Abstract
In two-component signal transduction, response regulator proteins contain the catalytic machinery for their own covalent phosphorylation and can catalyze phosphotransfer from a partner sensor kinase or autophosphorylate using various small molecule phosphodonors. Although response regulator autophosphorylation is physiologically relevant and a powerful experimental tool, the kinetic determinants of the autophosphorylation reaction and how those determinants might vary for different response regulators and phosphodonors are largely unknown. We characterized the autophosphorylation kinetics of 21 variants of the model response regulator Escherichia coli CheY that contained substitutions primarily at nonconserved active site positions D+2 (CheY residue 59) and T+2 (CheY residue 89), two residues C-terminal to conserved D57 and T87, respectively. Overall, the CheY variants exhibited a >105-fold range of rate constants (kphos/KS) for reaction with phosphoramidate, acetyl phosphate, or monophosphoimidazole, with the great majority of rates enhanced over wild type CheY. Although phosphodonor preference varied substantially, nearly all the CheY variants reacted faster with phosphoramidate than acetyl phosphate. Correlation between increased positive charge of the D+2/T+2 side chains and faster rates indicated electrostatic interactions are a kinetic determinant. Moreover, sensitivities of rate constants to ionic strength indicated that both long-range and localized electrostatic interactions influence autophosphorylation kinetics. Increased nonpolar surface area of the D+2/T+2 side chains also correlated with enhanced autophosphorylation rate, especially for reaction with phosphoramidate and monophosphoimidazole. Computer docking suggested that highly accelerated monophosphoimidazole autophosphorylation rates for CheY variants with a tyrosine at position T+2 likely reflect structural mimicry of phosphotransfer from the sensor kinase histidyl phosphate.
Keywords: CheY, response regulator, two-component signal transduction, acetyl phosphate, phosphoramidate, monophosphoimidazole
Two-component signal transduction systems are ubiquitous in bacteria and control a variety of cellular processes1–3. Despite their amazing functional diversity, two-component systems share a fundamental signaling scheme based on the transfer of phosphoryl groups between conserved protein domains. Signal transduction involves autophosphorylation of a sensor kinase on a histidyl residue, typically modulated by environmental conditions. The phosphoryl group is then transferred to an aspartyl residue on a partner response regulator protein, which alters the ability of the response regulator to execute an output response. This central His to Asp phosphotransfer event is catalyzed by a conserved active site on the receiver domain of the response regulator. It has been recognized for more than twenty years that receiver domains can also catalyze their own phosphorylation using a variety of small molecule phosphodonors4. Response regulator autophosphorylation with the metabolic intermediate acetyl phosphate contributes to the regulation of multiple two-component systems in vivo5, 6 and the use of small molecule phosphodonors in vitro has been instrumental in facilitating functional analysis of numerous response regulators7–9. Because autophosphorylation of the response regulator and phosphotransfer from the sensor kinase to the response regulator both proceed through the same basic phosphorus substitution chemistry, mechanistic insights into the catalysis of response regulator autophosphorylation will enhance understanding of the more complex phosphotransfer reaction between proteins.
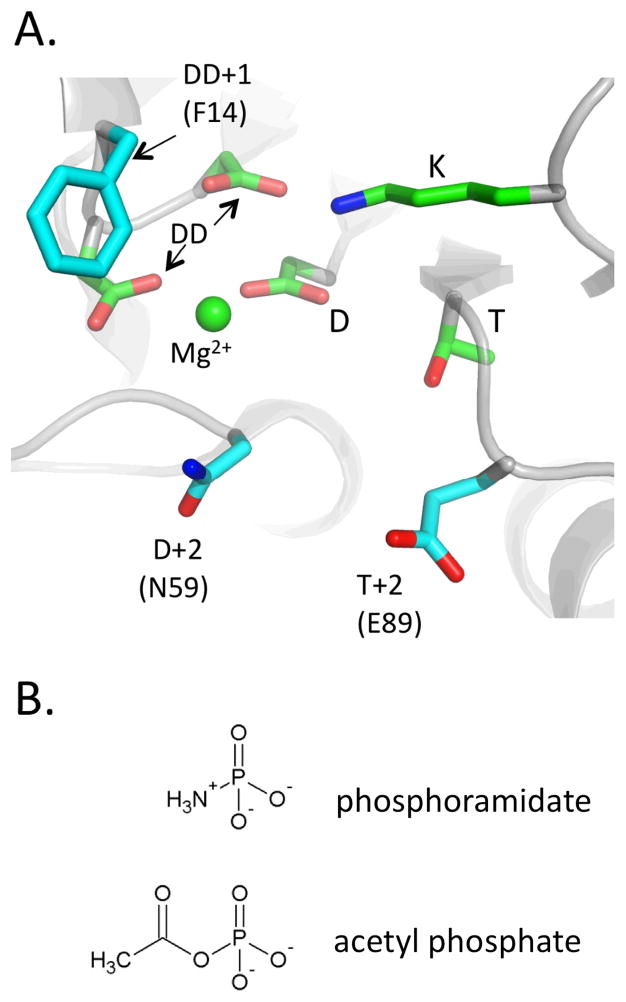
Response regulator receiver domains have a conserved (β/α)5 fold with the conserved active site located on the β/α loops that cluster on one face of the domain10 (Figure 1A). The phosphorylatable aspartate (D) is positioned centrally in the active site and is surrounded by a threonine/serine (T), a lysine (K), and two additional acid residues (DD), which coordinate a magnesium ion. The Mg2+, threonine/serine, and lysine interact with the three phosphoryl oxygens in the phosphorylated forms of receivers10, 11. Based on transition state analogue structures of related phosphatases within the haloacid dehalogenase superfamily12, the same interactions are also likely to occur in the response regulator transition state. Autophosphorylation proceeds by nucleophilic attack of the aspartyl carboxylate on the phosphodonor phosphorus atom in a substitution reaction. The receiver domain active site also catalyzes the subsequent hydrolysis of the aspartyl phosphate in a self-catalyzed dephosphorylation reaction.
Figure 1.
Features of the receiver domain active site (A) and chemical structures of phosphodonor molecules (B). (A) The active site of E. coli CheY (PDBid 1FQW) with residues conserved amongst all receiver domains colored green and the three nonconserved residues studied here colored cyan. Conserved residues are labeled as outlined in the text: DD is the metal binding pair (CheY D12 and D13), D is the phosphorylated aspartate (CheY D57), and T and K are the conserved threonine/serine and lysine (CheY T87 and K109) residues. Nonconserved residues are labeled according to their sequence relationship to the closest conserved residue with their identities in E. coli CheY in parentheses. (B) Prototypes of the two chemical classes of phosphodonors. Phosphoramidate and acetyl phosphate represent the phosphoramidate and acyl phosphate chemical classes, respectively. Charges shown are predominant at pH 7.0. CheY reacts poorly with the unprotonated form of PAM (pKa~8.0)18.
Known small molecule phosphodonors for receiver domains fall into two chemical classes (Figure 1B). Phosphoramidates (R2NH+-PO32−) contain a phosphorus-nitrogen bond and include monophosphoimidazole (MPI), a near-identical model of the phosphohistidine side chain in sensor kinases. The class also includes the compound phosphoramidate (PAM; NH3+-PO32−), often used to phosphorylate response regulators in vitro, and the smallest compound in the class. The other phosphodonor class is acyl phosphates (RCO2-PO32−), mixed anhydrides that include the physiologically relevant acetyl phosphate6. Both the compounds PAM and acetyl phosphate (AcP; CH3CO2-PO32−), well-characterized representatives of the two chemical classes, are relatively reactive to nucleophilic substitution. Both have high free energies of hydrolysis and undergo limited hydrolysis at neutral pH13, 14. However, PAM and AcP differ in net charge, charge distribution, and size (Figure 1B).
Much of the kinetic and mechanistic characterization of receiver domain autophosphorylation has come from studies of the Escherichia coli/Salmonella chemotaxis response regulator CheY15–18. CheY consists only of a receiver domain and has a unique active site tryptophan residue that serves as a fluorescence probe for phosphorylation4. The rates of CheY autophosphorylation with PAM and AcP are similar16, 18 but much slower than phosphotransfer from the CheA kinase19. The disparity in rates of CheY phosphorylation appears to be partially due to weak binding between CheY and small molecule phosphodonors15. Plots of the pseudo-first order autophosphorylation rate constant versus PAM or AcP concentration are linear up to at least 100 mM phosphodonor with no sign of saturation, indicating binding constants between CheY and phosphodonor of ≫ 100 mM15, 17. CheY does not use general acid catalysis in autophosphorylation with either PAM or AcP18. Although autophosphorylation kinetic data for other response regulators is limited, the available data suggest that different response regulators react with different kinetics20 as well as preferences for different phosphodonors. For example, the response regulators CheB4 from E. coli and Spo0F21 from Bacillus subtilis do not autophosphorylate with AcP but react readily with PAM. However, the structural features that determine the autophosphorylation kinetics of different receiver domains and how these features exert their modulatory effects are not currently known. E. coli CheY serves as a highly experimentally accessible model system to explore features of receiver domains that modulate functional differences between response regulators, a family of signaling proteins with tens of thousands of known members3, 22.
