Abstract
Humans have approximately 400 intact odorant receptors, but each individual has a unique set of genetic variations that lead to variation in olfactory perception. We used a heterologous assay to determine how often genetic polymorphisms in odorant receptors alter receptor function. We identified agonists for 18 odorant receptors and found that 63% of the odorant receptors we examined had polymorphisms that altered in vitro function. On average, two individuals differ functionally at over 30% of their odorant receptor alleles. To show that these in vitro results are relevant to olfactory perception, we verified that variations in OR10G4 genotype explain over 15% of the observed variation in perceived intensity and over 10% of the observed variation in perceived valence for the high affinity in vitro agonist guaiacol, but do not explain phenotypic variation for the lower affinity agonists vanillin and ethyl vanillin.
The human genome contains approximately 800 odorant receptor genes that have been shown to exhibit high genetic variability1–3. In addition, humans exhibit considerable variation in the perception of odorants4, 5 and variation in an odorant receptor predicts perception in four cases: loss of function in OR11H7P, OR2J3, OR5A1, and OR7D4 leads to elevated detection thresholds for the respective agonists isovaleric acid6, cis-3-hexen-1-ol7, β-ionone8, and androstenone9. These results suggest that although the olfactory system uses a combinatorial code where multiple receptors encode a given odorant, a single receptor can have a large influence on the perception of an odorant.
Understanding the role of a single receptor requires functional data for receptor/odorant pairs. Matching mammalian odorant receptors to ligands has seen limited success, and the picture is even worse when considering human odorant receptors; ligands have been published for only 22 of the approximately 400 intact human odorant receptors6, 8–17. This lack of data is a critical bottleneck in the field; matching ligands to odorant receptors is essential for understanding the olfactory system at all levels and is building viable models of olfaction.
Using a high-throughput system for functional testing of odorant receptors18, we can now elucidate the role of missense single nucleotide polymorphisms in odorant receptor function. Here we identify ligands for several orphan odorant receptors, determine the prevalence and functional consequences of missense mutations in odorant receptors, and measure the effect of these functional changes on human olfactory perception.
Results
High-throughput screening of human odorant receptors
To identify agonists for a variety of odorant receptors, we cloned a library of 511 human odorant receptors for a high-throughput heterologous screen. These clones represent 394 (94%) of the 418 intact odorant receptor genes, and 428,793 (47%) of their 912,912 intact odorant receptor alleles present in the 1000 Genomes Project. Some odorant receptors were represented by multiple nonsynonymous alleles in the screen.
We screened the odorant receptor library with a panel of 73 odorants that have been used in previous psychophysical testing9, 19 and used a cyclic adenosine monophosphate (cAMP)-mediated luciferase assay to measure receptor activity20 (Supplementary Fig. 1). In the primary screen we stimulated at a concentration of 100 μM. We selected 1572 odorant/receptor pairs from this primary screen for a secondary screen in which each odorant receptor was tested against a no-odor control as well as 1, 10 and 100 μM concentrations of the odorant in triplicate. For 425 odorant/receptor pairs, at least one concentration of the odorant produced significantly higher activation than the no-odor control. These odorant/receptor pairs included 190 clones representing 160 unique odorant receptors.
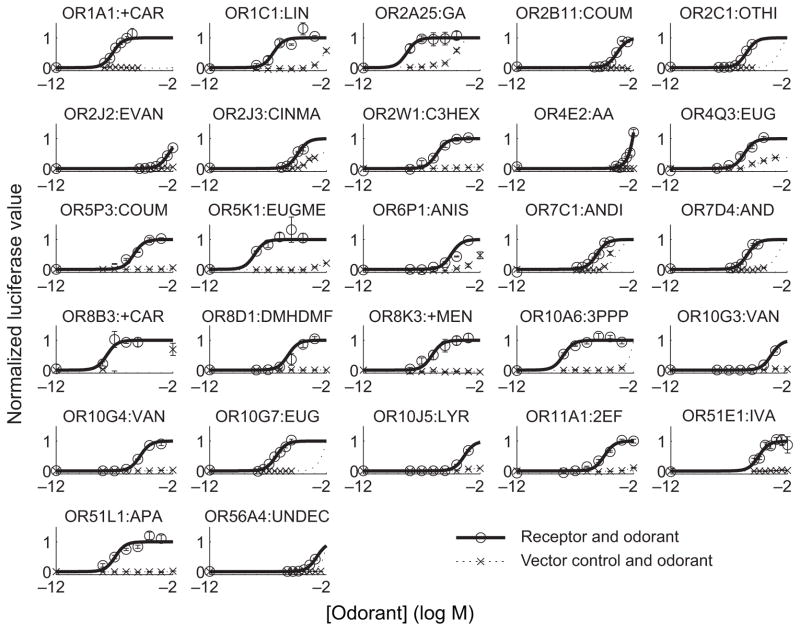
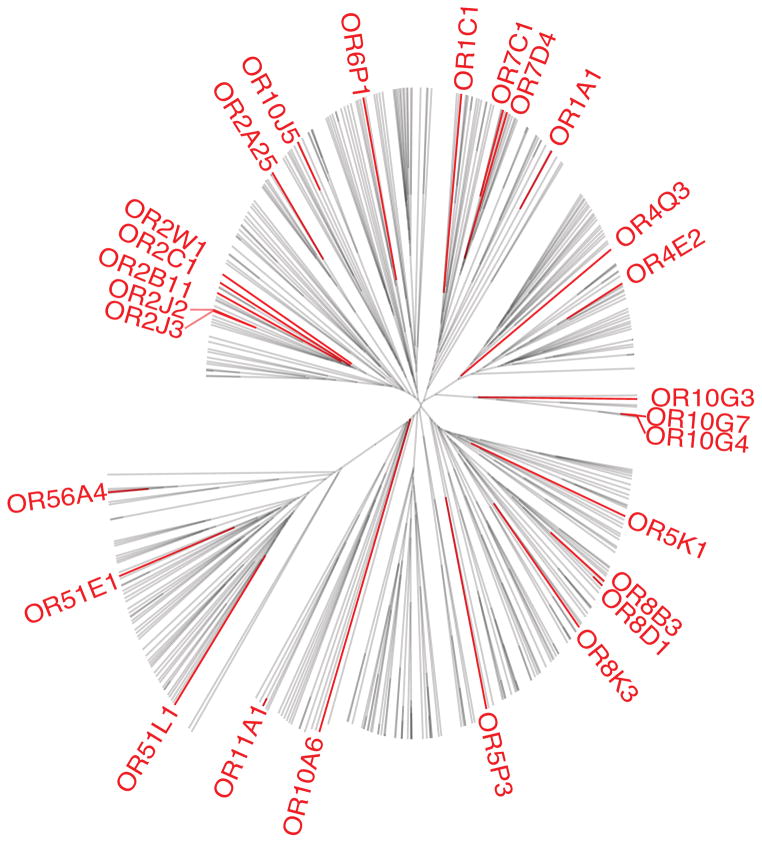
We then constructed dose-response curves for at least one putative agonist of 160 odorant receptors. 27 odorant receptors showed a significant response to at least one agonist, including nine that have previously been shown to respond to at least one agonist in the published literature9, 16, 17 (Fig. 1). For the other 18 odorant receptors we identified new agonists. This nearly doubles the total number of published human odorant receptors with known agonists, bringing the total to 406, 8–17. The receptors identified by this method are spread throughout 9 of the 13 gene families of odorant receptors21 (Fig. 2), suggesting that our assay is useful for examining ligand-receptor interactions across a wide variety of odorant receptors.
