Abstract
Lung cancer remains one of the leading causes for cancer-related death in developed countries. In lung adenocarcinomas, EGFR mutations and EML4-ALK fusions are associated with response to EGFR and ALK inhibition. By contrast, therapeutically exploitable genetic alterations have been lacking in squamous-cell lung cancer. We conducted a systematic search for alterations that are therapeutically amenable and performed high-resolution gene-copy number analyses in a set of 232 lung cancer specimens. We identified frequent and focal FGFR1 amplification in squamous-cell lung cancer (n=155), but not in other lung cancer subtypes, and confirmed its presence in an independent cohort of squamous-cell lung cancer samples employing FISH (22% of cases). Using cell-based screening with the FGFR inhibitor (PD173074) in a large (n=83) panel of lung cancer cell lines, we demonstrated that this compound inhibited growth (p=0.0002) and induced apoptosis (p=0.008) specifically in those lung cancer cells carrying amplified FGFR1. We validated the dependency on FGFR1 of FGFR1-amplified cell lines by knockdown of FGFR1 and by ectopic expression of a resistance allele of FGFR1 (FGFR1V561M), which rescued FGFR1-amplified cells from PD173074-mediated cytotoxicity. Finally we showed that inhibition of FGFR1 with a small molecule led to significant tumor shrinkage in vivo. Focal FGFR1 amplification is common in squamous-cell lung cancer and associated with tumor growth and survival, suggesting that FGFR inhibitors may be a viable therapeutic option in this cohort of patients.
The identification of focal and recurrent amplification of FGFR1 in squamous-cell lung cancer represents the first therapeutically amenable target in this histological type of lung cancer that is strongly associated with smoking and resistance to targeted lung cancer drugs.
Introduction
Oncogenic protein kinases are frequently potential targets for cancer treatment. Examples include ERBB2 amplification in breast cancer, associated with clinical response to antibodies targeting ERBB2 (1), and KIT or PDGFRA mutations in gastrointestinal stromal tumors, which lead to sensitivity to the KIT/ABL/PDGFR inhibitor imatinib (2). In lung adenocarcinoma, patients with EGFR-mutant tumors (3–5) experience tumor shrinkage and prolongation in progression-free survival when treated with EGFR inhibitors (6). Furthermore, EML4-ALK gene fusion-positive lung cancers can be effectively treated with ALK inhibitors (7, 8). However, these alterations almost exclusively occur in the rare adenocarcinomas of patients who never smoked, but are uncommon in squamous-cell lung cancer, which is almost invariably associated with smoking (9). Although previous studies have reported recurrent genetic alterations in squamous-cell lung cancer (10), no therapeutically tractable targets have so far been identified. Thus, therapeutic options for squamous-cell lung cancer patients remain scarce, as molecularly targeted drugs such as erlotinib, gefitinib, pemetrexed and cetuximab are either poorly active (6, 11) or contraindicated (e.g. bevacizumab)(12). These observations emphasize the need for new “druggable” targets in squamous-cell lung cancer patients.
Results
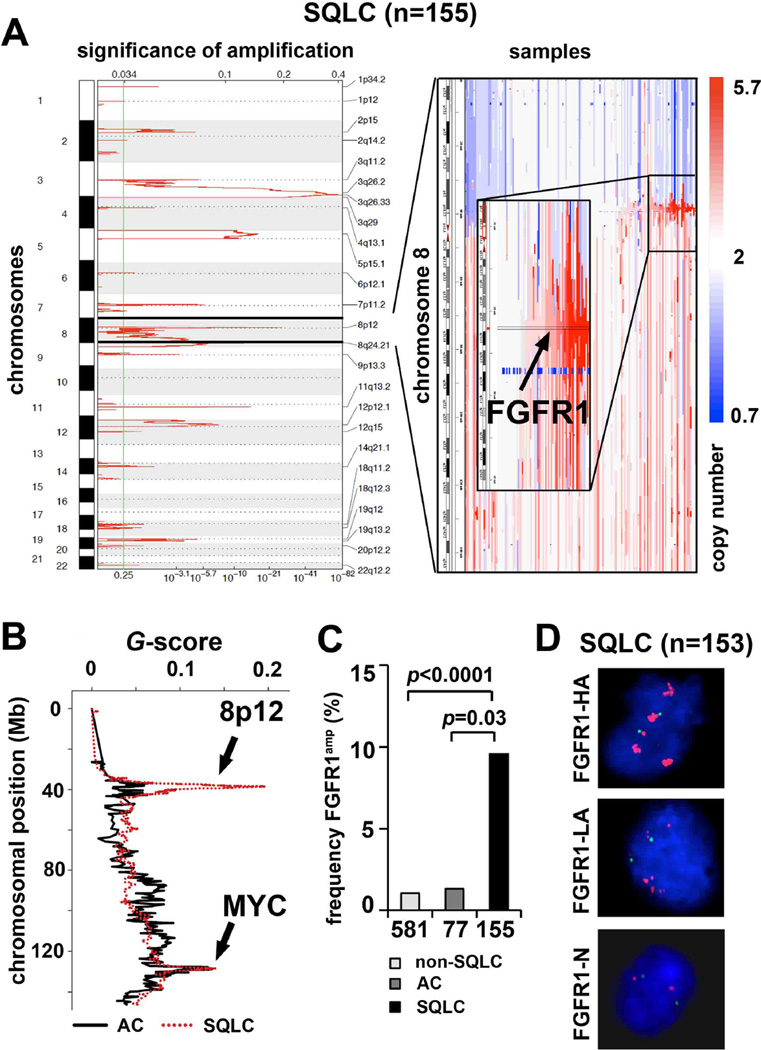
To identify therapeutically relevant genome alterations in squamous-cell lung cancer, we analyzed 155 primary squamous-cell lung cancer specimens using Affymetrix 6.0 SNP arrays, yielding high-resolution genomic profiles (median inter-marker distance <1 kb). To separate driver lesions from random noise, we applied the GISTIC algorithm (13, 14). We identified 25 significant amplification peaks, including the previously described amplification of SOX2 on chromosome 3q26.33 (Figure 1A and Supplementary Table 1) (10) and 26 significant deletions (Supplementary Figure 1 and Supplementary Table 1). The second most significant amplification (q=8.82*10−28) peak was identified on 8p12 and included FGFR1 as well as FLJ43582 in each sample called as amplified (Figure 1A). This region spanned 133kb (Supplementary Table 1), and was amplified at high amplitude (≥ 4 copies) in 15 of 155 squamous-cell lung cancer specimens (9.7%) (Figure 1A). Of note, 11 of the FGFR1-amplified tumors were from smokers and none of these were from never-smokers (Supplementary Table 2). Ten of the 15 FGFR1-amplified tumors also harbored a mutation in TP53 (Supplementary Table 2). Patients with FGFR1-amplified tumors (copy number > 9 in FISH analysis) had a non-significant trend toward inferior survival compared to patients whose tumors lacked FGFR1 amplifications (copy number = 2 in FISH analysis) (Supplementary Figure 2). We next analyzed copy-number alterations in lung adenocarcinoma specimens (n=77) and found no significant (q>0.25) amplification (≥ 4 copies; 1.3%) at 8p12 (Figure 1B). Finally, we analyzed a publicly available lung cancer SNP-array dataset (14) for the presence of FGFR1 amplifications (≥ 4 copies) and found it to occur in 6 out of 581 (1%) non-squamous cell lung cancers (Figure 1C). Thus, FGFR1 amplification is significantly enriched in squamous-cell lung cancer when compared to our own adenocarcinoma dataset (p=0.03) (Supplementary Table 3) and when compared to a published dataset of non-squamous-cell lung cancer (p<0.0001) (Figure 1C). Fluorescence in-situ hybridization (FISH) using an 8p12-specific probe on an independent set of 153 squamous-cell lung cancers confirmed the presence of frequent high-level amplification of FGFR1 in 34 of 153 patients (22%) (Figure 1D and Supplementary Table 4), 27 of whom were current smokers and none of whom were non-smokers. We note that FISH is not sensitive to the admixture of non-tumoral cells. Thus, focal amplification of FGFR1 is likely to be more frequent in squamous-cell lung cancer than estimated by SNP arrays (Supplementary Table 4) (15). We also sequenced the FGFR1 gene (16) in 94 squamous-cell lung cancers and 94 adenocarcinomas and found one mutation (FGFR1P578H) in the adenocarcinoma cohort, indicating that FGFR1 mutations might play an only minor role and might not be driver alterations in the pathogenesis of lung cancer.
