Abstract
Many biological complexes are naturally low in abundance and pose a significant challenge to their structural and functional studies. Here we describe a new method that utilizes strong oxidation and chemical linkage to introduce a high density of bioactive ligands onto nanometer-thick carbon films and enable selective enrichment of individual macromolecular complexes at subnanogram levels. The introduced ligands are physically separated. Ni-NTA, Protein G and DNA/RNA oligonucleotides were covalently linked to the carbon surface. They embody negligible mass and their stability makes the functionalized films able to survive long-term storage and tolerate variations in pH, temperature, salts, detergents, and solvents. We demonstrated the application of the new method to the electron microscopic imaging of the substrate-bound C3PO, an RNA-processing enzyme important for the RNA interference pathway. On the ssRNA-linked carbon surface, the formation of C3PO oligomers at subnanomolar concentrations likely mimics their assembly onto ssRNA substrates presented by their native partners. Interestingly, the 3D reconstructions by negative stain EM reveal a side port in the C3PO/ssRNA complex, and the 15 Å cryoEM map showed extra density right above the side port, which probably represents the ssRNA. These results suggest a new way for ssRNAs to interact with the active sites of the complex. Together our data demonstrate that the surface-engineered carbon films are suitable for selectively enriching low-abundance biological complexes at nanomolar level and for developing novel applications on a large number of surface-presented molecules.
Keywords: Chemical functionalization, carbon films, biological ligands, single particle cryoEM, C3PO/RNA complex, ChemiC
INTRODUCTION
Cells function by a fine interplay of macromolecular complexes (Alberts et al., 2007). Quantitative studies of the structures and functions of these complexes at molecular, cellular, and system levels derive deeper and broader insights in almost every frontier of biomedical research. However, many of these complexes have low copy numbers inside cells. Different techniques have been developed to enrich them for experimental investigations (Dong et al., 2008). We are interested in engineering a selection surface that allows affinity-based retention of macromolecular complexes in high density. Such a modification will introduce very little mass to the surface of the substrate, and thus very low background noise, but will be capable of selecting target complexes of low natural abundance or with a low expression level in heterologous systems. The enriched molecules would be suitable for many single molecule studies and for assembling multi-component biological processes in a controlled manner.
To develop such a method in a quantitative manner, we need to examine the selective enrichment of macromolecules on a substrate at the nanometer scale, and need to use electron microscopy to visualize them and quantify their surface density. Nanometer-thick carbon or graphene films appear to be the ideal substrates because of their mechanical strength and chemical stability. In the meantime, it is quite clear that the new substrates will be useful for preparing cryoEM specimens.
The current sample preparation method adopted for cryoEM was pioneered by Dübochet and colleagues (Adrian et al., 1984; Dubochet et al., 1982). It has, in the past three decades, generated great success in cryoEM study of biological complexes that are available at microgram or milligram levels, and helped resolve atomic-resolution structures of a few high-symmetry particles (Yu et al., 2008; Zhang et al., 2008). However, this powerful technology usually leads to the loss of more than 95% of the biological preparations. Alternative ways have been designed to deal with this issue. Double-stranded DNA-scaffolds (Selmi et al., 2011), 2D streptavidin crystals (Wang et al., 2008), and lipid monolayers (Kelly et al., 2008; Kelly et al., 2010) are three examples. But these three methods all introduce significant amounts of biomass into the final specimens and need specific conditions to maintain the integrity of the introduced materials. In contrast, the carbon-based engineering method to be described next introduces negligible biomass, and can tolerate a broad range of pH variation, harsh solvents (including some organic ones), high/low temperatures, and detergents. We developed procedures to introduce reactive carboxylate groups on the surface of 3–5 nm thick carbon films at a high surface density. We introduced new procedures to keep the chemically modified carbon films stable and clean for biological applications. We optimized reaction conditions to introduce Ni-NTA, DNA/RNA oligos, and Protein G to the surfaces of these carbon films. We quantified the surface density and binding affinity of these introduced groups separately. These surface-presented biological ligands retain their activities and are suitable for selective enrichment of target biological complexes.
To demonstrate the practical application of the new surface engineering method for single molecule studies, we used it to study single-stranded (ss)RNA-induced assembly of C3PO, component 3 Promoter of RNA Interference Silencing Complex (RISC), by single particle electron microscopy. C3PO is a newly characterized ribonuclease and its active complex is believed to contain six translin subunits and two TRAX subunits that form a hetero-octameric barrel with two active sites located inside (Ye et al., 2011). The endonucleolytic activity of C3PO is important for activating duplex small-interfering (si)RNA-initiated RISC, the effector complex of RNA interference (RNAi). In an active RISC, a single-stranded siRNA (called guide strand) directs Argonaute 2 (Ago2) endonuclease to cleave complementary mRNAs (Liu and Paroo, 2010; Miyoshi et al., 2005). While it has been shown by several published studies that C3PO is active only in the hetero-octameric form, it remains unclear how a substrate ssRNA is able to reach the interior of a C3PO barrel where its active sites are located (Tian et al., 2011; Ye et al., 2011). Recent crystallographic and EM studies have resolved several structures of C3PO in the absence of ssRNA. The crystal structure of the full-length human C3PO exhibits an asymmetric hetero-octamer made of six translin and two TRAX subunits (6:2 translin/TRAX)(Ye et al., 2011), which was proposed to be the active conformation based on mutagenesis and functional studies in solution. While the crystal structure of a truncated Drosophila C3PO mutant was hexameric [4:2 translin/TRAX; see (Tian et al., 2011)], the EM reconstruction of its full-length version appeared octameric (6:2 or 5:3 translin/TRAX). Intriguingly, in both cases, the RNA-binding sites and the catalytic residues for the C3PO RNA-processing activity are located at the interior surface of the octamer. It was proposed that C3PO might cleave short ssRNAs within its fully enclosed barrel. However, a challenging question is how an ssRNA is recruited to the interior of a C3PO complex. Our new carbon-based engineering technology makes it possible to present individual RNA or DNA molecules at spatially separated sites, similar to the presentation of the passenger RNA strands on the surface of individual Ago2/nicked dsRNA complexes. We were able to use these anchored ssRNAs to guide the assembly of C3PO complexes. It is possible that the C3PO complexes assembled on individual RNA oligos will recapitulate the properties of their in vivo assemblies on inactive Ago2 complexes. Single particle reconstruction of C3PO by negative-stain EM showed an olive-shaped structure, which resembles the asymmetric octamer (6:2 translin/TRAX) of an RNA-free human C3PO. A clear difference is that on one side, the EM map has a sizable opening, which is large enough for ssRNA molecules to bind or pass through. A cryoEM map at 15 Å resolution showed extra density above the side port, which likely came from the ssRNA bound to the C3PO complex laterally. Our results suggest that the enclosed octameric barrel of an RNA-free C3PO needs significant rearrangements in order to create such a lateral opening and allow an ssRNA to reach the enzymatic active sites from outside. The successful study of C3PO on the functionalized carbon films demonstrates the potential applications of our new technology to the structural and functional studies of many other important biological complexes.
MATERIALS and METHODS
Grid Preparation ---- ChemiC-coated copper grids
Copper grids were purchased from EMS. They were pre-cleaned with chloroform, 1.0% SDS and 100% ethanol. After air drying, they were stored at room temperature on a filter paper inside a covered petri dish. Immediately prior to use, both sides of the grids were negatively glow-discharged for 1.5 minutes (EMS 100 Glow Discharge Unit).
Carbon films were thermally evaporated onto freshly cleaved mica sheets from a pair of sharpened graphite carbon rods (Ted Pella, CA) that were heated to melting temperature at a high vacuum of 2.0 × 10−7 Torr inside a Denton Explorer 14 unit. The carbon films on mica sheets were stored at room temperature inside petri dishes for varying amount of time before being used.
To coat the copper grids, a carbon film on a piece of mica sheet was floated off in a water trough, and slowly settled onto the glow-discharged grids inside the trough. The grids were then slowly dried at 50°C overnight. Prior to chemical modification, the carbon-coated grids were heated to 200°C in air for 10 minutes. We found that this treatment was critical because it allowed the carbon films to adhere very well to the grid surface so that delamination of carbon films was minimized during subsequent steps.
The carbon films on the grids were first oxidized by floating them on top of droplets of 50 µl solution made of 0.40 M KMnO4 and 0.20 M NaOH on a piece of parafilm. After 1.5 hr oxidation, the grids were thoroughly washed in a sodium bisulfite solution to remove manganese oxides that were formed during the strong oxidation. We recognized that the sodium bisulfite washing was extremely critical in order to remove electron dense artifacts deposited on the carbon films during the oxidation. We examined the cleanness of the carbon films by EM before moving to the next step. For simplicity, we will call these chemically reactive carbon films the “ChemiC films”.
For covalent conjugation reactions, the carboxylate groups on the ChemiC films were first converted into amine-reactive esters by using a zero-length cross-linker, 1-ethyl-3-(3-dimethyl-aminopropyl) carbodiimide hydrochloride (EDC, Pierce), in the presence of N-hydroxyl-sulfosuccinimide (sulfo-NHS, Pierce). More specifically, the grids were immersed for 10 minutes in a solution of 5.0 mM EDC, 5.0 mM sulfo-NHS and 0.10 M MES at pH 5.0. In order to quantify the density of amine-reactive groups on the carbon surface, primary amine (−NH2)-containing Quantum Dots (QD 605, amino-modified from Invitrogen) at 1.0 nM in 50 mM borate buffer (pH 8.3) were used as a quality-control assay. We counted the number of QDs in randomly selected areas, and calculated the averaged surface density.
Ni-NTA-ChemiC films
For selective enrichment of His-tagged proteins to the carbon films, we prepared ChemiC films that were charged with Ni-NTA. 100 µM nitrilotriacetic acid (NTA)-NH2 (AB-NTA from Dojindo Molecular Technologies) was reacted with the NHS-modified ChemiC films for 1 hour in a 50 mM borate buffer, pH 8.5. The surface NTA groups were charged for 30 minutes with 100 µM NiCl2 in water. The resulted Ni-NTA-ChemiC grids were tested for their stability under different pH, temperature, solvents, detergents and salts.
Protein G-ChemiC films
Similar to the production of the Ni-NTA-ChemiC films, the NHS-modified ChemiC films were incubated with 100 nM recombinant Protein G (ID Labs, Inc.) in a solution containing 20 mM HEPES, 100 mM NaCl, pH 7.2 for 1 – 2 hours. The grids were washed with the same buffer prior to use. We checked the chemical stability of the introduced Protein G on the ChemiC films after one week storage at room temperature.