Figure 1.
Dose response curves of the most common functional allele for 27 receptors. Circles and solid lines represent the response of the odorant receptor to the odorant in the title of each pane, X’s and dotted lines represent the response of the vector-transfected control to the odorant in the title of each pane. Error bars are standard error. See Table S1 for odor abbreviations.
Figure 2.
Unrooted tree based on similarity of amino acid properties. 27 odorant receptors with agonists are highlighted in red, and represent 9 of the 13 odorant receptor gene families. Grantham’s amino acid property scales were used to quantify receptor similarity30 and distances were calculated using the unweighted pair group method with arithmetic mean (UPGMA).
Genetic variation in odorant receptors
We identified agonists for seven odorant receptors that segregate between intact and disrupted forms (Table 1), bringing the total number of segregating pseudogenes with known agonists to eight6. Combined with psychophysics in a genotyped population, these odorant receptor-agonist pairs can be used to probe the role of a single odorant receptor in olfactory perception.
Table 1.
Seven segregating pseudogenes with agonists. The frequency of the disrupted allele in the 1000 Genomes Project22 is listed. In cases where the variant allele alters a highly-conserved domain in the protein, the conserved amino acid that varies is underscored.
| Odorant receptor name | Frequency of pseudogene allele | Result | Agonist |
|---|---|---|---|
| OR2B11 | 43% | 8 amino acid protein | Cinnamaldehyde |
| OR4E2 | 30% | MAYDRY domain | Amyl acetate |
| OR8K3 | 24% | MAYDRY domain | (+)-Menthol |
| OR10A6 | 22% | PMLNPLIY domain | 3-phenyl propyl propionate |
| OR2C1 | 4% | 272 amino acid protein | Octanethiol |
| OR4Q3 | 1.50% | 159 amino acid protein | Eugenol |
| OR10G7 | 1.40% | 191 amino acid protein | Eugenol |
In addition to segregating pseudogenes and missense variation in conserved amino acid residues, a segregating missense variation that alters non-conserved amino acid residues of odorant receptors can also account for a portion of the variance in odor perception7–9. How many of the odorant receptors with intact open reading frames have functionally different variants, adding to the already considerable amount of variation in the human odorant receptor repertoire? We found a median of 5 alleles with a minor allele frequency (MAF) greater than 1% across 418 odorant receptors in the 1000 Genomes Project. 18 odorant receptors had only one allele with an MAF over 1% across the 2184 haplotypes. In contrast, OR51A2 had 19 different variants with an MAF over 1%. The odorant receptors for which we identified agonists did not exhibit a significantly different number of polymorphisms than odorant receptors without identified agonists (median alleles = 5 for both sets, Mann-Whitney U-test, Z = 0.77, p = 0.44, 2-sided).
To test how variability in amino acid sequence affected odorant receptor activation by odorants, we targeted odorant receptors with at least one known agonist and cloned alleles from pooled genomic DNA with the goal of representing the majority of protein-coding alleles seen in the 1000 Genomes Project. For 16 odorant receptors we successfully cloned 51 alleles, representing an average of 27,118 (77%) of their 34,944 alleles present in the 1000 Genomes Project. One mechanism through which genetic polymorphisms could influence receptor function is by altering cell-surface expression. We assessed the cell surface expression of these 51 cloned alleles using live-cell immunostaining against the N-terminal Rho tag followed by Fluorescent Activated Cell Sorting (FACS). Relative surface expression among each set of variants does not correlate with either relative potency (Spearman rho=0.04, p=0.82, Supplementary Fig. 2a) or relative efficacy (Spearman rho=0.13, p=0.45, Supplementary Fig. 2b) of the variants in the functional assay. While a complete lack of surface expression eliminates receptor responses to known agonists, a high level of surface expression does not reliably confer additional sensitivity. A small amount of cell surface expression is sufficient to confer functional responses. In summary, FACS does not provide enough resolution to determine if functional variation is due to cell-surface expression defects.
Functional consequences of genetic variation
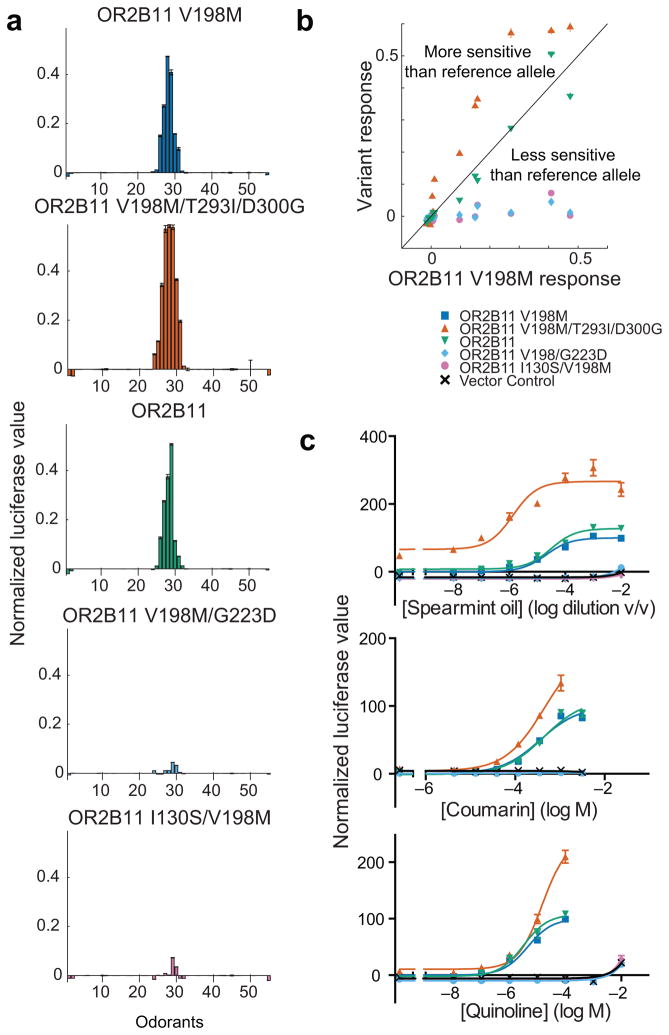
We screened 46 of the alleles used in the FACS analysis against 55 odorants chosen quantitatively to span the physicochemical space17 (Supplementary Fig. 3). Across odorants the absolute magnitudes of response varied, but the relative responses of variant alleles remained consistent (Fig. 3a,b, Supplementary Fig. 4). In other words, if a variant is hypersensitive to one agonist, that variant tends to be hypersensitive to all agonists. We found no case of a genetic change that resulted in a change in odor tuning (Supplementary Fig. 4), but our odorant library design was chosen to span odorant space and was therefore not ideal for identifying more subtle changes.