This study represents the first step towards a long-term goal of elucidating how various amino acids at nonconserved active site positions modulate the rates of response regulator autophosphorylation and phosphotransfer from the kinase. Examination of the active conformation of CheY (PDBid 1FQW), believed to be the ground state for the autophosphorylation reaction17, 20, 23, reveals that nonconserved residues D+2, T+1, and T+2 [CheY residues 59, 88, and 89, respectively; one or two residues C-terminal to the conserved D or T), form the surface of the active site where the phosphodonor molecule would be expected to approach and dock. In contrast, the conserved Asp, Thr/Ser, Lys, and divalent cation are located deeper within the CheY active site (Figure 1A). Thus, the conserved portions of the CheY active site are positioned to interact with the phosphoryl group of the phosphodonor, whereas the nonconserved portions are positioned to interact with the leaving group. In this work, we focus primarily on the effect of residues at positions D+2 and T+2 on autophosphorylation because response regulators exhibit far more amino acid sequence diversity at D+2 and T+2 than at T+1, which is Ala or Gly in more than 70% of response regulators22, 24. Furthermore, previous work from this laboratory examining the related self-catalyzed dephosphorylation reaction, demonstrated that substitutions at D+2 and T+2 impact the rates of E. coli CheY and B. subtilis Spo0F autodephosphorylation and that a given residue often has similar effects on the reaction rates in the two response regulators25, 26.
We show here that a set of 21 CheY mutants exhibited greater than a 105-fold span of autophosphorylation rates with AcP, PAM, or MPI with up to 103-fold rate enhancements over wild type CheY. The great majority of the CheY variants reacted faster with PAM and MPI than with AcP. Statistical correlations between autophosphorylation rates and side chain properties, aided by direct biochemical analysis and molecular modeling, allowed identification and characterization of several mechanisms for rate modulation and phosphodonor preference including attractive electrostatic interactions and steric complementarity between CheY and the phosphodonor.
EXPERIMENTAL PROCEDURES
Site directed mutagenesis and protein purification
Site directed mutagenesis of the pRS3 plasmid27, which carries the Escherichia coli cheY and cheZ genes under regulation of the Serratia marcescens trp promoter, was carried out using Quikchange methodology (Agilent Technologies). Resultant plasmids were transformed into the E. coli ΔcheY strain KO641recA for CheY expression and purification as described26.
Chemicals
Acetyl phosphate (AcP) (potassium lithium salt) was from Sigma-Aldrich. The potassium salt of phosphoramidate (PAM) was synthesized as described28. The PAM was > 95% pure as assessed by 31P NMR with the remaining phosphorus present as inorganic phosphate. The calcium salt of monophosphoimidazole (MPI) was synthesized according to published protocols29. To prepare the sodium salt of MPI (Na2MPI), an aqueous solution of CaMPI was flowed through a column containing Chelex resin in the sodium form (Biorad) that had been washed in water, followed by flash freezing and lyophilization.
Fluorescence spectroscopy
Time courses for reaction of CheY (wild type and mutant) with small molecule phosphodonors were measured by stopped-flow tryptophan fluorescence, essentially as described17. A rapid mixing device (Applied Photophysics RX2000) with a dead time of 8 ms was used to react equal volumes of solutions containing CheY and phosphodonor (AcP, PAM, or MPI) while maintaining constant temperature at 25 ± 0.5 °C with a circulating water bath. Tryptophan fluorescence (excitation 295 nm, emission 346 nm) was recorded at 20 ms intervals using a Perkin-Elmer LS-50B Luminescence Spectrometer with FL Winlab 1.1 software. The final reaction concentrations were 2.5 μM CheY and 0.5–30 mM phosphodonor. The CheY solutions were in 100 mM Hepes pH 7.0, 10 mM MgCl2 (total ionic strength 130 mM). The phosphodonor solutions were also in 100 mM Hepes pH 7.0, 10 mM MgCl2 with appropriate amounts of KCl to maintain a constant ionic strength of 330 mM in spite of differing phosphodonor concentrations. Thus mixing of equal volumes of the CheY and phosphodonor solutions yielded a final reaction ionic strength of 230 mM. A minimum of four phosphodonor concentrations were tested for each CheY variant and a series of three to five time courses were recorded for each phosphodonor concentration. The range of phosphodonor concentrations was adjusted for each CheY variant to give kobs values between ~0.05 and 2.0 s−1. The entire process was carried out in duplicate for each CheY variant.
Reactions resulted in an exponential loss of tryptophan fluorescence due to the phosphorylation of CheY Asp57. The time courses were fit to an equation for a single exponential decay using either Excel or Prism software to obtain a pseudo-first order rate constant (kobs) that reflects the rate of accumulation of phosphorylated CheY. Plots of kobs (averaged from three to five replicates) versus phosphodonor concentration were linear for all the CheY variants and phosphodonor concentrations used in this study, consistent with the previously derived relationship15–17:
| (eqn. 1) |
where KS is the dissociation equilibrium constant for noncovalent complex formation between CheY and phosphodonor, kphos is the rate constant for phosphotransfer within the noncovalent complex, and kdephos is the rate constant for CheY autodephosphorylation. Values for kphos/KS, the apparent bimolecular rate constant for autophosphorylation, were determined from the slopes of the linear plots.
Ionic strength experiments
For several of the CheY variants, kphos/KS values were determined at four different ionic strengths ranging from 0.23 M to 1.63 M. Time courses were measured by stopped-flow fluorescence as described above. Both the CheY and phosphodonor reaction solutions contained 100 mM Hepes pH 7.0, 10 mM MgCl2, and KCl was added to both the protein and phosphodonor solutions to achieve the desired ionic strength. For each CheY variant and ionic strength condition, time courses were recorded at four different phosphodonor concentrations. Plots of kobs versus phosphonor concentration were linear and the slope gave kphos/KS values, exactly as described above for rate constants measured at ambient ionic strength. Each experimental condition (CheY variant/ionic strength) was carried out in duplicate.
Quantitative analysis of the effects of ionic strength on CheY autophosphorylation kinetics was based on a published method30. For enzymatic reactions where the formation of Coulombic interactions (long-range electrostatic interactions) are a kinetic determinant [e.g. formation of ion pairs between enzyme (E) and substrate (S)], increasing solution ionic strength slows the reaction due to charge screening effects of solution counterions. Based on polyelectrolyte theory and supported empirically by the behavior of multiple enzymes30–32, plots of the log10 of kcat/Km (or individual kinetic parameters) versus the log10 of the concentration of monovalent salt are linear over a large range of ionic strength. The slope of the resultant line is −n′, where n′ approximates the number of counterions released from charged residues or substrate ions upon complex formation, which is usually reflective of the number of charges involved in salt bridges between enzyme and substrate in the E·S complex (see Supplemental text for additional details). For our studies, kphos/KS values for CheY autophosphorylation measured at different ionic strengths were first corrected for small differences in Mg2+ binding affinities at different ionic strengths (see Supplemental text). Then, for each CheY and phosphodonor pair, the log10 of the corrected kphos/KS values were plotted versus the log10 of the total ionic strength of the reaction solution. The data were fit using linear regression (Prism) and slopes (−n′ values) determined.
Docking MPI into CheY variant active site
Atomic coordinates for MPI were generated using the prodrg server (http:/davapc1.bioch.dundee.ac.uk/prodrg/). Water and glycerol molecules and sulfate ions were removed from PDBid 3FFW25, the Protein Data Bank (PDB) file for the X-ray crystal structure of the CheY triple mutant F14Q/N59Q/E89Y complexed with BeF3− and Mn2+. Using the Docking Wizard utility within the PyMOL Molecular Graphics System, Version 1.1. (Schrödinger, LLC), the three phosphoryl oxygen atoms in MPI were superimposed on the three fluorine atoms in the BeF3− anion bound to chain B of the edited pdb 3FFW, with the imidazole oriented toward solvent. Keeping the phosphoryl oxygens stationary, the torsional angle within MPI was rotated manually to minimize apparent steric overlap between the MPI and CheY active site atoms and to allow solvent accessibility of the carbon atom corresponding to Cγ in phosphohistidine, as would be expected for the sensor kinase phosphohistidine phosphodonor. Hydrogen atoms were added to the resultant complex using Reduce33 and a dot image representing positive and negative interatomic interactions in the docked structure was generated using Probe34.
Structure-based estimation of non-polar surface areas
The non-polar surface areas of residues at positions D+2 (CheY residue 59) and T+2 (CheY residue 89) were estimated based on X-ray crystal structures. Of the eight amino acids found at positions D+2 or T+2 in the 19 tested CheY mutants that did not contain a substitution at DD+1 (CheY residue 14), five amino acids at D+2 and seven amino acids at T+2 are present in structures of CheY variants in the Protein Data Bank (PDBids 1FQW, 1MIH, 3F7N, 3FFT, 3FFW, 3FFX, 3FGZ, 3RVJ, 3RVL, 3RVN, 3RVP)11, 25, 35 (C.A. Starbird, R.M. Immormino, R.E. Silversmith, R.B. Bourret, unpublished). The conformation of the remaining four amino acids (Ala, Leu, and Glu at D+2 plus Ala at T+2) were conservatively modeled using the BeF3− bound structure of wild type CheY (PDBid 1FQW)11 as a scaffold. For example, the Ala side chains were modeled by terminating the wild type side chains at the beta carbon. The resulting structures or models were then stripped of solvent molecules, except the divalent metal, protonated using Reduce and the van der Waals (VDW) surface for the residues surrounding the site of phosphorylation was calculated with Probe33, 34. The dot density for the VDW surface calculated by Probe was left at the default value of 16 dots per square angstrom. To determine if dots on the VDW surface belonged to the active site pocket or were external, a computer program was written that casts rays from a point in the center of the pocket (the nitrogen linked to the phosphorus of MPI docked to the CheY active site) outward to the dots on the VDW surface. Dots for which the rays could be drawn without passing through the VDW surface were assumed to be part of the active site. The active site surface dots belonging to non-polar side chain atoms (carbon or hydrogen) were counted and divided by 16 to give the non-polar surface areas for residues D+2 or T+2 in square angstroms. In cases where there was more than one crystal structure with the same residue at position D+2 or T+2, we used the average of the calculated surface areas for the residue from each structure. The total non-polar surface areas for residues D+2 and T+2 were then determined by adding the non-polar surface area values for particular amino acids at D+2 and T+2.