Preparation of QD–His10 peptide and QD-HA peptide complexes
To quantify the surface density of Ni-NTA groups, a His10 peptide (GGGHHHHHHHHHH) with a biotin added to the N-terminus was synthesized at the Protein Chemistry Technology Center in our institution. The peptides were used to fully saturate all the binding sites of the QD-streptavidin 605 (Invitrogen) by mixing them at 10:1 molar ratio in 0.10 M PBS, pH 6.5. After end-over-end rotation for 30 minutes at room temperature, excess peptides were separated by ultracentrifugation at 400,000 × g for one hour. A bright orange pellet of the QD-peptide conjugates was clearly visible at the bottom of the centrifuge tube and allowed the reliable removal of unbound peptides. The centrifugation step was repeated twice after washing and resuspension in the PBS buffer. The formation of QD-His10 conjugates was confirmed by labeling TALON Co-beads with them and examining the QD fluorescence on the Co-beads. The Quantum Dots without the peptides failed to make the Co-beads fluoresce.
In a similar fashion, an N-terminally biotinylated peptide (PSPSL-YPYDVPDYA-PDPKFE) with the HA epitope (underlined) was conjugated with QD-streptavidin. To test the accessibility of the HA peptides, the QD-HA conjugates were mixed with monoclonal anti-HA antibodies (Covance) in a 1:2 molar ratio. Successful formation of the QD-HA peptide/antibody complexes was confirmed by incubating Protein G-coated agarose beads with the complexes and examining the QD fluorescence. The QD-HA complex without the anti-HA antibody failed to introduce fluorescence to the agarose beads.
Expression and purification of valosin-containing protein (VCP)
His-tagged VCP was purified from transformed BL21 cells obtained from Dr. George de Martino’s lab (UT Southwestern). The cells were cultured to OD0.7–0.8, and then induced at 37 °C with 1.0 mM IPTG for 4 hours. Afterwards, the bacteria were collected by centrifugation, and lysed by sonication for 15 minutes in a buffer containing 150 mM KCl, 100 mM Tris pH 7.4, 5.0 mM MgCl2, 1.0 mM ATP, 2.0 mM β-mercaptoethanol (β-ME) and protease inhibitors. The cell lysate was centrifuged at 100,000×g. The supernatant was collected and run through a Co-NTA column. The Co-beads were washed with the same buffer supplemented with 20 mM imidazole (pH7.4). The protein was eluted with 350 mM imidazole. Further purification was done by size-exclusion chromatography in a Superose 6 column using a buffer made of 150 mM KCl, 50 mM Tris-HCl pH7.4, 2.0 mM MgCl2, 0.50 mM MgATP and 1.0 mM β-ME (Fig. S1).
To prepare the cryo-EM specimen of VCP, 5.0 µL of protein solution at 0.10 nM was loaded onto a Ni-NTA-ChemiC grid for 30 minutes in a wet chamber at room temperature. The grid was then transferred into a FEI Mark III Vitrobot, blotted with a piece of filter paper for ~6 seconds, and plunge-frozen into liquid ethane. The frozen grids were kept in liquid N2 until EM examination.
On the surface of a 3.05 mm grid, a 5.0 µL drop is about 0.7 mm thick. For a protein of 600 kDa, the measured 3D diffusion constant is ~35 µm2/s (Walters et al., 1984). It takes the protein molecule ~10 minutes to move 0.35 mm by random walk. We incubated for 30 minutes and thus there was enough time for all molecules to have a chance to reach the surface of a ChemiC grid.
Covalent linkages of oligonucleotide ligands (both DNA and RNA)
After EDC/NHS reaction, the ChemiC grids were incubated with 200 nM 5’-NH2-terminated oligonucleotides in 10 mM HEPES pH 7.5 for 1.0 hour at room temperature, and washed clean with nuclease-free water prior to use. In order to assay the number of attached oligos, we prepared grids with two test oligos, 5’-amine-C6-CTCCTCCTTCCTTC-3’ with or without a biotin at the 3’ end, and detected the presence of the oligos by using 10 nM QD-streptavidin in the borate buffer as described above. The grids were washed with nuclease-free water. The surface density of QD on these grids was quantified by EM.
C3PO purification and assembly on oligo-ChemiC films
Wild-type C3PO utilized in our experiments were purified as previously described (Ye et al., 2011). The avid affinity of C3PO for let-7 ssRNA was exploited in order to retain the oligomers on the surface of the oligo-ChemiC films. An oligonucleotide with a 5’ phosphorylation and 3’-amine-C6-dT modification (5’-pUGAGGAAGUAGG-UUGUAUAGU-dT-C6-NH2 3’) in 20 mM HEPES pH 7.5 was first covalently linked to NHS-activated ChemiC films. After a thorough wash with RNase free water, a 10 µL drop of 0.10 nM C3PO in a buffer of 150 mM NaCl and 10 mM Tris, pH 8.0 was loaded onto the grid and incubated in a wet chamber for 30 min. Excess solution was blotted away with a piece of filter paper and the grid was immediately stained with two drops of 2.0% phosphotungstic acid (PTA), pH 8.0 plus 0.50% trehalose, and dried in air. The trehalose was added to minimize the air-drying effect on the sample.
For cryo-specimens, C3PO was assembled on let-7 ssRNA grids in a wet chamber, blotted for ~7 seconds and plunge-frozen in liquid ethane inside an FEI Mark III Vitrobot. The grids were stored in liquid N2 until EM examination.
As control, 100 nM C3PO was mixed with ssDNAs (5’-TGAGGTAGTAGG-TTGTATAGT-3’) in a 1:2 molar ratio in a buffer containing 10 mM Tris pH 8.0 and 150 mM NaCl, loaded onto a glow-discharged carbon-coated grid and stained with 2.0% PTA pH 8.0 plus 0.50% trehalose.
Electron microscopy and image analysis
Negatively-stained C3PO complexes on regular glow-discharged grids or on let-7 ssRNA-ChemiC grids were examined in a JEOL JEM 2200FSFEG TEM operated at 200 kV accelerating voltage. EM images were taken under low dose condition (~20 electrons/Å2 at the specimen level) on Kodak SO-163 films with a nominal magnification of 60,000 × and defocus levels varying from −1.0 to −2.0 µm. The calibrated magnification was 61,198 ×. The EM negatives were screened on a SIRA optical diffractometer and those with obvious astigmatism or drift were eliminated. Good films were scanned in a Zeiss SCAI scanner at a step size of 14.0 µm, corresponding to a calibrated 2.29 Å/pixel at the specimen level. Individual C3PO particles were interactively selected and boxed using EMAN Boxer program (Ludtke et al., 1999). For the C3PO on ChemiC grids, a total of 12,587 particles were selected. The raw dataset was phase-flipped and band-pass filtered to suppress the low-frequency noise before it was subjected to multiple rounds of multivariate statistical analysis and hierarchical classification in IMAGIC (van Heel et al., 1996). The 2nd and 3rd eigenimages calculated from the translationally centered raw images showed strong C2 symmetry. Because of the band-pass filtering and the high signal-to-noise ratio in the negative-stain EM, the symmetry in the eigenimages probably reflects the particle property within the resolution range we are working in. Stable classification was achieved after six rounds. An initial model was generated using angular reconstitution without imposing symmetry (C1). A stable initial model was obtained by sequentially adding more and more class averages, and was refined by multiple rounds of projection matching and back projection. Once an initial model was obtained, its projections were used to align the raw dataset, before another round of MRA, classification, angular reconstitution and model refinement. After 3–4 rounds, the C1 model starts to be fairly stable with clear features suggesting C2 symmetry. To further improve the map, projection matching and model refinement was conducted by using programs written in SPIDER (Frank et al., 1996). After 6 rounds of refinement (C1 map as in Fig. S2a), the two-fold (C2) axis of the map was defined in Chimera, and the C2 symmetry was imposed. The last 3 rounds of refinement were executed with the C2 symmetry imposed along the Z-axis. The projections from the map, the class averages at the same orientation, and the typical raw images in the classes were compared, and were found to agree well with each other. The volume was corrected for the CTF envelope function by the sum of the CTF function calculated for all the particles used in the final calculation. To estimate the resolution of the 3D reconstruction, the dataset was divided into two roughly equal halves, and two independent maps were calculated from the two datasets, refined, aligned and then used for calculating Fourier Shell Correlation (FSC). A threshold value of 0.50 led to an estimated resolution of 21 Å (Fig. S2b).
To determine the handedness of the 3D reconstruction, three sets of tilted pairs of EM films (0° and 20°) were taken from negatively stained C3PO complexes. Because of the small size of the C3PO, we did not use the procedure developed by Henderson et al. (Henderson et al., 2011). Instead we did the following as outlined by Belnap et al. (Belnap et al., 1997). Using WEB, we first selected the corresponding pairs of particles from the two micrographs taken from almost the same fields in the untilted and tilted specimens, and then determined the relative rotation angle (θ’) between the tilt pairs as well as the angles between the tilt axis and Y-axis in both micrographs. The two micrographs of the tilted pairs were rotated such that in both cases the rotational axes run along the Y-axes. The EM micrographs were scanned such that in the default SPIDER coordinate system, the Z-axis is parallel to the direction of the electron beam. Under this condition, the rotational angle from the untilted to the tilted position is positive when the rotation is clockwise and is viewed along the Y-axis towards the origin. By using multi-reference alignment (AP SH) in SPIDER, the Euler angles of the untilted particles were determined against the projections of the 3D reconstruction. For every pair, the 3D model was first reoriented by applying the Euler angles so that it would reproduce the image of the untilted particle. The resultant 3D model was then rotated along Y-axis (which is the tilt axis) by θ’ and –θ’. Projections were calculated for these two positions and were compared with the image of the tilted particle. The same comparison was done using a mirrored model (along Y-axis) of the 3D reconstruction. For the model without mirror operation, ~60% of the tilted particles were assigned to the model projections at θ’. For the mirrored model, ~60% was assigned to projections at – θ’. The results from the analysis of 70 pairs of particle images suggested that the 3D model without mirror operation likely has the correct handedness (Fig. S3), even though 60% versus 40% is a slightly lower ratio. The reason behind the weaker power of discrimination seen here was likely the small size of the C3PO particle (~240 kDa), whose projections at 0º and 20º may not be strikingly different such that the noise in the images made it difficult for a reliable distinction between the two orientations (Henderson et al., 2011). Nevertheless, our main conclusion about the side port in the RNA-bound C3PO complex is not affected by the left uncertainty in the chirality determination.
To verify the 3D map for C3PO/ssRNA complex, we did a separate independent reconstruction for the C3PO/ssDNA mixture in suspension. The C3PO at 100 nM and ssDNA at 200 nM were mixed and immediately loaded onto the glow-discharged carbon-coated grids for negative staining. A dataset of 6,350 particles were built. The same procedure was used to build the initial model in IMAGIC and refine the 3D reconstruction in SPIDER to ~22 Å. The dataset also showed good C2 symmetry (Fig. S4a). The projections from the 3D reconstruction, the class averages, and the raw images in the classes agreed well with each other (Fig. S4b).
