistent with this, treatment of cancer cells with HDAC inhibitors induces growth arrest, differentiation, and apoptosis (57–61). Class I and Class II HDAC inhibitors increase the expression of p21, an inhibitor of the cyclin-cdk complex that is required for cell cycle progression (62). The contribution of individual HDACs in the regulation of p21 expression was measured by knockdown of HDACs in tumor cells. The p21 promoter contains six conserved Sp1/Sp3 binding sites that control the transcriptional activation of p21. Mutation or deletion of these sites leads to a marked reduction in p21 reporter activity. A positive effect on transcription is mediated by the association of p53 or E2F1 with Sp1 (63, 64), and negative regulation is achieved by binding of Sp1 to HDACs.
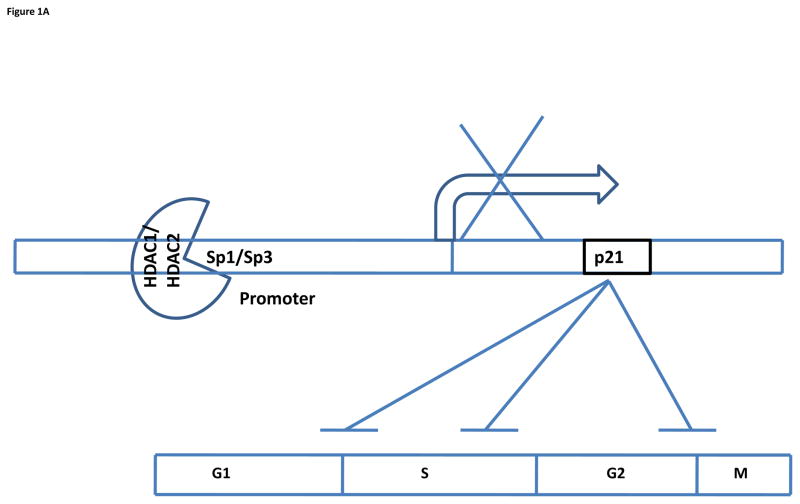
The regulation of p21 expression by HDAC1 has been demonstrated in U2OS cells. In U2OS cells, HDAC1 competes with p53 for binding to Sp1 and is recruited by Sp1 to the p21 promoter consequently regulating Sp1-dependent activation of p21 (64). HDAC2 also binds to the Sp1/Sp3 transcription factor to regulate p21 expression (65) (Figure 1A).
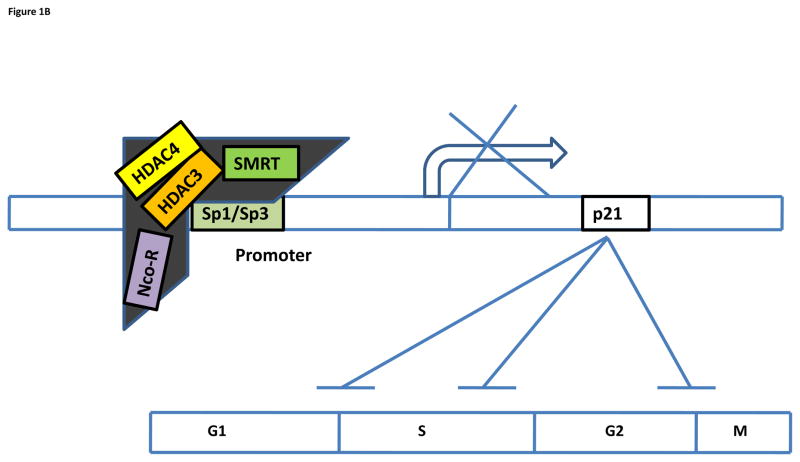
Figure 1.
(A) Repression of p21 gene transcription by HDAC1/2. p21 inhibits the cyclin-cdk complex at the indicated phases (G1/S, S, and G2/M) to block cell cycle progression. HDAC1/2 binds to Sp1/Sp3 and is recruited to the promoter site of p21, leading to hypoacetylation of the promoter region and inhibition of p21 transcription. (B) Repression of p21 gene transcription by HDAC3/4. HDAC3/4 is recruited to the promoter site by binding to the Sp1/3 transcription factor as part of a larger repressor complex, NCo-R-SMRT, to inhibit p21 transcription.