Figure 1. FGFR1 is amplified in squamous-cell lung cancer.
(A) Left panel: Significant (14) (FDR-value; x-axis) amplifications across all chromosomes (y-axis) in squamous-cell lung cancer (SQLC; n=155) as assessed by GISTIC. Right panel: Copy-number alterations (blue=deletion; white=copy number-neutral; red=amplification) at chromosome 8 (y-axis) across all SQLC samples (x-axis). Samples are ordered according to focal amplification of FGFR1. (B) Significant (G-score; y-axis) copy number changes in adenocarcinoma (AC; n=77), (black line) and SQLC (red dotted line) at chromosome 8. The q-value for the presence of 8p12 amplification is 8.82*10−28 for squamous-cell lung cancer and greater than 0.25 for adenocarcinoma. The chromosomal positions of FGFR1 (8p12) and MYC are highlighted (black arrows) (C) Frequency of FGFR1 amplification (% of samples ≥ copy number 4; y-axis) in non-SQLC lung cancer from a published dataset (14), AC and SQLC. P-values indicate statistical significance. (D) FISH analysis (green = control; red = FGFR1) of 153 SQLC samples (FGFR1-HA: copy number >9; FGFR1-LA: copy number >2 <9; FGFR1-N: copy number 2). Presented are example images from the three different FGFR1 amplification groups.
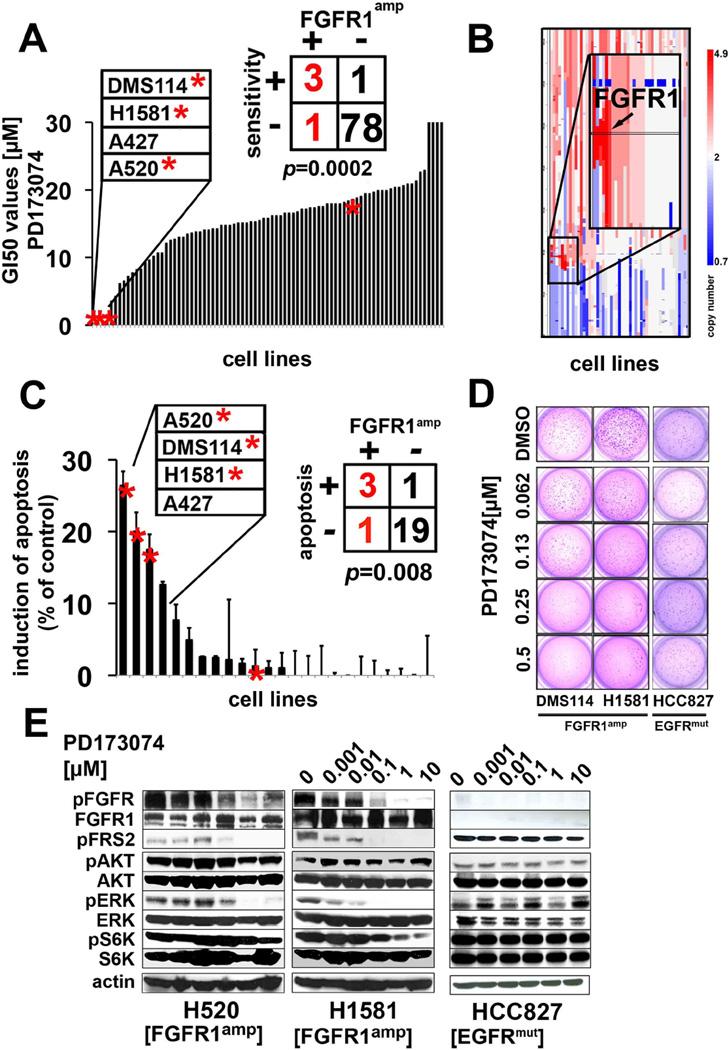
Next, we performed high-throughput cell-line screening (17, 18) to determine the activity of the non-isoform-specific FGFR inhibitor PD173074 (19) in a collection of 83 lung cancer cell lines (Supplementary Table 5) (17, 20). Of all cell lines tested, four had a half-maximal growth inhibitory concentration (GI50 values) below 1.0 µM (Figure 2A); remarkably, three of the four sensitive lung cancer cell lines exhibited focal amplification at 8p12 by 6.0 SNP-array analysis (Figure 2B) suggesting that FGFR1 amplifications are significantly (p=0.0002) associated with FGFR inhibitor activity (Figure 2A). As expected, FGFR1-amplified cells expressed higher levels of total FGFR1 protein (Supplementary Figure 3). Interestingly, one (H520) of the three FGFR1-amplified cell lines that were sensitive to PD173074 was derived from a squamous-cell lung cancer patient (Supplementary Table 5). We next tested whether amplification of FGFR1 could be linked with sensitivity to FGFR inhibition in an unbiased fashion. Application of a K-nearest neighbor-based analysis, followed by leave-one-out cross validation (17), revealed FGFR1 amplification to be the only genetic predictor of PD173074 sensitivity that retained significance following Bonferroni-based multiple-testing correction (p<0.05; Supplementary Table 6). Previous reports indicated that expression of FGFR ligands might contribute to the sensitivity to FGFR inhibitors in lung cancer (21). We did not observe elevated levels of FGF2 in the FGFR1-amplified cell lines (Supplementary Figure 4A), nor did we observe a difference in the expression of FGFR ligands between patients harboring FGFR1 amplification and those without FGFR1 amplification (Supplementary Figure 4B). However, FGFR1-amplified cells showed robust phosphorylation of FGFR, suggesting ligand-independent activation, which was further enhanced upon addition of exogenous FGF2 or FGF9 (Supplementary Figure 4C), compatible with paracrine activation of FGFR1 in FGFR1-amplified cells. We next measured induction of apoptosis in FGFR1-amplified cells after treatment with PD173074 and found a significant (p=0.008) enrichment of FGFR1-amplified lung cancer cells in the group of sensitive cells (Figure 2C and Supplementary Table 7). Furthermore, FGFR inhibition led to decreased colony formation of FGFR1-amplified but not of EGFR-mutant cells in soft agar (Figure 2D), further enforcing the notion that amplification of FGFR1 drives proliferation of these lung cancer cell lines. Treatment with PD173074 reduced the levels of phosphorylated FGFR1 (Supplementary Figure 5) and of the adaptor molecule FRS2 in a dose-dependent manner only in FGFR1-amplified cells, but not in the EGFR-mutant cell line HCC827 (Figure 2E). We also observed inhibition of phosphorylation of ERK but not of AKT and S6, indicating that the MAPK-pathway, and not the PI3K-pathway, is the major signaling pathway engaged by amplified FGFR1 (Figure 2E).