RESULTS
Nonconserved active site residues strongly influence CheY autophosphorylation kinetics
To assess the influence of variable active site residues in modulating catalysis of response regulator autophosphorylation, we measured pre-steady state kinetics for the reaction of a large set of CheY variants with AcP or PAM, representatives of the two chemical classes of small molecule phosphodonors (Figure 1B). In all, 21 single, double, and triple mutants of CheY were analyzed. The great majority (19 out of 21) were single or double mutants with substitutions at position D+2 (CheY N59, two residues from the phosphorylated D57) and/or T+2 (CheY E89, two residues from the conserved active site T87) (Figure 1A). The remaining two CheY variants were triple mutants with an additional substitution at position DD+1 (CheY F14, the residue after the conserved D12/D13 pair). Specific substitutions were chosen to give a variety of chemical properties and many also mimicked residues found at the corresponding positions in other well-studied receiver domains. For all of the reactions, stopped-flow fluorescence time courses that monitor the accumulation of phosphorylated CheY gave excellent fits to a single exponential decay (R2 > 0.99 with the great majority > 0.999), from which apparent first order rate constants (kobs) were obtained. Plots of kobs versus phosphodonor concentration were linear for all combinations of CheY and phosphodonor, as has been previously observed for reactions of wild type CheY with AcP and PAM15, 17. Thus, there was no detectable binding between phosphodonor and CheY (KS ≫ [phosphodonor]) under the conditions used here. The slopes of the resultant lines gave the effective bimolecular rate constants (kphos/KS) and are listed in Table 1.
Table 1.
Rate constants for autophosphorylation of wild type and mutant CheY with PAM or AcPa
| CheY variant designation | Residue at position
|
kphos/KS (M−1s−1) b
|
PAM preferencec | |||
|---|---|---|---|---|---|---|
| 14 DD+1 |
59 D+2 |
89 T+2 |
PAM | AcP | ||
| wild type | F | N | E | 10 ± 0.50 | 11 ± 0.84 | 0.91 |
| D+2 single mutants | ||||||
| RE | F | R | E | 54 ± 1.6d | 7.6 ± 0.0d | 7.1 |
| KE | F | K | E | 32 ± 1.3 | 5.8 ± 0.35 | 5.5 |
| ME | F | M | E | 26 ± 4.8 | 1.3 ± 0.61 | 20 |
| LE | F | L | E | 12 ± 0.68 | 0.97 ± 0.12 | 12 |
| AE | F | A | E | 9.0 ± 0.21d | 1.3 ± 0.21d | 6.9 |
| DE | F | D | E | 3.7 ± 0.55 | <0.2e | >19 |
| EE | F | E | E | 2.7 ± 0.20 | <0.2e | >13 |
| T+2 single mutants | ||||||
| NY | F | N | Y | 240 ± 34 | 13 ± 1.9 | 18 |
| NL | F | N | L | 140 ± 21 | 33 ± 2.8 | 4.2 |
| NR | F | N | R | 93 ± 18 | 62 ± 5.5 | 1.5 |
| NK | F | N | K | 60 ± 7.3 | 42 ± 0.90 | 1.4 |
| NH | F | N | H | 30 ± 6.8 | 12 ± 2.1 | 2.5 |
| NQ | F | N | Q | 19 ± 0.28d | 36 ± 1.8d | 0.53 |
| NA | F | N | A | 10 ± 0.85d | 18 ± 2.0d | 0.55 |
| D+2/T+2 double mutants | ||||||
| KY | F | K | Y | 370 ± 1.4 | 23 ± 1.5 | 16 |
| MR | F | M | R | 66 ± 1.3 | 18 ± 0.49 | 3.7 |
| MK | F | M | K | 97 ± 4.5 | 16 ± 1.1 | 6.1 |
| EH | F | E | H | 37 ± 2.1 | 2.3 ± 0.00 | 16 |
| DL | F | D | L | 71 ± 4.9 | 0.27 ± 0.058 | 260 |
| DD+1/D+2/T+2 triple mutants | ||||||
| *KY | Q | K | Y | 330 ± 4.7 | 25 ± 0.35 | 13 |
| *MR | E | M | R | 46 ± 0.71 | 9.5 ± 1.3 | 4.8 |
Reaction conditions were 25 °C, pH 7.0, 230 mM ionic strength, 10 mM MgCl2.
Rate constants represent the average and standard deviation of at least two independent experiments.
Ratio of kphos/KS values for PAM and AcP.
A kphos/KS value of 0.2 M−1s−1 is the estimated lower limit of detection for the assay.
This set of CheY mutants and phosphodonors gave kphos/KS values that spanned greater than an 1,800-fold range in magnitude (Table 1), demonstrating that the residues at positions D+2 and T+2 in receiver domain active sites can have a profound impact on autophosphorylation kinetics. Under these reaction conditions (pH 7.0, 230 mM ionic strength), wild type CheY gave similar rates for reaction with AcP (11 M−1s−1) and PAM (10 M−1s−1). In contrast, all other CheY variants exhibited different rates for reaction with PAM and AcP, indicating that the substitutions affected aspects of the autophosphorylation reaction that are different for the two classes of phosphodonors.
Autophosphorylation of CheY D+2 single mutants
Replacement of the wild type Asn at CheY position D+2 with amino acids that varied in size and polarity gave autophosphorylation rates that spanned a >270-fold range. For reaction with PAM, the substitutions resulted in both rate increases (up to five-fold) and decreases (up to four-fold) relative to wild type CheY. In contrast, the same substitutions all gave reduced rates for AcP. Strikingly, with the exception of wild type CheY, CheY variants with Glu at position T+2 and different amino acids at D+2 displayed the same rank order (D+2 residues: R/K>M/L/A>D/E) for reaction with both phosphodonors. This ranking suggests a correlation between increased positive charge at D+2 and faster autophosphorylation rates. Although an acidic side chain at D+2 was detrimental to reaction with both phosphodonors, the effect was especially severe for AcP, where CheY DE and CheY EE gave no detectable accumulation of CheY-P (Table 1). Taking into account the previously measured autodephosphorylation rates of CheY DE and CheY EE26, both variants were determined to possess kphos/KS values for reaction with AcP below the lower limit measureable (Table 1). Thus, negative charge at both CheY D+2 and T+2 is extremely detrimental, if not prohibitive, for CheY autophosphorylation with AcP.
Autophosphorylation of CheY T+2 single mutants
The single amino acid replacements of glutamate at CheY T+2 overwhelmingly gave enhanced rates of autophosphorylation with both phosphodonors, but especially for PAM. Notably, CheY NY exhibited a 24-fold rate enhancement with PAM whereas the same substitution gave a rate indistinguishable from wild type CheY for reaction with AcP. The rank order for CheY proteins varying at T+2 for reaction with AcP (T+2 residues: R/K>Q/L/A>H/Y/E) was similar to that observed for D+2 variants, suggesting that positive charge at T+2 also enhances autophosphorylation rate. However, the rank order for the reaction of this set of CheY proteins with PAM was different (T+2 residues: Y/L>R/K>H/Q>A/E), suggesting that large hydrophobic residues at T+2 may be advantageous for reaction with PAM, thus implicating other kinetic determinants besides side chain charge.
Autophosphorylation of CheY DD+1/D+2/T+2 triple mutants
The two CheY triple mutants included in this study, CheY *KY and CheY *MR (Table 1), contained a substitution at DD+1 (CheY Phe14) in addition to D+2 and T+2 and were designed to mimic the active sites of B. subtilis Spo0F and E. coli PhoB, respectively. Both CheY triple mutants gave rates that were similar to the corresponding D+2/T+2 double mutants. Although a very small data set, these results suggest that, as is the case with autodephosphorylation25, 26, residue DD+1 does not appear to modulate autophosphorylation rates.
Variation in phosphodonor preference
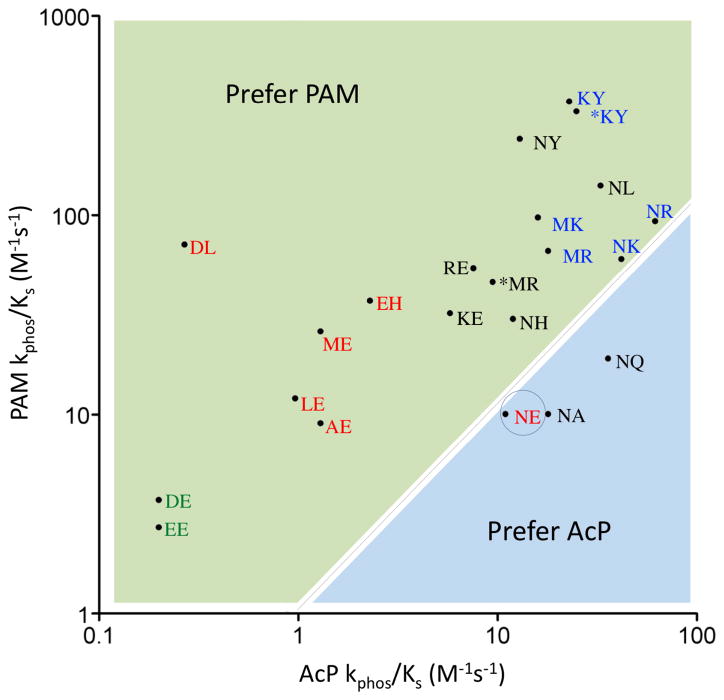
Wild type CheY displays nearly identical kphos/KS values for reactions with AcP and PAM at a standard ionic strength of 230 mM (Table 1) and thus has essentially no preference for one phosphodonor compared to the other (PAM preference = ~1). In contrast, there was a greater than 500-fold variation in preference for PAM over AcP for the mutant set studied here with preferences ranging from 0.5 to 260 (Table 1). A plot of log (kphos/KS PAM) versus log (kphos/KS AcP) (Figure 2) graphically illustrates the preference of all but two of the CheY mutants for PAM, as well as the diversity of magnitudes of the preference for PAM. For the two CheY variants (CheY NA and CheY NQ) that preferred AcP, the preference was weak (only two-fold).