Wilson et al. have tested the role of HDAC3 in colon cancer. HDAC3 expression is restricted to the proliferative compartment of the mouse small intestine and colon and regulates proliferation by repressing p21 expression (66). Like other Class I HDACs, HDAC3 represses p21 transcription in a Sp1/Sp3-dependent manner. HDAC3 is recruited to the Sp1/3 sites of the p21 promoter as part of the N-CoR/SMRT repressor complex that also contains HDAC4 and HDAC5, indicating that Class I and Class II HDACs act together in repressing p21 gene transcription (67–70) (Figure 1B). A similar role for HDAC3 in the repression of E2F-dependent gene transcription has been observed in primary differentiated neurons (71).
Likewise, a role for HDAC4 in controlling colon cancer cell proliferation through the repression of p21 transcription has been reported. HDAC4 forms part of the HDAC4-HDAC3-N-CoR/SMRT corepressor complex that represses p21 transcription by associating with Sp1 at the proximal p21 promoter and repressing Sp1 transcription. The importance of HDAC4 in the corepressor complex is highlighted by the fact that downregulation of HDAC4 markedly reduces HDAC3 association at the p21 promoter site. This is indicative of a scaffolding role for HDAC4 in mediating the repression of p21, since the deacetylase activity of the repressor complex is contributed by HDAC3 (72).
The involvement of HDAC3 in the control of the G1-S transition in resting T cells has been revealed by its association with pro-IL-16 (73). Pro-IL-16 is a PDZ domain-containing protein that is expressed in T cells and functions to repress Skp2 transcription, leading to increased levels of p27 and a subsequent block in the G0/G1 phase of the cell cycle (74). Pro-IL-16 recruits HDAC3 to the Skp2 promoter by binding to the GA binding protein β1 subunit (GABP) transcription factor in quiescent cells, resulting in the repression of Skp2 (73). Skp2 is a component of the SCF ubiquitin ligase and mediates the degradation of p27 (75,76). Thus, pro-IL-16 maintains T cells in a G0/G1 arrested state by recruiting the GABP-HDAC3 complex to the Skp2 promoter prior to T-cell activation. Following T-cell activation, pro-IL-16 is cleaved by caspases, thereby relieving the repression of Skp2 and promoting cell cycle progression.
HDAC7 is expressed during the early stages of embryogenesis and is important for the regulation of endothelial cell cycle progression. Overexpression, as well as knockdown, of HDAC7 in human umbilical vein endothelial cells (HuVECs) results in the downregulation of cyclin D1, suggesting that cell cycle progression is intricately dependent on the level of HDAC7. Knockdown of HDAC7 also causes an increase in Rb protein levels, leading to the inhibition of E2F activity and cell cycle elongation at the G1 phase (77). HDAC7 participates in the repression of gene transcription via recruitment of corepressors N-CoR and SMRT to the nucleus. This indicates that HDAC7 downregulates cyclin D1 expression as part of a larger repressor complex.
HDACs and p53
p53 was the first reported nonhistone target of p300/CBP (78). p53 is a tumor suppressor transcription factor that is activated in response to genotoxic stress. Activation of p53 halts cell cycle progression. During cell cycle progression in unperturbed conditions, p53 levels are maintained at low levels by the E3 ligase, MDM2 (79). Modification of p53 by phosphorylation and acetylation has been observed to increase p53 protein stability and activity. Acetylation of p53 at six lysine residues, including K320, K373, and K382 by CBP increases the stability of p53 (80). In addition, HDAC inhibitor, TSA increases p53 stability (80). This suggested a possible role for HDACs in activating the p53 pathway. The relevant HDACs in p53 regulation were subsequently identified. Class I HDACs and Class III HDACs have been implicated in the regulation of p53 function. HDAC1, but not HDACs 2, 3, 4, 5, or 7, is capable of deacetylating p53 on K382, K320, and K373. Deacetylaton of p53 by HDAC1 is mediated by MDM2, since HDAC1 has no appreciable effect in the absence of MDM2 in regulating p53 function. Binding, as well as deacetylation, of p53 is enhanced in the presence of MDM2. Thus, MDM2 and HDAC1 cooperatively suppress p300-mediated acetylation of p53 thus affecting p53 stability and activity (80).