Figure 2. FGFR1 amplification and sensitivity to FGFR inhibition.
(A) GI50-values (y-axis) of PD173074 across 83 lung cancer cell lines (x-axis). FGFR1-amplified (copy number≥4) cell lines are marked with asterisks. (B) Copy number alterations (x-axis, blue=deletion; white= copy number 2; red= amplification) on chromosome 8 with a zoom in on 8p12 (FGFR1 locus is highlighted) across all cell lines (y-axis). (C) Induction of apoptosis (difference between PD173074 at 1µM and DMSO control after 72h; y-axis) across 24 cell lines (x-axis; asterisks denote FGFR1 amplification copy number≥4) as measured by flowcytometry (after Annexin V/PI staining). (D) FGFR1-amplified cell lines were plated in soft agar and treated either with DMSO (control) or decreasing concentrations of PD173074. (E) Phosphorylation of FGFR and of downstream molecules in FGFR1-amplified (H1581, H520) and in FGFR1 wildtype (EGFR-mutant) cells (HCC827) after treatment with PD173074 as assessed by immunoblotting.
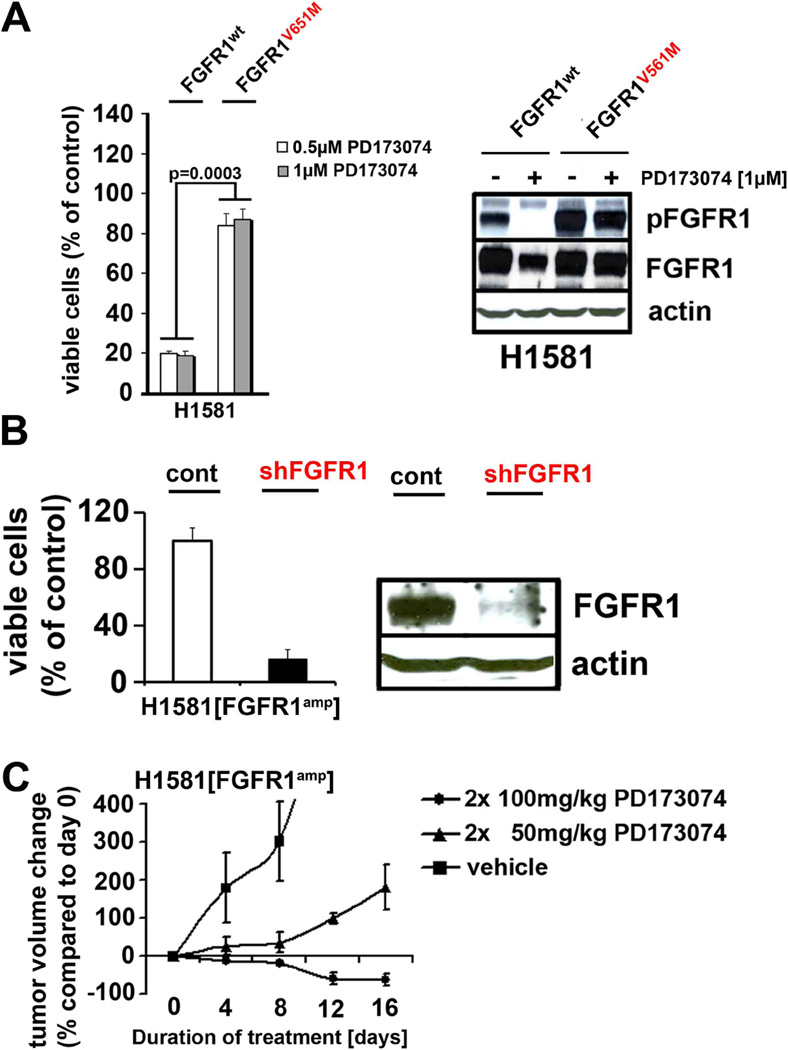
In order to validate FGFR1 as the critical target of PD173074 in FGFR1-amplified lung cancer cells, we ectopically expressed the V561M mutation (22) at the gatekeeper position of FGFR1 (FGFR1V561M), preventing access of the compound to the hinge region of the kinase (23) (Supplementary Figure 6). Expression of FGFR1V561M in FGFR1-amplified lung cancer cells abolished PD173074-mediated cytotoxicity and dephosphorylation of FGFR (Figure 3A), consistent with the notion that FGFR1 is the critical target of PD173074 in FGFR1-amplified lung cancer cells. Furthermore, in a panel of 105 biochemically-screened kinases FGFR1 was one of only two kinases strongly inhibited by PD173074 (Supplementary Table 8), recapitulating previous reports (22).
Figure 3. FGFR1-amplified cells are dependent on FGFR1 in vitro and in vivo.
