Summary
T cell exhaustion is common during chronic infections. While CD4+ T cells are critical for controlling viral load during chronic viral infections, less is known about their differentiation and transcriptional program. We defined the phenotypic, functional and molecular profiles of exhausted CD4+ T cells. Global transcriptional analysis demonstrated a molecular profile distinct from effector or memory CD4+ T cells and also from exhausted CD8+ T cells, though some common features of CD4+ and CD8+ T cell exhaustion were revealed. We have demonstrated unappreciated roles for transcription factors (TFs) including Helios, type I interferon (IFN-I) signaling and a diverse set of co-inhibitory and costimulatory molecules during CD4+ T cell exhaustion. Moreover, the signature of CD4+ T cell exhaustion was found to be distinct from that of other CD4+ T cell lineage subsets and was associated with TF heterogeneity. This study provides a framework for therapeutic interventions targeting exhausted CD4+ T cells.
Introduction
Following acute infections, memory T cells form that persist long-term and can rapidly perform effector functions and expand upon reinfection (Jameson and Masopust, 2009). In contrast, during many persisting infections, T cells become ‘exhausted’, a state characterized by poor effector functions and high expression of multiple inhibitory receptors (Wherry, 2011). CD8 T cell exhaustion occurs in mice during chronic lymphocytic choriomeningitis virus (LCMV) and other chronic infections in mice, in primates infected with simian immunodeficiency virus (SIV) and in humans infected with HIV, hepatitis B virus (HBV), hepatitis C virus (HCV) and other pathogens, as well as in cancer (Wherry, 2011). In recent years the pathways involved in CD8+ T cell exhaustion have begun to be defined. In contrast, while CD4+ T cells play a pivotal role in chronic infection and cancer, the effect of persisting infection on their function and differentiation remains less well understood.
Robust and functional CD4+ T cell responses are a critical feature of effective antiviral immunity and can prevent CD8+ T cell exhaustion during chronic viral infections. For example, CD4+ T cell depletion during chronic LCMV infection leads to lifelong uncontrolled viremia (Matloubian et al., 1994). Similarly, during HIV infection, the progression to AIDS is temporally associated with (and defined by) loss of CD4+ T cells. During HCV infection, a robust early CD4+ T cell response is important for clearing the infection and chronic infection is accompanied by low or absent CD4+ T cell responses while resolution is associated with vigorous CD4+ T cell responses (Schulze Zur Wiesch et al., 2012).
While CD4+ T cell production of interferon-γ (IFN-γ), tumor necrosis factorα (TNF-α) and interleukin-2 (IL-2) is decreased during chronic infection, suggesting similar defects to exhausted CD8+ T cells (Wherry, 2011), other functions such as production of IL-10 and IL-21 are increased (Brooks et al., 2006; Ejrnaes et al., 2006; Elsaesser et al., 2009; Frohlich et al., 2009). IL-21 is important for CD8+ T cells in this setting, but could also influence B cells. In chronic LCMV and HCV infections, large amounts of T-dependent virus-specific antibodies are produced, suggesting that at least some aspects of CD4+ T cell help to B cells remain intact (Bartosch et al., 2003; Buchmeier et al., 1980) and virus-specific CD4+ T cells transferred to chronically infected mice remain capable of providing help to B cells for at least 50 days (Oxenius et al., 1998). Thus, whether CD4+ T cells become ‘exhausted’ during chronic infection or develop down an alternate path of differentiation remains unclear. Moreover, the extent to which the program of exhaustion of CD4+ and CD8+ T cells overlaps during chronic viral infection is unknown. Together, these studies indicate that while CD4+ T cells develop some functional defects, they may gain and/or sustain other properties during chronic infection suggesting that the impact of chronic infection on exhaustion of CD4+ and CD8+ T cells might be different.
To begin to define the molecular pathways involved in CD4+ T cell dysfunction during chronic infection, genome-wide transcriptional profiling was performed. A common signature of exhaustion shared between virus-specific CD4+ and CD8+ T cells was revealed, as well as unique aspects of CD4+ T cell exhaustion. CD4+ T cell exhaustion was defined by a distinct pattern of inhibitory and costimulatory molecule expression, cell cycle changes and a unique TF profile. In addition, exhausted CD4+ T cells showed a loss of a strong T helper-1 (Th1) cell-associated transcriptional signature but not an obvious skewing towards Th2, Th17, T follicular helper (Tfh) or inducible regulatory T (iTreg) cell fates. These studies provide insights into the molecular mechanisms of CD4+ versus CD8+ T cell dysfunction during chronic infection and define the nature of CD4+ T cell exhaustion versus memory. Specific genes and pathways identified here represent potential therapeutic targets for interventions aimed at restoring function from exhausted CD4 T cells. Moreover, the broader signatures of CD4+ T cell exhaustion might aid in identifying this type of T cell dysfunction in other settings to predict disease progression or therapeutic efficacy of vaccines or other interventions.
Results
Virus-specific CD4+ T cells have altered effector functions during chronic infection
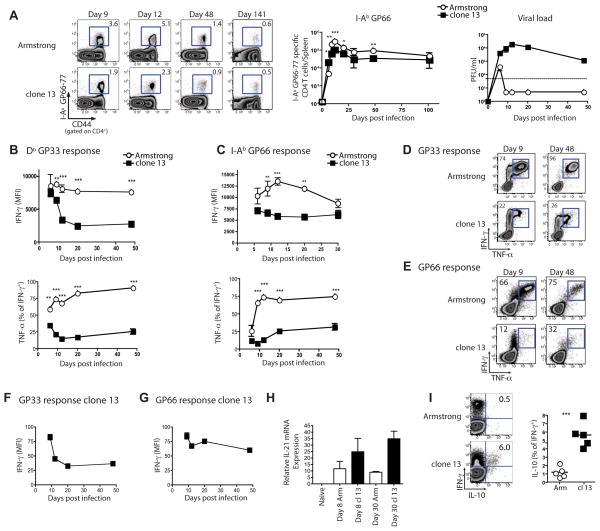
Infection with LCMV Armstrong (Arm) results in an acute infection while clone 13 causes a chronic infection (Ahmed et al., 1984; Wherry et al., 2003). To define how chronic infection affects CD4+ T cell differentiation, we examined LCMV-specific CD4+ T cell responses after either Arm or clone 13 infection (Figure 1). LCMV-specific CD4+ T cells underwent robust clonal expansion and persisted long-term following Arm or clone 13 infection (Figure 1A), though the magnitude of the response was lower during clone 13 infection.
Figure 1. Kinetics of cytokine production by virus-specific CD4+ and CD8+ T cells during chronic LCMV infection.
(A) Representative flow plots of I-AbGP66 tetramer staining on the indicated days p.i. with Arm or clone 13. Numbers show percent of CD4+ T cells specific for GP66. Total number of I-AbGP66 tetramer+ CD4+ T cells in spleen during acute (open symbols) or chronic (closed symbols) LCMV infection (middle). Viral load in the serum over time p.i. with either Arm (open symbols) or clone 13 (filled symbols) (right). (B) MFI of IFN-γ in response to GP33 stimulation (top) and the percentage of IFN-γ producing cells co-producing TNF-α (bottom). (C) CD4+ T cell responses to GP66-77 as described in (B). (D and E) Representative flow plots are shown for cytokine co-production in response to GP33 (D) or GP66 peptide (E). (F and G) MFI of IFN-γ as a percentage of d6 IFN-γ MFI for CD8 (F) and CD4+ T cells (G). (H) IL-21 mRNA expression on d8 and 30 p.i. with Arm or clone 13 in sorted I-AbGP66 tetramer+ CD4+ T cells. (I) Representative plots of IFN-γ and IL-10 production by GP66-specific CD4+ T cells on d30 p.i. and summary of the percent of IFNγ-producers making IL-10. Data are representative over at least 3 independent experiments with 2–4 mice/group. Error bars indicated standard deviation. Figure 1, see also Figure S1.
To compare dysfunction between virus-specific CD4+ and CD8+ T cell responses, we measured IFN-γ, TNF-α and IL-2 ex vivo by intracellular cytokine staining (ICS). During clone 13 infection, CD8+ T cells initially developed effector functions but become progressively dysfunctional (Figure 1B and D, Figure S1, and (Wherry, 2011)). LCMV-specific CD4+ T cells also displayed poor co-production of cytokines or polyfunctionality after infection with clone 13 ((Figure 1C and E; Figure S1; (Brooks et al., 2005; Fuller and Zajac, 2003; Oxenius et al., 1998)). It was unclear whether this CD4+ T cell dysfunction was progressive, as was the CD8+ T cell response, or arose early during infection. To examine this issue we compared IFN-γ production/cell as a percentage of that observed on day 6 (d6). These analyses revealed that IFN-γ production by CD8+ T cells gradually waned while IFN-γ production by CD4+ T cells changed little from the low production at d6 post infection (p.i.) (Figure 1F and G). Thus, endogenous virus-specific CD4+ T cell responses during clone 13 infection display early and sustained dysfunction compared to memory CD4+ T cells in agreement with previous data using TCR transgenic cells (Brooks et al., 2005).