In addition to HDAC1, Sirt1, a Class III HDAC, has been reported to bind and deacetylate p53 and effect its activity (81, 82). Sirt1 has been shown to deacetylate p53 on K382, resulting in the suppression of radiation-induced p53 acetylation and impaired transcriptional activity (81). Recently, the ability of Sirt3 to deacetylate p53 in the mitochondrial compartment has been reported in EJ bladder carcinoma cells that are naïve for p53 function. This study revealed a novel mechanism of sirtuin regulation of mitochondrial p53 (83). However, this study utilized an inducible system for p53 expression, and the in vivo interaction and deacetylation of p53 by Sirt3 in the mitochondria needs to be confirmed.
Conclusions and perspectives
A summary of HDAC regulation of the cell cycle is presented in Table 1. HDACs act to promote cell cycle progression and proliferation by inhibiting the expression and the activities of important cell cycle modulators, including p53, pRb, E2F, p21, and p27 resulting in increased proliferation (27). Hence, inhibitors of HDACs have been explored as potential anti-cancer agents. Much of the knowledge obtained about the regulation of cell cycle progression by HDACs stems from the use of HDAC inhibitors. Treatment of cells with HDAC inhibitors has profound cellular effects, including cell cycle arrest, differentiation, and apoptosis. HDAC inhibitors have shown tremendous potential in the treatment of cancer and are currently being employed in clinical trials. However, a better understanding of the biological function of each HDAC will enable the development of a more efficient HDACi treatment with fewer undesirable side effects. For instance, the treatment of patients with HDACi has undesirable hematological side effects. This is due to the fact that treatment with HDAC inhibitors results in a loss of HDAC 1 and 2 activity, which leads to hematopoietic defects since HDAC1 and HDAC2 have a critical role in hematopoiesis (53). Despite having overlapping functions due to their ability to heterodimerize with each other, increased expression of HDAC2 in HDAC1 null cells cannot compensate for the loss of HDAC1 function, indicating that the deacetylases have distinct functions (52). Although a certain level of redundancy is shared by HDACs, the differences in localization and targets of HDACs indicate that they have specialized cell-type specific functions. Hence, in addition to their role in tumorigenesis, a comprehensive understanding of the functions of HDACs in the cell cycle with respect to the cell environment is necessary for effective design and use of HDAC inhibitors.
Table 1.
Summary of HDAC regulation of cell cycle. HDACs promote cell cycle progression by repressing protein function via repression of gene transcription at the promoter level, and by direct deacetylation of key cell cycle regulators.
| Deacetylation of histones at promoter region | |
|---|---|
| Promoter/gene | HDAC |
| p21 | HDAC1, HDAC2, HDAC3, HDAC4 |
| Skp2 | HDAC3 |
| E2F/Rb | HDAC1, HDAC2, HDAC3 |
| Deacetylation of non-histone proteins | |
| Protein | HDAC |
| E2F | HDAC1 |
| p53 | HDAC1, Sirt1 |
Acknowledgments
We apologize to all investigators whose work was not cited in this article due to space limitations. HDAC-related research in our laboratory is supported by grants from the National Institutes of Health (GM081650), the American Heart Association (0755298B), and an endowment from the Kaul Foundation.