Figure 2.
Phosphodonor preference (PAM versus AcP) for the set of CheY variants studied. For each CheY variant, the position of the point reflects the logarithms of the kphos/KS values (Table 1) for reaction with AcP (abscissa) or PAM (ordinate). CheY variants are designated by the amino acids at positions D+2 and T+2, respectively with wild type CheY (NE) circled. *KY and *MR have additional substitutions at DD+1 (F14Q and F14E) to mimic the Spo0F and PhoB response regulators respectively. Designations are color coded to reflect the net charge of residues at positions DD+1, D+2, and T+2 with net charge = +1 (blue), 0 (black), −1 (red), and −2 (green). The black dotted line (slope = 1) separates the plot into regions that prefer PAM (light green) or prefer AcP (light blue).
MPI further enhances kinetics for some CheY variants
PAM and AcP differ in both chemical linkage (and implicit charge), and size (Figure 1B). To probe the mechanistic basis for the general preference for PAM over AcP observed here, we measured the kinetics of autophosphorylation with MPI (monophosphoimidazole) for several CheY mutants that strongly preferred PAM over AcP. MPI has the same N-P chemical linkage (and thus similar charge distribution) as PAM but is larger than AcP. If size were the dominant deterring factor for AcP, then the reaction rates would be expected to be PAM > AcP > MPI. However, rates for reaction with MPI were all at least as fast as with PAM (Table 2), with several mutants exhibiting extremely large rate enhancements. Thus, preference for PAM over AcP is not due to phosphodonor size but must be due to chemical linkage or charge.
Table 2.
Rate constants for autophosphorylation of selected CheY variants with MPI, PAM, or AcPa
| Residue at position
|
kphos/KS (M−1s−1)
|
|||
|---|---|---|---|---|
| D+2 | T+2 | MPIb | PAMc | AcPc |
| K | Yd | 43,000 ± 120 | 330 | 25 |
| N | Y | 3,900 ± 470 | 240 | 13 |
| E | H | 440 ± 2.8 | 37 | 2.3 |
| M | E | 51 ± 7.0 | 26 | 1.3 |
| D | E | 2.5 ± 1.5 | 3.7 | < 0.2 |
| N | Ee | 43 ± 2.8 | 10 | 11 |
Reaction conditions were 25 °C, pH 7.0, 230 mM ionic strength, 10 mM MgCl2.
Values are the average and standard deviation of two independent measurements.
Variant is CheY *KY, the triple mutant with a glutamine at DD+1 (see Table 1)
Wild type CheY.
Several of the CheY variants demonstrated large preferences for MPI over PAM. In particular, CheY *KY and CheY NY, both of which exhibited rates with PAM > 20-fold faster than wild type CheY, reacted with MPI another 16-130-fold faster than their rates with PAM. The kphos/KS value for reaction of CheY *KY with MPI (43,000 M−1s−1) represents the fastest autophosphorylation rate of this mutant set and is 4,300-fold faster than the reaction of wild type CheY with AcP or PAM. In addition to CheY *KY and CheY NY, CheY EH also exhibited a further rate enhancement of >10-fold with MPI relative to its rate with PAM. Thus, instead of being a deterrent, the added bulk of MPI appeared to substantially increase autophosphorylation rates for three CheY variants relative to their rates with PAM. In contrast, CheY DE and CheY ME exhibited rates for MPI that were similar to their rates with PAM, indicating that MPI did not offer anything deleterious or advantageous over PAM for two CheY mutants. Thus the large rate enhancements for MPI over PAM appeared to correlate with the presence of an aromatic group at the T+2 position.
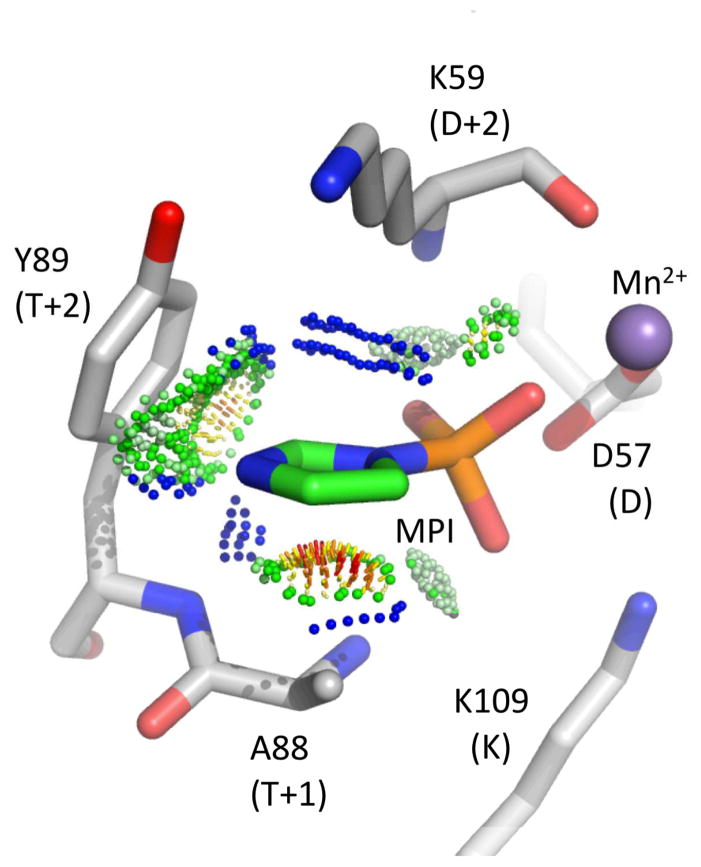
Docking MPI into CheY *KY structure implicates role of steric complementarity
To explore the structural basis of the large rate enhancements for several CheY variants - especially CheY *KY and CheY NY - with MPI, we docked the MPI molecule into the active site of CheY *KY as described in Experimental Procedures. The X-ray crystal structure of CheY *KY (PDBid 3FFW)25 complexed with Mn2+ and the phosphoryl group analogue BeF3− was used as a model of the active conformation, the probable reactive species in autophosphorylation17, 20, 23. The docked orientation (Figure 3) positioned the imidazole ring of the MPI and the aromatic ring of the tyrosine at T+2 virtually perpendicular to one another in an optimal position for edge-to-face π-π interactions36. In addition, there were a large number of van der Waals interactions between the imidazole group of MPI and both the tyrosine side chain at T+2 and the lysine at D+2. It is likely that similar interactions also occur in the other two CheY variants (CheY NY and CheY EH, both of which have an aromatic group at position T+2) that show very fast reaction with MPI. Thus, the modeling suggested that enhanced binding between phosphodonor and CheY due to an enlarged sterically complementary hydrophobic binding surface and π-π interactions is a likely explanation for the greatly enhanced rates. The 10-fold increase in rate of CheY *KY over CheY NY may be due to a larger binding interface (K verus N) as well an additional positive charge (see below).
Figure 3.
Model of MPI docked into the active site of CheY *KY·Mn2+· BeF3− (PDBid 3FFW)25. Green and blue probe dots represent favorable van der Waals interactions whereas orange and red dots represent steric clashes. The BeF3− ion present in the original structure is not shown for clarity.