In contrast, IL-21 expression was higher in exhausted CD4+ T cells (Figure 1H and (Elsaesser et al., 2009; Frohlich et al., 2009; Yi et al., 2009)) and a subpopulation of exhausted CD4+ T cells also co-produced IL-10 with IFN-γ (Figure 1I) (Brooks et al., 2006). Together these observations indicate that CD4+ T cells do not merely lose cytokine production, but rather have an altered functional profile, suggesting a change in the pattern of differentiation during chronic infection.
Comparing transcriptional profiles of CD4+ and CD8+ T cells from acute versus chronic infection
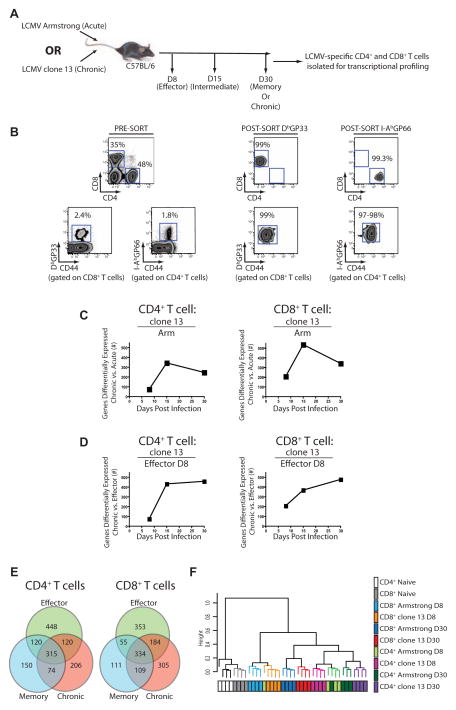
To examine the molecular mechanisms of altered function of CD4+ T cells during chronic infection, global transcriptional profiling was performed on tetramer+ CD4+ or CD8+ T cells at d8, 15 and 30 p.i. (Figure 2A and 2B). This approach allowed direct comparison of CD4+ and CD8+ T cells from mice with the same viral load. An initial evaluation confirmed changes known to occur in effector, memory or exhausted T cells (Figure S2) (Wherry et al., 2007). Pairwise comparisons of exhausted CD4+ T cells with naïve, effector or memory CD4+ T cells revealed a total of 715 transcripts differentially expressed (>2-fold) in exhausted compared to naïve CD4+ T cells (Table S1). To compare the CD4+ T cell responses during clone 13 versus Arm infection over time, we calculated the number of differentially expressed genes unique to CD4+ T cells that responded to either infection. Early in the response (d8), few genes were uniquely changed in either infection suggesting that, initially, effector CD4+ T cells have a similar transcriptional profile in both infections. However, gene expression of the CD4+ T cells responding to the two infections diverged as the chronic infection evolved (Figure 2C). A similar pattern was observed for CD8+ T cells suggesting that this divergence was common for both CD4+ and CD8+ T cells.
Figure 2. Transcriptional profiling of LCMV-specific CD4+ and CD8+ T cells from mice infected with Arm or clone 13.
(A) Schematic of the experimental design. (B) Pre- and post-sort purities for DbGP33-specific CD8+ T cells and I-AbGP66-specific CD4+ T cells. (C) The number of differentially expressed genes for CD4+ (left) and CD8+ T cells (right) from clone 13 compared to Arm at each time p.i. (D) The total number of differentially expressed genes from virus-specific CD4+ or CD8+ T cells from clone 13 infected mice on d8, 15, and 30 p.i. compared to d8 effector T cells from Arm. (E) Differentially expressed genes between d8 effector T cells from Arm infected mice, memory T cells from d30 p.i. with Arm and exhausted T cells isolated from d30 p.i. with clone 13. (F) Hierarchical clustering of CD4+ and CD8+ T cell responses to acute and chronic infections. Genes differentially expressed by a covariance of 0.1 were analyzed using Spearman’s correlation. Microarray data are representative of 4 independent samples per time point. Figure 2, see also Figure S2 and Table S1.
One possibility was that T cells responding to chronic infection remain in a prolonged effector state. To test this possibility, we compared the differentially expressed genes in CD4+ T cells responding to chronic infection on different days p.i. with those of effector CD4+ T cells on d8 p.i. with Arm. The transcriptional profiles at d15 and 30 p.i of clone 13 infection were distinct from those of effector cells, suggesting that the CD4+ T cells responding to chronic infection were not simply arrested in an effector state (Figure 2D). A similar pattern was observed using the clone 13 d8 signature as the “denominator” (Figure S2). Indeed, exhausted CD4+ T cells had a distinct transcriptional profile compared to effector or memory CD4+ T cells (Figure 2E). A similar pattern was observed for exhausted CD8+ T cells. While exhausted CD4+ and CD8+ T cells were distinct from effector or memory T cells of the same lineage, unsupervised hierarchical clustering indicated that while exhausted and memory CD4+ T cells were clearly distinct, these cells were more closely related to one another than exhausted CD4+ T cells are to exhausted CD8+ T cells (Figure 2F). While CD4+ and CD8+ T cells remained as separate branches, within each lineage, samples clustered mainly by time point. Thus, the profile of exhausted CD4+ T cells is divergent from effector and memory CD4+ T cells, as well as exhausted CD8+ T cells.
Transcriptional profiles reveal categories of genes prominently associated with exhausted CD4+ T cells
To begin to define how cellular processes were impacted in exhausted CD4+ T cells, differentially expressed genes were assigned to putative functional categories (Table 1 and Table S2). This analysis revealed categories with substantial (>50%), moderate (35–50%) or low (<35%) numbers of genes shared between exhausted and memory CD4+ T cells (Table 1). Categories with substantial overlap included metabolism, costimulation, cytokines, chemokines, and adhesion molecules. Categories with moderate overlap included those involved in regulating cell signaling, inhibitory molecules and transcription. In this set, half of the transcripts had similar changes during chronic and acute infection while the remainder was unique to either infection. In this latter group, genes tended to be exhaustion-biased. The categories with low overlap included “IFN-induced” genes and genes involved in cell cycle and DNA repair and remodeling. Regulation of many genes in these categories was biased towards exhausted CD4+ T cells. An exception was the set of genes involved in DNA repair, which was biased towards memory. This analysis identifies potential cellular pathways of interest that distinguish functional memory CD4+ T cells from CD4+ T cells responding to chronic infection.
Table 1.
Percentage of differentially expressed genes that are unique or shared between CD4 T cells from clone 13 and Armstrong-infected mice
| Category | clone 13 | Shared | Armstrong |
|---|---|---|---|
| Cytokines/Chemokines | 6 | 76 | 18 |
| Adhesion/Integrins | 25 | 75 | 0 |
| Regulation of apoptosis/Cell death | 21 | 73 | 6 |
| Cytokine receptors/Chemokine receptors | 26 | 70 | 4 |
| Metabolism | 32 | 60 | 8 |
| Ubiquitin ligase activity/Proteasome | 25 | 58 | 17 |
| Costimulatory molecules | 27 | 55 | 18 |
| Vesicle/Membrane biology | 38 | 52 | 10 |
| Cell surface | 41 | 51 | 8 |
|
| |||
| Co-inhibitory molecules | 38 | 50 | 13 |
| Cell signaling | 46 | 49 | 5 |
| Regulation of transcription | 32 | 48 | 20 |
| Solute carriers | 27 | 47 | 27 |
| Phosphatases | 45 | 45 | 9 |
| Cytoskeleton | 47 | 35 | 18 |
| IFN-induced | 53 | 33 | 13 |
| Antigen Receptors/MHC | 15 | 31 | 54 |
|
| |||
| Regulation of cell proliferation/Cell cycle | 60 | 29 | 11 |
| DNA repair/Replication | 21 | 29 | 50 |
| RNA binding/Ribosomal | 29 | 29 | 43 |
| DNA structure | 59 | 18 | 24 |
Numbers are the percent of total genes in category that are either biased towards CD4 T cells from clone 13 infection, towards memory CD4 T cells, or differentially expressed by both (compared to naive T cells).