(A) Left panel: Viability (PD173074 treatment as compared to DMSO control) of FGFR1-amplified cells expressing wildtype or mutant (V561M) FGFR1 treated with PD173074 (0.5µM white bars; 1.0µM grey bars). Right panel: phosphorylation of FGFR in the FGFR1V561M and FGFR1wt cells detected by immunoblotting. (B) Upper panel: Viability (PD173074 treatment as compared to DMSO control; y-axis) of H1581 cells after transduction with control shRNA or shRNA targeting FGFR1. Right panel: Silencing of FGFR1 in H1581 cells was confirmed by immunoblotting. (C) In mice engrafted with H1581 cells either treated with vehicle or PD173074 (dosage as indicated; y-axis), tumor volume was measured over time (x-axis).
The high analytical resolution of the 6.0 SNP arrays, together with the large size of our data set, limited the number of candidate genes in the 8p12 amplicon to only two genes, FGFR1 and FLJ43582. A previous report analyzing the 8p12 locus in lung cancer applying lower-resolution techniques suggested WHSC1L1 to be the relevant oncogene in the 8p12 amplicon (24). To test whether genes other than FGFR1 drive tumorigenesis in the 8p12-amplified tumors, we silenced the genes WHSC1L1 (24) and FLJ43582 using five different shRNA constructs in the 8p12-amplified lung cancer cell line H1581. Although silencing of either one of these genes did not inhibit cellular viability (Supplementary Figure 7), silencing of FGFR1 strongly reduced the viability of the FGFR1-amplified lung cancer cells (Figure 3B). In light of the focality of the 8p12 amplicon (including FGFR1 and FLJ43582) and the lack of effect of shRNA-mediated knockdown of either FLJ43582 and WHSC1L1 in FGFR1-amplified cells, our data suggests that FGFR1 is the relevant target in these cells. Of note, the cell line H1703, which bears a copy-number gain at 8p12 and that had been reported to depend on WHSC1L1 (24) was not sensitive to FGFR inhibition (Supplementary Figure 8). By contrast, H1703 cells depend on PDGFRA for their survival (25) due to amplification (copy number > 2.8) of the gene encoding this kinase (26, 27). Thus, our data suggests that the gene targeted by the 8p12 amplicon is primarily FGFR1 and its amplification induces FGFR1 dependency.
Finally, treatment with 100mg/kg twice a day of PD173074 resulted in tumor shrinkage in mice engrafted with FGFR1-amplified cells (Figure 3C). This reduction in tumor size was paralleled by reduction in the levels of phospho-ERK but not of phospho-AKT in immunohistochemical analyses of explanted tumors, validating our in-vitro findings that MAPK-signaling is the key pathway engaged by amplified FGFR1 (Supplementary Figure 9A). Treatment at 50mg/kg twice a day resulted in only a minimal exposure when compared to the gavage of 100mg/kg twice a day due to the short half-life of the compound in vivo (Supplementary Figure 9B). Thus, although we cannot formally exclude inhibitory effects on VEGFR2, the observed tumor regression is likely to be mediated by inhibition of FGFR1. By contrast, xenografted EGFR-mutant H1975 cells did not show signs of regression upon PD173074 treatment (Supplementary Figure 9C). Thus, FGFR1 amplification leads to FGFR1 dependency in vivo.
Discussion
Here, we have identified frequent high-level amplification of FGFR1 in squamous-cell lung cancer of smokers that sensitizes the tumors to FGFR1 inhibition. Previous studies in lung cancer cohorts of mixed subtypes and low technological resolution (24, 28) or small size (10) have reported occasional amplification of the 8p locus in lung cancer. However, the large size of our sample set was necessary to reveal the high prevalence of this amplicon in squamous-cell lung cancer (approximately 10%) in comparison to other lung cancer subtypes (1%). Given the insensitivity of FISH analyses to admixture of non-tumoral cells, the true prevalence of this amplification is likely to still be substantively underestimated by SNP arrays and to be up to 20%. We conclude that FGFR1 amplification is one of the hallmark alterations in squamous-cell lung cancer, similar to amplification of SOX2. These two alterations were almost completely mutually exclusive (Supplementary Table 9), suggesting an epistatic relationship. Furthermore, FGFR1 amplification induced a strong FGFR1 dependency that could be exploited therapeutically, resulting in induction of apoptosis. Thus, FGFR1 amplification represents an opportunity for targeted therapy in squamous-cell lung cancer. We therefore suggest that FGFR1 inhibitors, which are currently in clinical testing in tumor types bearing genetic alterations in FGFR genes (29–31), should be evaluated in patients with FGFR1-amplified squamous-cell lung cancer.
Materials and Methods
Genomic analyses
The tumor specimens analyzed in this study have been collected under local Institutional Review Board approval. All patients gave written informed consent. Genomic DNA was hybridized to Affymetrix 6.0 SNP arrays following manufacturer’s instructions. Raw signal intensities were normalized and modeled using a Gaussian-mixture model. Background-corrected intensities were normalized across all arrays of one batch using quantile normalization. Raw copy numbers were calculated by dividing the normalized tumor-derived signal intensities by the mean signal intensities derived from the normal samples hybridized in the same batch. Raw copy number data were segmented using circular binary segmentation and visualized in the integrated genome viewer (IGV) (32). GISTIC was performed as described previously (13, 14). The human genome build hg18 was utilized. Dideoxy sequencing was performed on whole-genome amplified DNA of primary tumors. Cell lines were sequenced using cDNA. All primer sequences are available on request. All raw data are publically available (GEO; GSE25016).
Tissue microarray construction
Tissue microarray slides were obtained from Formalin-fixed paraffin-embedded lung squamous-cell carcinoma samples. The tissue microarrays contained samples of a total of 172 patients from the University Hospital Zurich, each of these samples was present in duplicate cores, each core 0.6 mm in diameter (33). A second tissue microarray of 22 patients from Weill Cornell Medical Center was obtained with each sample present in triplicate cores, each core 0.6 mm in diameter. Subsequently, 153 samples were used for FISH analysis.
Gene expression
After RNA isolation biotin labeled cRNA preparation was performed using Epicentre TargetAmp™ Kit (Epicentre Biotechnologies) and Biotin-16-UTP (10 mM; Roche Molecular Biochemicals) or Illumina TotalPrep RNA Amplification Kit (Ambion). Biotin labeled cRNA (1.5 µg) was hybridized to Sentrix® whole genome bead chips WG6 version 2, (Illumina) and scanned on the Illumina® BeadStation 500×. For data collection, we used Illumina BeadStudio 3.1.1.0 software.†Gene pattern analysis platform (34) was used to visualize the normalized data.