Figure 3.
Functional testing of odorant receptor variants. (a) Sensitivity-ordered tuning curves for 5 variant alleles of OR2B11 tested against the 55 representative odorants at 100 μM. If a given odorant did not significantly activate any of the variant receptors above the no-odor control (2-tailed t-test, α=0.05/55), that odorant’s response was set to zero across all variants. Odorants were ordered along the x-axis according to the response they elicited from the OR2B11 reference allele (see Fig. S3 for odor names). Error bars are standard error over three replicates. (b) The responses of the four variant alleles to the 55 representative odorants at 100 μM are plotted against the OR2B11 reference allele’s responses. The black line represents the unit slope line. (c) Dose response curves for the OR2B11 alleles for three different odorants. Y-axis represents the luciferase value normalized to the reference allele. Error bars are standard error over three replicates.
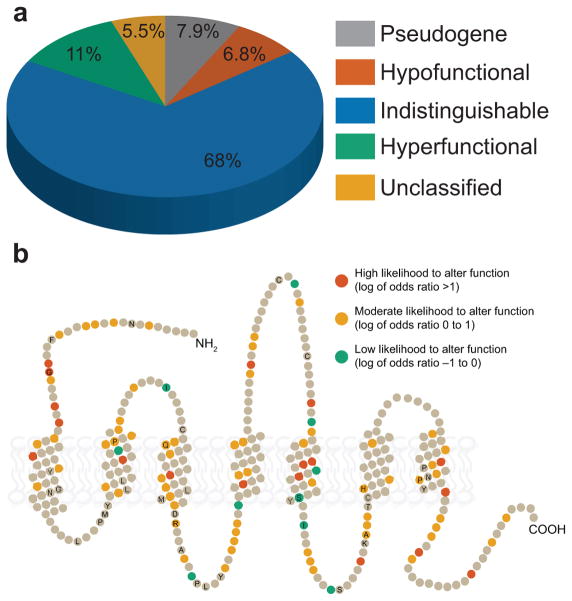
We then examined how the variant allele responses compared across a range of concentrations by constructing a dose response curve from 10 nM to 10 mM (Fig. 3c, Supplementary Fig. 5). Here, we included the 15 odorant receptors tested against all 55 odorants as well as 12 additional odorant receptors. We typically used only a single agonist, as our results from using a broad set of odorants suggested that the differences between alleles using one odorant were highly correlated to differences between alleles using different odorants. We fit the data to a sigmoid curve and compared the variant alleles using an extra sums-of-squares test. A pair of alleles was classified as hyper/hypofunctional if one allele in the pair had both a lower potency (EC50) and a lower efficacy (maximum value). Comparing one allele to all other alleles of the same odorant receptor from the 1000 Genomes Project revealed that 11% of the alleles were hyperfunctional, 68% were indistinguishable and 6.8% were hypofunctional. 7.9% of the alleles were pseudogenes and for 5.5% of the alleles potency and efficacy did not change concordantly, so we could not clearly classify them as hypo or hyperfunctional (Fig. 4a). 63% (17/27) of the odorant receptors we examined had polymorphisms that altered in vitro function. Residues that are polymorphic across alleles with measured function are shown in Figure 4b. There is no obvious pattern to the amino acids that change function; they are found all over the protein. The odds that a residue altered function in our assay did not correlate with evolutionary conservation (GERP score, r = −0.04, p = 0.83), predictions from SIFT (r = 0.05, p = 0.80), or predictions from PolyPhen (r = −0.05, p = 0.81).
Figure 4.
Summary of functional variation. (a) The type of functional differences among 27 odorant receptors of 1092 participants from the 1000 Genomes Project. Note that pseudogenes account for a small portion of the variability relative to missense variations. (b) Snake plot of a typical odorant receptor showing residues where SNPs alter the function of the receptor. Amino acid residues that did not vary between any of the minor alleles and their reference allele are shown in gray. The remaining residues are colored according to the odds that they alter function given our current dose-response data. Amino acid positions conserved in at least 90% of the receptors are labeled with their single-letter amino acid code.
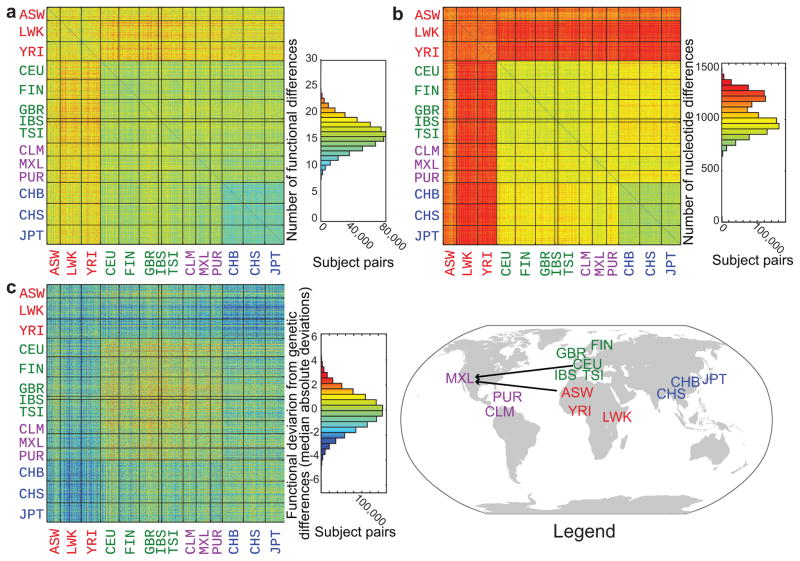
To quantify functional differences across the 1000 Genomes Project population we assigned in vitro results to each participant according to their allele type. We had in vitro results for 46,561 (79%) of the 58,968 alleles (27 odorant receptors x 1092 subjects x 2 alleles). When we conservatively classified all pairwise comparisons including those involving untested alleles as functionally identical, we saw an average of 16 functional differences in dose response out of 54 possible functional differences (27 odorant receptors tested in dose-response x 2 alleles, Fig. 5a, histogram). When we classified all pairwise comparisons including an untested allele as functionally different, we saw an average of 22 functional differences in dose response out of 54 possible functional differences. These results were consistent if we excluded the 500 related participants. In other words, two individuals differ functionally at over 30% (16/54) of their odorant receptor alleles. Pairs where both participants had Asian ancestry (CHB, CHS, and JPT) were more functionally similar than pairs where neither participant had Asian ancestry (median Asian = 13; median non-Asian = 17; Mann-Whitney U-test, z=127, p < 0.0001, 2-sided). Pairs where both participants had African ancestry (ASW, LWK, and YRI) were more functionally different than pairs where neither participant had African ancestry (median African = 16; median non-African = 15; Mann-Whitney U-test, z=29 p < 0.0001, 2-sided)22, in line with those populations having a greater genetic diversity (Fig. 5b). However, when taking genetic diversity into account, pairs where both participants had African ancestry (ASW, LWK, and YRI) were more functionally similar than pairs where neither participant had African ancestry (median African = −0.83; median non-African = 0.36; Mann-Whitney U-test, z=149, p < 0.0001, 2-sided) (Fig. 5c). This shows that, although there is greater genetic variability among Africans, much of this diversity does not translate into functional differences relative to other groups.