The IFN-responsive pathway is associated with CD4+ T cell exhaustion
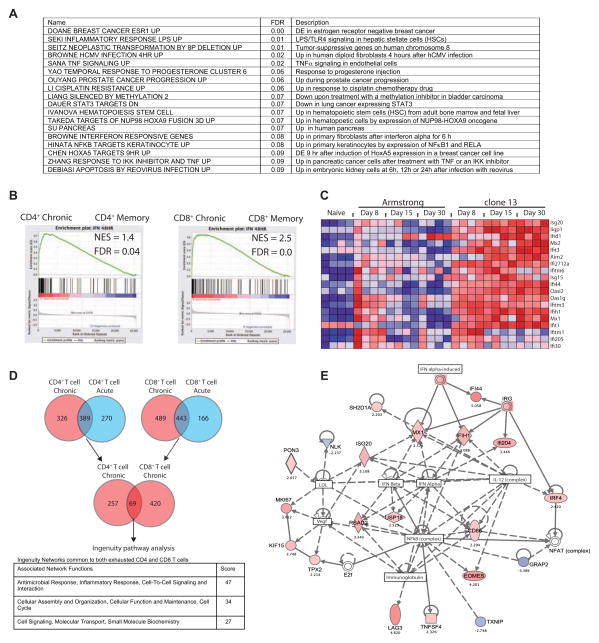
To define additional biological pathways associated with CD4+ T cell exhaustion, we used gene set enrichment analysis (GSEA) and a curated panel of gene sets from the Molecular Signatures Database (MSigDB; the C2 set) that contains information about biological pathways, disease phenotypes and gene sets from the biomedical literature. Several gene sets enriched in exhausted, compared to effector and memory CD4+ T cells (false discovery rate (FDR) < 0.1) were associated with inflammation including type I IFN (IFN-I) signaling (Figure 3A). We next used a signature of IFN-I induced transcriptional changes (Agarwal et al., 2009) and GSEA to test whether this biologically defined type I IFN signature was enriched in memory versus exhausted CD4+ or CD8+ T cells. Indeed, both CD4+ and CD8+ T cells from chronically infected mice showed a strong enrichment for type I IFN induced transcriptional changes (Figure 3B).
Figure 3. Biological pathways enriched in exhausted CD4+ T cells.
(A) Enriched gene sets in exhausted versus effector or memory CD4+ T cells using GSEA analysis and the MSigDB C2 curated gene sets. (B) GSEA of exhausted versus memory CD4+ or CD8+ T cells with an IFN-I transcriptional signature (Agarwal et al., 2009). (C) Heatmap of IFN responsive genes differentially expressed by exhausted CD4+ T cells (over naïve). (D) The core exhausted signature shared in CD4+ and CD8+ T cells (see also Table S3). Networks identified and their score from Ingenuity pathway analysis are shown. (E) Ingenuity pathway analysis of the main network associated with both exhausted CD4+ and CD8+ T cells. Figure 3, see also Figure S3 and Table S2 and S3.
The presence of a strong type I IFN signature at d30 of chronic infection was intriguing since circulating type I IFN is not detectable at this time (Lee et al., 2009; Zuniga et al., 2008). Many of the IFN-induced genes upregulated in exhausted CD4+ T cells (Table S2) were also present in the early stages (e.g. d6-8) of both infections (Figure 3C). As virus was cleared in Arm infection, however, expression of these genes returned to baseline. In contrast, expression of the majority of these genes was high and sustained in T cells responding to clone 13 infection (Figure 3C and Figure S3A). Since circulating type I IFN peaks and resolves with similar kinetics in Arm and clone 13 infections (Lee et al., 2009; Zuniga et al., 2008), these observations suggest a disconnect between detectable circulating type I IFN protein and activation of the downstream IFN dependent transcriptional programs in CD4+ T cells during acute versus chronic infections.
We next examined phosphorylated signal transducer and activator of transcription-1 (pStat1) directly ex vivo and after stimulation with IFN-β or IFN-γ. Phosphorylated Stat1 was detectable directly ex vivo in total CD4+ T cells from chronically infected mice, but not in CD4+ T cells from Arm immune mice (Figure S3B and S3C). However, the ability of CD4+ T cells from chronically infected mice to increase pStat1 in response to IFN-I or IFN-γ was blunted compared to the pStat1 induction in CD4+ T cells from Arm immune mice. These observations suggest that exhausted CD4+ T cells are subjected to ongoing chronic Stat1 activation and desensitization of this pathway or express negative regulators of IFN signaling, such as Socs proteins. Of note, Socs1 and Socs3 mRNA was upregulated in CD4+ T cells during clone 13 infection (Figure S3D).
To further investigate pathways similarly regulated in exhausted CD4+ and CD8+ T cells, we analyzed the differentially expressed genes common to exhausted CD4+ and CD8+ T cells (but unchanged in memory T cells) (Table S3) using Ingenuity Pathway Analysis (IPA) (Figure 3D). Three networks were identified (scores > 2 have at least 99% confidence of not being generated by chance). The network with the highest score contained genes involved in antimicrobial responses, inflammatory responses and cell-to-cell signaling (Figure 3E). This analysis revealed an association of the IFN pathway with transcriptional pathways including NF-κB, NFAT and Eomes, as well as the inhibitory receptor LAG-3 and the adaptor protein SH2D1A (Sap).
Expression of distinct patterns of inhibitory receptors in exhausted CD4+ and CD8+ T cells
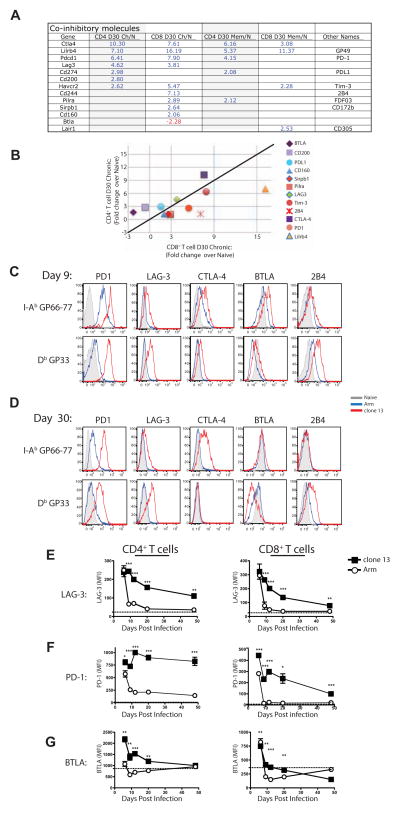
A key feature of CD8+ T cell exhaustion is sustained expression of multiple inhibitory receptors (Blackburn et al., 2009). While virus-specific CD4+ T cells also express inhibitory receptors during chronic infections (reviewed in (Odorizzi and Wherry, 2012)), it is unclear whether these cells express the same repertoire of inhibitory receptors as exhausted CD8+ T cells. Both virus-specific CD4+ and CD8+ T cells upregulated inhibitory receptors during clone 13 infection (Figure 4A). However, the specific inhibitory receptors upregulated and the degree of expression differed greatly between CD4+ and CD8+ T cells. Inhibitory receptor genes biased towards exhausted CD8+ T cells including Cd244, Havcr2 (encoding Tim3) and Lilrb4. Other inhibitory receptors, however, were biased toward expression in CD4+ T cells including Ctla4, Cd200 and Btla (Figure 4A and B). Protein expression confirmed the transcriptional profiles, including CD8+ T cell biased 2B4 expression, mutual, though slightly CD8+ biased LAG-3 expression and CD4+ T cell biased CTLA-4 (Figure 4C–E). PD-1 was highly expressed by CD4+ T cells during chronic infection and this high expression was sustained through at least d50 p.i. (Figure 4C, D and F and Figure S4 and data not shown) even when PD-1 expression by exhausted CD8 T cells in the same mice declined (Figure 4F). The expression of BTLA also differed between CD4+ and CD8+ T cells. CD4+ T cells had increased expression of BTLA during clone 13 infection until ~ d20 p.i. and then sustained BTLA expression at least as high as naïve T cells (Figure 4G). Virus-specific CD8+ T cells, in contrast, expressed lower BTLA and by d30 p.i. these CD8+ T cells had less BTLA than naïve T cells.
Figure 4. Inhibitory receptor expression by exhausted CD4+ and CD8+ T cells.
(A) Genes encoding coinhibitory receptors differentially expressed by memory and exhausted virus-specific T cells. (B) Fold change in expression of coinhibitory receptors by exhausted T cells at d30 p.i. compared to naïve T cells. (C and D) Representative histograms showing expression of coinhibitory receptors (PD-1, LAG-3, CTLA-4, BTLA, and 2B4) on LCMV-specific CD4+ and CD8+ T cells on d9 (C) or d30 (D). Histograms depict naïve T cells (CD44Lo; grey) or virus-specific T cells from Arm (blue) or clone 13 (red) infection. (E, F and G) MFI of LAG-3 (E), PD-1 (F) and BTLA (G) expression by LCMV-specific CD4+ (left) or CD8+ (right) T cells. Data are representative of 3–8 independent experiments with at least 3 mice per group at each time point. Error bars represent standard deviation. Figure 4, see also Figure S4 and Table S2.