FGFR1 amplification FISH assay
A fluorescence in-situ hybridization (FISH) assay was used to detect the FGFR1 amplification at the chromosomal level on the tissue microarrays. We performed fluorescence signal detection, with two probes on chromosome 8. The reference probe is located on a stable region of chromosome 8p23.2 and selected based on SNP array analysis. Only samples where the control BAC was detectable were used for the determination of the copy number of FGFR1. The target probe is located on the FGFR1 locus spanning 8p11.23 to 8p11.22. We used the digoxigenin labelled BAC clones CTD 2523O9 producing a green signal as reference probe. The target probe was labelled with biotin to produce a red signal using RP11-148D21 BAC clones (Invitrogen). Deparaffinized sections were pre-treated with a 100 mM Tris and 50 mM EDTA solution at 92.8C° for 15 min. and digested with Digest-All III (dilution 1:2) at 37°C for 14 min.; FGFR1 FISH probes were denatured at 73°C for 5 min. and immediately placed on ice. Subsequently, the tissue sections and FGFR1 FISH probes were co-denatured at 94°C for 3 min. and hybridized overnight at 37°C. Post hybridization washing was done with 2× SSC at 75°C for 5 min., and the fluorescence detection was carried out using streptavidin-Alexa-594 conjugates (dilution 1:200) and anti-digoxigenin-FITC (dilution 1:200). Slides were then counterstained with 4’,6-Diamidin-2’ phenylindoldihydrochlorid (DAPI) and mounted.†The samples were analyzed under a 63× oil immersion objective using a fluorescence microscope (Zeiss) equipped with appropriate filters, a charge-coupled device camera and the FISH imaging and capturing software Metafer 4 (Metasystems). The evaluation of the tests was done independently by three experienced evaluators (R.M, S.M. and S.P.). At least 100 nuclei per case were evaluated. The thresholds for assigning a sample to the FGFR1 “high amplification” group was copy number nine. All samples that had a copy number below nine and above two were assigned to the group of “low amplification” cohort. All the remaining samples were assigned “normal”.
Cell lines and reagents
Cell lines were obtained from ATCC, DSMZ, or from own and other cell culture collections and were maintained as described previously. PD173074 was purchased from commercial suppliers, dissolved in DMSO or vehicle solution and stored at −20°C.
Cell-line screening
Cell-line screening was performed as previously described (17) with various concentrations of PD173074. Viability was determined after 96h by measuring cellular ATP content (CellTiter-Glo, Promega). Half-maximal inhibitory concentrations (GI50) were determined using the statistical data analysis software “R” with the package “ic50”.
Apoptosis
For determination of apoptosis, cells were seeded in six-well plates, incubated for 24h, treated with either DMSO (control) or 1.0 µM PD173074 for 72 h and stained with annexinV and propidium iodide (PI). Finally the cells were analyzed on a FACS Canto FlowCytometer (BD Biosciences). The difference between the relative percentage of annexin V/PI positive cells treated with DMSO and cells treated with PD173074 was determined (induction of apoptosis rate).
Lentiviral RNAi and retroviral expression
The V561M mutation was introduced into FGFR1 cloned in pBABE-Puro by site-directed mutagenesis. Replication-incompetent retroviruses were produced by cotransfection with the pCL-ampho plasmid in HEK 293T cells. Hairpin shRNA targeting the different genes were ordered from Sigma. All sequences are given in a Supplementary Table (Supplementary Table 10). Replication-incompetent lentiviruses were produced from pLKO.1-Puro based vectors by contransfection with Δ8.9 and pMGD2 in 293T cells as described previously (35). After transduction cells were selected with puromycin (1.5µg/ml) and five days after selection cells were counted using trypan blue.
Western blotting
The following antibodies were used for immunoblotting: β-actin (MPBioscience), phospho-FGFR (Y653, Y654), pFRS2 (Tyr196), p-AKT (S473), pS6, S6, AKT, p-ERK, ERK (Cell Signaling Technology), total-FGFR1 (Santa Cruz), anti-rabbit-HRP, anti-mouse-HRP-antibody (Millipore).
Soft-agar assay
Cells were suspended in growth media containing 10% FCS and 0.6% agar and plated in triplicate on 50µl solidified growth medium (10% FCS; 1.0% agar). Growth medium containing indicated compound concentrations was added on top. Colonies were analyzed using the Scanalyzer imaging system (Lemnatec, Germany).
Xenograft mouse models
All animal procedures were approved by the local animal protection committee and the local authorities. 5*106 tumor cells were injected subcutaneously into male nude mice. After the tumors reached a size of at least 50mm3, the animals were treated twice daily by oral gavage with PD173074 (15 mg/ml for 50mg/kg or 30mg/ml for 100mg/kg schedule) solved in vehicle (sodium lactate) or vehicle detergent alone. Tumor size was monitored measuring perpendicular diameters as described previously (17). For the determination of tumor growth under treatment with PD173074, each experiment presented in the figures compromises the measurement of 5 different tumors.
Statistical analyses
Tests for statistical significance were either two-tailed t-tests or Fishers’s exact tests. Prediction of compound activity was performed using the KNN algorithm as described previously (17). Multiple hypothesis testing was performed employing the statistical data analysis software “R” using p-value adjustment.
Supplementary Material
Acknowledgements
We thank Drs. Mathew Meyerson and Adam Bass for sharing upublished data and Drs. Christian Reinhardt and Axel Ullrich for discussion. Funding: This work was supported by the Deutsche Krebshilfe (grant 107954 to RKT), by the German Ministry of Science and Education (BMBF) as part of the NGFNplus program (grant 01GS08100 to RKT and 01GS08101 to JW and PN), by the Max Planck Society (M.I.F.A.NEUR8061 to RKT), by the Deutsche Forschungsgemeinschaft (DFG) through SFB (TP6 to RKT and RTU; TP5 to LH and RB), the Ministry for Innovation, Science, Research and Technology of the State of Nordrhein-Westfalen (MIWT, 4000- 12 09 to RKT and BK) and by an anonymous foundation to RKT. EB was supported by the PNES INCA grant 2008. RKT reports the following potential sources of conflict of interest: consulting and lecture fees (Sequenom, Sanofi-Aventis, Merck, Roche, Infinity, Boehringer, Astra-Zeneca, Atlas-Biolabs); research support (Novartis, AstraZeneca).
Footnotes
List of Supplementary Material
Supplementary Figure 1: Significant deletions are observed in squamous-cell lung cancer.
Supplementary Figure 2: FGFR1 amplification has no significant impact on overall survival of SQLC patients.
Supplementary Figure 3: FGFR1 amplification correlates with FGFR1 protein expression.
Supplementary Figure 4: Expression of FGFR ligands does not correlate with FGFR1 amplification status.
Supplementary Figure 5: Treatment of FGFR1-amplified cell line H520 with PD173074 leads to dephosphorylation of FGFR1 as measured by immunoprecipitation assay.
Supplementary Figure 6: PD173074 binds inside the ATP-binding pocket of FGFR1.