Statistical correlations between D+2/T+2 side chain properties and CheY autophosphorylation rates
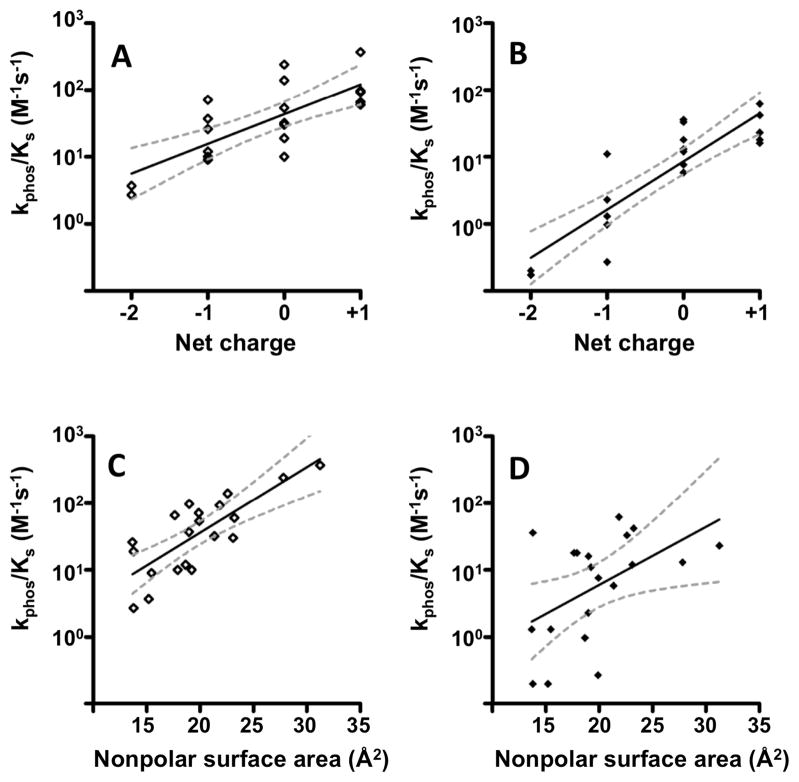
The trends discussed above suggested that increased positive charge of D+2/T+2 side chains or the presence of an aromatic side chain could be kinetic determinants that enhanced CheY autophosphorylation. To test these trends more rigorously, and to gain insight into which factors may dominate for each of the two classes of phosphodonors, we carried out statistical analysis of rates for wild type CheY plus all 19 CheY single or double mutants for reaction with AcP or PAM. Plots were generated relating net electrostatic charge or nonpolar surface area of residues D+2 and T+2 versus log (kphos/KS) for reaction with AcP or PAM (Figure 4AB). There was a significant Pearson correlation between net charge and log (kphos/KS) for both PAM and AcP (p = 0.0001 and p < 0.0001, respectively, supporting the notion that increased positive charge enhances rate for both AcP and PAM. Furthermore, the magnitudes of the slopes of the linear regression lines (0.44 for PAM and 0.72 for AcP; Figure 4AB) suggest that the charge effect was greater for AcP (−2 charge) than PAM (−1 charge), consistent with the observation that CheY EE and CheY DE gave no detectable reaction with AcP but reacted with PAM just modestly slower than with wild type CheY. The two classes of phosphodonors also exhibited differences in correlations between log (kphos/KS) and the nonpolar surface area of the active site “pocket” formed by the T+2 and D+2 side chains. For PAM, there was a strong Pearson correlation (p < 0.0001) and the linear regression analysis showed a tight 95% confidence band whereas for acetyl phosphate, the Pearson correlation was weak (but still significant; p = 0.0251) and the linear regression exhibited a wide 95% confidence band (Figure 4CD). Thus, CheY has different kinetic determinants for reaction with PAM or AcP. Overall, CheY autophosphorylation with PAM is more positively impacted by increasing nonpolar surface area and less positively impacted by increased charge than reaction with AcP.
Figure 4.
Correlations between net charge (A and B) or non-polar surface area (C and D) at positions D+2 and T+2 and the measured rate constants (kphos/KS, from Table 1) for autophosphorylation reactions with PAM (A and C) or AcP (B and D). Each point represents a single CheY variant. Net charge was calculated assuming histidyl residues are neutral. Non-polar surface area was calculated as described in Experimental Procedures. For each plot, the black dashed line is the linear regression best-fit line and the gray dashed lines represent the 95% confidence bands. R2 values were 0.57, 0.76, 0.61, and 0.25 for panels A, B, C, and D, respectively.
Ionic strength effects on CheY autophosphorylation kinetics
The correlation between increased net charge of active site residues and faster autophosphorylation observed here (Figure 4AB) implicates a role for electrostatic forces in driving CheY autophosphorylation kinetics. Previous studies demonstrated that CheY autophosphorylation rates decrease with increasing ionic strength16. Rates that are sensitive to ionic strength in this manner generally reflect bimolecular reaction steps where two separated oppositely charged species come together to form an ionic interaction (long-range electrostatic interactions)30, such as the association of a charged enzyme active site with an oppositely charged substrate30, 37. Solution counterions act to shield the charged groups and prevent the interaction. To assess whether the rate differences observed for CheY D+2/T+2 substitutions (Table 1) were due to the same electrostatic interactions that lead to the ionic strength dependence, we measured the ionic strength dependences of autophosphorylation rates for a subset of CheY variants and quantitatively analyzed the results.
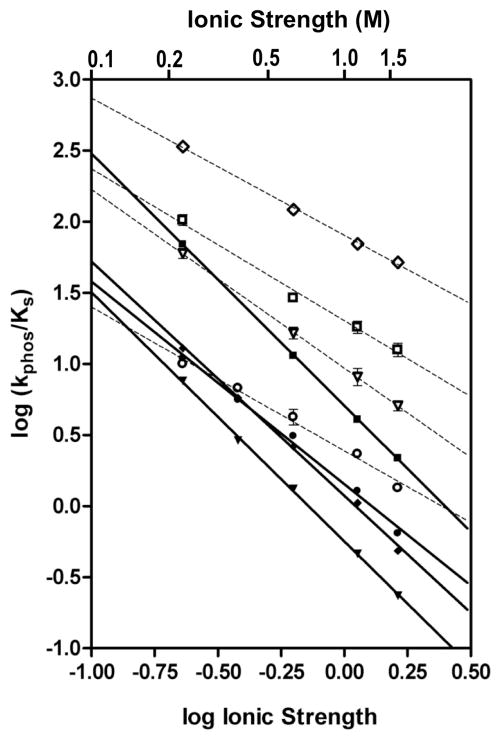
Plots of log (ionic strength) versus log (kphos/KS) for autophosphorylation of CheY NE, CheY RE, CheY NR, and CheY NY with either PAM or AcP approximated linearity (Figure 5), as predicted by polyelectrolyte theory30. In this analysis, for enzymes where the rate of dissociation of the E·S complex is much faster than the chemical reaction step as occurs with CheY (see Supplemental Information), the absolute value of the slope of the linear relationship (n′; Table 3) reflects the number of counterions displaced in the reaction step that is ionic strength sensitive. A priori, we might expect that, if the amino acid substitutions and ionic strength affected the same long-range electrostatic interaction(s), then the kinetic differences between the mutants would diminish as ionic strength increased and the rates would eventually converge when all long-range electrostatic interactions are shielded by counterions. This would be reflected in different values of n′ such that the variants with the fastest rates (at low ionic strength) would have the largest ionic strength dependences (highest n′). However, the slopes for the various CheY variants were only modestly sensitive to the amino acid substitution, but instead, largely dependent on the phosphodonor (Table 3). n′ values for all four CheY variants for reaction with PAM were close to unity (ranging from 0.97– 1.3), which matches the −1 net charge of the PAM zwitterion (Figure 1B). In contrast, the n′ values for reaction with AcP (net charge is −2, Figure 1B) ranged from 1.4–1.8 and, for each CheY variant, n′ for AcP was between 1.4 and 1.7-fold greater than for PAM. The value of n′ reflects the extent of the Coulombic interactions in the E·S complex and approximates the number of counterions released from charged residues and substrate ions upon complex formation, which is reflective of the number of charges involved in salt bridges between enzyme and substrate in the E·S complex. The same qualitative result- that the CheY amino acid substitutions had small effects on the ionic strength sensitivity and that reactions with AcP had a 1.4–1.7 -fold higher ionic strength sensitivity than PAM- was achieved with an alternative method of plotting the data whereby log (kphos/KS) was plotted versus a term that included the square root of the ionic strength38.
Figure 5.
Ionic strength dependence of autophosphorylation rates for a subset of CheY variants. Closed symbols are for reaction with AcP and open symbols are for reaction with PAM. CheY variants are CheY NE (circles), CheY NY (diamonds), CheY RE (inverted triangles) and CheY NR (squares), where the two letters designate the amino acid at position D+2 and T+2 (CheY positions 59 and 89). Error bars represent standard deviation from duplicate experiments.
Table 3.
n ′ values derived from plots of log (ionic strength) versus log (kphos/KS)
| Residue at position
|
n′ a
|
||
|---|---|---|---|
| D+2 | T+2 | AcP | PAM |
| N | Eb | 1.4 ± 0.04 | 1.0 ± 0.05 |
| R | E | 1.8 ± 0.02 | 1.3 ± 0.06 |
| N | R | 1.8 ± 0.01 | 1.1 ± 0.06 |
| N | Y | 1.6 ± 0.03 | 0.97 ± 0.04 |
n′ is the absolute value of the slope from Figure 5 and represents the number of counterions displaced on the protein or substrate in the reaction step(s) represented by kphos/KS.
Wild type CheY
The relative insensitivity of the n′ values to amino acid substitution indicates that ionic strength and substitutions at D+2 and T+2 affect two different electrostatic phenomena. The correlation of the n′ values with the −1 and −2 charges of the phosphodonors supports the notion that the ionic strength dependence reflects the binding of counterions to the phosphodonor anion to disrupt ion pair formation between the phosphodonor and a species that is common to all the mutant reactions, such as the CheY active site (see Discussion). Furthermore, because the amino acid substitutions had only small effects on the ionic strength sensitivities, the electrostatic component of the kinetic effects exerted by the substituted side chains likely do not involve long-range electrostatic interactions (i.e. the coming together of separate charges to form an ion pair). Instead, the side chains likely exert their effects via more localized electrostatic interactions that would not be susceptible to the effects of bulk solvent.
DISCUSSION
Although response regulator autophosphorylation was discovered more than twenty years ago4 and has since been established to have physiological relevance for multiple response regulators5, 6, the reaction kinetics have been rigorously characterized only for CheY15– 18. Prior to this study, the ranges of reaction rates and phosphodonor discrimination possible with the response regulator scaffold were unknown. As summarized in this Discussion, there are now specific proposed mechanisms for modulation of the catalysis of CheY phosphorylation that can be tested for generality in other response regulators, as well as in sensor kinase/response regulator pairs.
Specifically, this study provided substantial insight into the precise mechanisms by which electrostatic and steric interactions influence the kinetics of CheY autophosphorylation. We demonstrated that two types of electrostatic interactions affect reaction kinetics: long range (getting the phosphodonor into the active site) and short range (interactions between the active site and the phosphodonor once the enzyme/substrate complex has formed). Furthermore, the “direction” of long- and short-range electrostatic interactions is the same - both are enhanced by positive charge in the CheY active site. With regard to steric effects, the generally faster reaction rates observed with PAM compared to AcP were not attributable to the smaller size of PAM. Instead, the data provided strong evidence that aromatic/bulky hydrophobic groups in the CheY active site can interact directly with the imidazole portion of MPI (or the nitrogen atom of PAM) to increase binding energy and thus enhance phosphorylation rate.