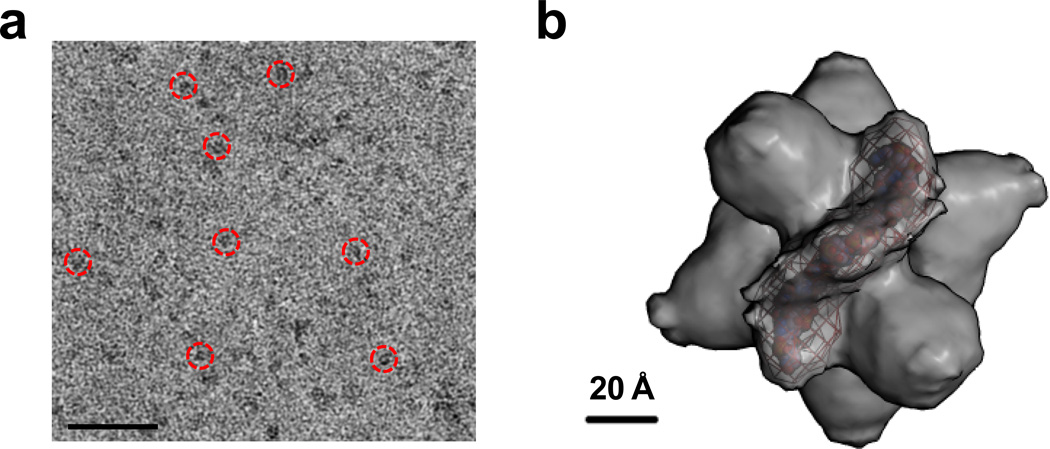
To demonstrate the feasibility of performing 3D reconstructions from cryoEM images obtained using the new ChemiC films, C3PO complexes formed on let-7 ssRNA-anchored ChemiC grids were frozen and imaged under low dose conditions with a defocus level of −1.5 to −2.5 microns in the JEOL2200FS FEG microscope. Because the ChemiC films retained all the molecules within a layer of solution that was ~10 nm above the carbon surface, the vitrified ice of the cryo-specimens could be made as thin as 30–40 nm, which would decrease the noise due to the electron scattering by ice when compared with the conventional 100–150 nm thick ice. With in-column energy filtering of 15 eV, we were able to visualize well the C3PO/RNA complex in the images (Fig. 7a). Particles were selected manually and a dataset of ~39,000 particles were built for analysis. After phase-flipping and band-pass filtering, the images were subjected to multivariate statistical analysis in IMAGIC, revealing two-fold symmetry in the 2nd and 3rd eigenimages (Fig. S5a). The map of negatively stained C3PO on let-7 ssRNA grids was filtered to 30 Å, and then used as an initial model for the refinement. The projections from the map, the class averages and the raw images aligned to the projections were found to agree well with each other (Fig. S5b). At the 0.5 threshold of the Fourier shell correlation, the estimated resolution was ~15 Å (Fig. S5c). Even thought it was a bit low, it is reasonable with the consideration of the small size of the complex. A negative B-factor of −800 Å2 was applied to sharpen the map. The putative RNA density was segmented and presented in Fig. 7b.
Figure 7. cryoEM map of C3PO/ssRNA complexes on ChemiC films.
(a). cryoEM images of the complexes. The ice thickness was empirically tested to achieve good contrast for the ~240 kDa complex. The proteins are black for good visibility. The red circles highlight a few particles. Scale bar = 100 nm. (b). The cryoEM map of the complex at a nominal 15 Å resolution. The putative RNA density was segmented and displayed as purple mesh. A model (ball structure) of the 21-nt let-7 ssRNA was generated in Coot (Emsley et al., 2010) and inserted into the volume enclosed by the mesh.
RESULTS
Chemical functionalization of ultrathin carbon films (ChemiC films)
Ultrathin carbon films that are a few nanometers thick have seen extensive use in electron microscopy. They exhibit superior thermal and electrical conductivity as well as high mechanical strength at microscopic scales. It is possible to make these films cover tens of centimeters on various substrates, such as glass slides, mica sheets, silicon wafers, etc. We used chemical oxidation to break the C=C bonds in the graphite patches and introduced reactive groups to the top layer of the carbon films. Specific ligands were then linked covalently to these reactive groups (Fig.1a). We used electron microscopy to examine the quality of the chemically functionalized carbon films, developed assays to measure the surface density of the introduced ligands and obtained projection images of individual biological complexes. Because of the high density on 2D surfaces, these ligands could serve as high-affinity binders to enrich biological complexes at (sub)nanomolar concentrations into spatially discrete spots, where specific biological reactions can be reconstituted.
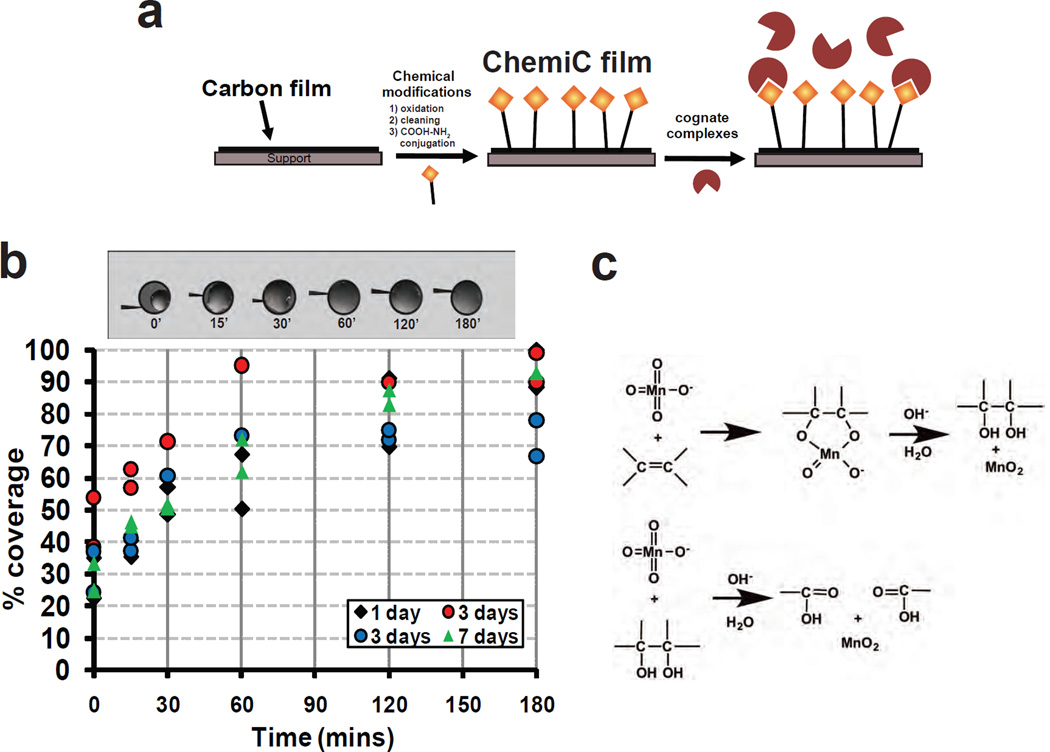
Figure 1. Chemical modification of carbon films for attaching specific complexes.
(a) Overall functionalization scheme. In order to attach specific complexes, the carbon films will be first chemically modified to generate functional groups suitable for conjugating specific ligands. This affinity-based approach will allow the selective enrichment of the complexes on the surface and enable the study of low-abundance complexes.
(b) ChemiC films remain hydrophilic for an extended period of time. The top shows the images of water droplets on the 3.05-mm discs of carbon films that were oxidized for 0, 15, 30, 60, 120 and 180 minutes. 1.0 µL water was loaded to the surface, and the spreading of the water was measured with ImageJ and expressed as percentage of the total area of the disc. Four sets of grids were treated at the same time. The %coverage of the water droplets was measured after the grids were stored at room temperature for 1 (black diamonds), 3 (red and blue circles) and 7 (green triangles) days. Two-hour oxidization was found to be enough for reaching pretty good wettability of the treated carbon films.
(c) Tentative chemical mechanism for carbon oxidation by permanganate. The C=C double bonds are mediated by sp2 electrons in the graphite carbon films. Permanganate ions conjugates with the C=C by electrophilic attack at the right orientations. Hydrolysis of the intermediate led to the formation of –OH, and the break of the –CH(OH)--CH(OH)- linkage (through sp2 electrons) leads to the opening of the hexameric carbon ring in graphite. Further oxidation gives rise to –CHO and –COOH groups. The oxidation reaction produces MnO2 deposits on the carbon surface, which needs to be removed thoroughly through bisulfite reaction.
To accomplish our general scheme and make it easy to implement reproducibly, we first demonstrated that it is possible to oxidize the surface of carbon films, and the oxidized films remain hydrophilic for a long period of time. We used the extent of spreading of one-microliter water droplets on 3.05 mm diameter films as a direct measure of surface hydrophilicity (Fig. 1b). After 60-minute or longer reaction, the oxidized carbon films became highly hydrophilic, and allowed the water droplets to spread and cover 60–95% of the surface area. At room temperature, the treated films remained hydrophilic after 7 days, suggesting that the oxidization introduced hydrophilic groups permanently to the carbon surface. Without oxidization, the carbon films would become hydrophobic after a day at room temperature.
From previous chemical analysis (Aitchison et al., 2007; Hummers and Offeman, 1958), the oxidization introduces hydroxyls, aldehydes and carboxylates onto the graphite patches in the carbon films. Fig. 1c presents a putative scheme of the chemical reactions. The introduced groups are highly reactive for covalent linkage of biological ligands. We will take advantage of the carboxylate groups, and introduce specific ligands that are used extensively in biomedical research. We tried to measure the FTIR signatures from these surface groups, but found out that the signals were below the instrument sensitivity.