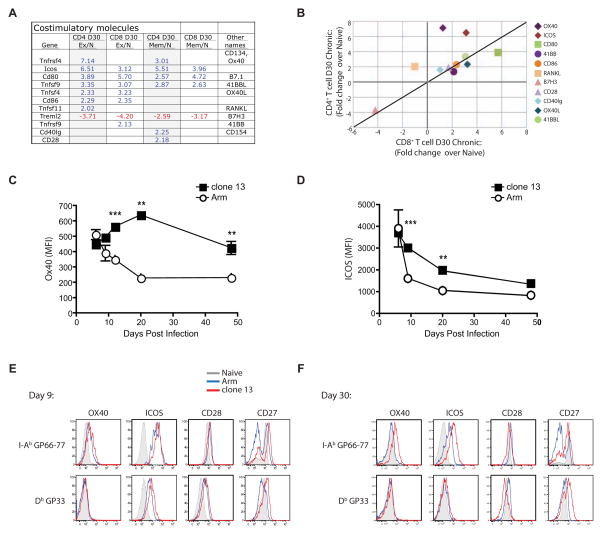
Differential expression of costimulatory receptors in exhausted CD4+ and CD8+ T cells
Given the expression of inhibitory receptors during chronic infection, one might predict that costimulatory receptors would be downregulated. However, expression of several costimulatory molecule mRNAs was increased in exhausted CD4+ and CD8+ T cells (Figure 5A and B). While Tfnrsf4 (encoding Ox40) and Icos were biased toward expression by CD4+ T cells, only Cd80 was clearly biased toward CD8+ T cells (Figure 5B). Protein expression of OX40 and ICOS showed a strong bias toward expression by CD4+ T cells responding to clone 13 infection in the spleen (Figure 5C and D) and other tissues (Figure S4). Exhausted CD4+ T cells also expressed higher CD27 by flow cytometry compared to memory CD4+ T cells (Figure 5F). CD28 was the only costimulatory molecule examined that was slightly decreased on both exhausted CD4+ and CD8+ T cells (Figure 5E and F). These data suggest that therapies combining inhibitory receptor blockade and costimulatory receptor engagement may be beneficial.
Figure 5. Costimulatory molecule expression by T cells during Arm versus clone 13 infection.
(A) Genes encoding costimulatory molecules differentially expressed in LCMV-specific CD4+ and CD8+ T cells at d30 p.i. (B) Fold changes over naïve for exhausted CD4+ and CD8+ T cells at d30 p.i. (C and D) MFI of OX40 (C) and ICOS (D) by I-AbGP66-specific CD4+ T cells over time. (E and F) Representative histograms showing expression of ICOS, OX40, CD28 and CD27 at d9 (E) and d30 (F) p.i. Histograms depict naïve T cells (CD44Lo; grey) or virus-specific T cells from Arm (blue) or clone 13 (red) infection. Data are representative of 4–7 independent experiments with at least 3 mice per group at each time point. Error bars represent standard deviation. Figure 5, see also Figure S4 and Table S2
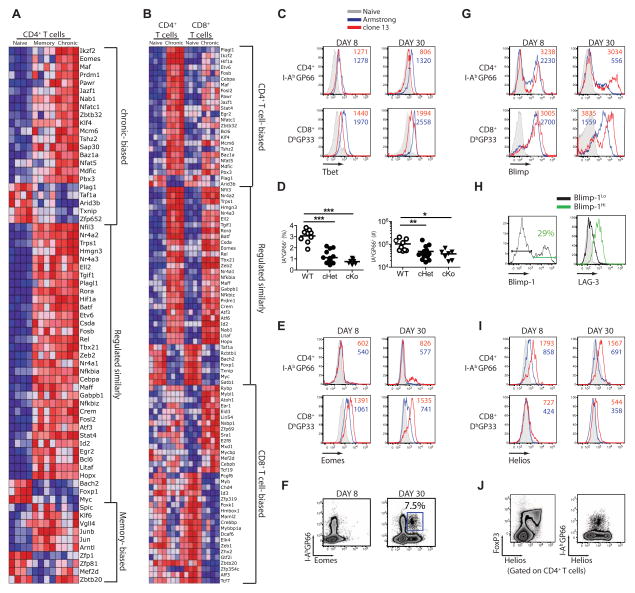
TFs involved in CD4+ T cell exhaustion
Several TFs including Blimp-1, Batf, T-bet, Eomes and NFAT have been implicated in CD8+ T cell exhaustion during chronic infection (Agnellini et al., 2007; Kao et al., 2011; Paley et al., 2012; Quigley et al., 2010; Shin et al., 2009). The TFs involved in CD4+ T cell dysfunction during chronic infection, however, are poorly understood. Exhausted CD4+ T cells differentially expressed a number of TFs compared to functional memory CD4+ T cells (Figure 6A and Table S2). First, there was a core set of TFs similarly regulated in memory and exhausted compared to naïve CD4+ T cells. This set included Fosb and Id2. In addition, there was a smaller set of TFs whose change in expression was more associated with memory, including Klf6, Jun and Junb. Finally, there were several TFs such as Eomes, Prdm1 (Blimp-1) and Ikzf2 (Helios) associated with exhausted CD4+ T cells (Figure 6A).
Figure 6. TF profile of memory and exhausted CD4+ and CD8+ T cells.
(A) Heatmap of TFs differentially expressed by either memory or exhausted virus-specific CD4+ T cells. (B) Heatmap of TFs differentially expressed by exhausted virus-specific CD4+ or CD8+ T cells. (C) Histograms show protein expression of T-bet by LCMV-specific CD4+ and CD8+ T cells with Arm (blue), clone 13 (red) infection, or in CD4+CD44lo cells (grey). (D) I-AbGP66-specific CD4+ T cell frequency (left) and total numbers per spleen (right) in WT, Tbx21flox/+XCd4creor Tbx21flox/floxXCd4cre mice at d30 p.i. with LCMV clone 13. (E) Eomes expression by LCMV-specific CD4+ and CD8+ T cells from Arm (blue), clone 13 (red) infection, or CD4+CD44lo cells (grey). Numbers show MFI in cells from clone 13 (red) or Armstrong-infected mice (blue). (F) Representative flow plots for Eomes expression on d8 and d30 p.i. with clone 13. Number shows the percent of I-AbGP66+ CD4+ T cells expressing Eomes. (G) Representative histograms of YFP expression by LCMV-specific CD4+ T cells and CD8+ T cells from Blimp-1-YFP reporter mice at d8 and d30 p.i. with LCMV clone 13. Numbers show the MFI of YFP expression in cells from clone 13 (red) or Armstrong-infected mice (blue). (H) At d30 p.i. with clone 13, IAbGP66-specific CD4+ T cells from Blimp-1-YFP mice were gated on Blimp-1Hi or Blimp-1Lo and examined for LAG-3 expression. Number shows the percent of I-AbGP66+ CD4+ T cells expressing Blimp-1. (I) Helios expression by LCMV-specific CD4+ or CD8+ T cells. Gated on I-AbGP66+ CD4+ T cells from Arm (Red) or clone 13 (blue) infection. (J) Representative flow plots of Helios versus FoxP3 or I-AbGP66 tetramer for CD4+ T cells at d30 p.i. with clone 13. Data are representative of 2–4 independent experiments with at least 3 mice per group at each time point. Figure 6, see also Figure S5.
The expression pattern of key transcription factors can, at least partially, define cellular identity and provide clues into the regulation of cellular function and differentiation. We therefore investigated the unique and overlapping TF expression patterns between exhausted CD4+ and CD8+ T cells (Figure 6B). A core set of TFs was changed in a similar manner in virus-specific exhausted CD4+ and CD8+ T cells, including Batf, Tbx21, Prdm1 and Eomes. In addition, both exhausted CD4+ and CD8+ T cells had changes for separate sets of TFs. Notable CD8+ T cell-biased changes in TF expression included downregulation of Tcf7 while CD4+-biased changes included increased Ikzf2 (Helios) and Klf4 (Figure 6B). Thus, while there is clearly a core program of T cell dysfunction during chronic infection shared by both lineages, there were also unique transcriptional changes for CD4+ versus CD8+ T cells during chronic infection.
We next examined protein expression for TFs of interest. As expected, T-bet was upregulated upon activation, but its expression was lower in exhausted CD4+ and CD8+ T cells compared to memory T cells (Figure 6C). T-bet is needed to sustain CD8+ T cell responses during chronic infection (Kao et al., 2011; Paley et al., 2012) and T-bet represses PD-1 expression in both CD8+ and CD4+ T cells (Kao et al., 2011). In this regard, it is worth noting that T-bet expression was lower in exhausted CD4+ T cells and PD-1 was higher compared to exhausted CD8+ T cells. We previously demonstrated a role for T-bet in sustaining exhausted CD8+ T cell responses (Kao et al., 2011; Paley et al., 2012). To directly test whether T-bet was required for exhausted CD4+ T cells, we examined LCMV-specific CD4+ T cells during clone 13 infection in WT mice or mice conditionally deficient in one or both copies of Tbx21 only in T cells (Tbx21flox/floxXCd4cre). In the full (Tbx21flox/floxXCd4cre) or partial (Tbx21flox/+XCd4cre) absence of T-bet there was a significant reduction in the LCMV-specific CD4+ T cells compared to WT mice at d30 p.i. (Figure 6D). T-bet was necessary in a cell intrinsic manner since the tetramer+ response of Tbx21−/− CD4+ T cells was substantially lower than that of WT CD4+ T cells in mixed bone marrow chimeras (Figure S5). Thus, while T-bet expression is reduced in CD4+ T cells during LCMV clone 13 infection, low amounts of T-bet are still important to sustain both CD4+ and CD8+ T cell responses during chronic infection.