Supplementary Figure 7: Knockdown of genes adjacent to FGFR1 on 8p12.
Supplementary Figure 8: PD173074 is not active in the PDGFRA- and FGFR1-amplified cell line H1703.
Supplementary Figure 9: PD173074 shows anti-tumor activity in vivo.
Supplementary Table 1: Significant amplifications and deletions are noted in a subset of 155 SQLC samples.
Supplementary Table 2: Clinical features and co-occurent mutations of FGFR1-amplified SQLC samples.
Supplementary Table 3: Significant amplifications and deletions are noted in a subset of 77 adenocarcinoma samples.
Supplementary Table 4: FGFR1 amplification is detected using FISH on tumor microarrays.
Supplementary Table 5: GI50 values are not associated with mutation status across the lung cancer cell line panel.
Supplementary Table 6: KNN algorithm-based scoring predicts PD173074 sensitivity.
Supplementary Table 7: PD173074 induces apoptosis in FGFR1-amplified cell lines.
Supplementary Table 8: PD173074 has specific activity against two kinases.
Supplementary Table 9: FGFR1 and SOX2 amplification in squamous-cell lung carcinoma.
Supplementary Table 10: Sequences of all shRNA constructs that were used in the study.
Authors contributions: J.W. and M.L.S designed and performed experiments, analyzed data and wrote the manuscript, D.S. designed experiments and analyzed data. M.P., T.Z., F.L. and D.R. analyzed data, J.M.H., R.T.U., R.M., S.M., F.F., S.H., M.K., J.S., F.G., I.D., S.Q., L.H., H.B-W., I.B. and J.A. performed experiments and discussed data, A.S., H.M., P.W., S.A., Z.W., M.C., G.W., P.R., B.S., E.B., C.B., P.L., S.S., L.T.B., W.E.R., C.L., I.P., J.S., J.C., H.G., W.T., H.S., E.T., E.S., D.H., F.C., C.L., S.D., M.H., R.B., W.P., B.K., M.B., R.B., K.E., E.S., J.W. and P.N., contributed critical tumor specimens and contributed to the discussion of the data. S.P. reviewed tumor histology, analyzed FISH data and wrote the manuscript. R.K.T. conceived the project, designed experiments, analyzed data and wrote the manuscript.
References and Notes
- 1.Slamon DJ, Leyland-Jones B, Shak S, Fuchs H, Paton V, Bajamonde A, Fleming T, Eiermann W, Wolter J, Pegram M, Baselga J, Norton L. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med. 2001;344:783–792. doi: 10.1056/NEJM200103153441101. [DOI] [PubMed] [Google Scholar]
- 2.Heinrich MC, Corless CL, Demetri GD, Blanke CD, von Mehren M, Joensuu H, McGreevey LS, Chen CJ, Van den Abbeele AD, Druker BJ, Kiese B, Eisenberg B, Roberts PJ, Singer S, Fletcher CD, Silberman S, Dimitrijevic S, Fletcher JA. Kinase mutations and imatinib response in patients with metastatic gastrointestinal stromal tumor. J Clin Oncol. 2003;21:4342–4349. doi: 10.1200/JCO.2003.04.190. [DOI] [PubMed] [Google Scholar]
- 3.Pao W, Miller V, Zakowski M, Doherty J, Politi K, Sarkaria I, Singh B, Heelan R, Rusch V, Fulton L, Mardis E, Kupfer D, Wilson R, Kris M, Varmus H. EGF receptor gene mutations are common in lung cancers from "never smokers" and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci U S A. 2004;101:13306–13311. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, Boggon TJ, Naoki K, Sasaki H, Fujii Y, Eck MJ, Sellers WR, Johnson BE, Meyerson M. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 5.Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat SM, Supko JG, Haluska FG, Louis DN, Christiani DC, Settleman J, Haber DA. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 6.Mok TS, Wu YL, Thongprasert S, Yang CH, Chu DT, Saijo N, Sunpaweravong P, Han B, Margono B, Ichinose Y, Nishiwaki Y, Ohe Y, Yang JJ, Chewaskulyong B, Jiang H, Duffield EL, Watkins CL, Armour AA, Fukuoka M. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361:947–957. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- 7.Soda M, Choi YL, Enomoto M, Takada S, Yamashita Y, Ishikawa S, Fujiwara S, Watanabe H, Kurashina K, Hatanaka H, Bando M, Ohno S, Ishikawa Y, Aburatani H, Niki T, Sohara Y, Sugiyama Y, Mano H. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448:561–566. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 8.Kwak EL, Bang YJ, Camidge DR, Shaw AT, Solomon B, Maki RG, Ou SH, Dezube BJ, Janne PA, Costa DB, Varella-Garcia M, Kim WH, Lynch TJ, Fidias P, Stubbs H, Engelman JA, Sequist LV, Tan W, Gandhi L, Mino-Kenudson M, Wei GC, Shreeve SM, Ratain MJ, Settleman J, Christensen JG, Haber DA, Wilner K, Salgia R, Shapiro GI, Clark JW, Iafrate AJ. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med. 363:1693–1703. doi: 10.1056/NEJMoa1006448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Khuder SA. Effect of cigarette smoking on major histological types of lung cancer: a meta-analysis. Lung Cancer. 2001;31:139–148. doi: 10.1016/s0169-5002(00)00181-1. [DOI] [PubMed] [Google Scholar]
- 10.Bass AJ, Watanabe H, Mermel CH, Yu S, Perner S, Verhaak RG, Kim SY, Wardwell L, Tamayo P, Gat-Viks I, Ramos AH, Woo MS, Weir BA, Getz G, Beroukhim R, O'Kelly M, Dutt A, Rozenblatt-Rosen O, Dziunycz P, Komisarof J, Chirieac LR, Lafargue CJ, Scheble V, Wilbertz T, Ma C, Rao S, Nakagawa H, Stairs DB, Lin L, Giordano TJ, Wagner P, Minna JD, Gazdar AF, Zhu CQ, Brose MS, Cecconello I, R U, Jr, Marie SK, Dahl O, Shivdasani RA, Tsao MS, Rubin MA, Wong KK, Regev A, Hahn WC, Beer DG, Rustgi AK, Meyerson M. SOX2 is an amplified lineage-survival oncogene in lung and esophageal squamous cell carcinomas. Nat Genet. 2009;41:1238–1242. doi: 10.1038/ng.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pirker R, Pereira JR, Szczesna A, von Pawel J, Krzakowski M, Ramlau R, Vynnychenko I, Park K, Yu CT, Ganul V, Roh JK, Bajetta E, O'Byrne K, de Marinis F, Eberhardt W, Goddemeier T, Emig M, Gatzemeier U. Cetuximab plus chemotherapy in patients with advanced non-small-cell lung cancer (FLEX): an open-label randomised phase III trial. Lancet. 2009;373:1525–1531. doi: 10.1016/S0140-6736(09)60569-9. [DOI] [PubMed] [Google Scholar]