Phosphoramidates and acyl phosphates have different kinetic determinants
Amino acid substitutions at positions D+2 and T+2 modulated CheY autophosphorylation rates by two to three orders of magnitude for each of the three phosphodonors tested (Tables 1,2). Multiple lines of evidence support the notion that CheY and other receiver domains exist as an equilibrium between multiple conformational states39, 40 and that the activated conformation(s) autophosphorylate at an enhanced rate17, 23. Thus, a rate change as a result of substitution might reflect the effect of the substitution on the equilibrium between active and inactive conformational states17, 20. However, simply changing the fraction of CheY in the activated conformation would manifest in rates changing in concert for all phosphodonors. In contrast, a central observation here was that the effect of substitutions at D+2 and T+2 on CheY autophosphorylation kinetics was phosphodonor dependent (Tables 1, 2; Figure 2). With the inclusion of MPI, kinetic preferences for different phosphodonors for a single CheY variant were as high as 1,700-fold. Thus, conformational state does not appear to be a dominant source of rate modulation for this mutant set. Furthermore, we probed the effect of conformational change on two of the CheY variants with enhanced rates of autophosphorylation. Both variants (CheY NR and NL) exhibited additional rate enhancements, similar in magnitude to that shown by wild type CheY, when pushed into an activated conformation by binding to the FliM1–16 peptide (Table S2).
If conformation is not playing a dominant role in autophosphorylation rate modulation, then what is? Correlations between measured rate constants and chemical properties of the D+2/T+2 substitutions allowed us to propose several kinetic determinants to account for the observed rate differences. Specifically, increased positive charge and increased nonpolar surface area of the D+2/T+2 side chains both correlated with enhanced autophosphorylation rates (Figure 4). The analysis further revealed that the different phosphodonor classes were affected differentially by the kinetic determinants. Increased positive charge enhanced autophosphorylation rate for both AcP and PAM but did so to a greater degree for AcP. Moreover, because reaction with AcP displayed a stronger dependence on ionic strength than with PAM (Figure 5), the preference for PAM over AcP for any mutant would be dependent on ionic strength. Increased nonpolar surface area strongly correlated with faster rate for reaction with PAM but the correlation for reaction with AcP was weak. Reaction with MPI showed yet a larger impact of nonpolar surface area than PAM. Plot of log kphos/KS versus nonpolar surface area for the small set of mutants for which we measured reaction rates with MPI gave a linear regression line with a slope more than two-fold higher than with PAM (data not shown). The differential impacts of kinetic determinants on reaction with different phosphodonors underlies the variation in phosphodonor preference.
Electrostatic interactions modulate CheY autophosphorylation at multiple levels
The correlation between charge of the D+2/T+2 side chains and reaction rate implicated a role for electrostatic interactions in the differing autophosphorylation kinetics of the CheY variants. However, the similar ionic strength sensitivities of the CheY variants suggested at least two electrostatic phenomena: one that was ionic strength-sensitive and one that was ionic strength-insensitive but sensitive to the charge at D+2/ T+2. Furthermore, quantitative analysis (Figure 5, Table 3) revealed that the magnitude of the ionic strength sensitivity (n′) was greater for AcP (net charge of −2) than for PAM (net charge of −1). n′ values reflect the number of counterions released from charged species in order to form a noncovalent complex.
Ionic strength inhibits long-range electrostatic interactions between a positively charged active site and negatively charged substrate
Taken together, the observations summarized in the paragraph above provide compelling evidence that the ionic strength sensitive step involves direct interaction of the phosphodonor anion with a positively charged species that is common to the reactions for all the CheY variants. Calculation of the electrostatic surface potential of the CheY·Mg2+ active site reveals a positively charged surface centered on the Mg2+ and conserved lysine residue, which more than neutralizes the three conserved aspartyl residues (Adaptive Poisson-Boltzmann Solver software41; PDBid 2CHE42 or 1FQW11). Structures of phosphatases within the haloacid dehalogenase superfamily (which have virtually identical active sites and catalyze the same chemistry as receiver domains) complexed with substrate analogues show two salt bridges between phosphoryl group oxygen atoms and Mg2+ and the conserved Lys12, 43. Therefore, a likely candidate for the ionic strength sensitive step in CheY autophosphorylation is the initial binding of phosphodonor to CheY, which would involve formation of salt bridges between phosphoryl group oxygen anions and the Mg2+ and lysyl cations. This suggests that the inhibitory effect of increasing ionic strength on kphos/KS would reflect a higher KS (weaker binding) rather than a decrease in kphos and is consistent with the notion that ionic strength generally affects long-range electrostatic interactions. We also considered and rejected an alternate possibility that noncovalent complexes formed between Mg2+ and phosphodonor function as the reactive species in autophosphorylation and that ionic strength disrupts complex formation (see Supplemental Information).
The magnitudes of the n′ values measured here deserve further comment. The agreement between n′ values for reactions with PAM (~1) and the net charge of PAM (−1) was striking as n′ should approximate the number of counterions released from charged residues or substrate ions that interact. However, PAM is a zwitterion (Figure 1B) and the observation that PAM behaves more like a −1 charge than a −2 suggests that the positively charged nitrogen atom inhibits the ability of the K+ counterions to bind to phosphoryl oxygens on PAM. Although n′ values for reaction with AcP were 1.4 – 1.8-fold higher than for PAM, they did not reach the theoretical value of ~2 expected for release of two counterions from AcP. This is similar to studies with RNase A in which n′ increased linearly with substrate charge but with a slope of only ~0.2 (n′/charge). The ratio of n′/charge of <1 was attributed to uneven counterion density on the substrate30, which would be expected for AcP.
D+2/T+2 residues affect localized electrostatic interactions
The electrostatic interactions implicated by the correlation between positive charge of D+2/ T+2 and enhanced rate were virtually insensitive to ionic strength. Thus, these interactions are likely to be more localized and not involve two separated and solvated charged species coming together to form an ion pair. The locations of the D+2 and T+2 side chains in the active site further implicate that these interactions likely involve energetics at the leaving group region of the autophosphorylation reaction. The D+2 and T+2 side chains are positioned near the leaving group atom bonded to the phosphorus, which is different for the phosphodonor classes. The leaving group for PAM is NH3 (neutral) and the nitrogen atom carries a partial positive charge in the transition state. In contrast, the leaving group for AcP is acetate (−1 charge) and the oxygen atom has a partial negative charge in the transition state. Thus, it is reasonable that positive charge at D+2/T+2 may help stabilize the transition state of AcP more so than PAM via electrostatic attractive forces between positively charged side chain(s) and the leaving group atom. This is consistent with the observation that the slopes of the linear correlation between net positive charge and rate was higher and the correlations were statistically stronger for AcP than for PAM (Figure 4AB). Similar roles of stabilizing negative charges in the transition state have been proposed for arginine residues that are prevalent in active sites of phosphotransfer enzymes44. Thus, electrostatic interactions due to D+2/T+2 substitutions likely affect the kphos component of kphos/KS.
Non-polar surface area and steric complementarity
The overall preference of PAM over AcP for this CheY mutant set is consistent with accounts in the literature of receiver domains that react readily with PAM but do not react with AcP4, 21. PAM and AcP have similar kinetics of hydrolysis in the absence of catalyst13, 14 so the preference for PAM is not due to an innately higher reactivity of PAM. In the mutant set studied here, reaction with PAM improved as the nonpolar surface enlarged but AcP did not appear to benefit similarly. This was particularly evident in three CheY variants with tyrosine residues at T+2 (CheY NY, CheY KY, and CheY *KY), which displayed greatly enhanced rates with PAM that were not achievable with AcP. CheY NY and CheY *KY underwent dramatic further rate enhancements with MPI (Table 2) and docking showed a strong likelihood of steric complementarity between MPI and both the D+2 and T+2 side chains (Figure 3). Why didn’t AcP similarly benefit from the added nonpolar surface area? Interestingly, docking of AcP into the active site of CheY *KY (as shown in Figure 3 for MPI) showed that the planar -COCH3 group linked to the phosphoryl group in AcP clashed with multiple atoms of the D+2 and T+2 side chain in all possible rotamers. In MPI, the planar imidazole is one atom closer to the phosphoryl group and so fits more snuggly in the active site. These trends likely reflect a tradeoff between enhanced surface for potential interaction with the limitation that the increased bulk does not introduce steric clash.
Finally, in addition to steric complementarity, the observed correlation between nonpolar surface area and autophosphorylation rate also likely reflects energetic contributions of providing a more hydrophobic environment for catalysis of the phosphotransfer reaction. A more hydrophobic active site pocket would result in strengthening the multiple types of electrostatic interactions that drive the reaction (described above) due to a decrease in the local dielectric constant45.