Optimizing carboxylate-amine reaction on the surface of the ChemiC films
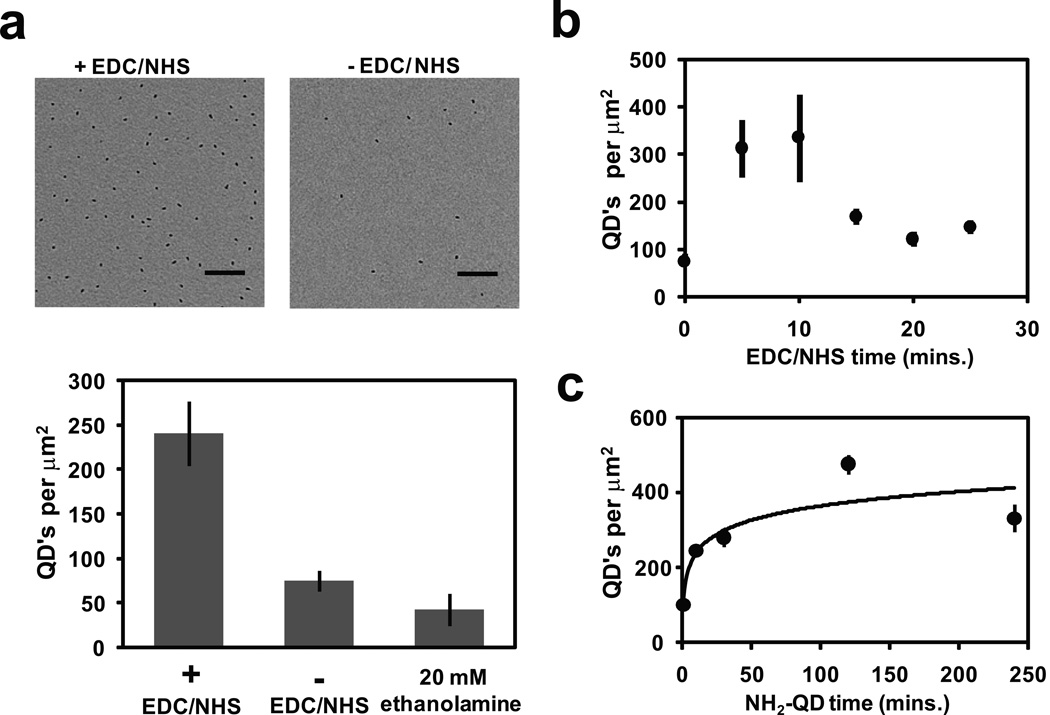
After oxidization and proper cleaning, the ChemiC films are suitable to react preferentially with primary amines, which will allow an extensive list of biological ligands to be introduced under regular laboratory settings. We developed a reproducible procedure to clean the oxidized carbon surface and remove all electron-dense artifacts (see Methods). We optimized the reaction conditions so that it will allow a quantitative control of the surface densities of reactive carboxylates as well as amide formation. Our reaction is based on the EDC/NHS conjugation scheme (Hermanson, 2008). For the quantification of introduced ligands, we used amine-Quantum Dots (QD’s) to estimate the surface density of reactive groups by EM. The QD density on the carbon surface will provide a lower limit to the surface density of reactive groups.
When we compared the reactions with and without sulfo-NHS, the surface treated with NHS had much higher surface density of QD’s, suggesting that the reaction needs NHS-based catalysis for high efficiency (Fig. 2a). When ethanolamine was used to block all reactive sites, the surface QD density decreased to background levels, which supported that the reaction is amine-specific. These two aspects together demonstrate that the EDC/NHS reaction on the ChemiC films is successful and can be quantitatively controlled by manipulating the reaction conditions.
Figure 2. Quantification of amine-reactive esters using amine-Quantum Dots.
(a) Top: Two images show the QDs on the carbon surface when the linkage was done with (left image) or without (right) the EDC/NHS treatment. Scale bar = 100 nm. Bottom: Quantification of the surface density of the linked QDs (error bars from S.E.M., n=7). As a negative control, 20 mM ethanolamine was introduced to block all the reactions, and the remaining QDs reflect the nonspecific binding to the blocked ChemiC surface.
(b)The reaction varies with the incubation time of 5.0 mM EDC and 5.0 mM sulfo-NHS. The oxidization time was fixed at two hours, and the surface density was measured by direct counting under the EM (error bars represent S.E.M., n=7).
(c) The surface density of linked QDs as a function of incubation time with NH2-QD. The oxidization was fixed at two hours and the treatment with EDC/NHS was 10 minutes (error bars are S.E.M., n=7).
With the QD assay, we next optimized the parameters for the EDC/NHS treatment. At room temperature, the 5–10 minute treatment yielded the highest QD density (Fig. 2b). Longer treatment actually decreased the surface density of the reactive carboxylates, which in turn led to the decreased QD density.
In a similar fashion, the amide-based conjugation was optimized, and 2–4 hours was found sufficient to saturate almost all carboxylate groups on the surface (Fig. 2c). The estimated carboxylate density on the carbon surfaces is at least 400 per µm2. At this density, each anchored ligand (QD in this case) on average occupies a 50 × 50 nm2 surface area. This level of density is suitable for anchoring large multi-Mega-Dalton molecular complexes into individual spaces without mutual interference.
Ni-NTA-ChemiC films suitable for selective retention of His-tagged proteins
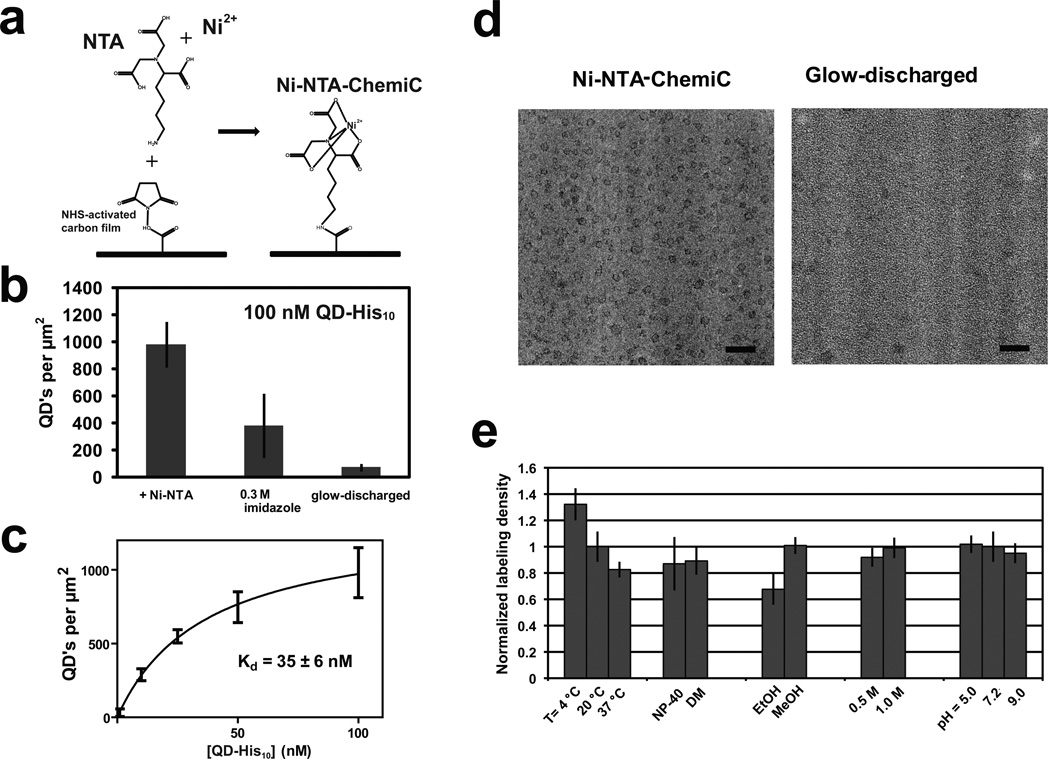
His-tags (6–12 consecutive Histidine residues) have seen extensive applications in protein purification and anchorage. Introducing Ni-NTA groups therefore came naturally to our design (Fig. 3a). To detect His-tag binding, we applied His-tagged QDs (see Methods). With 100 nM QD-His10 all Ni-NTA sites should be saturated, and the measured surface density of Ni-NTA sites was ~1000 per µm2, higher than the estimate from the direct conjugation of amine-QD (Fig.3b). In consideration of the size of the QD, the conjugation of amine-NTA was probably much more efficient than that of the amine-QD. The bulky QD might also suffer from steric hindrance in the conjugation of amine-QD to those carboxylates that happened to be in a disfavored orientation. We therefore think that the 1000 per µm2 is a more faithful estimate of the surface density of small biological ligands. The surface binding affinity of QD-His10 is ~35 nM (Fig. 3c), close to the estimated affinity of His-tags to Ni-NTA groups on the surface of affinity beads (Block et al., 2009).
Figure 3. Binding of His-tagged complexes to Ni-NTA-ChemiC films.
(a) Ni-NTA groups are covalently linked to make the Ni-NTA-ChemiC films.
(b) Enrichment of QD-His10 peptide complexes to the Ni-NTA-ChemiC films. The highest density is ~1000 per µm2 (error bar from S.E.M., n=7). 0.30 M imidazole was able to elute the majority of the bound QD-His10 peptide complex. As a control, the glow-discharged carbon films were loaded with the same amount of QD-His10 peptide complexes, and blotted for direct counting under EM. The residual amount is about 50 per µm2. All grids were incubated in 5.0µL of QD-His10 peptide solution for 15 minutes, washed three times with buffer, and dried in air before EM examination.
(c) The surface density as a function of the QD-His10 peptide concentration used in the reaction (error bars are S.E.M, n=7). Fitting with a binding function, B = Bmax / (1 + kd / [L]), gives a kd = 35 ± 6.0nM.
(d) CryoEM images of His-tagged VCP on Ni-NTA-ChemiC (left) showed significant enrichment in comparison to the same quantities of complexes loaded to the glow-discharged carbon surfaces (right). The ChemiC grids were incubated with 5.0 µl of the protein at 0.10 nM for 30 minutes. The total amount of protein per grid was ~0.3 nanograms. Scale bar = 50 nm.
(e) Tolerance of the Ni-NTA-ChemiC for variations in temperature, detergents, organic solvents, salts and pH. The grids were labeled containing with 5 nM QD-His10 peptide complex at different temperatures and with different detergents (0.5 mM NP-40 and 5 .0 mM DM) in buffer containing 20 mM HEPES pH 7.2, 150 mM NaCl. They were also immersed in different solvents (20% ethanol and 20% methanol), buffers with different salt concentrations (20 mM HEPES pH 7.2 with 0.5 M or 1.0 M NaCl), and buffers at different pH levels (0.1 M MES pH 5.0, 20 mM HEPES pH 7.2, 150 NaCl and 50 mM Borate buffer pH 9.0) for 30 minutes. After washing with water, they were labeled with 10 nM QD-His10 peptide complex. The labeling density is normalized to room temperature (20°C) and pH 7.2 conditions which was ~ 250 QD’s per µm2. Error bars are S.E.M., n=10.
For efficient binding, we would like to enhance the apparent surface affinity of Ni-NTA. It is expected that complexes with multiple copies of His-tags and ligands containing two or three Ni-NTA moieties should interact with sub-nM affinity. To test this idea, we imaged the hexameric His-tagged VCP AAA+ ATPase on the Ni-NTA-ChemiC grids under cryo-condition (Fig. S1). Six His-tags made it possible for the VCP molecules at 0.10 nM (5.0 µL) to be enriched onto the Ni-NTA-ChemiC surface. After 30-min incubation, direct blotting and plunge-freezing, a high density of VCP complexes on the surface were observed under cryoEM (Fig. 3d). From the measured surface density, ~45% of the 0.50 femtomoles of VCP complexes loaded on each grid were retained. Under the same conditions, the density of the His-tagged VCP loaded on freshly glow-discharged carbon films, both negatively- and positively-charged, was much lower (Fig. 3d).