Figure 5.
Functional differences between participants. The number of functional differences (a), nucleotide differences (b), and z-scored functional differences minus z-scored nucleotide differences (c) among 27 odorant receptors of 1092 participants from the 1000 Genomes Project. The colors of the squares represent the number of differences between participants. Participant populations are labeled on the axes and separated by black grid lines. The histograms of the number of differences show the color key used in the main figure. The legend displays ethnic groups from (a–c) at the place of geographic origin; arrows point to the location of sample collection. ASW, African ancestry in Southwest USA; CEU, Utah residents with Northern and Western European ancestry from the CEPH collection; CHB, Han Chinese in Beijing; CHS, Han Chinese South; CLM, Colombian in Medellin, Colombia; FIN, Finnish; GBR, British individuals from England and Scotland; IBS, Iberian populations in Spain; JPT, Japanese in Tokyo; LWK, Luhya in Webuya, Kenya; MXL, Mexican ancestry in Los Angeles, California; PUR, Puerto Rican; TSI, Tuscanians in Italy; YRI, Yoruba in Ibadan, Nigeria.
Perceptual consequences of genetic variation
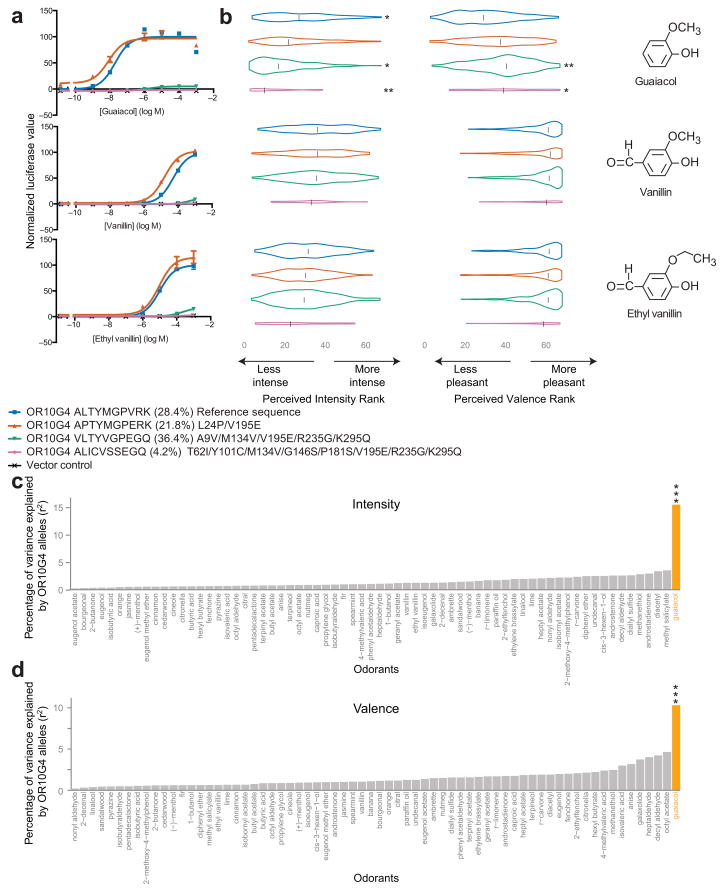
We have so far shown that genetic changes are widespread in the human population and that these genetic changes result in widespread in vitro functional changes. We next set out to determine if the observed in vitro functional changes lead to the predicted perceptual consequences. We selected an odorant receptor, OR10G4, for further testing because we had genomic DNA of subjects that had been tested for their perception of three OR10G4 agonists19, and because functional and non-functional OR10G4 alleles were common in the 1000 Genomes Project22. We successfully obtained OR10G4 sequences from 308 of the 391 participants who had rated their perceived intensity and valence for guaiacol, vanillin, and ethyl vanillin. We then examined the effect of each OR10G4 allele on the perceptual phenotypes (Fig. 6).
Figure 6.
OR10G4 allele effects on perceived intensity and valence. (a) Concentration response curves of OR10G4 alleles with a frequency greater than 4% in the participant population. Error bars are standard errors of 3 replicates. Y-axis values are normalized to the baseline response of the reference allele. (b) Perceived intensity and valence rank for three in vitro OR10G4 agonists by allele of OR10G4. Each participant is represented twice—once for the maternal and once for the paternal allele. The width of each violin is proportional to the number of participants assigning a given rank. The black line inside the violin denotes the median rank. The amino acid changes are relative to the hg19 reference sequence. The frequency listed is the allele frequency in the 308 participants. All unlisted alleles occurred with a frequency lower than 4%. Asterisks signify that the allele had a significant effect in the regression model, and are only shown for regression models that were overall significantly different from a constant model; one asterisk signifies p < 0.05, two asterisks signify p < 0.01. (c,d) Percentage of perceptual variance (r2) in intensity (c) and valence (d) ranking explained by OR10G4 allele types. Each odor was analyzed using the multiple linear regression model outlined in the main text. Three asterisks signifies p < 0.001 after false-discovery rate (FDR) correction. For all other odorants, p > 0.05 after FDR correction.