- 12.Sandler A, Gray R, Perry MC, Brahmer J, Schiller JH, Dowlati A, Lilenbaum R, Johnson DH. Paclitaxel-carboplatin alone or with bevacizumab for non-small-cell lung cancer. N Engl J Med. 2006;355:2542–2550. doi: 10.1056/NEJMoa061884. [DOI] [PubMed] [Google Scholar]
- 13.Beroukhim R, Getz G, Nghiemphu L, Barretina J, Hsueh T, Linhart D, Vivanco I, Lee JC, Huang JH, Alexander S, Du J, Kau T, Thomas RK, Shah K, Soto H, Perner S, Prensner J, Debiasi RM, Demichelis F, Hatton C, Rubin MA, Garraway LA, Nelson SF, Liau L, Mischel PS, Cloughesy TF, Meyerson M, Golub TA, Lander ES, Mellinghoff IK, Sellers WR. Assessing the significance of chromosomal aberrations in cancer: Methodology and application to glioma. Proc Natl Acad Sci U S A. 2007;104:20007–20012. doi: 10.1073/pnas.0710052104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, Barretina J, Boehm JS, Dobson J, Urashima M, Mc Henry KT, Pinchback RM, Ligon AH, Cho YJ, Haery L, Greulich H, Reich M, Winckler W, Lawrence MS, Weir BA, Tanaka KE, Chiang DY, Bass AJ, Loo A, Hoffman C, Prensner J, Liefeld T, Gao Q, Yecies D, Signoretti S, Maher E, Kaye FJ, Sasaki H, Tepper JE, Fletcher JA, Tabernero J, Baselga J, Tsao MS, Demichelis F, Rubin MA, Janne PA, Daly MJ, Nucera C, Levine RL, Ebert BL, Gabriel S, Rustgi AK, Antonescu CR, Ladanyi M, Letai A, Garraway LA, Loda M, Beer DG, True LD, Okamoto A, Pomeroy SL, Singer S, Golub TR, Lander ES, Getz G, Sellers WR, Meyerson M. The landscape of somatic copy-number alteration across human cancers. Nature. 2009;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weir BA, Woo MS, Getz G, Perner S, Ding L, Beroukhim R, Lin WM, Province MA, Kraja A, Johnson LA, Shah K, Sato M, Thomas RK, Barletta JA, Borecki IB, Broderick S, Chang AC, Chiang DY, Chirieac LR, Cho J, Fujii Y, Gazdar AF, Giordano T, Greulich H, Hanna M, Johnson BE, Kris MG, Lash A, Lin L, Lindeman N, Mardis ER, McPherson JD, Minna JD, Morgan MB, Nadel M, Orringer MB, Osborne JR, Ozenberger B, Ramos AH, Robinson J, Roth JA, Rusch V, Sasaki H, Shepherd F, Sougnez C, Spitz MR, Tsao MS, Twomey D, Verhaak RG, Weinstock GM, Wheeler DA, Winckler W, Yoshizawa A, Yu S, Zakowski MF, Zhang Q, Beer DG, Wistuba, Watson MA, Garraway LA, Ladanyi M, Travis WD, Pao W, Rubin MA, Gabriel SB, Gibbs RA, Varmus HE, Wilson RK, Lander ES, Meyerson M. Characterizing the cancer genome in lung adenocarcinoma. Nature. 2007;450:893–898. doi: 10.1038/nature06358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Greenman C, Stephens P, Smith R, Dalgliesh GL, Hunter C, Bignell G, Davies H, Teague J, Butler A, Stevens C, Edkins S, O'Meara S, Vastrik I, Schmidt EE, Avis T, Barthorpe S, Bhamra G, Buck G, Choudhury B, Clements J, Cole J, Dicks E, Forbes S, Gray K, Halliday K, Harrison R, Hills K, Hinton J, Jenkinson A, Jones D, Menzies A, Mironenko T, Perry J, Raine K, Richardson D, Shepherd R, Small A, Tofts C, Varian J, Webb T, West S, Widaa S, Yates A, Cahill DP, Louis DN, Goldstraw P, Nicholson AG, Brasseur F, Looijenga L, Weber BL, Chiew YE, DeFazio A, Greaves MF, Green AR, Campbell P, Birney E, Easton DF, Chenevix-Trench G, Tan MH, Khoo SK, Teh BT, Yuen ST, Leung SY, Wooster R, Futreal PA, Stratton MR. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sos ML, Michel K, Zander T, Weiss J, Frommolt P, Peifer M, Li D, Ullrich R, Koker M, Fischer F, Shimamura T, Rauh D, Mermel C, Fischer S, Stuckrath I, Heynck S, Beroukhim R, Lin W, Winckler W, Shah K, LaFramboise T, Moriarty WF, Hanna M, Tolosi L, Rahnenfuhrer J, Verhaak R, Chiang D, Getz G, Hellmich M, Wolf J, Girard L, Peyton M, Weir BA, Chen TH, Greulich H, Barretina J, Shapiro GI, Garraway LA, Gazdar AF, Minna JD, Meyerson M, Wong KK, Thomas RK. Predicting drug susceptibility of non-small cell lung cancers based on genetic lesions. J Clin Invest. 2009;119:1727–1740. doi: 10.1172/JCI37127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McDermott U, Sharma SV, Dowell L, Greninger P, Montagut C, Lamb J, Archibald H, Raudales R, Tam A, Lee D, Rothenberg SM, Supko JG, Sordella R, Ulkus LE, Iafrate AJ, Maheswaran S, Njauw CN, Tsao H, Drew L, Hanke JH, Ma XJ, Erlander MG, Gray NS, Haber DA, Settleman J. Identification of genotype-correlated sensitivity to selective kinase inhibitors by using high-throughput tumor cell line profiling. Proc Natl Acad Sci U S A. 2007;104:19936–19941. doi: 10.1073/pnas.0707498104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mohammadi M, Froum S, Hamby JM, Schroeder MC, Panek RL, Lu GH, Eliseenkova AV, Green D, Schlessinger J, Hubbard SR. Crystal structure of an angiogenesis inhibitor bound to the FGF receptor tyrosine kinase domain. EMBO J. 1998;17:5896–5904. doi: 10.1093/emboj/17.20.5896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sos ML, Fischer S, Ullrich R, Peifer M, Heuckmann JM, Koker M, Heynck S, Stuckrath I, Weiss J, Fischer F, Michel K, Goel A, Regales L, Politi KA, Perera S, Getlik M, Heukamp LC, Ansen S, Zander T, Beroukhim R, Kashkar H, Shokat KM, Sellers WR, Rauh D, Orr C, Hoeflich KP, Friedman L, Wong KK, Pao W, Thomas RK. Identifying genotype-dependent efficacy of single and combined PI3K- and MAPK-pathway inhibition in cancer. Proc Natl Acad Sci U S A. 2009;106:18351–18356. doi: 10.1073/pnas.0907325106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marek L, Ware KE, Fritzsche A, Hercule P, Helton WR, Smith JE, McDermott LA, Coldren CD, Nemenoff RA, Merrick DT, Helfrich BA, Bunn PA, Jr, Heasley LE. Fibroblast growth factor (FGF) and FGF receptor-mediated autocrine signaling in non-small-cell lung cancer cells. Mol Pharmacol. 