Eomesodermin (Eomes) is expressed by CD8+ T cells after Arm or clone 13 infection. While mRNA coding for this TF was not upregulated in effector or memory CD4+ T cells (Figure 6A, B and Table S2), Eomes was increased in exhausted CD4+ T cells. Flow cytometric analysis revealed expression of Eomes by only a subset of exhausted CD4+ T cells (Figure 6E and F). These observations suggest a potential unexpected role for Eomes in CD4+ T cells during chronic viral infection, but also point to heterogeneity in the exhausted CD4+ T cell population (see below).
During clone 13 infection, high Blimp-1 (encoded by Prdm1) in CD8+ T cells promotes expression of inhibitory receptors and fosters exhaustion (Shin et al., 2009). Prdm1 mRNA was also upregulated by exhausted CD4+ T cells. There was again substantial heterogeneity in Blimp-1 expression by flow cytometry using a Blimp-1-YFP reporter mouse with only a subset of CD4+ T cells expressing high Blimp-1 after Arm or clone 13 infection (Figure 6G). During chronic viral infection, the Blimp-1hi subset of LCMV-specific CD4+ T cells also had high expression of the inhibitory receptor LAG-3 at d30 p.i. (Figure 6H) consistent with observations in exhausted CD8+ T cells (Shin et al., 2009).
The Ikaros family TF Helios (gene Ikzf2) was one of the most differentially expressed TFs in exhausted CD4+ T cells compared to memory CD4+ T cells or exhausted CD8+ T cells. During clone 13 infection, virus-specific CD4+ T cells expressed more Helios mRNA and protein than their effector and memory CD4+ counterparts (Table S2 and Figure 6I). While Helios is highly expressed by a subpopulation of Treg cells (Getnet et al., 2010), none of the exhausted CD4+ T cells expressed FoxP3 (Figure S5; and see below) and virus-specific CD4+ T cells expressed intermediate amounts of Helios compared to the high amounts in Foxp3+ CD4+ T cells (Figure 6J and data not shown). Thus, while both T-bet and Blimp-1 appear to have similar expression patterns in both exhausted CD4+ and CD8+ T cells, other TFs such as Helios are more biased to exhausted CD4+ T cells.
Exhausted CD4+ T cells are heterogeneous and distinct from other CD4+ T cell effector subsets
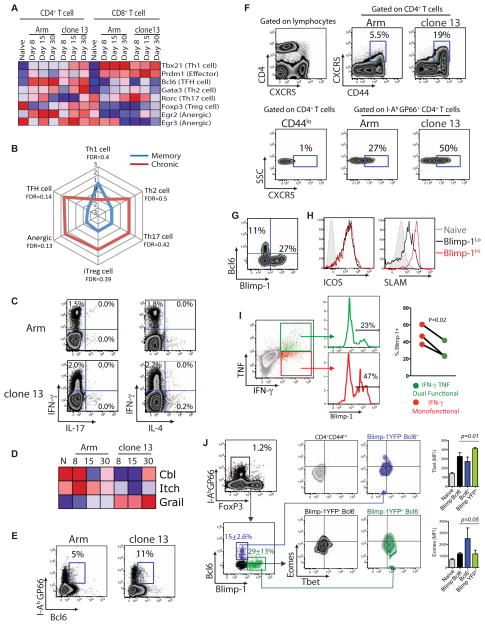
CD4+ T cells can differentiate into distinct lineages including Th1, Th2, Th17, iTreg and Tfh cells. LCMV Arm infection induces Th1 cells producing IFN-γ. One possible impact of chronic viral infection is skewing of CD4+ T cells to another lineage. Indeed, while no clear signature of lineage specific TFs was apparent, increased expression of GATA-3 and Bcl6 (Figure 7A) or the cytokines IL-10 and IL-21 (Figure 1) suggested that lineage skewing might occur. The signatures for Th2, Th17, iTreg, nTreg, Tfh cell subsets and anergic cells defined in previous studies (Safford et al., 2005; Wei et al., 2009; Yusuf et al., 2011) were all slightly biased toward exhausted CD4+ T cells compared to memory CD4+ T cells using GSEA, but no enrichment reached statistical significance (Figure 7B). Indeed, when RT-PCR was used to measure expression of selected TFs, many lineage-defining TFs showed only moderately increased expression in exhausted CD4+ T cell populations compared to memory CD4+ T cells (Figure S6). The major pattern to emerge, however, was the absence of enrichment of a clear Th1 signature that was seen for memory CD4+ T cells.
Figure 7. T helper cell lineage differentiation during clone 13 infection.
(A) Heatmap of expression of TFs associated with distinct T helper cell lineages in LCMV-specific CD4+ and CD8+ T cells. (B) GSEA of exhausted versus memory CD4+ T cells with signatures from the specified Th cell subsets. Normalized enrichment scores (NES) are plotted. Red = clone 13. Blue = Arm. Using an FDR score of 0.01 there was no significant enrichment of any subset for exhausted CD4+ T cells. (C) Representative flow plots show IL-4 or IL-17 production following peptide stimulation and ICS (gated on CD4+ T cells). Numbers show the percent of CD4+ T cells expressing cytokines. (D) Heatmap of Cbl, Itch and Grail expression by LCMV-specific CD4+ T cells. (E) Representative flow plots of Bcl6 expression by CD4+ T cells at d30 p.i. with Arm or clone 13. Numbers show the percent of I-AbGP66+ CD4+ T cells expressing Bcl6. (F) CXCR5 expression by total and IAbGP66+ CD4+ T cells. Numbers show the percentage of cells expressing CXCR5. (G) Representative plots of Blimp-1-YFP versus Bcl6 expression in I-AbGP66+ CD4+ T cells at d30 p.i. with clone 13. (H) LCMV-specific CD4+ T cells from Blimp-1-YFP mice were examined for expression of ICOS and SLAM at d30 p.i. with clone 13. I-AbGP66+ CD4+ T cells were gated for low or high Blimp-1 expression then examined for ICOS or SLAM expression. (I) Blimp-1-YFP mice were infected with LCMV clone 13 and on d30 p.i. ICS was performed with GP66 peptide stimulation followed by staining for IFN-γ and TNF. Dual functional (IFN-γ+TNF+) and monofunctional (IFN-γ+ only) virus-specific CD4+ T cells were then examined for Blimp-1-YFP expression. P-value determined using a paired T-test. (J) Representative flow plots of TF expression in I-AbGP66+ CD4+ T cells at d30 post infection with LCMV clone 13. Foxp3- I-AbGP66+ CD4+ T cells were separated into 3 distinct populations (Blimp-1+Bcl6−, Blimp-1−Bcl6−, Blimp-1+Bcl6+) then examined for expression of Eomes and Tbet. Numbers represent the percentage of cells expressing the marker with standard deviation. Statistical relevance was determined using two-way ANOVA. Data are representative of 2–5 independent experiments with at least 3 mice per group at each time point. Error bars represent standard deviation. Figure 7, see also Figure S6.
Another possibility was that multiple distinct subpopulations existed during chronic viral infection with each contributing modestly to the transcriptional profile leading to an unfocused enrichment pattern. However, neither IL-4 nor IL-17 protein expression was increased suggesting that virus-specific CD4+ T cells from chronic infection had not become full-fledged Th2 or Th17 cells (Figure 7C). In addition, essentially all of the I-AbGP66 tetramer+ CD4+ T cells during either Arm or clone 13 infection lacked expression of Foxp3 (Figure S5 and see below). We also examined potential skewing towards anergic T cells or Tfh cells. Anergic CD4+ T cells express ubiquitin protein ligases cbl-b, itch and GRAIL (Mueller, 2004). Only GRAIL was upregulated by virus-specific CD4+ T cells from chronic infection (Figure 7D). However, GRAIL is also upregulated by Th2 cells from schistosome-infected mice and is required for their hyporesponsiveness (Taylor et al., 2009). Thus, while some molecular components may be shared, CD4+ T cell anergy and CD4+ T cell dysfunction during chronic viral infection may not be entirely overlapping processes.