Highly accelerated reactions between CheY variants and MPI may reflect structural mimicry of phosphotransfer from sensor kinases
In two-component systems, the phosphohistidyl containing domain that transfers directly to the receiver can be either a ‘dimerization and histidine phosphotransfer’ (DHp) domain (present in canonical sensor kinases) or a ‘histidine-containing phosphotransfer’ (Hpt) domain (commonly found in phosphorelays and chemotaxis systems), both of which are four-helix bundles. Phosphotransfer from both DHp46, 47 and Hpt19, 48 domains to response regulators occurs rapidly with rate constants of ~106 M−1s−1 or greater. Whereas previously determined autophosphorylation rates for wild type CheY were ~105-times slower than transfer from cognate proteins, the rate constant for CheY *KY autophosphorylation with MPI (4.3 × 104 M−1s−1) measured here approaches rates seen with the protein domains. Could interactions between the imidazole side chain and D+2/T+2 side chain atoms predicted by docking also occur in complexes between phosphorylated histidine kinases and response regulators? In the docked structure (Figure 3), the monophosphoimidazole carbon atom corresponding to histidyl Cγ is facing solvent, allowing plenty of room for the rest of the histidyl side chain. Assessment of about 14,000 response regulator sequences showed that about 35% have an aromatic residue (F/Y/W/H) at position T+222 so these interactions could be common. Indeed, in the co-crystal structure of the Hpt/response regulator pair YPD1/SLN1-R1 (PDBid 2R25)49, there is a phenylalanine at the SLN1-R1 T+2 position that interacts with the conserved histidyl imidazole in YPD1, providing evidence that very similar interactions may help mediate binding between the Hpt domain and the response regulator. In the HK853/RR468 complex between a kinase DHp domain and a cognate receiver, the imidazole group of the conserved His forms one end of a sandwich of van der Waals interactions with receiver domain residues D+2 (Met) and T+2 (Lys)50. In the low resolution complex between the DHp domain of the TrrA kinase and the TrrA response regulator (PDBid 3AOR)51, TrrA T+2 Y82 interacts with ThkA H547 via parallel π-π stacking interactions, but there is uncertainty in this exact alignment due to weak electron density in this region. Thus there may be multiple modes of interaction between D+2/T+2 side chains with the histidyl group, including direct interaction between an aromatic response regulator T+2 side chain and the conserved histidyl side chain from the partner protein. It is likely that these interactions not only help stabilize the kinase/receiver complex but could also strengthen the electrostatic interactions within the transition state by providing a more hydrophobic environment.
The roles of response regulator nonconserved active site residues D+2 and T+2
The influence of residues D+2 and T+2 on CheY autophosphorylation kinetics established here represents yet another example of the impact of these two positions on receiver domain phosphorylation reactions. Located at the ‘gateway’ of the active site, residues D+2 and T+2 also play roles in sensor kinase recognition and binding50, 52, 53, receiver domain autodephosphorylation kinetics25, 26, and phosphatase binding and catalysis35, 54, 55. Bioinformatics analysis of 1,555 receiver domain sequences52 reveals strong evolutionary covariation between residues at D+2 and T+2 (Michael Laub, Pers. Comm.), consistent with a functional role for these positions in differentiating response regulator receiver domain functions.
Although amino acid substitutions at D+2 and T+2 had large effects on the kinetics and specificity of CheY autophosphorylation, other nonconserved active site positions likely also modulate response regulator autophosphorylation. For example, residue T+1 is hypothesized to control access to the phosphorylation site24. Identification of all major determinants of response regulator phosphorylation chemistry and characterization of the interactions between such factors will require additional investigation.
Supplementary Material
Acknowledgments
Funding
Research reported in this publication was supported by the National Institute of General Medical Sciences of the National Institutes of Health under award number R01GM050860. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
We thank Dr. Ashalla Freeman, Dr. Yael Pazy-Benhar, Rachel Creager-Allen, and Stephani Page for helpful discussions and Dr. Mike Laub (MIT) for sharing covariation analysis results. We also thank Dr. Rick Stewart (University of Maryland) and Dr. Gideon Schreiber (Weizmann Institute, Israel) for thoughtful discussion regarding analysis of the ionic strength effects.
ABBREVIATIONS
- AcP
acetyl phosphate
- PAM
phosphoramidate
- MPI
monophosphoimidazole
- DD
the two conserved acid residues on the receiver domain β1α1 loop that chelate the Mg2+ (D12 and D13 for E. coli CheY)
- D
the conserved phosphorylated aspartate on β3α3 (D57 for CheY)
- T
the conserved threonine/serine on β4α4 (T87 for CheY)
- K
the conserved lysine on β5α5 (K109 for CheY)
- DD+1, D+2, and T+2
the nonconserved residues positioned one or two residues carboxyl terminal to DD, D, or T respectively (F14, N59, and E89 for CheY)
- CheY XY
CheY double mutant where X and Y are the identities of the residues at positions D+2 (residue 59 in CheY) and T+2 (residue 89 in CheY), respectively
- CheY *XY
CheY triple mutant where X and Y are the identities of the residues at positions D+2 (59) and T+2 (89), respectively, with an additional substitution at DD+1
Footnotes
Supplemental materials for this manuscript include Table S1 (Mg2+ binding constants for select CheY variants), Table S2 (rate constants in the presence of FliM peptide) an addendum to Experimental Procedures (describing corrections made to autophosphorylation rates at different ionic strengths due to differences in Mg2+ affinity and additional detail describing the method of quantitation of ionic strength effects), and an addendum to the Discussion describing consideration of a possible role for soluble complexes between acetyl phosphate and Mg2+ in autophosphorylation rate modulation. Supplemental materials may be accessed free of charge at http://pubs.acs.org.
References
- 1.Bourret RB, Silversmith RE. Two-component signal transduction. Curr Opin Microbiol. 2010;13:113–115. doi: 10.1016/j.mib.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Capra EJ, Laub MT. Evolution of two-component signal transduction systems. Annu Rev Microbiol. 2012;66:325–347. doi: 10.1146/annurev-micro-092611-150039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Galperin MY. Diversity of structure and function of response regulator output domains. Curr Opin Microbiol. 2010;13:150–159. doi: 10.1016/j.mib.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lukat GS, McCleary WR, Stock AM, Stock JB. Phosphorylation of bacterial response regulator proteins by low molecular weight phospho-donors. Proc Natl Acad Sci U S A. 1992;89:718–722. doi: 10.1073/pnas.89.2.718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wolfe AJ. The acetate switch. Microbiol Mol Biol Rev. 2005;69:12–50. doi: 10.1128/MMBR.69.1.12-50.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wolfe AJ. Physiologically relevant small phosphodonors link metabolism to signal transduction. Curr Opin Microbiol. 2010;13:204–209. doi: 10.1016/j.mib.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bourret RB, Thomas SA, Page SC, Creager-Allen RL, Moore AM, Silversmith RE. Measurement of response regulator autodephosphorylation rates spanning six orders of magnitude. Methods Enzymol. 2010;471:89–114. doi: 10.1016/S0076-6879(10)71006-5. [DOI] [PubMed] [Google Scholar]
- 8.Buckler DR, Stock AM. Synthesis of [32P]phosphoramidate for use as a low molecular weight phosphodonor reagent. Anal Biochem. 2000;283:222–227. doi: 10.1006/abio.2000.4639. [DOI] [PubMed] [Google Scholar]
- 9.Gao R, Tao Y, Stock AM. System-level mapping of Escherichia coli response regulator dimerization with FRET hybrids. Mol Microbiol. 2008;69:1358–1372. doi: 10.1111/j.1365-2958.2008.06355.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bourret RB. Receiver domain structure and function in response regulator proteins. Curr Opin Microbiol. 2010;13:142–149. doi: 10.1016/j.mib.2010.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee SY, Cho HS, Pelton JG, Yan D, Berry EA, Wemmer DE. Crystal structure of activated CheY. Comparison with other activated receiver domains. J Biol Chem. 2001;276:16425–16431. doi: 10.1074/jbc.M101002200. [DOI] [PubMed] [Google Scholar]
- 12.Wang W, Cho HS, Kim R, Jancarik J, Yokota H, Nguyen HH, Grigoriev IV, Wemmer DE, Kim SH. Structural characterization of the reaction pathway in phosphoserine phosphatase: crystallographic “snapshots” of intermediate states. J Mol Biol. 2002;319:421–431. doi: 10.1016/S0022-2836(02)00324-8. [DOI] [PubMed] [Google Scholar]
- 13.Chanley JD, Feageson E. A study of hydrolysis of phosphoramides. II Solvolysis of phosphoramidic acid and comparison with phosphate esters. J Am Chem Soc. 1963;85:1181–1190. [Google Scholar]
- 14.Koshland DE. Efffect of catalysts on the hydrolysis of acetyl phosphate. Nucleophilic displacement mechanisms in enzymic reactions. J Am Chem Soc. 1952;74:2286–2296. [Google Scholar]
- 15.Da Re SS, Deville-Bonne D, Tolstykh T, Veron M, Stock JB. Kinetics of CheY phosphorylation by small molecule phosphodonors. FEBS Lett. 1999;457:323–326. doi: 10.1016/s0014-5793(99)01057-1. [DOI] [PubMed] [Google Scholar]