To test the robustness of the Ni-NTA-ChemiC films, we treated them at three different temperatures (4, 20, 37 degrees C), two different detergents (0.5 mM NP-40 and 5.0 mM n-decyl-β-D-maltopyranoside (DM)), two different organic solvents (20% ethanol and 20% methanol), two different salt concentrations (0.5 and 1.0 M NaCl), and three different pHs (5.0, 7.2, and 9.0). After the treatment, the grids were washed and labeled with 5.0 nM QD-His10. The quantified surface densities of the QDs suggested that Ni-NTA ChemiC films were able to tolerate these insults very well (Fig. 3e). For long-term storage, we tested instead the more sensitive Protein G grids (see next).
CryoEM imaging of low-abundance macromolecular complexes has been difficult because the blotting/plunging method usually leads to the loss of more than 95% of the sample. Without affinity-driven interactions with the carbon film, it is necessary to start with concentrated molecules in a small volume. In contrast, the functionalized films can retain more than 40% of the loaded molecules at subnanomolar concentrations. The method described here is therefore particularly suitable for imaging molecular complexes that are available at nM concentrations in small volumes. It opens a new avenue to the 3D reconstructions of low-abundance molecular complexes that are currently evading structural studies even by the less mass-demanding cryoEM techniques.
ChemiC films presenting Protein G for antibody capture
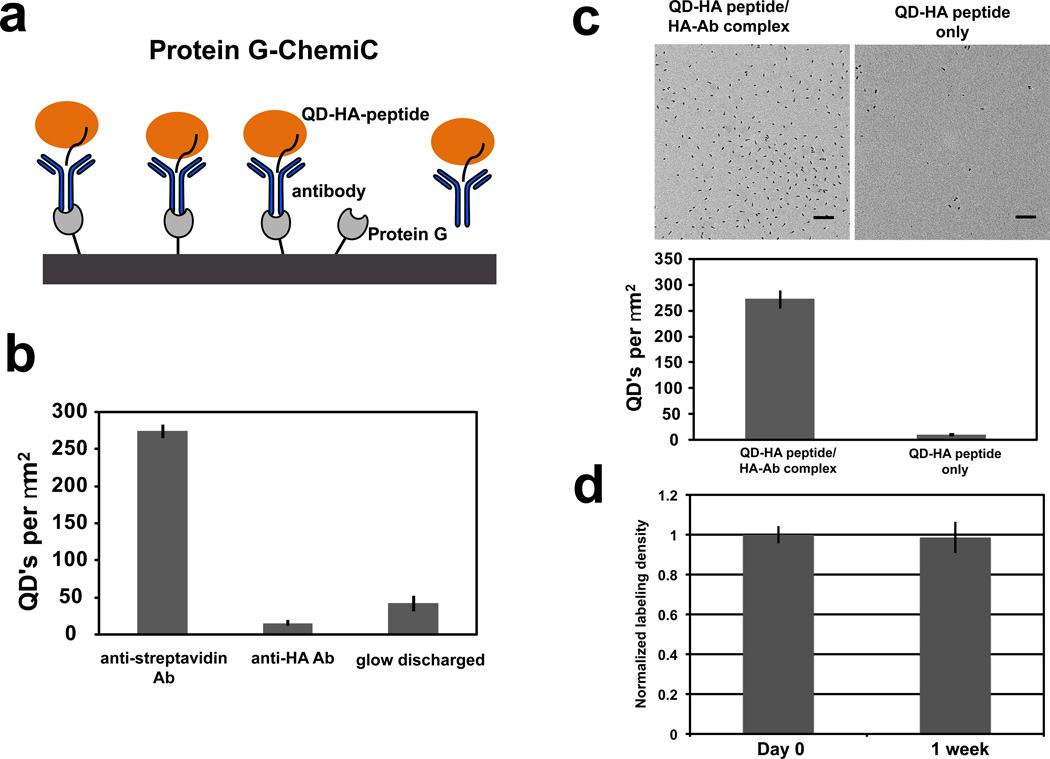
Antibody-antigen interactions can be exploited on carbon films using covalently bound Protein G (Fig. 4a). The EDC/NHS treated surface was reacted with the primary amines of Protein G. To verify the presence of Protein G, we labeled the surface with anti-streptavidin IgG antibodies and then incubated it with QD-streptavidin. Anti-hemagglutinin (HA) IgGs were used as a control for specificity. A glow-discharged grid was included as a control for conventional methods. Minimal labeling was observed in both glow-discharged and anti-HA antibody-bound grids (Fig. 4b), suggesting that the Protein G linked at the carbon surface is capable of capturing IgG antibodies, and the captured IgGs retain their specificity. The selectivity of the surface-presented Protein G molecules was further validated through a quantitative reaction with streptavidin-coated Quantum Dots that were conjugated to biotinylated HA-peptides and bound to anti-HA antibodies (see Methods). As control, a QD-HA peptide complex was used. The Protein G-ChemiC surface efficiently captured the Ab-containing complexes, resulting in approximately a 20-fold increase in labeling density (Fig. 4c).
Figure 4. Protein G-ChemiC films for antibody-based capture of specific complexes.
(a) Recombinant Protein G (grey) is covalently linked to the surface and can be used to capture antibodies (blue) through Fc binding. The bound antibodies can recognize the target proteins (orange) containing specific epitopes (black line). An antibody-containing complex may be readily bound to the Protein G surface.
(b) Protein G-ChemiC grids first incubated with anti-streptavidin or anti-HA antibodies, and then labeled with 10 nM QD-streptavidin after BSA blocking (0.1 mg/mL). Only those treated with the anti-streptavidin antibodies showed specific binding of QD-streptavidin. Glow-discharged surface has very low binding (error bars are S.E.M., n=7).
(c) Typical TEM images of Protein G grids blocked with BSA and labeled with 10 nM QD-HA peptide with and without anti-HA antibodies (top). More than 20-fold higher density of Ab-containing QD complexes demonstrated efficient binding to the Protein G-ChemiC surface (bottom). Error bars are S.E.M., n = 10. Scale bar = 100 nm.
(d) The Protein G-ChemiC grid is suitable for long-term storage. Protein G is known to be stable after lyophilization. The grids were rinsed with water which diluted the salts and buffer to low ionic strength, dried in air, and then left in room temperature for a week before they were washed with the buffer and tested for anti-body capture. The captured antibodies were quantified by the QD-HA peptide complexes. The Protein G grids with anti-HA antibody are compared with the same blocked with mouse IgG as control prior to labeling with 10 nM QD-HA peptide.. Error bars represent S.E.M., n = 10.
Because Protein G is a relatively stable protein, we tested its stability on the surface of the ChemiC films over a week. The Protein G-grids were prepared in a buffer, rinsed with water before it was dried out for long-term storage. We compared the binding of the QD-HA-peptide to the Protein G grids that were saturated with anti-HA antibody or mouse IgG as negative control. The HA-Ab labeled Protein G grids showed at least 10 fold increase in labeling compared to negative controls. The data showed that these grids are fairly stable over a long period of time (Fig. 4d).
ChemiC films presenting oligonucleotides (oligos)
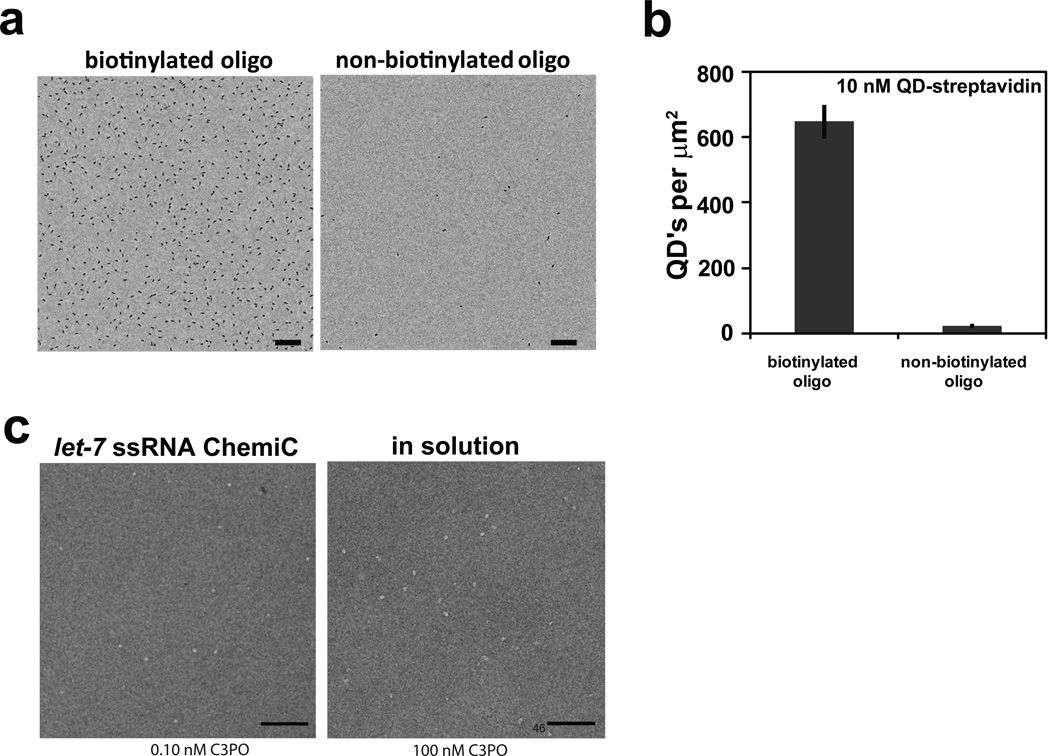
The same conjugation scheme can be extended to a battery of biological ligands. We tested reactions that allowed the surface-anchorage of DNA/RNA oligos to the ChemiC films. Surface-anchored DNA or RNA oligos were introduced by using 5’ or 3’-amines. Due to the multiple primary and secondary amines in the nucleotide bases, it initially appeared possible that the oligos could be anchored through a base. Considering the freedom of the 5’ and 3’ ends, we believed that the reaction was probably more favorable to the terminal amines. For quantification purposes, we used an oligo containing a 5’-amine and a 3’-biotin for chemical conjugation, and quantified its surface density with streptavidin-coated QDs (Fig. 5a, 5b). Because the average distance between two neighboring anchored sites is ~50 nm, the length of the oligos should preferably be less than 25 nm in order to minimize mutual interference between neighboring sites. This dimensional limit is equivalent to ~50 bases, and should not impose any limit to many applications.
Figure 5. Anchorage of DNA and RNA-oligos on ChemiC films.
(a) ChemiC films were conjugated with 1.0 µM of ssDNA, which contains 5’ -amine and 3’–biotin. 10 nM QD-streptavidin was used to quantify the oligos. An EM image (left) shows the high density of oligos on the surface. The surface density of the QD was counted after the films were incubated with 10 nM QD-streptavidin conjugates and washed with buffer. As a control, the oligos of the same sequence but without 3’-biotin was used in the reaction and a typical image is presented to the right side. Scale bar = 100 nm.