Recent work suggests a bias towards Tfh during chronic viral infections (Fahey et al., 2011). A number of markers have been used to identify Tfh cells including high expression of ICOS, OX40, PD-1, IL-21, CXCR5, Bcl6, GL7 and low Blimp-1 and SLAM (Johnston et al., 2009). While several of these markers are also features of exhausted CD4+ T cells (Figures 1 and 6) and might suggest a possible Tfh skewing, some can also reflect recent T cell activation. Indeed, while exhausted CD4+ T cells expressed ICOS, OX40 and PD-1, the majority of these cells were Bcl6Lo (Figure 7E). We, therefore, examined the expression of CXCR5 to further investigate the differentiation into Tfh cells. As previously described (Fahey et al., 2011), a higher proportion of LCMV-specific CD4+ T cells isolated from chronically infected mice expressed CXCR5 compared to cells isolated from mice previously infected with LCMV Arm (Figure 7F). Thus, a higher proportion of LCMV-specific CD4+ T cells in chronically infected mice expressed several key Tfh markers including CXCR5 and Bcl6. To interrogate this issue in more detail, we examined co-expression of Blimp-1 and Bcl6. Consistent with their antagonistic roles (Johnston et al., 2009), the expression of Blimp-1 and Bcl6 in exhausted CD4+ T cells was largely mutually exclusive: Blimp-1hi cells did not express Bcl6 (Figure 7G). SLAM expression was low on some of the Blimp-1lo CD4+ T cells consistent with a subset of these Blimp-1lo cells expressing Bcl6 (Figure 7H) and representing true Tfh. In contrast, both Blimp-1lo and Blimp-1hi cells had high expression expressed of ICOS (Figure 7H) suggesting that ICOS is an unreliable marker of Tfh during chronic LCMV infection.
We next investigated whether the heterogeneity in expression of TFs was related to CD4+ T cell function. On 30 p.i. ICS following peptide stimulation revealed IFN-γ and TNF dual cytokine producing and IFN-γ producing “monofunctional” populations of virus-specific CD4+ T cells consistent with Figure 1. To determine whether expression of Blimp-1 was related to CD4+ T cell function we analyzed YFP expression as a proxy for Blimp-1 in Blimp-1-YFP reporter mice. This analysis demonstrated that virus-specific CD4+ T cells that co-produced IFN-γ and TNF had substantially lower expression of Blimp-1 compared to IFN-γ monofunctional CD4+ T cells that contained an increased proportion of Blimp-1-YFPHi cells. “Reverse” gating also demonstrated that Blimp-1Hi cells contained fewer polyfunctional cells than the Blimp-1lo population (data not shown). These data link TF heterogeneity to function in virus-specific CD4+ T cells from chronic infection and suggest an association between elevated Blimp-1 expression and decreased polyfunctionality. Clearly, however, Blimp-1 expression does not fully account for full spectrum of heterogeneity in virus-specific CD4+ T cells during chronic infection and other layers of control (e.g. via other TFs, inhibitory receptors, etc) are likely to exist.
We next investigated the single-cell co-expression patterns of Blimp-1-YFP, FoxP3, Bcl6, T-bet and Eomes in exhausted CD4+ T cells on d30 p.i. by flow cytometry (Figure 7J). FoxP3 was not expressed by I-AbGP66-specific CD4+ T cells (Figure 7J and Figure S5). However, three distinct subpopulations were defined by Bcl6 and Blimp-1 expression (Bilmp-1+Bcl6−, Blimp-1−Bcl6+ and Blimp-1−Bcl6−). Within these subsets, T-bet expression was highest in the Blimp-1+Bcl6− subset, but not different between the Blimp-1−Bcl6+ and double negative subsets (P=0.01, two-way ANOVA). In contrast, while some co-expression of Blimp-1 (i.e. YFP) and Eomes was observed, there were also clear populations of Blimp-1+Eomes−, Blimp-1−Eomes+ and double negative cells (Figure 7J and Figure S6). Thus, virus-specific CD4+ T cell populations during chronic LCMV infection were comprised of multiple distinct subsets of cells based on expression of key TFs. Together these studies reveal key insights into the biology of CD4 T cells responding to chronic viral infection. While there was clearly a shared program with exhausted CD8 T cells such as a signature of ongoing inflammation, CD4 T cells during chronic infection displayed distinct features including an altered profile of inhibitory and costimulatory molecules, a distinct pattern of transcription factors expression and the existence of multiple distinct subpopulations that may have different function. Future studies are necessary to understand the development and potential to re-invigorate function in these subsets of antiviral CD4 T cells during chronic infection.
Discussion
A major question during chronic infections is whether CD4+ and CD8+ T cell exhaustion is due to the same molecular pathways. Here we addressed this issue and our results demonstrate that “exhausted” CD4+ T cells differ substantially from effector and memory CD4+ T cells. While there was clearly a core program conserved between exhausted CD4+ and CD8+ T cells, exhausted CD4+ T cells differed in several key respects. We identified cell cycle regulation, DNA repair, IFN-I signaling, altered inhibitory and costimulatory receptor expression as well as diverse TF expression patterns among the key changes in exhausted CD4+ T cells. TFs and inhibitory receptors are centrally involved in both CD4+ and CD8+ T cell exhaustion, but the repertoire of inhibitory and transcriptional pathways used was distinct. These data suggest that exhausted CD4+ T cells share some transcriptional “modules” with exhausted CD8+ T cells, but also use other gene expression programs that distinguish them functionally and phenotypically.
We identified a core set of TFs similarly regulated in exhausted CD4+ and CD8+ T cells. In particular, Blimp-1, T-bet and Eomes have all been implicated in CD8+ T cell responses to infection. Like in CD8+ T cells (Shin et al., 2009), high Blimp-1 was associated with increased inhibitory receptor expression and decreased polyfunctionality for exhausted CD4+ T cells, though whether Blimp-1 is causal or just associated with these changes will require future investigation. T-bet was also a positive regulator of CD4+ T cell responses during chronic LCMV infection, reminiscent of its role in sustaining exhausted CD8+ T cells (Kao et al., 2011; Paley et al., 2012). A small subset of exhausted CD4+ T cells also expressed Eomes in contrast to lack of this TF memory CD4+ T cells. Finally, the Ikaros family member Helios was strongly biased towards expression in exhausted CD4+ T cells. Helios can modulate the function of Ikaros by affecting its intracellular localization (Hahm et al., 1998) perhaps impacting Ikaros-dependent expression of cytokines including IFN-γ, TNF-α and IL-2 (Quirion et al., 2009). These studies also revealed substantial heterogeneity in TF expression suggesting that the pool of “exhausted” CD4+ T cells likely represents a mixture of subsets of cells. A single term (exhausted) describing these cells may under-represent the true diversity of this population. Future studies using TF reporters should allow detailed interrogation of function and lineage relationships between these subpopulations and also facilitate assessment of how expression of individual TFs specifically relates to functional and phenotypic heterogeneity.
Exhausted CD4+ and CD8+ T cells upregulated IFN-responsive genes during chronic infection suggesting that T cells were continuing to respond to IFN signals after ~1 month of chronic infection. Serum IFN protein peaks within 48 hours after both Arm and clone 13 infection and falls to baseline within one week (Lee et al., 2009; Zuniga et al., 2008), though low IFN mRNA expression has been detected later during chronic LCMV infection (Lee et al., 2009). These observations suggest that exhausted T cells are either able to sustain this IFN-responsive transcription program with minimal continued IFN-I stimulation or that other signals could substitute for IFN-I. Of note, two recent reports demonstrate an immunoregulatory role for type I IFN during chronic LCMV infection and suggest an important role for CD4+ T cells in coordinating this effect (Teijaro et al., 2013; Wilson et al., 2013). IFN responsive genes are also a negative prognostic indicator during human Mycobacterium tuberculosis infection (Berry et al., 2010) and are associated with pathogenic SIV infection (Bosinger et al., 2009). Thus, sustained expression of an IFN-I-induced transcriptional program might be a common feature of many chronic diseases, though future studies are necessary to determine the consequences of prolonged expression of this set of genes in CD4+ T cells.
Exhausted CD4+ T cells also expressed a different repertoire of inhibitory receptors compared to exhausted CD8+ T cells. CD4+ T cell biased expression of inhibitory molecules like BTLA, CTLA-4, CD200 and even PD-1 at later time points, as well as the lack of the CD8+ T cell biased molecules including 2B4 and Pilra, suggests qualitative differences in the negative regulatory circuits for CD4+ and CD8+ T cells during the same chronic infection. These differences might reflect ligand availability, different APC interactions or hardwired transcriptional differences. Surprisingly, with the exception of downregulation of CD28, many costimulatory receptors were increased. Together, these observations suggest that distinct combinations of inhibitory and costimulatory pathways could lead to selective targeting of exhausted CD4+ and/or CD8+ T cells. Indeed, ligating 4-1BB in combination with blockade of PD-1 leads to a dramatic rescue of exhausted CD8+ T cells (Vezys et al., 2011).