References
- 1.Isenberg I. Histones. Annu Rev Biochem. 1979;48:159–91. doi: 10.1146/annurev.bi.48.070179.001111. [DOI] [PubMed] [Google Scholar]
- 2.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–5. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 3.Wolffe AP, Hayes JJ. Chromatin disruption and modification. Nucleic Acids Res. 1999;27:711–20. doi: 10.1093/nar/27.3.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sterner DE, Berger SL. Acetylation of histones and transcription-related factors. Microbiol Mol Biol Rev. 2000;64:435–459. doi: 10.1128/mmbr.64.2.435-459.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–40. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 6.Marks PA, Richon VM, Rifkind RA. Histone deacetylase inhibitors: inducers of differentiation or apoptosis of transformed cells. J Natl Cancer Inst. 2000;92:1210–6. doi: 10.1093/jnci/92.15.1210. [DOI] [PubMed] [Google Scholar]
- 7.Marks PA, Rifkind RA, Richon VM, Breslow R. Inhibitors of histone deacetylase are potentially effective anticancer agents. Clin Cancer Res. 2001;7:759–60. [PubMed] [Google Scholar]
- 8.Marks PA. The clinical development of histone deacetylase inhibitors as targeted anticancer drugs. Expert Opin Investig Drugs. 2010;19:1049–1066. doi: 10.1517/13543784.2010.510514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Taunton J, Hassig CA, Schreiber SL. A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science. 1996;272:408–411. doi: 10.1126/science.272.5260.408. [DOI] [PubMed] [Google Scholar]
- 10.Yang XJ, Seto E. Collaborative spirit of histone deacetylases in regulating chromatin structure and gene expression. Curr Opin Genet Dev. 2003;13:143–153. doi: 10.1016/s0959-437x(03)00015-7. [DOI] [PubMed] [Google Scholar]
- 11.Gregoretti IV, Lee YM, Goodson HV. Molecular evolution of the histone deacetylase family: functional implications of phylogenetic analysis. J Mol Biol. 2004;338:17–31. doi: 10.1016/j.jmb.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 12.Yang XJ, Seto E. The Rpd3/Hda1 family of lysine deacetylases: from bacteria and yeast to mice and men. Nat Rev Mol Cell Biol. 2008;9:206–18. doi: 10.1038/nrm2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peserico A, Simone C. Physical and functional HAT/HDAC interplay regulates protein acetylation balance. J Biomed Biotechnol. doi: 10.1155/2011/371832. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vidal M, Gaber RF. RPD3 encodes a second factor required to achieve maximum positive and negative transcriptional states in Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:6317–27. doi: 10.1128/mcb.11.12.6317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McKinsey TA, Zhang CL, Lu J, Olson EN. Signal-dependent nuclear export of a histone deacetylase regulates muscle differentiation. Nature. 2000;408:106–11. doi: 10.1038/35040593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang XJ, Gregoire S. Class II histone deacetylases: from sequence to function, regulation, and clinical implication. Mol Cell Biol. 2005;25:2873–84. doi: 10.1128/MCB.25.8.2873-2884.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cress WD, Seto E. Histone deacetylases, transcriptional control, and cancer. J Cell Physiol. 2000;184:1–16. doi: 10.1002/(SICI)1097-4652(200007)184:1<1::AID-JCP1>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 18.Ahringer J. NuRD and SIN3 histone deacetylase complexes in development. Trends Genet. 2000;16:351–6. doi: 10.1016/s0168-9525(00)02066-7. [DOI] [PubMed] [Google Scholar]
- 19.Denu JM. The Sir 2 family of protein deacetylases. Curr Opin Chem Biol. 2005;9:431–40. doi: 10.1016/j.cbpa.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 20.Michan S, Sinclair D. Sirtuins in mammals: insights into their biological function. Biochem J. 2007;404:1–13. doi: 10.1042/BJ20070140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Donmez G, Guarente L. Aging and disease: connections to sirtuins. Aging Cell. 2010;9:285–290. doi: 10.1111/j.1474-9726.2010.00548.x. [DOI] [PubMed] [Google Scholar]