(b) Quantification of the surface density of the oligos on the ssDNA-ChemiC films generated in (a). The QD-streptavidin density reflects the minimal density of the oligos (~650 per µm2). The nonspecific binding of the QD-streptavidin to the oligo-ChemiC was fairly low (less than 40 per µm2). Error bars represent S.E.M., n = 11.
(c) Left: Negative-stain EM image of 0.10 nM C3PO loaded on the let-7 ssRNA-ChemiC films. Oligomeric particles in red circles are typical C3PO oligomers (likely to be octamers). Right: 100 nM C3PO mixed with ssDNA (5’-TGAGGTAGTA-GGTTGTATAGT-3’) at 1:2 molar ratio in solution before loading on glow-discharged carbon-coated grids. C3PO/ssDNA complexes at high concentrations are primarily octameric (red circles highlight typical oligomers) with a distinct hollow interior similar to the C3PO complexes assembled on anchored let-7 ssRNAs. Scale bar = 100 nm.
The versatility of the carboxyl-amine conjugation bestows the capability of introducing many other ligands. We are establishing conditions for presenting mono-and poly-saccharides, specific amino acids, glutathione, dsDNAs, dsRNAs, microRNAs, lipid analogs, etc. In the next section, we will demonstrate the practical application of the ChemiC grids to addressing a biological problem.
C3PO assembled on let-7 ssRNA-ChemiC films
Structural studies of C3PO as dimers, tetramers, hexamers and octamers raised a fundamental question about what oligomeric state the native C3PO takes inside a cell (Parizotto et al., 2013; Tian et al., 2011), and how C3PO recruits short ssRNAs for degradation. Crystallographic studies were all conducted with high concentrations (0.2–0.4 mM) of proteins (Tian et al., 2011; Ye et al., 2011), orders of magnitude higher than the native concentration.
Inside a cell, the C3PO recognizes the nicked passenger stand RNA on the surface of the RISC-dsRNA complex. It is reasonable to think that the passenger strand RNA must have one end protruding out as a short single-stranded fragment because C3PO does not act on duplex RNAs. With a similar consideration, the surface anchored ssRNAs on the thin carbon films mimic, to certain extent, the native scenario, and should be in a configuration that is recognizable by freely floating C3PO. To do that, C3PO protein at 0.10 nM was incubated with the let-7 ssRNA-ChemiC grids for ~30 minutes before the protein complexes were negatively stained for EM imaging. We introduced 0.50 % trehalose in the stain solutions to minimize the potential air-drying effects. In a cell of 20 µm in diameter, 300 molecules in the cytosol would give rise to a concentration of ~0.12 nM. The concentration regime we used here is likely close to the native concentration of low-abundance macromolecular complexes. In a parallel experiment, 100 nM C3PO incubated with let-7 ssDNA in solution (1000 × higher in concentration on glow-discharged carbon films) showed similar oligomers with distinct central hollowness in many images (Fig. 5c). The reaction conditions on the ssRNA-ChemiC films therefore seem to be applicable to low-abundance complexes inside a cell. These data suggest that the anchored ssRNAs are capable of guiding the assembly of C3PO into oligomers, which may represent the in vivo behavior of C3PO. Mg2+ was found to be able to activate the C3PO hydrolysis of the anchored RNAs (data not shown), suggesting that the C3PO assembled on the ssRNAs are in a conformation that is very close to the catalytically competent state.
C3PO assembled on ssRNA-ChemiC films shows a side port
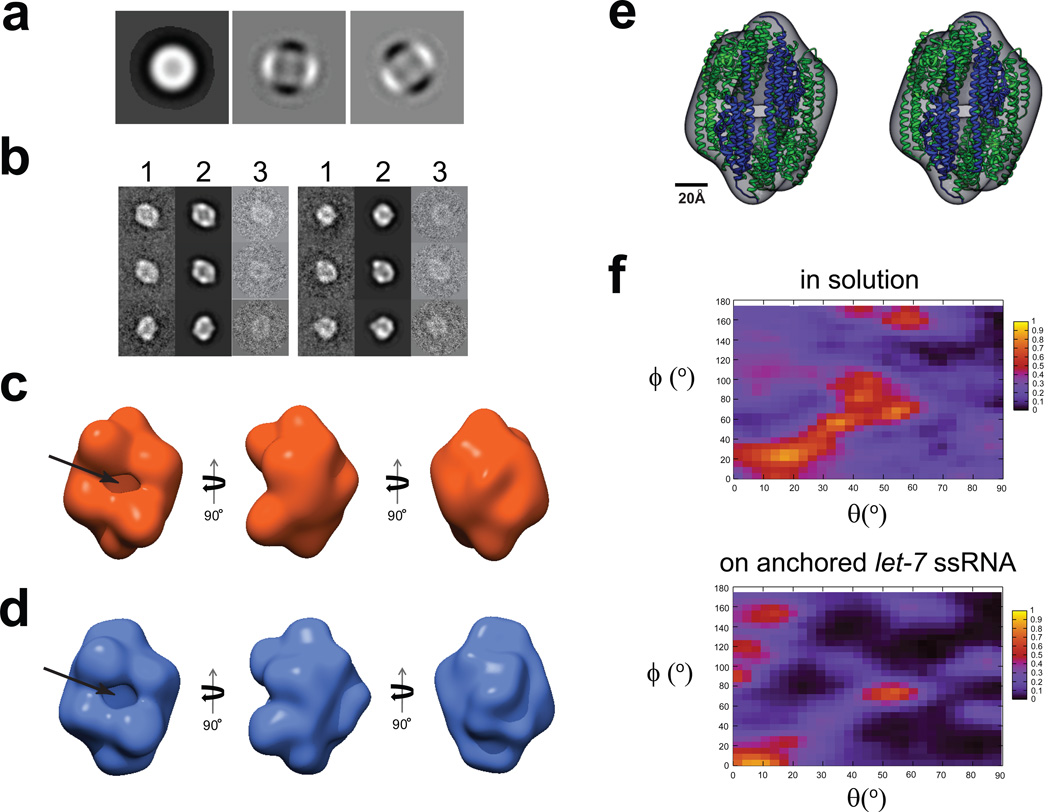
X-ray crystallographic studies of octameric C3PO revealed a football-shaped structure that encloses the active endonuclease sites inside the internal cavity. This structural model raises a serious question on how the ssRNA substrates reach the interior active sites. To address this question, we built a large dataset of single-molecule images taken from negatively stained C3PO complexes that were assembled on the ssRNA-ChemiC films. Based on the 2nd and 3rd eigenimages out of multivariate statistical analysis, the particles have clear two-fold (C2) symmetry (Fig. 6a). Our reconstruction started with C1 symmetry, but showed apparent C2 symmetry [Fig. S2a; see (Jiang et al., 2004)]. We calculated a 3D structure at an estimated resolution of 21 Å (Fig. 6b & 6c and Fig. S2b; details in Methods). The small mass size of C3PO led us to first determine the structure of the negatively stained samples in order to obtain a reliable model from images of high signal-to-noise ratio. The handedness of the structure was tested against 70 pairs of particles from tilted and untilted images. Because of the small size of the C3PO particles, we used a different procedure for handedness validation (Fig. S3 and Methods).
Figure 6. 3D reconstructions of C3PO assembled on ssRNA-ChemiC films.
(a) First three eigenimages after the multivariate statistical analysis of the center-aligned raw dataset. Strong 2-fold symmetry is evident from the second and third eigenimages, which were also seen in the dataset of the C3PO/DNA complexes in solution (Figure S4a).
(b) Representative class sums (1), projections from the 3D reconstruction (2) at the corresponding orientations, and the corresponding raw data (3) agree well with each other, and clearly reveal a football-shaped structure with a hollow interior. The matching among these three sets suggests that the 3D reconstruction does reflect the features in the raw images of the C3PO/ssRNA complexes on the ssRNA-ChemiC films.
(c) Three different views of the 3D reconstruction of C3PO/anchored let-7 ssRNA complexes. The side port is ~15 × 25 Å2. The volume is surfaced to enclose a mass of ~240 kDa, the expected mass of a C3PO octamer. If we threshold the density to reflect a density of a hexamer (180 kDa), the densities started to show disconnections, suggesting the 3D map likely represents an octamer.
(d) Three views of the 3D reconstruction calculated from negatively stained C3PO/ssDNA complexes in solution. This control reconstruction was obtained completely independently of the map in (c). It shows very similar features. The size of the side port is almost the same.
(e) Stereo view of the X-ray structure of the human C3PO octamer (ribbons; PDB code: 3PJA; blue for TRAX and green for translin subunits) docked into the 3D model (semi-transparent surface map in grey) of C3PO on the ssRNA-ChemiC-coated grids. The octameric structural model was positioned such that the two TRAX subunits are at the side port. The crystal structure shows a small gap at the TRAX dimer interface, which was proposed to become wider in order to admit the ssRNA substrates.
(f) Heat maps showing the orientational distribution for individual C3PO particles bound with ssDNA in solution (top) and that for C3PO complexes assembled on ssRNA-ChemiC films (bottom). The angles θand ϕfollow the standard definition in SPIDER.
The most striking feature is that the RNA-guided C3PO assembly contains a side port, whose lateral dimensions are large enough for ssRNAs or ssDNAs to interact (Fig. 6c). At the current condition, we were not able to recognize the density of the ssRNA. The apparent hypothesis is that ssRNAs sit at the lateral opening in order to reach the active sites of the C3PO oligomer.
As a comparison, we reconstructed the C3PO structure after the proteins at 100 nM were mixed with ssDNAs in solution and then negatively stained for imaging (Fig. 6d and Fig. S4). The two independently calculated structures exhibit similar overall architecture with the side port. In negative-stain EM, the average density of RNA nucleotides is significantly lower than the protein density (Penczek et al., 1992). Further, lack of sequence specificity in the C3PO binding to ssRNA would make the RNA molecules poorly registered on the C3PO octamer, possibly decreasing its density level. It is therefore not surprising that the negative-stain map at the current threshold level has no visible RNA density in the side opening or inside the C3PO barrel.
Even though the limited resolution of negative-stain EM does not allow us to distinguish TRAX from translin in the 3D reconstructions, the clear two-fold symmetry and the comparison with the octameric X-ray structure (Fig.6a, 6e and Fig. S4a) suggest that the RNA-guided C3PO assembly is probably octameric with two TRAX molecules lining the side port. Such an arrangement leaves the active sites right next to the side port so that even ssRNAs bound at the opening may be able to reach the active sites. This mode of action does not require the deep insertion of the ssRNAs into the internal cavity, and offers a possible explanation for the fragmentation of circular RNAs by active C3PO (Ye et al., 2011). Further experiments will be needed to evaluate this mechanism.