2009;75:196–207. doi: 10.1124/mol.108.049544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou W, Hur W, McDermott U, Dutt A, Xian W, Ficarro SB, Zhang J, Sharma SV, Brugge J, Meyerson M, Settleman J, Gray NS. A structure-guided approach to creating covalent FGFR inhibitors. Chem Biol. 17:285–295. doi: 10.1016/j.chembiol.2010.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blencke S, Zech B, Engkvist O, Greff Z, Orfi L, Horvath Z, Keri G, Ullrich A, Daub H. Characterization of a conserved structural determinant controlling protein kinase sensitivity to selective inhibitors. Chem Biol. 2004;11:691–701. doi: 10.1016/j.chembiol.2004.02.029. [DOI] [PubMed] [Google Scholar]
- 24.Tonon G, Wong KK, Maulik G, Brennan C, Feng B, Zhang Y, Khatry DB, Protopopov A, You MJ, Aguirre AJ, Martin ES, Yang Z, Ji H, Chin L, Depinho RA. High-resolution genomic profiles of human lung cancer. Proc Natl Acad Sci U S A. 2005;102:9625–9630. doi: 10.1073/pnas.0504126102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rikova K, Guo A, Zeng Q, Possemato A, Yu J, Haack H, Nardone J, Lee K, Reeves C, Li Y, Hu Y, Tan Z, Stokes M, Sullivan L, Mitchell J, Wetzel R, Macneill J, Ren JM, Yuan J, Bakalarski CE, Villen J, Kornhauser JM, Smith B, Li D, Zhou X, Gygi SP, Gu TL, Polakiewicz RD, Rush J, Comb MJ. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell. 2007;131:1190–1203. doi: 10.1016/j.cell.2007.11.025. [DOI] [PubMed] [Google Scholar]
- 26.McDermott U, Ames RY, Iafrate AJ, Maheswaran S, Stubbs H, Greninger P, McCutcheon K, Milano R, Tam A, Lee DY, Lucien L, Brannigan BW, Ulkus LE, Ma XJ, Erlander MG, Haber DA, Sharma SV, Settleman J. Ligand-dependent platelet-derived growth factor receptor (PDGFR)-alpha activation sensitizes rare lung cancer and sarcoma cells to PDGFR kinase inhibitors. Cancer Res. 2009;69:3937–3946. doi: 10.1158/0008-5472.CAN-08-4327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ramos AH, Dutt A, Mermel C, Perner S, Cho J, Lafargue CJ, Johnson LA, Stiedl AC, Tanaka KE, Bass AJ, Barretina J, Weir BA, Beroukhim R, Thomas RK, Minna JD, Chirieac LR, Lindeman NI, Giordano T, Beer DG, Wagner P, Wistuba, Rubin MA, Meyerson M. Amplification of chromosomal segment 4q12 in non-small cell lung cancer. Cancer Biol Ther. 2009;8:2042–2050. doi: 10.4161/cbt.8.21.9764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao X, Weir BA, LaFramboise T, Lin M, Beroukhim R, Garraway L, Beheshti J, Lee JC, Naoki K, Richards WG, Sugarbaker D, Chen F, Rubin MA, Janne PA, Girard L, Minna J, Christiani D, Li C, Sellers WR, Meyerson M. Homozygous deletions and chromosome amplifications in human lung carcinomas revealed by single nucleotide polymorphism array analysis. Cancer Res. 2005;65:5561–5570. doi: 10.1158/0008-5472.CAN-04-4603. [DOI] [PubMed] [Google Scholar]
- 29.Reis-Filho JS, Simpson PT, Turner NC, Lambros MB, Jones C, Mackay A, Grigoriadis A, Sarrio D, Savage K, Dexter T, Iravani M, Fenwick K, Weber B, Hardisson D, Schmitt FC, Palacios J, Lakhani SR, Ashworth A. FGFR1 emerges as a potential therapeutic target for lobular breast carcinomas. Clin Cancer Res. 2006;12:6652–6662. doi: 10.1158/1078-0432.CCR-06-1164. [DOI] [PubMed] [Google Scholar]
- 30.Turner N, Grose R. Fibroblast growth factor signalling: from development to cancer. Nat Rev Cancer. 2009;10:116–129. doi: 10.1038/nrc2780. [DOI] [PubMed] [Google Scholar]
- 31.Turner N, Pearson A, Sharpe R, Lambros M, Geyer F, Lopez-Garcia MA, Natrajan R, Marchio C, Iorns E, Mackay A, Gillett C, Grigoriadis A, Tutt A, Reis-Filho JS, Ashworth A. FGFR1 amplification drives endocrine therapy resistance and is a therapeutic target in breast cancer. Cancer Res. 2010;70:2085–2094. doi: 10.1158/0008-5472.CAN-09-3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.2010 www.broadinstitute.org/igv/, [Google Scholar]
- 33.Perner S, Wagner PL, Soltermann A, LaFargue C, Tischler V, Weir BA, Weder W, Meyerson M, Giordano TJ, Moch H, Rubin MA. TTF1 expression in non-small cell lung carcinoma: association with TTF1 gene amplification and improved survival. J Pathol. 2009;217:65–72. doi: 10.1002/path.2443. [DOI] [PubMed] [Google Scholar]
- 34.2010 http://www.broadinstitute.org/cancer/software/genepattern/ [Google Scholar]
- 35.Sos ML, Rode HB, Heynck S, Peifer M, Fischer F, Kluter S, Pawar VG, Reuter C, Heuckmann JM, Weiss J, Ruddigkeit L, Rabiller M, Koker M, Simard JR, Getlik M, Yuza Y, Chen TH, Greulich H, Thomas RK, Rauh D. Chemogenomic profiling provides insights into the limited activity of irreversible EGFR Inhibitors in tumor cells expressing the T790M EGFR resistance mutation. Cancer Res. 2010;70:868–874. doi: 10.1158/0008-5472.CAN-09-3106. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.