- 16.Mayover TL, Halkides CJ, Stewart RC. Kinetic characterization of CheY phosphorylation reactions: comparison of P-CheA and small-molecule phosphodonors. Biochemistry. 1999;38:2259–2271. doi: 10.1021/bi981707p. [DOI] [PubMed] [Google Scholar]
- 17.Schuster M, Silversmith RE, Bourret RB. Conformational coupling in the chemotaxis response regulator CheY. Proc Natl Acad Sci U S A. 2001;98:6003–6008. doi: 10.1073/pnas.101571298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Silversmith RE, Appleby JL, Bourret RB. Catalytic mechanism of phosphorylation and dephosphorylation of CheY: kinetic characterization of imidazole phosphates as phosphodonors and the role of acid catalysis. Biochemistry. 1997;36:14965–14974. doi: 10.1021/bi9715573. [DOI] [PubMed] [Google Scholar]
- 19.Stewart RC, Jahreis K, Parkinson JS. Rapid phosphotransfer to CheY from a CheA protein lacking the CheY-binding domain. Biochemistry. 2000;39:13157–13165. doi: 10.1021/bi001100k. [DOI] [PubMed] [Google Scholar]
- 20.Barbieri CM, Mack TR, Robinson VL, Miller MT, Stock AM. Regulation of response regulator autophosphorylation through interdomain contacts. J Biol Chem. 2010;285:32325–32335. doi: 10.1074/jbc.M110.157164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zapf JW, Hoch JA, Whiteley JM. A phosphotransferase activity of the Bacillus subtilis sporulation protein Spo0F that employs phosphoramidate substrates. Biochemistry. 1996;35:2926–2933. doi: 10.1021/bi9519361. [DOI] [PubMed] [Google Scholar]
- 22.Ulrich LE, Zhulin IB. The MiST2 database: a comprehensive genomics resource on microbial signal transduction. Nuc Acids Res. 2010;38:D401–407. doi: 10.1093/nar/gkp940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ames SK, Frankema N, Kenney LJ. C-terminal DNA binding stimulates N-terminal phosphorylation of the outer membrane protein regulator OmpR from Escherichia coli. Proc Natl Acad Sci U S A. 1999;96:11792–11797. doi: 10.1073/pnas.96.21.11792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Volz K. Structural conservation in the CheY superfamily. Biochemistry. 1993;32:11741–11753. doi: 10.1021/bi00095a001. [DOI] [PubMed] [Google Scholar]
- 25.Pazy Y, Wollish AC, Thomas SA, Miller PJ, Collins EJ, Bourret RB, Silversmith RE. Matching biochemical reaction kinetics to the timescales of life: structural determinants that influence the autodephosphorylation rate of response regulator proteins. J Mol Biol. 2009;392:1205–1220. doi: 10.1016/j.jmb.2009.07.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thomas SA, Brewster JA, Bourret RB. Two variable active site residues modulate response regulator phosphoryl group stability. Mol Microbiol. 2008;69:453–465. doi: 10.1111/j.1365-2958.2008.06296.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boesch KC, Silversmith RE, Bourret RB. Isolation and characterization of nonchemotactic CheZ mutants of Escherichia coli. J Bacteriol. 2000;182:3544–3552. doi: 10.1128/jb.182.12.3544-3552.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sheridan RC, McCullough JF, Wakefield ZT, Allcock HR, Walsh EJ. Phospharamidic acid and its salts. Inorg Synth. 1971;13:23–26. [Google Scholar]
- 29.Rathlev T, Rosenberg T. Non-enzymic formation and rupture of phosphorus to nitrogen linkages in phosphoramido derivatives. Arch Biochem Biophys. 1956;65:319–339. doi: 10.1016/0003-9861(56)90197-7. [DOI] [PubMed] [Google Scholar]
- 30.Park C, Raines RT. Quantitative analysis of the effect of salt concentration on enzymatic catalysis. J Am Chem Soc. 2001;123:11472–11479. doi: 10.1021/ja0164834. [DOI] [PubMed] [Google Scholar]
- 31.Hicks SN, Smiley RD, Hamilton JB, Howell EE. Role of ionic interactions in ligand binding and catalysis of R67 dihydrofolate reductase. Biochemistry. 2003;42:10569–10578. doi: 10.1021/bi034643d. [DOI] [PubMed] [Google Scholar]
- 32.Plantinga MJ, Korennykh AV, Piccirilli JA, Correll CC. Electrostatic interactions guide the active site face of a structure-specific ribonuclease to its RNA substrate. Biochemistry. 2008;47:8912–8908. doi: 10.1021/bi800592g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Word JM, Lovell SC, Richardson JS, Richardson DC. Asparagine and glutamine: using hydrogen atom contacts in the choice of side-chain amide orientation. J Mol Biol. 1999;285:1735–1747. doi: 10.1006/jmbi.1998.2401. [DOI] [PubMed] [Google Scholar]
- 34.Chen VB, Arendall WB, 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Silversmith RE, Guanga GP, Betts L, Chu C, Zhao R, Bourret RB. CheZ-mediated dephosphorylation of the Escherichia coli chemotaxis response regulator CheY: role for CheY glutamate 89. J Bacteriol. 2003;185:1495–1502. doi: 10.1128/JB.185.5.1495-1502.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meyer EA, Castellano RK, Diederich F. Interactions with aromatic rings in chemical and biological recognition. Angew Chem Int Ed. 2003;42:1210–1250. doi: 10.1002/anie.200390319. [DOI] [PubMed] [Google Scholar]
- 37.Sines JJ, Allison SA, McCammon JA. Point charge distributions and electrostatic steering in enzyme/substrate encounter: Brownian dynamics of modified copper/zinc superoxide dismutases. Biochemistry. 1990;29:9403–9412. doi: 10.1021/bi00492a014. [DOI] [PubMed] [Google Scholar]
- 38.Schreiber G, Fersht AR. Rapid, electrostatically assisted association of proteins. Nat Struct Biol. 1996;3:427–431. doi: 10.1038/nsb0596-427. [DOI] [PubMed] [Google Scholar]
- 39.McDonald LR, Boyer JA, Lee AL. Segmental motions, not a two-state concerted switch, underlie allostery in CheY. Structure. 2012;20:1363–1373. doi: 10.1016/j.str.2012.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stock AM, Guhaniyogi J. A new perspective on response regulator activation. J Bacteriol. 2006;188:7328–7330. doi: 10.1128/JB.01268-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci U S A. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stock AM, Martinez-Hackert E, Rasmussen BF, West AH, Stock JB, Ringe D, Petsko GA. Structure of the Mg(2+)-bound form of CheY and mechanism of phosphoryl transfer in bacterial chemotaxis. Biochemistry. 1993;32:13375–13380. doi: 10.1021/bi00212a001. [DOI] [PubMed] [Google Scholar]
- 43.Roberts A, Lee SY, McCullagh E, Silversmith RE, Wemmer DE. YbiV from Escherichia coli K12 is a HAD phosphatase. Proteins. 2005;58:790–801. doi: 10.1002/prot.20267. [DOI] [PubMed] [Google Scholar]
- 44.Lassila JK, Zalatan JG, Herschlag D. Biological phosphoryl-transfer reactions: understanding mechanism and catalysis. Annu Rev Biochem. 2011;80:669–702. doi: 10.1146/annurev-biochem-060409-092741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Janin J, Chothia C. Stability and specificity of protein-protein interactions: the case of the trypsin-trypsin inhibitor complexes. J Mol Biol. 1976;100:197–211. doi: 10.1016/s0022-2836(76)80148-9. [DOI] [PubMed] [Google Scholar]
- 46.Groban ES, Clarke EJ, Salis HM, Miller SM, Voigt CA. Kinetic buffering of cross talk between bacterial two-component sensors. J Mol Biol. 2009;390:380–393. doi: 10.1016/j.jmb.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Skerker JM, Prasol MS, Perchuk BS, Biondi EG, Laub MT. Two-component signal transduction pathways regulating growth and cell cycle progression in a bacterium: a system-level analysis. PLoS Biol. 2005;3:e334. doi: 10.1371/journal.pbio.0030334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Janiak-Spens F, Cook PF, West AH. Kinetic analysis of YPD1-dependent phosphotransfer reactions in the yeast osmoregulatory phosphorelay system. Biochemistry. 2005;44:377–386. doi: 10.1021/bi048433s. [DOI] [PubMed] [Google Scholar]
- 49.Zhao X, Copeland DM, Soares AS, West AH. Crystal structure of a complex between the phosphorelay protein YPD1 and the response regulator domain of SLN1 bound to a phosphoryl analog. J Mol Biol. 2008;375:1141–1151. doi: 10.1016/j.jmb.2007.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Casino P, Rubio V, Marina A. Structural insight into partner specificity and phosphoryl transfer in two-component signal transduction. Cell. 2009;139:325–336. doi: 10.1016/j.cell.2009.08.032. [DOI] [PubMed] [Google Scholar]
- 51.Yamada S, Akiyama S, Sugimoto H, Kumita H, Ito K, Fujisawa T, Nakamura H, Shiro Y. The signaling pathway in histidine kinase and the response regulator complex revealed by X-ray crystallography and solution scattering. J Mol Biol. 2006;362:123–139. doi: 10.1016/j.jmb.2006.07.012. [DOI] [PubMed] [Google Scholar]
- 52.Skerker JM, Perchuk BS, Siryaporn A, Lubin EA, Ashenberg O, Goulian M, Laub MT. Rewiring the specificity of two-component signal transduction systems. Cell. 2008;133:1043–1054. doi: 10.1016/j.cell.2008.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Szurmant H, Hoch JA. Interaction fidelity in two-component signaling. Curr Opin Microbiol. 2010;13:190–197. doi: 10.1016/j.mib.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pazy Y, Motaleb MA, Guarnieri MT, Charon NW, Zhao R, Silversmith RE. Identical phosphatase mechanisms achieved through distinct modes of binding phosphoprotein substrate. Proc Natl Acad Sci U S A. 2010;107:1924–1929. doi: 10.1073/pnas.0911185107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhao R, Collins EJ, Bourret RB, Silversmith RE. Structure and catalytic mechanism of the E. coli chemotaxis phosphatase CheZ. Nat Struct Biol. 2002;9:570–575. doi: 10.1038/nsb816. [DOI] [PubMed] [Google Scholar]
- 56.Silversmith RE, Smith JG, Guanga GP, Les JT, Bourret RB. Alteration of a nonconserved active site residue in the chemotaxis response regulator CheY affects phosphorylation and interaction with CheZ. J Biol Chem. 2001;276:18478–18484. doi: 10.1074/jbc.M011418200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.