To test whether the surface-presented ssRNAs lend significant constraints to the orientational freedom of the C3PO oligomers, we compared the Euler angle distribution of individual images from the two C3PO datasets with RNAs either anchored to ChemiC films or free in solution. The distribution of Euler angles assigned by projection matching shows very similar patterns (Fig. 6f). The molecules on the ChemiC surface have even less significant peaks in the distribution statistic, suggesting that the RNAs above the carbon films might have slightly decreased the chance of C3PO oligomers to take the four preferred orientations that resulted from the C3PO-carbon film interactions (top panel, Fig. 6f). Our data alleviate the concern that the anchored ssRNAs might restrict the rotational freedom of the assembled C3PO oligomers and interfere with the single molecule imaging and analysis.
Our cryoEM map from a small dataset of frozen C3PO/RNA complexes on the ChemiC grids demonstrated that the ChemiC grids are suitable for preparing biological complexes for 3D electron microscopy. The map revealed an extra density right above the side port (Fig. 7b). The extra density very likely comes from the ssRNA. It is thicker than a rigid 21-nt ssRNA model (Fig. 7b), probably because of the flexibility in the nucleotides and the lack of sequence-specificity for C3PO-ssRNA interaction. The protein part still has a side port and a hollow interior, and it takes a similar shape as the negative-stain map with better recognition of the individual domains. Our results showcased that with the ChemiC films a 240 kDa protein complex can be well recognized under cryoEM and suitable for 3D reconstruction. It is a surprise that the ssRNA binds on top of the side port instead of being enclosed inside the C3PO barrel. We need better resolutions to elucidate the molecular mechanism of C3PO enzymatic action on ssRNAs (Llaguno et al., manuscript in preparation).
DISCUSSION
The chemical modification and functionalization of nanometer-thick carbon films represents a robust and simple strategy to enrich biological complexes on an inert surface that has high mechanical stability, good electrical and heat conductivity, superior uniformity in two dimensions, and low electron and photon scattering power. Even though we had to use EM techniques to develop this technology and have applied it to the 3D electron microscopy, the versatility in presenting various biological ligands on the ChemiC films makes our strategy suitable for many other applications. Our results by using ssRNA-ChemiC films demonstrate the ssRNA-guided assembly of C3PO at subnanogram levels, and for the first time showed a side port in the C3PO that is suitable for RNA-protein interaction. Our cryoEM study further found that the ssRNA binds to the location of the side port, suggesting that the side port is where the C3PO acts on the ssRNA. Our studies suggest that the ChemiC films with bioactive ligands can be exploited to reconstitute biochemical reactions, and may be useful for various single molecule studies (Cerf et al., 2011; Fowler et al., 2009).
When compared with other methods that have been developed for cryoEM imaging of biological complexes, ChemiC films appear much simpler to produce and more robust. The lipid monolayer based method is limited by the available lipid-conjugated ligands (Kelly et al., 2008). Transfer of a monolayer from the air-water interface to the surface of a grid poses a significant technical challenge. The monolayer does not permit the use of detergents or other harsh solvents. The network of duplex DNAs formed on a grid surface was reported to allow the selective anchorage of G-protein coupled receptors (Selmi et al., 2011). The significant mass of DNA strands in the background certainly would increase noise in EM images. The third method relies on 2D crystals of streptavidin where the background of the crystal could be subtracted through Fourier filtering (Wang et al., 2008). For this method, the target complexes need to be biotinylated, and only in good crystalline areas could images be reliably filtered. Recently graphene films supported by holey carbon-films were proposed to be a better support for single particle cryoEM (Pantelic et al., 2011a; Pantelic et al., 2011b). Hydrophobicity of the graphene surface and the expense in making large-dimension graphene films poses challenges to wide applications (Obraztsov, 2009; Park and Ruoff, 2009). In principle, functionalized multilayered graphene films would be equivalent to or even better than our surface-engineered carbon films. We have been trying to apply the same functionalization strategy to 3–4 layered graphene films in the lab, but are still searching for a good method to reliably transfer the graphene films to EM grids (unpublished observations). With more recent advancement in producing multilayered centimeter-sized graphene films, it will eventually become feasible (Obraztsov, 2009; Park and Ruoff, 2009). We expect that the same surface engineering techniques will be suitable for producing functionalized graphene films.
ChemiC films are suitable for imaging biological complexes
The ChemiC films can present a large number of ligands at a high surface density. On the size of a 3.0 × 3.0 mm2 patch that is enough for covering one EM grid, the total number of ligands is ~109–1010. These ligands are well separated. They can have minimal mass, or they can be functional proteins. Short peptides, small proteins, oligonucleotides, lipids, sugars, and other chemical moieties may all be introduced. On the other hand, the discrete ligands serve as isolated reaction centers. Biological functions, such as the ribosome-mediated protein translation, the telomerase-mediated telomere length maintenance, the spliceosome, the exocyst (TerBush et al., 1996), etc. may all be assembled at these centers. Due to the spatial separation of the anchored ligands, there is minimal interference between neighboring reconstituted units. In addition, the high density of the ligands on the ChemiC films increases the effective affinity of the ligands within the small volume around the surface, which may serve as an absorptive trap and retain the cognate complexes at even sub-nM concentrations. The strong binding of the complexes also prevents the adsorbed complexes from detaching during the fast blotting/freezing step when these complexes are prepared for cryoEM imaging.
Similarly, it is expected that the local enrichment of the complexes can be further improved when two or more copies of the ligands are linked together in one reaction center. For example, if tris-Ni-NTA is anchored to the ChemiC films (Huang et al., 2009), the apparent affinity to the His-tagged complexes would be much higher and the selectivity will be even more stringent. This design would work especially well for those that contain multiple copies of His-tags such that the multi-dentate interaction between bis- (or tris-) Ni-NTA and His-tags on the ChemiC grids collectively increase binding affinity. The same idea may be extended to other applications so that ligands with merely sub-µM affinity could be engineered to enhance significantly the efficacy of selective enrichment on the surface.
The high affinity of Protein G to IgG antibodies is suitable for capturing a wide range of biological complexes on the ChemiC grids. The targeted pull-down of a protein using specific antibodies will be useful for determining the structures of antibody-bound proteins. For example, epidermal growth factor receptor (EGFR) in cancer cells is a leading target for targeted therapy with antibodies. Structural studies of the IgG-bound EGFR may pave a unique venue to determining the structure of mature full-length receptors, which has been a lingering challenge for a long time (Arkhipov et al., 2013; Wang et al., 2011). The same is true for a long list of eukaryotic membrane proteins whose structural and functional studies have been prohibitively difficult because of the difficulty in producing them in large quantities. The ChemiC technology therefore has the potential to break new grounds in these lagging-behind areas.
let-7 ssRNA-ChemiC films as substrates for C3PO assembly
The similarity between the 3D map of C3PO assembled on anchored ssRNAs and that of the molecules mixed with oligos in solution suggests the ChemiC technology can be used for structural interrogation of many protein-ligand interactions. The similar orientational distributions for both datasets suggest that the anchored ssRNAs did not introduce strong orientational bias. The 21-nucleotide let-7 ssRNA with a six-carbon linker (~8 nm in length) together with the relatively small size of the complex (~240 kDa) should provide sufficient rotational freedom so that most, if not all, orientations in the hemispherical orientational space are sampled. We therefore conclude that the anchored ssRNAs impose no obvious constraint on the orientational freedom of the C3PO oligomers. Importantly, when a low concentration of C3PO molecules (0.1 nM, octamer equivalent) were incubated on the let-7 ssRNA-ChemiC grids, the majority of the proteins were assembled into individual octamers because of the stability of the C3PO/RNA complex in the absence of Mg2+ ions. The surface density of the anchored octamers is high enough so that more than 40% of all loaded 300 million octamers (0.10 nM in 5.0 µl) were retained on each grid.
The side port in the two 3D EM maps and the RNA density in the cryoEM map suggest a new way for the ssRNAs to access the active sites of the C3PO complex. The sugar-embedded C3PO and the small size of C3PO oligomer made it unlikely that the side port is an air-drying artifact. The cryoEM map further showed the empty barrel with RNA-bound at the side. The size of the side port and the possible domain rearrangement in the RNA-bound form of C3PO suggest that the eight subunits need to rearrange themselves from the RNA-free octamer structure seen by X-ray crystallography, and assemble into the active RNA-bound state. Further studies are being performed to understand how a ssRNA interacts with the C3PO complex and at what stage the protein portion of the complex transits from the laterally open structure to an enclosed barrel.
Potential applications of ChemiC films in other areas
The nanometer-thick carbon films up to 200 cm2 could be deposited on an inert surface. The procedures described above can be adapted to functionalize such a surface with different chemical moieties at suitable densities. The substrates could be modified or sandwiched against a multi-well array. Such an array may be employed to present millions of individually isolated molecules for various purposes. Individual DNAs, RNAs, antibodies, proteins, lipids, or sugars can all be presented as discrete moieties with no mutual interference (Blixt et al., 2004; Groves and Boxer, 2002; Haab et al., 2001; Ramachandran et al., 2004). The apparent concentration on the surface is high enough such that selective binding can occur with high efficiency. These arrays will likely be useful for various studies, not limited to biological imaging.
Conclusions
Chemical engineering of the ChemiC films is successfully developed and is being applied to a group of biological ligands. It was used as a platform to mimic the interaction of an oligonucleotide with its cognate protein (ssRNA to C3PO), and revealed the novel side port for the RNA-guided C3PO assembly. It will likely see more extensive use not only for imaging individual biological molecules but also for utilizing the surface presentation of various molecules in biomedical research and bioengineering.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr. George de Martino (UT Southwestern Medical Center) for sharing the VCP construct and Dr. Puey Ounjai for initiating the water spreading assay. Special thanks go to Drs. Fred Sigworth, Jose Rizo, Sandy Schmid and Woodring Wright for reading and commenting on the manuscript. This work was mainly supported by an NIGMS EUREKA award (R01GM088745 to Q.-X. J.), and partially by an NIH grant (R01GM93271 to Q.X.J.), a Welch Foundation Grant (I1684 to Q-X J.), a CPRIT grant (RP120474 to Q.-XJ.), and an AHA National Innovative Award (12IRG9400019 to Q.-X.J.). M.C.L. was partly supported by a NIH-NCI Cancer Biology Training Grant (5T32CA124334-04 under the direction of Dr. Jerry Shay at UT Southwestern Medical Center). This work was performed in laboratories constructed with support from NIH grant C06RR30414 (Dr. Jerry Shay as the PI).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Data Deposition: The cryoEM map for the C3PO complex assembled on ssRNA-ChemiC films is deposited to the EM Data Bank with an access number (XXXX).