During chronic infection, CD4+ T cells increased expression of mRNA encoding several TFs implicated in the development of different Th subsets including Tbx21 (Th1 cell), Gata3 (Th2 cell), Rora (Th17 cell) and Bcl6 (Tfh cell). While protein expression of some of these TFs was uniform, others such as Blimp-1, Eomes and Bcl6 were only expressed by subsets of CD4+ T cells, suggesting heterogeneity. Blimp-1 and Bcl6 were examined given the expression of a number of Tfh cell-like markers by exhausted CD4+ T cells. Bcl6 was only expressed by a subset of these CD4+ T cells and this expression was inversely related to Blimp-1. While Bcl6 staining can be challenging in T cells, combined analyses with Blimp-1 and CXCR5 expression, suggest it is unlikely that all LCMV-specific CD4+ T cells become true germinal center Tfh cell during chronic infection despite uniformly high expression of some Tfh cell markers like ICOS, OX40 and PD-1. Moreover, the exhausted CD4+ T cells population contained at least partially non-overlapping subsets of Bcl6+, Blimp-1+ and Eomes+ cells. This observation is consistent with GSEA that demonstrated little enrichment for alternative lineage profiles and suggests a model where exhausted CD4+ T cell populations are heterogeneous containing multiple distinct subpopulations based on expression of key TFs. Future studies should help clarify the precise how these TF-defined subsets relate to the phenotypic and functional diversity of virus-specific CD4+ T cells during chronic infection.
In summary, this work reveals pathways associated with CD4+ T cell exhaustion and suggests new approaches for therapeutic interventions. The altered pathways identified in CD4+ T cells from chronic infection not only allowed us to gain insight into the unique differentiation of “exhausted” CD4+ T cells, but also revealed shared regulators with exhausted CD8+ T cells. The coinhibitory, costimulatory, inflammatory and transcriptional pathways identified suggest that exhausted CD4+ and CD8+ T cell subsets are subject to different molecular control and suggest that these T cell populations could be selectively targeted therapeutically.
Experimental Procedures
Animals and viruses
C57Bl/6J mice were purchased from Jackson Laboratories (Bar Harbor, Maine). Tbx21flox mice were crossed with Cd4Cre mice (Kao et al., 2011). Blimp-1YFP reporter mice were from E. Meffre (Yale University, New Haven, CT). All mice were used in accordance with Institutional Animal Care and Use Committee (IACUC) guidelines. LCMV Arm and LCMV clone 13 were grown in BHK cells and titered on Vero cells (Wherry et al., 2003). Mice were infected with either Arm (2×105 PFU) i.p. or clone 13 (2×106 PFU) i.v.
Flow cytometry, sorting and intracellular cytokine staining
Lymphocytes were isolated from spleen, stained and analyzed by flow cytometry as described (Wherry et al., 2003). Virus-specific CD8+ or CD4+ T cells were examined using MHC class I or class II tetramers. MHC class I peptide tetramers were made and used as described (Wherry et al., 2003). MHC class II tetramer was obtained from the NIH Tetramer Core Facility (Emory University, Atlanta, GA). For a list of antibodies used see Supplemental Experimental Procedures. For ICS, 106 splenocytes were cultured in the presence or absence of peptide (0.2μg/ml for CD8 peptides and 2μg/ml for the CD4+ peptide) and brefeldin A for 5h at 37°C. Staining was carried out using the BD cytofix/cytoperm kit. Samples were collected using an LSR II flow cytometer (BD).
Gene Expression Profiling
Microarray analysis was performed on four independent samples of FACS-purified T cells from the spleen, sorted based on CD8, CD4, CD44, and tetramer. Cells were sorted directly into TRIzol LS (Invitrogen, Carlsbad, CA) and samples processed and hybridized at the University of Pennsylvania microarray facility using Affymetrix mouse Gene 1.0 ST arrays. Quality-control checks were performed with R (BioConductor). The data were preprocessed with RMA (Robust Multichip Average) normalization. Technical replicates were averaged for fold-change. Genes were considered differentially expressed if > 2-fold different.
Molecular signatures and Gene set enrichment analysis (GSEA)
Molecular signatures were generated using the class neighbors function in GenePattern. Genes were ranked using the signal-to-noise ratio (SNR) and a permutation test used to calculate statistical significance. Gene Set Enrichment Analysis (GSEA) was performed using the Broad Institute program (http://www.broadinstitute.org/gsea/index.jsp). GSEA used gene sets from the Molecular Signature Database v2.5 (Subramanian et al., 2005) or published gene expression arrays for Th17, iTreg, Th1 cells, Th2 cells (Wei et al., 2009), anergic T cells (Safford et al., 2005), TFH cells (Yusuf et al., 2011). Normalized enrichment score (NES) and q value were calculated by permutation testing.
Statistical analysis
Non-array data were analyzed using a two-tailed Student’s t-test. P-value of ≤0.05 was considered significant.
Supplementary Material
Highlights.
Exhausted CD4+ T cells have a distinct molecular profile
Exhausted CD4+ and CD8+ T cells have a diverse inhibitory receptor profile
A unique TF signature is associated with CD4+ T cell exhaustion
The molecular profile of exhausted CD4+ T cells is distinct from other Th subsets
Acknowledgments
We thank J. Faust, D.J. Hussey and D.E. Ambrose for FACS sorting. Thanks to members of the Wherry lab, G. Perona-Wright and I. Stylianou for reading of the manuscript. This work was supported by grants from the NIH (AI071309, AI083022, AI082630, AI095608, AI078897, and HHSN266200500030C) to E.J.W and the American Heart Association postdoctoral fellowship (to A.C.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agarwal P, Raghavan A, Nandiwada SL, Curtsinger JM, Bohjanen PR, Mueller DL, Mescher MF. Gene regulation and chromatin remodeling by IL-12 and type I IFN in programming for CD8 T cell effector function and memory. J Immunol. 2009;183:1695–1704. doi: 10.4049/jimmunol.0900592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agnellini P, Wolint P, Rehr M, Cahenzli J, Karrer U, Oxenius A. Impaired NFAT nuclear translocation results in split exhaustion of virus-specific CD8+ T cell functions during chronic viral infection. Proc Natl Acad Sci U S A. 2007;104:4565–4570. doi: 10.1073/pnas.0610335104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed R, Salmi A, Butler LD, Chiller JM, Oldstone MB. Selection of genetic variants of lymphocytic choriomeningitis virus in spleens of persistently infected mice. Role in suppression of cytotoxic T lymphocyte response and viral persistence. J Exp Med. 1984;160:521–540. doi: 10.1084/jem.160.2.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartosch B, Bukh J, Meunier JC, Granier C, Engle RE, Blackwelder WC, Emerson SU, Cosset FL, Purcell RH. In vitro assay for neutralizing antibody to hepatitis C virus: evidence for broadly conserved neutralization epitopes. Proc Natl Acad Sci U S A. 2003;100:14199–14204. doi: 10.1073/pnas.2335981100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry MP, Graham CM, McNab FW, Xu Z, Bloch SA, Oni T, Wilkinson KA, Banchereau R, Skinner J, Wilkinson RJ, et al. An interferon-inducible neutrophil-driven blood transcriptional signature in human tuberculosis. Nature. 2010;466:973–977. doi: 10.1038/nature09247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn SD, Shin H, Haining WN, Zou T, Workman CJ, Polley A, Betts MR, Freeman GJ, Vignali DA, Wherry EJ. Coregulation of CD8+ T cell exhaustion by multiple inhibitory receptors during chronic viral infection. Nature immunology. 2009;10:29–37. doi: 10.1038/ni.1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosinger SE, Li Q, Gordon SN, Klatt NR, Duan L, Xu L, Francella N, Sidahmed A, Smith AJ, Cramer EM, et al. Global genomic analysis reveals rapid control of a robust innate response in SIV-infected sooty mangabeys. J Clin Invest. 2009;119:3556–3572. doi: 10.1172/JCI40115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks DG, Teyton L, Oldstone MB, McGavern DB. Intrinsic functional dysregulation of CD4 T cells occurs rapidly following persistent viral infection. J Virol. 2005;79:10514–10527. doi: 10.1128/JVI.79.16.10514-10527.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks DG, Trifilo MJ, Edelmann KH, Teyton L, McGavern DB, Oldstone MB. Interleukin-10 determines viral clearance or persistence in vivo. Nat Med. 2006;12:1301–1309. doi: 10.1038/nm1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchmeier MJ, Welsh RM, Dutko FJ, Oldstone MB. The virology and immunobiology of lymphocytic choriomeningitis virus infection. Adv Immunol. 1980;30:275–331. doi: 10.1016/s0065-2776(08)60197-2. [DOI] [PubMed] [Google Scholar]