There were four OR10G4 alleles with an MAF greater than 4% in the participant population: the reference allele (ALTYMGPVRK), and three variant alleles that differ from the reference allele by two (APTYMGPERK), five (VLTYVGPEGQ), or eight (ALICVSSEGQ) amino acids. The APTYMGPERK allele was more sensitive to guaiacol than the reference allele, but the effect was small (log EC50 ALTYMGPVRK = −7.4, log EC50 APTYMGPERK = −7.7, sum of squares test, F(3,42) = 6.38, p < 0.002). The VLTYVGPEGQ allele had a much lower affinity to the three odorants than the reference allele, but still showed significant responses (log EC50 = −5.5, sum of squares test against reference, F(3,42) = 459, p < 0.001; sum of squares test against vector control, F(3,42) = 149, p < 0.001). The ALICVSSEGQ allele was not significantly different from the control cells transfected with vector only (sum of squares test against vector control, F(3,42) = 2.2, p = 0.11) (Fig. 6a). We generated odorant receptors with each of the SNPs in a reference background and found that no single SNP accounted for the functional impairment in the VLTYVGPEGQ and ALICVSSEGQ alleles, suggesting that multiple residues interact to cause the decrease in affinity (Supplementary Fig. 6).
Multiple regression analysis was used to test if OR10G4 allele-type significantly predicted participants’ perception of the three in vitro agonists. The predictors, allele counts (0,1,or 2) for the four alleles with MAF > 4% in the participant population, were regressed against the odor rating rank. OR10G4 allele type predicted 15.4% of the variance in perceived intensity of guaiacol (r2 = 0.165, adjusted r2 = 0.154, compared to constant model, F(4,303) = 15.0, p < 0.001 after false discovery rate (FDR) correction). The model estimated that subjects with none of the major alleles would rank the intensity of guaiacol 24th relative to the other tested odors. Each copy of the ALTYMGPVRK allele is associated with an increase in perceived intensity (decreased rank) of guaiacol by 2.1 ranks (β = 2.10, p < 0.04), and each copy of the VLTYVGPEGQ and ALICVSSEGQ alleles is associated with a decrease in perceived intensity by 2.4 and 4.3 ranks respectively (β = −2.39, p < 0.02; β = −4.34, p < 0.005). The APTYMGPERK allele was not significantly associated with the intensity rank (β= 1.01, p = 0.32).
In addition to intensity, OR10G4 allele type predicted 10.3% of the variance in perceived valence of guaiacol (r2 = 0.115, adjusted r2 = 0.103, compared to constant model, F(4,303) = 9.85, p < 0.001 after false discovery rate (FDR) correction). The model estimated that subjects with none of the major alleles would rank the valence of guaiacol 29th relative to the other tested odors. Each copy of the VLTYVGPEGQ and ALICVSSEGQ alleles is associated with an increase in perceived valence (increased rank) of guaiacol by 3.3 and 3.7 ranks respectively (β = 3.33, p < 0.002; β = 3.71, p < 0.03), but the ALTYMGPVRK and APTYMGPERK alleles were not significantly associated with the valence rank (β = −0.69, p = 0.52; β = 1.88, p = 0.08).
In contrast to guaiacol, neither perceived intensity nor valence of vanillin and ethyl vanillin were predicted by OR10G4 allele-type (vanillin intensity–compared to constant model, F(4,303) = 0.95, uncorrected p = 0.44; ethyl vanillin intensity–compared to constant model, F(4,303) = 0.95, uncorrected p = 0.44; vanillin valence–compared to constant model, F(4,303) = 0.84, uncorrected p = 0.50; ethyl vanillin valence–compared to constant model, F(4,303) = 0.50, uncorrected p = 0.74). As further controls, the 308 participants were also psychophysically tested for their intensity and valence perception of 63 odors that are not known to be OR10G4 agonists, as well as two solvents. Of the 68 compounds, only guaiacol intensity and valence were significantly correlated with OR10G4 allele type (Fig. 6c,d).
Discussion
Here we have identified 27 odorant receptors with known agonists that have functionally different alleles that segregate in the human population and demonstrated that this segregation is relevant to human odorant perception. This nearly doubles the number of human odorant receptors with a known agonist, and is the first investigation of the functional role of genetic variation in a large set of odorant receptors. Pairing odorants and odorant receptors and verifying the functional consequences of segregating polymorphisms in vitro allows us to address previously inaccessible questions regarding how activation of an individual odorant receptor alters olfactory perception. This promises to be a rich future field of study, as we do not currently know how the odorant receptor array codes for odor threshold, intensity, or character. Understanding how the functional alteration of an odorant receptor affects the neural code is a crucial first step in a model of olfactory perception.
Each pair of individuals had, on average, differences in 16–22 out of a possible 54 alleles (27 odorant receptor genes with dose-response data x 2 alleles per subject). If we extrapolate to the approximately 400 intact odorant receptors, we would expect each pair of individuals to differ at somewhere between 237–326 of the 800 alleles. This suggests that odor detection at the peripheral level is highly variable. Variation at the peripheral level leads to variability in odor perception across individuals in several cases; in addition to the OR10G4/guaiacol association demonstrated here, four olfactory perceptual phenotypes have previously been linked to a single odorant receptor genes6–9 and five additional olfactory phenotypes have been linked to regions of the genome containing more than one receptor23–25. Each individual, therefore, has a highly personalized set of olfactory receptors that affects his or her perception of odors.
We chose to focus only on SNPs in the coding regions of the odorant receptors due to the lack of an efficient assay for testing the effects of noncoding polymorphisms on expression. That said, there is considerable variation in noncoding regions, which can lead to altered gene transcription26 and even changes in sensory perception27. Similarly, we did not examine copy number variation, which is widespread in human odorant receptors28, 29. Thus, our data underestimate the potential extent of variation in each individual’s expressed odorant receptor repertoire.
Our study did not find any evidence suggesting SNPs that alter in vitro function are restricted to a particular domain of the receptor, deviate from neutral evolution, or are predicted by two popular computational alogrithms. Note, however, that our study was not designed to carefully detect changes due to a particular SNP; because we did not generate every possible combination of SNPs for the majority of odorant receptors, SNP-specific alterations may be confounded by linkage in the tested alleles.
Although we found that OR10G4 has at least three in vitro agonists, the OR10G4 allele type only predicted perceived intensity and valence for guaiacol. The dose-response curves in Figure 6a show that guaiacol is a more potent agonist than either vanillin or ethyl vanillin. Although more data is needed, one possible interpretation is that the intensity and valence of odorants that only weakly activate a receptor will not be altered by functional variation in the receptor. Indeed, this is similar to the association between OR7D4 and androstenone9. In that case, both of the major alleles respond to androstenone in vitro, but the WM allele is much less potent than the RT allele. As with OR7D4, participants with the lower affinity in vitro allele find the odor to be less intense and more pleasant. This suggests that not all functional variation in vitro will lead to perceptual variation, but the exact rules determining how much of this variation is compensated for at later stages of processing will require further investigation.
OR10G4 explains 15.4% of the variance in guaiacol intensity, which is lower than the 39% of androstenone intensity variation explained by OR7D4 genotype. The reason for this lower explanatory value is unclear. One possibility is that more odorant receptors play a role in the perception of guaiacol than in the perception of androstenone, therefore reducing the influence of a single odorant receptor on the percept. Another is that confounding variables, such as culture and genetic background may have differential effects on the two phenotypes.