- 22.Canto C, Auwerx J. Caloric restriction, SIRT1 and logevity. Trends Endocrinol Metab. 2009;20:325–331. doi: 10.1016/j.tem.2009.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Finkel T, Deng CX, Mostoslavsky R. Recent progress in the biology and physiology of sirtuins. Nature. 460:587–591. doi: 10.1038/nature08197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Michishita E, Park JY, Burneskis JM, Barrett JC, Horikawa I. Evolutionarily conserved and nonconserved cellular localizations and functions of human SIRT proteins. Mol Biol Cell. 2005;16:4623–35. doi: 10.1091/mbc.E05-01-0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schwer B, North BJ, Frye RA, Ott M, Verdin E. The human silent information regulator (Sir)2 homologue hSIRT3 is a mitochondrial nicotinamide adenine dinucleotide-dependent deacetylase. J Cell Biol. 2002;158:647–57. doi: 10.1083/jcb.200205057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yamamoto H, Schoonjans K, Auwerx J. Sirtuin functions in health and disease. Mol Endocrinol. 2007;21:1745–55. doi: 10.1210/me.2007-0079. [DOI] [PubMed] [Google Scholar]
- 27.Glozak MA, Sengupta N, Zhang X, Seto E. Acetylation and deacetylation of non-histone proteins. Gene. 2005;363:15–23. doi: 10.1016/j.gene.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 28.Nurse P. Ordering S phase and M phase in the cell cycle. Cell. 1994;79:547–50. doi: 10.1016/0092-8674(94)90539-8. [DOI] [PubMed] [Google Scholar]
- 29.Sherr CJ, Roberts JM. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 1999;13:1501–12. doi: 10.1101/gad.13.12.1501. [DOI] [PubMed] [Google Scholar]
- 30.Pearce AK, Humphrey TC. Integrating stress-response and cell-cycle checkpoint pathways. Trends Cell Biol. 2001;11:426–33. doi: 10.1016/s0962-8924(01)02119-5. [DOI] [PubMed] [Google Scholar]
- 31.Abraham RT. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev. 2001;15:2177–96. doi: 10.1101/gad.914401. [DOI] [PubMed] [Google Scholar]
- 32.Bartek J, Lukas J. Chk1 and Chk2 kinases in checkpoint control and cancer. Cancer Cell. 2003;3:421–9. doi: 10.1016/s1535-6108(03)00110-7. [DOI] [PubMed] [Google Scholar]
- 33.Weinberg RA. The retinoblastoma protein and cell cycle control. Cell. 1995;81:323–30. doi: 10.1016/0092-8674(95)90385-2. [DOI] [PubMed] [Google Scholar]
- 34.Muller H, Helin K. The E2F transcription factors: key regulators of cell proliferation. Biochim Biophys Acta. 2000;1470:M1–12. doi: 10.1016/s0304-419x(99)00030-x. [DOI] [PubMed] [Google Scholar]
- 35.Stevaux O, Dyson NJ. A revised picture of the E2F transcriptional network and RB function. Curr Opin Cell Biol. 2002;14:684–91. doi: 10.1016/s0955-0674(02)00388-5. [DOI] [PubMed] [Google Scholar]
- 36.Trimarchi JM, Lees JA. Sibling rivalry in the E2F family. Nat Rev Mol Cell Biol. 2002;3:11–20. doi: 10.1038/nrm714. [DOI] [PubMed] [Google Scholar]
- 37.Leone G, DeGregori J, Yan Z, Jakoi L, Ishida S, Williams RS, Nevins JR. E2F3 activity is regulated during the cell cycle and is required for the induction of S phase. Genes Dev. 1998;12:2120–30. doi: 10.1101/gad.12.14.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.DeGregori J, Kowalik T, Nevins JR. Cellular targets for activation by the E2F1 transcription factor include DNA synthesis- and G1/S-regulatory genes. Mol Cell Biol. 1995;15:4215–24. doi: 10.1128/mcb.15.8.4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bagchi S, Weinmann R, Raychaudhuri P. The retinoblastoma protein copurifies with E2F-I, an E1A-regulated inhibitor of the transcription factor E2F. Cell. 1991;65:1063–72. doi: 10.1016/0092-8674(91)90558-g. [DOI] [PubMed] [Google Scholar]