SUPPLEMENTAL INFORMATION
Supplementary figures (Figures S1 to S5)
Contributions
Q.-X.J designed the experiments and started the initial test of the concept of chemical engineering. M.C.L. executed most of the experiments and brought together the complete procedure. H.X. and L.S. did the single particle reconstruction of the C3PO/ssDNA complex in solution. N.H. prepared the C3PO protein. Q.L. and H.Z. helped design the C3PO experiments on let-7 oligo. M.C.L. and Q.-X.J. wrote the manuscript with comments from all other co-authors.
Competing financial interests
The authors declare no competing financial interest.
References
- Adrian M, Dubochet J, Lepault J, McDowall AW. Cryo-electron microscopy of viruses. Nature. 1984;308:32–36. doi: 10.1038/308032a0. [DOI] [PubMed] [Google Scholar]
- Aitchison TJ, Ginic-Markovic M, Matisons JG, Simon GP, Fredericks PM. Purification, Cutting, and Sidewall Functionalization of Multiwalled Carbon Nanotubes Using Potassium Permanganate Solutions. The Journal of Physical Chemistry C. 2007;111:2440–2446. [Google Scholar]
- Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P. Molecular Biology of the Cell. 5th ed. New York: Galand Science; 2007. [Google Scholar]
- Arkhipov A, Shan Y, Das R, Endres Nicholas F, Eastwood Michael P, Wemmer David E, Kuriyan J, Shaw David E. Architecture and Membrane Interactions of the EGF Receptor. Cell. 2013;152:557–569. doi: 10.1016/j.cell.2012.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belnap DM, Olson NH, Baker TS. A method for establishing the handedness of biological macromolecules. Journal of Structural Biology. 1997;120:44–51. doi: 10.1006/jsbi.1997.3896. [DOI] [PubMed] [Google Scholar]
- Blixt O, Head S, Mondala T, Scanlan C, Huflejt ME, Alvarez R, Bryan MC, Fazio F, Calarese D, Stevens J, Razi N, Stevens DJ, Skehel JJ, van Die I, Burton DR, Wilson IA, Cummings R, Bovin N, Wong C-H, Paulson JC. Printed covalent glycan array for ligand profiling of diverse glycan binding proteins. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:17033–17038. doi: 10.1073/pnas.0407902101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Block H, Maertens B, Spriestersbach A, Brinker N, Kubicek J, Fabis R, Labahn J, Schäfer F. Chapter 27 Immobilized-Metal Affinity Chromatography (IMAC): A Review. In: Richard RB, Murray PD, editors. Methods in Enzymology. Academic Press; 2009. pp. 439–473. [DOI] [PubMed] [Google Scholar]
- Cerf A, Alava T, Barton RA, Craighead HG. Transfer-Printing of Single DNA Molecule Arrays on Graphene for High-Resolution Electron Imaging and Analysis. Nano Letters. 2011;11:4232–4238. doi: 10.1021/nl202219w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong M, Yang LL, Williams K, Fisher SJ, Hall SC, Biggin MD, Jin J, Witkowska HE. A "tagless" strategy for identification of stable protein complexes genome-wide by multidimensional orthogonal chromatographic separation and iTRAQ reagent tracking. Journal of proteome research. 2008;7:1836–1849. doi: 10.1021/pr700624e. [DOI] [PubMed] [Google Scholar]
- Dubochet J, Lepault J, Freeman R, Berriman JA, Homo JC. ELECTRON-MICROSCOPY OF FROZEN WATER AND AQUEOUS-SOLUTIONS. Journal of Microscopy-Oxford. 1982;128:219–237. [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler JD, Allen MJ, Tung VC, Yang Y, Kaner RB, Weiller BH. Practical Chemical Sensors from Chemically Derived Graphene. ACS Nano. 2009;3:301–306. doi: 10.1021/nn800593m. [DOI] [PubMed] [Google Scholar]
- Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- Groves JT, Boxer SG. Micropattern formation in supported lipid membranes. Accounts of Chemical Research. 2002;35:149–157. doi: 10.1021/ar950039m. [DOI] [PubMed] [Google Scholar]
- Haab BB, Dunham MJ, Brown PO. Protein microarrays for highly parallel detection and quantitation of specific proteins and antibodies in complex solutions. Genome Biology. 2001;2 doi: 10.1186/gb-2001-2-2-research0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson R, Chen S, Chen JZ, Grigorieff N, Passmore LA, Ciccarelli L, Rubinstein JL, Crowther RA, Stewart PL, Rosenthal PB. Tilt-Pair Analysis of Images from a Range of Different Specimens in Single-Particle Electron Cryomicroscopy. Journal of Molecular Biology. 2011;413:1028–1046. doi: 10.1016/j.jmb.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermanson GT. Bioconjugate techniques. Amsterdam [u.a: Academic Press; 2008. [Google Scholar]
- Huang Z, Hwang P, Watson DS, Cao L, Szoka FC. Tris-Nitrilotriacetic Acids of Subnanomolar Affinity Toward Hexahistidine Tagged Molecules. Bioconjugate Chemistry. 2009;20:1667–1672. doi: 10.1021/bc900309n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hummers WS, Offeman RE. Preparation of Graphitic Oxide. Journal of the American Chemical Society. 1958;80:1339–1339. [Google Scholar]
- Jiang QX, Wang DN, MacKinnon R. Electron microscopic analysis of KvAP voltage-dependent K+ channels in an open conformation. Nature. 2004;430(7001):806–810. doi: 10.1038/nature02735. [DOI] [PubMed] [Google Scholar]
- Kelly DF, Dukovski D, Walz T. Monolayer purification: A rapid method for isolating protein complexes for single-particle electron microscopy. Proceedings of the National Academy of Sciences. 2008;105:4703–4708. doi: 10.1073/pnas.0800867105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly DF, Dukovski D, Walz T. Strategy for the Use of Affinity Grids to Prepare Non-His-Tagged Macromolecular Complexes for Single-Particle Electron Microscopy. Journal of Molecular Biology. 2010;400:675–681. doi: 10.1016/j.jmb.2010.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Paroo Z. Biochemical principles of small RNA pathways. Annual review of biochemistry. 2010;79:295–319. doi: 10.1146/annurev.biochem.052208.151733. [DOI] [PubMed] [Google Scholar]
- Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- Miyoshi K, Tsukumo H, Nagami T, Siomi H, Siomi MC. Slicer function of Drosophila Argonautes and its involvement in RISC formation. Genes & development. 2005;19:2837–2848. doi: 10.1101/gad.1370605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obraztsov AN. Chemical vapour deposition: Making graphene on a large scale. Nature nanotechnology. 2009;4:212–213. doi: 10.1038/nnano.2009.67. [DOI] [PubMed] [Google Scholar]
- Pantelic RS, Suk JW, Hao YF, Ruoff RS, Stahlberg H. Oxidative Doping Renders Graphene Hydrophilic, Facilitating Its Use As a Support in Biological TEM. Nano Letters. 2011a;11:4319–4323. doi: 10.1021/nl202386p. [DOI] [PubMed] [Google Scholar]
- Pantelic RS, Suk JW, Magnuson CW, Meyer JC, Wachsmuth P, Kaiser U, Ruoff RS, Stahlberg H. Graphene: Substrate preparation and introduction. Journal of structural biology. 2011b;174:234–238. doi: 10.1016/j.jsb.2010.10.002. [DOI] [PubMed] [Google Scholar]
- Parizotto EA, Lowe ED, Parker JS. Structural basis for duplex RNA recognition and cleavage by Archaeoglobus fulgidus C3PO. Nat Struct Mol Biol. 2013;20:380–386. doi: 10.1038/nsmb.2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, Ruoff RS. Chemical methods for the production of graphenes. Nature nanotechnology. 2009;4:217–224. doi: 10.1038/nnano.2009.58. [DOI] [PubMed] [Google Scholar]
- Penczek P, Radermacher M, Frank J. Three-dimensional reconstruction of single particles embedded in ice. Ultramicroscopy. 1992;40:33–53. [PubMed] [Google Scholar]
- Ramachandran N, Hainsworth E, Bhullar B, Eisenstein S, Rosen B, Lau AY, Walter JC, LaBaer J. Self-Assembling Protein Microarrays. Science. 2004;305:86–90. doi: 10.1126/science.1097639. [DOI] [PubMed] [Google Scholar]
- Selmi DN, Adamson RJ, Attrill H, Goddard AD, Gilbert RJC, Watts A, Turberfield AJ. DNA-Templated Protein Arrays for Single-Molecule Imaging. Nano Letters. 2011;11:657–660. doi: 10.1021/nl1037769. [DOI] [PubMed] [Google Scholar]
- TerBush DR, Maurice T, Roth D, Novick P. The Exocyst is a multiprotein complex required for exocytosis in Saccharomyces cerevisiae. Embo Journal. 1996;15:6483–6494. [PMC free article] [PubMed] [Google Scholar]
- Tian Y, Simanshu DK, Ascano M, Diaz-Avalos R, Park AY, Juranek SA, Rice WJ, Yin Q, Robinson CV, Tuschl T, Patel DJ. Multimeric assembly and biochemical characterization of the Trax-translin endonuclease complex. Nat Struct Mol Biol. 2011;18:U658–U651. doi: 10.1038/nsmb.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Heel M, Harauz G, Orlova EV, Schmidt R, Schatz M. A new generation of the IMAGIC image processing system. J Struct Biol. 1996;116(1):17–24. doi: 10.1006/jsbi.1996.0004. 2837. [DOI] [PubMed] [Google Scholar]
- Walters RR, Graham JF, Moore RM, Anderson DJ. Protein diffusion coefficient measurements by laminar flow analysis: Method and applications. Analytical Biochemistry. 1984;140:190–195. doi: 10.1016/0003-2697(84)90152-0. [DOI] [PubMed] [Google Scholar]
- Wang L, Ounjai P, Sigworth FJ. Streptavidin crystals as nanostructured supports and image-calibration references for cryo-EM data collection. Journal of structural biology. 2008;164:190–198. doi: 10.1016/j.jsb.2008.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Longo PA, Tarrant MK, Kim K, Head S, Leahy DJ, Cole PA. Mechanistic insights into the activation of oncogenic forms of EGF receptor. Nature Structural & Molecular Biology. 2011;18:1388–1393. doi: 10.1038/nsmb.2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye XC, Huang NA, Liu Y, Paroo Z, Huerta C, Li P, Chen S, Liu QH, Zhang H. Structure of C3PO and mechanism of human RISC activation. Nat Struct Mol Biol. 2011;18:U650–U643. doi: 10.1038/nsmb.2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu XK, Jin L, Zhou ZH. 3.88 angstrom structure of cytoplasmic polyhedrosis virus by cryo-electron microscopy. Nature. 2008;453:U415–U473. doi: 10.1038/nature06893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Settembre E, Xu C, Dormitzer PR, Bellamy R, Harrison SC, Grigorieff N. Near-atomic resolution using electron cryomicroscopy and single-particle reconstruction. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:1867–1872. doi: 10.1073/pnas.0711623105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.