The role of a single odorant receptor in olfactory perception is currently unknown, in part because of the large search space for both odorants and odorant receptors and the redundant nature of the combinatorial code for odorant identity. By assigning ligands to odorant receptors, measuring the functional consequences of segregating polymorphisms in vitro, and linking in vitro function to human behavior, these data provide a solid platform from which to probe the effects of a single odorant receptor on olfactory perception.
METHODS
Cloning
Odorant receptor open reading frames were amplified from the genomic DNA of 20 participants from the International Hapmap Consortium using Phusion polymerase and subcloned into pCI expression vectors (Promega) containing the first 20 residues of human rhodopsin (Rho tag). The sequences of the cloned receptors were verified by sequencing (3100 Genetic Analyzer, Applied Biosystems).
Fluorescence-activated Cell Sorter (FACS) Analysis
We conducted FACS analysis on all tested clones for the 17 odorant receptors where we had more than one clone (Supplementary Fig. 5). Hana3A cells were maintained in minimal essetial medium (Sigma) containing 10% fetal bovine serum (Sigma), 500 ug/ml peniciilin-streptomycin (Invitrogen) and 6 ug/ml amphotericin B (Sigma). Cells were seeded in 35mm dishes (Falcon) and grown overnight at 37°C and 5% CO2. The following day, each dish was transfected using 4ul Lipofectamine 2000 (Invitrogen), 1200ng Rho-tagged odorant receptor, 300ng hRTP1S, and 20ng of EGFP to control for transfection efficiency. 24-hours post-transfection, cells were washed with PBS and detached from the dishes using Cellstripper (Cellgro). Primary incubation was carried out at 4°C using mouse monoclonal antibody anti-rhodopsin 4D231 (gift from R. Molday) diluted 1:50 in PBS containing 2% FBS, and 15mM NaN3 for 30 minutes. Cells were washed in PBS/FBS/NaN3, followed by secondary incubation with Phycoerythrin (PE)-conjugated donkey anti-mouse antibody (Jackson Immunologicals) diluted 1:100 in PBS/FBS/NaN3 for 30 minutes covered with aluminum foil. Cells were washed and resuspended in PBS/FBS/NaN3 containing 1:500 dilution of 7-Aminoactinomycin D (7AAD 1mg/ml; Calbiochem), a fluorescent, cell-impermeable DNA binding agent that selectively stains dead cells. Fluorescent cell sorting was conducted using a BD FACSCanto (BD Biosciences). Cells that were EGFP-negative and/or 7AAD-positive were removed from further analysis. Cell-surface expression is quantified as PE fluorescence intensity. Data collection and analysis were not randomized.
Luciferase assay
The Dual-Glo™ Luciferase Assay System (Promega) was used to measure receptor reponses as previously described20. Hana3A cells were transfected with 5 ng/well of RTP1S32, 5 ng/well of pRL-SV40, 10 ng/well of CRE-luciferase, 2.5 ng/well of M333, and 5 ng/well of odorant receptor. 1M odorant stocks are diluted in DMSO. 24 hours following transfection, transfection media was removed and replaced with the appropriate concentration of odor diluted from the 1M stocks in CD293 (Gibco). Four hours following odor stimulation luminescence was measured using a Polarstar Optima plate reader (BMG). All luminescence values were divided by the Renilla Luciferase activity to control for transfection efficiency in a given well. Data were analyzed with Microsoft Excel, GraphPad Prism 4, and MATLAB.
1000 Genomes Project data
Allele frequency in the human population was derived from the May 2011 phased release of the 1000 Genomes Project public data (ftp://ftp-trace.ncbi.nih.gov/1000genomes/ftp/release/20110521/)22. Variant calls were obtained from the public repository in vcf format using tabix34. A custom-written MATLAB script was used to translate the vcf file into 2184 full-length phased alleles (two alleles for each of the 1092 participants in the public database).
Human odorant receptor genotyping
Venous blood (8.5 ml) was collected from participants and genomic DNA was prepared with the Qiagen PAXgene blood DNA kit. For sequencing, human genomic DNAs were amplified with HotStar Taq (Qiagen) with primers upstream (5′-ACCTGGTTGATGCAGTTTCC-3′) and downstream (5′-AAACCTATTGATGAGAAATGAGTCAA-3′) of the OR10G4 open reading frame. The PCR products were then purified using Sephacryl S-400 (GE Healthcare) and sequenced with a 3100 or 3730 Genetic Analyzer (ABI Biosystems).
Procedures for olfactory psychophysics
All psychophysical data was obtained from Keller et al. (2012)19 and approved by the Rockefeller University Institutional Review Board. All subjects gave informed consent to participate and were financially compensated for their time and effort. Exclusion criteria for subjects were: allergies to odors or fragrances, a history of nasal illness, upper respiratory infection, seasonal allergy, prior endoscopic surgery on the nose, pre-existing medical condition that has caused a reduced sense of smell such as head injury, cancer therapy, radiation to head and neck, or alcoholism. Pregnant women and children under 18 were excluded from this study. Of the 308 subjects (138 male), 133 were Caucasian, 29 were Asian, and 77 were African-American. The median age was 35 years, with a range of 19 to 66. In brief, participants rated the intensity and valence of 66 odorants on a 7-point scale. The intensity scale was labeled with 1 as “extremely weak” and 7 as “extremely strong”. The valence scale was labeled with 1 as “extremely unpleasant” and 7 as “extremely pleasant”. Stimuli were presented in jars. For a detailed description of the psychophysical methods, see Keller et al.9. Three of these odorants, ethyl vanillin, vanillin and guaiacol, are in vitro agonists to OR10G4. We examined the ratings of the higher of two tested concentrations. Ethyl vanillin and vanillin were presented at a 1/200 dilution in propylene glycol, guaiacol was presented at a 1/1,000,000 dilution in paraffin oil. Our data collection and analysis was blind to genotype, as all sequencing was conducted after phenotyping of the human subjects was complete. Data collection and analysis were not randomized.