- 40.Lukas J, Bartkova J, Rohde M, Strauss M, Bartek J. Cyclin D1 is dispensable for G1 control in retinoblastoma gene-deficient cells independently of cdk4 activity. Mol Cell Biol. 1995;15:2600–11. doi: 10.1128/mcb.15.5.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brehm A, Miska EA, McCance DJ, Reid JL, Bannister AJ, Kouzarides T. Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature. 1998;391:597–601. doi: 10.1038/35404. [DOI] [PubMed] [Google Scholar]
- 42.Magnaghi-Jaulin L, Groisman R, Naguibneva I, Robin P, Lorain S, Le Villain JP, Troalen F, Trouche D, Harel-Bellan A. Retinoblastoma protein represses transcription by recruiting a histone deacetylase. Nature. 1998;391:601–5. doi: 10.1038/35410. [DOI] [PubMed] [Google Scholar]
- 43.Lai A, Kennedy BK, Barbie DA, Bertos NR, Yang XJ, Theberge MC, Tsai SC, Seto E, Zhang Y, Kuzmichev A, Lane WS, Reinberg D, Harlow E, Branton PE. RBP1 recruits the mSIN3-histone deacetylase complex to the pocket of retinoblastoma tumor suppressor family proteins found in limited discrete regions of the nucleus at growth arrest. Mol Cell Biol. 2001;21:2918–32. doi: 10.1128/MCB.21.8.2918-2932.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang H, Clark I, Nicholson PR, Herskowitz I, Stillman DJ. The Saccharomyces cerevisiae SIN3 gene, a negative regulator of HO, contains four paired amphipathic helix motifs. Mol Cell Biol. 1990;10:5927–36. doi: 10.1128/mcb.10.11.5927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ayer DE, Lawrence QA, Eisenman RN. Mad-Max transcriptional repression is mediated by ternary complex formation with mammalian homologs of yeast repressor Sin3. Cell. 1995;80:767–76. doi: 10.1016/0092-8674(95)90355-0. [DOI] [PubMed] [Google Scholar]
- 46.Dannenberg JH, David G, Zhong S, van der Torre J, Wong WH, Depinho RA. mSin3A corepressor regulates diverse transcriptional networks governing normal and neoplastic growth and survival. Genes Dev. 2005;19:1581–95. doi: 10.1101/gad.1286905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hassig CA, Fleischer TC, Billin AN, Schreiber SL, Ayer DE. Histone deacetylase activity is required for full transcriptional repression by mSin3A. Cell. 1997;89:341–7. doi: 10.1016/s0092-8674(00)80214-7. [DOI] [PubMed] [Google Scholar]
- 48.David G, Grandinetti KB, Finnerty PM, Simpson N, Chu GC, Depinho RA. Specific requirement of the chromatin modifier mSin3B in cell cycle exit and cellular differentiation. Proc Natl Acad Sci U S A. 2008;105:4168–72. doi: 10.1073/pnas.0710285105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Martinez-Balbas MA, Bauer UM, Nielsen SJ, Brehm A, Kouzarides T. Regulation of E2F1 activity by acetylation. EMBO J. 2000;19:662–71. doi: 10.1093/emboj/19.4.662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Marzio G, Wagener C, Gutierrez MI, Cartwright P, Helin K, Giacca M. E2F family members are differentially regulated by reversible acetylation. J Biol Chem. 2000;275:10887–92. doi: 10.1074/jbc.275.15.10887. [DOI] [PubMed] [Google Scholar]
- 51.Zupkovitz G, Grausenburger R, Brunmeir R, Senese S, Tischler J, Jurkin J, Rembold M, Meunier D, Egger G, Lagger S, Chiocca S, Propst F, Weitzer G, Seiser C. The cyclin-dependent kinase inhibitor p21 is a crucial target for histone deacetylase 1 as a regulator of cellular proliferation. Mol Cell Biol. 2010;30:1171–81. doi: 10.1128/MCB.01500-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lagger G, O’Carroll D, Rembold M, Khier H, Tischler J, Weitzer G, Schuettengruber B, Hauser C, Brunmeir R, Jenuwein T, Seiser C. Essential function of histone deacetylase 1 in proliferation control and CDK inhibitor repression. EMBO J. 2002;21:2672–81. doi: 10.1093/emboj/21.11.2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wilting RH, Yanover E, Heideman MR, Jacobs H, Horner J, van der Torre J, DePinho RA, Dannenberg JH. Overlapping functions of Hdac1 and Hdac2 in cell cycle regulation and haematopoiesis. EMBO J. 2010;29:2586–97. doi: 10.1038/emboj.2010.