- Ejrnaes M, Filippi CM, Martinic MM, Ling EM, Togher LM, Crotty S, von Herrath MG. Resolution of a chronic viral infection after interleukin-10 receptor blockade. J Exp Med. 2006;203:2461–2472. doi: 10.1084/jem.20061462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsaesser H, Sauer K, Brooks DG. IL-21 is required to control chronic viral infection. Science. 2009;324:1569–1572. doi: 10.1126/science.1174182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahey LM, Wilson EB, Elsaesser H, Fistonich CD, McGavern DB, Brooks DG. Viral persistence redirects CD4 T cell differentiation toward T follicular helper cells. J Exp Med. 2011;208:987–999. doi: 10.1084/jem.20101773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frohlich A, Kisielow J, Schmitz I, Freigang S, Shamshiev AT, Weber J, Marsland BJ, Oxenius A, Kopf M. IL-21R on T cells is critical for sustained functionality and control of chronic viral infection. Science. 2009;324:1576–1580. doi: 10.1126/science.1172815. [DOI] [PubMed] [Google Scholar]
- Fuller MJ, Zajac AJ. Ablation of CD8 and CD4 T cell responses by high viral loads. J Immunol. 2003;170:477–486. doi: 10.4049/jimmunol.170.1.477. [DOI] [PubMed] [Google Scholar]
- Getnet D, Grosso JF, Goldberg MV, Harris TJ, Yen HR, Bruno TC, Durham NM, Hipkiss EL, Pyle KJ, Wada S, et al. A role for the transcription factor Helios in human CD4(+)CD25(+) regulatory T cells. Mol Immunol. 2010;47:1595–1600. doi: 10.1016/j.molimm.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahm K, Cobb BS, McCarty AS, Brown KE, Klug CA, Lee R, Akashi K, Weissman IL, Fisher AG, Smale ST. Helios, a T cell-restricted Ikaros family member that quantitatively associates with Ikaros at centromeric heterochromatin. Genes & development. 1998;12:782–796. doi: 10.1101/gad.12.6.782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jameson SC, Masopust D. Diversity in T cell memory: an embarrassment of riches. Immunity. 2009;31:859–871. doi: 10.1016/j.immuni.2009.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston RJ, Poholek AC, DiToro D, Yusuf I, Eto D, Barnett B, Dent AL, Craft J, Crotty S. Bcl6 and Blimp-1 are reciprocal and antagonistic regulators of T follicular helper cell differentiation. Science. 2009;325:1006–1010. doi: 10.1126/science.1175870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao C, Oestreich KJ, Paley MA, Crawford A, Angelosanto JM, Ali MA, Intlekofer AM, Boss JM, Reiner SL, Weinmann AS, et al. Transcription factor T-bet represses expression of the inhibitory receptor PD-1 and sustains virus-specific CD8(+) T cell responses during chronic infection. Nat Immunol. 2011;12:663–671. doi: 10.1038/ni.2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee LN, Burke S, Montoya M, Borrow P. Multiple mechanisms contribute to impairment of type 1 interferon production during chronic lymphocytic choriomeningitis virus infection of mice. J Immunol. 2009;182:7178–7189. doi: 10.4049/jimmunol.0802526. [DOI] [PubMed] [Google Scholar]
- Matloubian M, Concepcion RJ, Ahmed R. CD4+ T cells are required to sustain CD8+ cytotoxic T-cell responses during chronic viral infection. J Virol. 1994;68:8056–8063. doi: 10.1128/jvi.68.12.8056-8063.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller DL. E3 ubiquitin ligases as T cell anergy factors. Nature immunology. 2004;5:883–890. doi: 10.1038/ni1106. [DOI] [PubMed] [Google Scholar]
- Odorizzi PM, Wherry EJ. Inhibitory receptors on lymphocytes: insights from infections. J Immunol. 2012;188:2957–2965. doi: 10.4049/jimmunol.1100038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oxenius A, Zinkernagel RM, Hengartner H. Comparison of activation versus induction of unresponsiveness of virus- specific CD4+ and CD8+ T cells upon acute versus persistent viral infection. Immunity. 1998;9:449–457. doi: 10.1016/s1074-7613(00)80628-7. [DOI] [PubMed] [Google Scholar]
- Paley MA, Kroy DC, Odorizzi PM, Johnnidis JB, Dolfi DV, Barnett BE, Bikoff EK, Robertson EJ, Lauer GM, Reiner SL, et al. Progenitor and terminal subsets of CD8+ T cells cooperate to contain chronic viral infection. Science. 2012;338:1220–1225. doi: 10.1126/science.1229620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quigley M, Pereyra F, Nilsson B, Porichis F, Fonseca C, Eichbaum Q, Julg B, Jesneck JL, Brosnahan K, Imam S, et al. Transcriptional analysis of HIV-specific CD8+ T cells shows that PD-1 inhibits T cell function by upregulating BATF. Nat Med. 2010;16:1147–1151. doi: 10.1038/nm.2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quirion MR, Gregory GD, Umetsu SE, Winandy S, Brown MA. Cutting edge: Ikaros is a regulator of Th2 cell differentiation. J Immunol. 2009;182:741–745. doi: 10.4049/jimmunol.182.2.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safford M, Collins S, Lutz MA, Allen A, Huang CT, Kowalski J, Blackford A, Horton MR, Drake C, Schwartz RH, et al. Egr-2 and Egr-3 are negative regulators of T cell activation. Nature immunology. 2005;6:472–480. doi: 10.1038/ni1193. [DOI] [PubMed] [Google Scholar]
- Schulze Zur Wiesch J, Ciuffreda D, Lewis-Ximenez L, Kasprowicz V, Nolan BE, Streeck H, Aneja J, Reyor LL, Allen TM, Lohse AW, et al. Broadly directed virus-specific CD4+ T cell responses are primed during acute hepatitis C infection, but rapidly disappear from human blood with viral persistence. J Exp Med. 2012;209:61–75. doi: 10.1084/jem.20100388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin H, Blackburn SD, Intlekofer AM, Kao C, Angelosanto JM, Reiner SL, Wherry EJ. A role for the transcriptional repressor Blimp-1 in CD8(+) T cell exhaustion during chronic viral infection. Immunity. 2009;31:309–320. doi: 10.1016/j.immuni.2009.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JJ, Krawczyk CM, Mohrs M, Pearce EJ. Th2 cell hyporesponsiveness during chronic murine schistosomiasis is cell intrinsic and linked to GRAIL expression. The Journal of clinical investigation. 2009;119:1019–1028. doi: 10.1172/JCI36534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teijaro JR, Ng C, Lee AM, Sullivan BM, Sheehan KC, Welch M, Schreiber RD, de la Torre JC, Oldstone MB. Persistent LCMV infection is controlled by blockade of type I interferon signaling. Science. 2013;340:207–211. doi: 10.1126/science.1235214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vezys V, Penaloza-Macmaster P, Barber DL, Ha SJ, Konieczny B, Freeman GJ, Mittler RS, Ahmed R. 4-1BB Signaling Synergizes with Programmed Death Ligand 1 Blockade To Augment CD8 T Cell Responses during Chronic Viral Infection. J Immunol. 2011 doi: 10.4049/jimmunol.1100077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei G, Wei L, Zhu J, Zang C, Hu-Li J, Yao Z, Cui K, Kanno Y, Roh TY, Watford WT, et al. Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity. 2009;30:155–167. doi: 10.1016/j.immuni.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wherry EJ. T cell exhaustion. Nat Immunol. 2011;12:492–499. doi: 10.1038/ni.2035. [DOI] [PubMed] [Google Scholar]
- Wherry EJ, Blattman JN, Murali-Krishna K, van der Most R, Ahmed R. Viral persistence alters CD8 T-cell immunodominance and tissue distribution and results in distinct stages of functional impairment. J Virol. 2003;77:4911–4927. doi: 10.1128/JVI.77.8.4911-4927.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wherry EJ, Ha SJ, Kaech SM, Haining WN, Sarkar S, Kalia V, Subramaniam S, Blattman JN, Barber DL, Ahmed R. Molecular signature of CD8+ T cell exhaustion during chronic viral infection. Immunity. 2007;27:670–684. doi: 10.1016/j.immuni.2007.09.006. [DOI] [PubMed] [Google Scholar]
- Wilson EB, Yamada DH, Elsaesser H, Herskovitz J, Deng J, Cheng G, Aronow BJ, Karp CL, Brooks AG. Blockade of Chronic Type I Interferon Signaling to Control Persistent LCMV Infection. Science. 2013;340:202–207. doi: 10.1126/science.1235208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi JS, Du M, Zajac AJ. A vital role for interleukin-21 in the control of a chronic viral infection. Science. 2009;324:1572–1576. doi: 10.1126/science.1175194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusuf I, Kageyama R, Monticelli L, Johnston RJ, Ditoro D, Hansen K, Barnett B, Crotty S. Germinal center T follicular helper cell IL-4 production is dependent on signaling lymphocytic activation molecule receptor (CD150) J Immunol. 2011;185:190–202. doi: 10.4049/jimmunol.0903505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuniga EI, Liou LY, Mack L, Mendoza M, Oldstone MB. Persistent virus infection inhibits type I interferon production by plasmacytoid dendritic cells to facilitate opportunistic infections. Cell Host Microbe. 2008;4:374–386. doi: 10.1016/j.chom.2008.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.