Statistical analysis
Screening procedure
We stimulated the entire odorant receptor library with 73 odorants used in previous psychophysical testing9. We applied the odorants at 100 μM (except for androstenone and androstadienone, which were both applied at 10 μM) and ranked odorant/receptor pairs by their activity above the no odor condition. We selected the top 5% of odorant/receptor pairs from this primary screen--some receptors were very promiscuous, so we tested only the top ten ligands for a given receptor. We then performed a secondary screen in which each odorant receptor was tested against a no-odor control as well as 1, 10 and 100 μM. Each comparison was performed in triplicate, where each measure was collected from separate wells, but each well contains cells from the same parent plate of cells. Statistical significance was assessed by 2-sided t-test comparing the 3 wells stimulated with odor with the 3 wells stimulated with media alone. As this was a screening procedure, the data distribution was assumed to be normal but this was not formally tested. In addition, the tests were uncorrected for multiple comparisons. We then constructed dose-response curves using concentrations ranging from 10 nM to 10 mM for the odor/receptor pairs that were significantly different from baseline in the secondary screen. Each odorant receptor-odorant dose was tested in triplicate, where each measure was collected from separate wells, but each well contains cells from the same parent plate of cells, and a vector-only control was included for each odorant. We fit the data to a sigmoidal curve. We counted an odorant as an agonist if the 95% confidence intervals of the top and bottom parameters did not overlap, the standard deviation of the fitted log EC50 was less than 1 log unit, and the extra sums-of-squares test confirmed that the odorant activated the receptor significantly more than the control, which was transfected with an empty vector. Data collection and analysis were not randomized.
Screening 55 odorants
To choose 55 odorants that quantitatively span chemical space we generated 20 physicochemical descriptors that predict 62% of the variance in mammalian odorant receptor responses17 for 2715 commonly used odorants. We then divided the 2715 odorants into 55 clusters using k-means clustering. For each cluster, we selected the odorant closest to the centroid of the cluster among odorants that are previously shown to activate at least one odorant receptor. If no such agonist was present in the cluster, we selected the odorant closest to the centroid of the cluster to maximize structural diversity. Each odorant was screened against each receptor variant at 100 μM in triplicate where each measure was collected from separate wells, but each well contains cells from the same parent plate of cells. We performed an ANOVA on the responses from the clones of each odorant receptor. We used 15 odorant receptors where we had more than one allele cloned with an allele frequency greater than 1% in the 1092 participants and the cloned alleles represented a large percentage of the 2184 alleles. For 13 odorant receptors, the cloned alleles represented more than 85% of the 2184 alleles. For OR2B11 the cloned alleles represented 37.5% of the alleles and for OR10G4 the cloned alleles represented 29.5% of the alleles. Data collection and analysis were not randomized.
Dose response curves
We tested odorant receptors with odorants ranging in concentration from 10 nM to 10 mM. All numerical results are reported as mean ± s.e.m. and represent data from a minimum of three replicates, where each measure was collected from separate wells, but each well contains cells from the same parent plate of cells. We fit the resulting data with a 3-parameter logistic model. We counted an odorant as an agonist if the 95% confidence intervals of the top and bottom parameters did not overlap, the standard deviation of the fitted log EC50 was less than 1 log unit, and the extra sums-of-squares test confirmed that the odorant activated the receptor significantly more than the vector-only transfected control.
For each pair of alleles, we determined if one model fit the data from both alleles better than two separate models using the extra sums-of-squares test. A pair of alleles was classified as hyper/hypofunctional if one allele in the pair had both a higher EC50 (lower efficacy) and a lower potency (dynamic range, or top-bottom). A pair of alleles was designated as “unclassified” if the potency and efficacy showed discordant changes (i.e. one allele was more sensitive, but had a lower efficacy).
To compare each pair of individuals, we took the four alleles from a single odorant receptor and removed any pairs of alleles that were indistinguishable according to the above criteria. Each remaining pair was counted as one functional difference. These values were summed across odorant receptors, with a maximum of 48 possible functional differences per pair of participants. Data collection and analysis were not randomized.
Odds that a SNP alters function
We aligned the nucleotide sequences of the odorant receptor variants to a multiple sequence alignment of 1425 intact mouse and human odorant receptors. For each SNP we calculated the ratio of the odds that a functional change (as defined above, relative to the most common functional variant) occurred in an allele with a non-synonymous amino acid to the odds that a functional change occurred in an allele with a synonymous amino acid. We used SNPnexus35 (Ensembl 63 build) to generate GERP, SIFT, and Polyphen scores.
Multiple linear regression model
Multiple regression analysis was used to test if the number of OR10G4 alleles significantly predicted participants’ perception of the three in vitro agonists. To determine the minimum sample size for this analysis, we performed a Monte-Carlo simulation using the data from Keller et al.9. We ranked each subject’s ratings of the odorants to control for differences in general olfactory acuity and usage for the rating scale across subjects. The predictors were allele counts (0,1,or 2) for the four alleles with MAF > 4% in the participant population. Data collection and analysis were not randomized.
Supplementary Material
Acknowledgments
This work was supported by R01 DC005782, R01 DC012095, R03 DC011373, R01 DC013339, and an NRSA postdoctoral fellowship F32 DC008932 to J.D.M. A portion of the work was performed using the Monell Chemosensory Receptor Signaling Core and Genotyping and DNA/RNA Analysis Core, which are supported, in part, by funding from the NIH-NIDCD Core Grant P30 DC011735. The FACS analysis was performed using the Duke Cancer Institute Flow Cytometry Core. The authors thank D. Marchuk for sharing equipment, Leslie B. Vosshall for supervising the collection of psychophysical data and DNA samples by A.K. in her laboratory, and R. Molday for 4D2 anti-rhodopsin antibody.
Footnotes
Author contributions
J.D.M. and H.M. conceived and designed the project. J.D.M., C.T., A.H.M., L.L.S., S.Z., L.L, T.Z., Y.R.L., H.Z., S.L., A.L. and K.A.A. performed research. A.K. collected the psychophysical data and provided DNA samples. J.D.M. carried out the analysis and wrote the paper with help from all authors. H.M. supervised the project.