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bhaskara S, Chyla BJ, Amann JM, Knutson SK, Cortez D, Sun ZW, Hiebert SW. Deletion of histone deacetylase 3 reveals critical roles in S phase progression and DNA damage control. Mol Cell. 2008;30:61–72. doi: 10.1016/j.molcel.2008.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Warbrick E. The puzzle of PCNA’s many partners. Bioessays. 2000;22:997–1006. doi: 10.1002/1521-1878(200011)22:11<997::AID-BIES6>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 56.Milutinovic S, Zhuang Q, Szyf M. Proliferating cell nuclear antigen associates with histone deacetylase activity, integrating DNA replication and chromatin modification. J Biol Chem. 2002;277:20974–8. doi: 10.1074/jbc.M202504200. [DOI] [PubMed] [Google Scholar]
- 57.Mariadason JM, Rickard KL, Barkla DH, Augenlicht LH, Gibson PR. Divergent phenotypic patterns and commitment to apoptosis of Caco-2 cells during spontaneous and butyrate-induced differentiation. J Cell Physiol. 2000;183:347–54. doi: 10.1002/(SICI)1097-4652(200006)183:3<347::AID-JCP7>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 58.Mariadason JM, Barkla DH, Gibson PR. Effect of short-chain fatty acids on paracellular permeability in Caco-2 intestinal epithelium model. Am J Physiol. 1997;272:G705–12. doi: 10.1152/ajpgi.1997.272.4.G705. [DOI] [PubMed] [Google Scholar]
- 59.Mariadason JM, Velcich A, Wilson AJ, Augenlicht LH, Gibson PR. Resistance to butyrate-induced cell differentiation and apoptosis during spontaneous Caco-2 cell differentiation. Gastroenterology. 2001;120:889–99. doi: 10.1053/gast.2001.22472. [DOI] [PubMed] [Google Scholar]
- 60.Heerdt BG, Houston MA, Augenlicht LH. Potentiation by specific short-chain fatty acids of differentiation and apoptosis in human colonic carcinoma cell lines. Cancer Res. 1994;54:3288–93. [PubMed] [Google Scholar]
- 61.Gurvich N, Tsygankova OM, Meinkoth JL, Klein PS. Histone deacetylase is a target of valproic acid-mediated cellular differentiation. Cancer Res. 2004;64:1079–86. doi: 10.1158/0008-5472.can-03-0799. [DOI] [PubMed] [Google Scholar]
- 62.Cheng M, Olivier P, Diehl JA, Fero M, Roussel MF, Roberts JM, Sherr CJ. The p21(Cip1) and p27(Kip1) CDK ‘inhibitors’ are essential activators of cyclin D-dependent kinases in murine fibroblasts. EMBO J. 1999;18:1571–83. doi: 10.1093/emboj/18.6.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Doetzlhofer A, Rotheneder H, Lagger G, Koranda M, Kurtev V, Brosch G, Wintersberger E, Seiser C. Histone deacetylase 1 can repress transcription by binding to Sp1. Mol Cell Biol. 1999;19:5504–11. doi: 10.1128/mcb.19.8.5504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lagger G, Doetzlhofer A, Schuettengruber B, Haidweger E, Simboeck E, Tischler J, Chiocca S, Suske G, Rotheneder H, Wintersberger E, Seiser C. The tumor suppressor p53 and histone deacetylase 1 are antagonistic regulators of the cyclin-dependent kinase inhibitor p21/WAF1/CIP1 gene. Mol Cell Biol. 2003;23:2669–79. doi: 10.1128/MCB.23.8.2669-2679.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Davie JR. Inhibition of histone deacetylase activity by butyrate. J Nutr. 2003;133:2485S–2493S. doi: 10.1093/jn/133.7.2485S. [DOI] [PubMed] [Google Scholar]
- 66.Wilson AJ, Byun DS, Popova N, Murray LB, L’Italien K, Sowa Y, Arango D, Velcich A, Augenlicht LH, Mariadason JM. Histone deacetylase 3 (HDAC3) and other class I HDACs regulate colon cell maturation and p21 expression and are deregulated in human colon cancer. J Biol Chem. 2006;281:13548–58. doi: 10.1074/jbc.M510023200. [DOI] [PubMed] [Google Scholar]
- 67.Fischle W, Dequiedt F, Hendzel MJ, Guenther MG, Lazar MA, Voelter W, Verdin E. Enzymatic activity associated with class II HDACs is dependent on a multiprotein complex containing HDAC3 and SMRT/N-CoR. Mol Cell. 2002;9:45–57. doi: 10.1016/s1097-2765(01)00429-4. [DOI] [PubMed] [Google Scholar]