References
- 1.Menashe I, Man O, Lancet D, Gilad Y. Different noses for different people. Nat Genet. 2003;34:143–144. doi: 10.1038/ng1160. [DOI] [PubMed] [Google Scholar]
- 2.Hasin-Brumshtein Y, Lancet D, Olender T. Human olfaction: from genomic variation to phenotypic diversity. Trends Genet. 2009;25:178–184. doi: 10.1016/j.tig.2009.02.002. [DOI] [PubMed] [Google Scholar]
- 3.Olender T, et al. Personal receptor repertoires: olfaction as a model. BMC Genomics. 2012;13:414. doi: 10.1186/1471-2164-13-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ayabe-Kanamura S, et al. Differences in perception of everyday odors: a Japanese-German cross-cultural study. Chem Senses. 1998;23:31–38. doi: 10.1093/chemse/23.1.31. [DOI] [PubMed] [Google Scholar]
- 5.Amoore JE. Chemical Senses and Flavor. Vol. 2. D. Reidel Publishing Company; Dordrecht-Holland: 1977. Specific anosmia and the concept of primary odors; pp. 267–281. [Google Scholar]
- 6.Menashe I, et al. Genetic elucidation of human hyperosmia to isovaleric acid. PLoS Biol. 2007;5:e284. doi: 10.1371/journal.pbio.0050284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McRae JF, et al. Genetic Variation in the Odorant Receptor OR2J3 Is Associated with the Ability to Detect the “Grassy” Smelling Odor, cis-3-hexen-1-ol. Chem Senses. 2012;37:585–593. doi: 10.1093/chemse/bjs049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jaeger SR, et al. A Mendelian Trait for Olfactory Sensitivity Affects Odor Experience and Food Selection. Curr Biol. 2013;23:1601–1605. doi: 10.1016/j.cub.2013.07.030. [DOI] [PubMed] [Google Scholar]
- 9.Keller A, Zhuang H, Chi Q, Vosshall LB, Matsunami H. Genetic variation in a human odorant receptor alters odour perception. Nature. 2007;449:468–472. doi: 10.1038/nature06162. [DOI] [PubMed] [Google Scholar]
- 10.Wetzel CH, et al. Specificity and sensitivity of a human olfactory receptor functionally expressed in human embryonic kidney 293 cells and Xenopus Laevis oocytes. J Neurosci. 1999;19:7426–7433. doi: 10.1523/JNEUROSCI.19-17-07426.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Spehr M, et al. Identification of a testicular odorant receptor mediating human sperm chemotaxis. Science. 2003;299:2054–2058. doi: 10.1126/science.1080376. [DOI] [PubMed] [Google Scholar]
- 12.Sanz G, Schlegel C, Pernollet JC, Briand L. Comparison of odorant specificity of two human olfactory receptors from different phylogenetic classes and evidence for antagonism. Chem Senses. 2005;30:69–80. doi: 10.1093/chemse/bji002. [DOI] [PubMed] [Google Scholar]
- 13.Matarazzo V, et al. Functional characterization of two human olfactory receptors expressed in the baculovirus Sf9 insect cell system. Chem Senses. 2005;30:195–207. doi: 10.1093/chemse/bji015. [DOI] [PubMed] [Google Scholar]
- 14.Jacquier V, Pick H, Vogel H. Characterization of an extended receptive ligand repertoire of the human olfactory receptor OR17-40 comprising structurally related compounds. J Neurochem. 2006;97:537–544. doi: 10.1111/j.1471-4159.2006.03771.x. [DOI] [PubMed] [Google Scholar]
- 15.Neuhaus EM, Mashukova A, Zhang W, Barbour J, Hatt H. A specific heat shock protein enhances the expression of mammalian olfactory receptor proteins. Chem Senses. 2006;31:445–452. doi: 10.1093/chemse/bjj049. [DOI] [PubMed] [Google Scholar]
- 16.Schmiedeberg K, et al. Structural determinants of odorant recognition by the human olfactory receptors OR1A1 and OR1A2. J Struct Biol. 2007;159:400–412. doi: 10.1016/j.jsb.2007.04.013. [DOI] [PubMed] [Google Scholar]
- 17.Saito H, Chi Q, Zhuang H, Matsunami H, Mainland JD. Odor coding by a Mammalian receptor repertoire. Sci Signal. 2009;2:ra9. doi: 10.1126/scisignal.2000016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saito H, Kubota M, Roberts RW, Chi Q, Matsunami H. RTP family members induce functional expression of mammalian odorant receptors. Cell. 2004;119:679–691. doi: 10.1016/j.cell.2004.11.021. [DOI] [PubMed] [Google Scholar]
- 19.Keller A, Hempstead M, Gomez IA, Gilbert AN, Vosshall LB. An olfactory demography of a diverse metropolitan population. BMC Neurosci. 2012;13:122. doi: 10.1186/1471-2202-13-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhuang H, Matsunami H. Evaluating cell-surface expression and measuring activation of mammalian odorant receptors in heterologous cells. Nat Protoc. 2008;3:1402–1413. doi: 10.1038/nprot.2008.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hayden S, et al. Ecological adaptation determines functional mammalian olfactory subgenomes. Genome Res. 2010;20:1–9. doi: 10.1101/gr.099416.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.The 1000 Genomes Project Consortium. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McRae JF, et al. Identification of Regions Associated with Variation in Sensitivity to Food- Related Odors in the Human Genome. Curr Biol. 2013;23:1596–1600. doi: 10.1016/j.cub.2013.07.031. [DOI] [PubMed] [Google Scholar]
- 24.Eriksson N, et al. Web-based, participant-driven studies yield novel genetic associations for common traits. PLoS Genet. 2010;6:e1000993. doi: 10.1371/journal.pgen.1000993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eriksson N, et al. A genetic variant near olfactory receptor genes influences cilantro preference. arXiv. 2012 arXiv:1209.2096 [q-bio.GN] [Google Scholar]
- 26.Zhang X, et al. Characterizing the expression of the human olfactory receptor gene family using a novel DNA microarray. Genome Biol. 2007;8:R86. doi: 10.1186/gb-2007-8-5-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fushan AA, Simons CT, Slack JP, Manichaikul A, Drayna D. Allelic Polymorphism within the TAS1R3 Promoter is Associated with Human Taste Sensitivity to Sucrose. Curr Biol. 2009 doi: 10.1016/j.cub.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Waszak SM, et al. Systematic inference of copy-number genotypes from personal genome sequencing data reveals extensive olfactory receptor gene content diversity. PLoS Comput Biol. 2010;6:e1000988. doi: 10.1371/journal.pcbi.1000988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nozawa M, Kawahara Y, Nei M. Genomic drift and copy number variation of sensory receptor genes in humans. Proc Natl Acad Sci U S A. 2007;104:20421–20426. doi: 10.1073/pnas.0709956104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grantham R. Amino acid difference formula to help explain protein evolution. Science. 1974;185:862–864. doi: 10.1126/science.185.4154.862. [DOI] [PubMed] [Google Scholar]
- 31.Laird DW, Molday RS. Evidence against the role of rhodopsin in rod outer segment binding to RPE cells. Invest Ophthalmol Vis Sci. 1988;29:419–428. [PubMed] [Google Scholar]
- 32.Zhuang H, Matsunami H. Synergism of accessory factors in functional expression of mammalian odorant receptors. J Biol Chem. 2007;282:15284–15293. doi: 10.1074/jbc.M700386200. [DOI] [PubMed] [Google Scholar]
- 33.Li YR, Matsunami H. Activation state of the m3 muscarinic acetylcholine receptor modulates mammalian odorant receptor signaling. Sci Signal. 2011;4:ra1. doi: 10.1126/scisignal.2001230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li H. Tabix: fast retrieval of sequence features from generic TAB-delimited files. Bioinformatics. 2011;27:718–719. doi: 10.1093/bioinformatics/btq671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chelala C, Khan A, Lemoine NR. SNPnexus: a web database for functional annotation of newly discovered and public domain single nucleotide polymorphisms. Bioinformatics. 2009;25:655–661. doi: 10.1093/bioinformatics/btn653. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.