- 68.Guenther MG, Lane WS, Fischle W, Verdin E, Lazar MA, Shiekhattar R. A core SMRT corepressor complex containing HDAC3 and TBL1, a WD40-repeat protein linked to deafness. Genes Dev. 2000;14:1048–57. [PMC free article] [PubMed] [Google Scholar]
- 69.Li J, Wang J, Nawaz Z, Liu JM, Qin J, Wong J. Both corepressor proteins SMRT and N-CoR exist in large protein complexes containing HDAC3. EMBO J. 2000;19:4342–50. doi: 10.1093/emboj/19.16.4342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Codina A, Love JD, Li Y, Lazar MA, Neuhaus D, Schwabe JW. Structural insights into the interaction and activation of histone deacetylase 3 by nuclear receptor corepressors. Proc Natl Acad Sci U S A. 2005;102:6009–14. doi: 10.1073/pnas.0500299102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Panteleeva I, Rouaux C, Larmet Y, Boutillier S, Loeffler JP, Boutillier AL. HDAC-3 participates in the repression of e2f-dependent gene transcription in primary differentiated neurons. Ann N Y Acad Sci. 2004;1030:656–60. doi: 10.1196/annals.1329.076. [DOI] [PubMed] [Google Scholar]
- 72.Wilson AJ, Byun DS, Nasser S, Murray LB, Ayyanar K, Arango D, Figueroa M, Melnick A, Kao GD, Augenlicht LH, Mariadason JM. HDAC4 promotes growth of colon cancer cells via repression of p21. Mol Biol Cell. 2008;19:4062–75. doi: 10.1091/mbc.E08-02-0139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang Y, Tuzova M, Xiao ZX, Cruikshank WW, Center DM. Pro-IL-16 recruits histone deacetylase 3 to the Skp2 core promoter through interaction with transcription factor GABP. J Immunol. 2008;180:402–8. doi: 10.4049/jimmunol.180.1.402. [DOI] [PubMed] [Google Scholar]
- 74.Center DM, Cruikshank WW, Zhang Y. Nuclear pro-IL-16 regulation of T cell proliferation: p27(KIP1)-dependent G0/G1 arrest mediated by inhibition of Skp2 transcription. J Immunol. 2004;172:1654–60. doi: 10.4049/jimmunol.172.3.1654. [DOI] [PubMed] [Google Scholar]
- 75.Carrano AC, Eytan E, Hershko A, Pagano M. SKP2 is required for ubiquitin-mediated degradation of the CDK inhibitor p27. Nat Cell Biol. 1999;1:193–9. doi: 10.1038/12013. [DOI] [PubMed] [Google Scholar]
- 76.Tsvetkov LM, Yeh KH, Lee SJ, Sun H, Zhang H. p27(Kip1) ubiquitination and degradation is regulated by the SCF(Skp2) complex through phosphorylated Thr187 in p27. Curr Biol. 1999;9:661–4. doi: 10.1016/s0960-9822(99)80290-5. [DOI] [PubMed] [Google Scholar]
- 77.Margariti A, Zampetaki A, Xiao Q, Zhou B, Karamariti E, Martin D, Yin X, Mayr M, Li H, Zhang Z, De Falco E, Hu Y, Cockerill G, Xu Q, Zeng L. Histone deacetylase 7 controls endothelial cell growth through modulation of beta-catenin. Circ Res. 2010;106:1202–11. doi: 10.1161/CIRCRESAHA.109.213165. [DOI] [PubMed] [Google Scholar]
- 78.Gu W, Roeder RG. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 79.Freedman DA, Wu L, Levine AJ. Functions of the MDM2 oncoprotein. Cell Mol Life Sci. 1999;55:96–107. doi: 10.1007/s000180050273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ito A, Kawaguchi Y, Lai CH, Kovacs JJ, Higashimoto Y, Appella E, Yao TP. MDM2-HDAC1-mediated deacetylation of p53 is required for its degradation. EMBO J. 2002;21:6236–45. doi: 10.1093/emboj/cdf616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Vaziri H, Dessain SK, Ng Eaton E, Imai SI, Frye RA, Pandita TK, Guarente L, Weinberg RA. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001;107:149–59. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- 82.Luo J, Nikolaev AY, Imai S, Chen D, Su F, Shiloh A, Guarente L, Gu W. Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell. 2001;107:137–148. doi: 10.1016/s0092-8674(01)00524-4. [DOI] [PubMed] [Google Scholar]
- 83.Li S, Banck M, Mujtaba S, Zhou MM, Sugrue MM, Walsh MJ. p53-induced growth arrest is regulated by the mitochondrial SirT3 deacetylase. PLoS One. 2010;5:e10486. doi: 10.1371/journal.pone.0010486. [DOI] [PMC free article] [PubMed] [Google Scholar]