Abstract
The ubiquitin-proteasome system (UPS) is the major intracellular degradation system, and its proper function is critical to the health and function of cardiac cells. Alterations in cardiac proteasomes have been linked to several pathological phenotypes, including cardiomyopathies, ischemia-reperfusion injury, heart failure, and hypertrophy. Defects in proteasome-dependent cellular protein homeostasis can be causal for the initiation and progression of certain cardiovascular diseases. Emerging evidence suggests that the UPS can specifically target proteins that govern pathological signaling pathways for degradation, thus altering downstream effectors and disease outcomes. Alterations in UPS-substrate interactions in disease occur, in part, due to direct modifications of 19S, 11S or 20S proteasome subunits. Post-translational modifications (PTMs) are one facet of this proteasomal regulation, with over 400 known phosphorylation sites, over 500 ubiquitination sites and 83 internal lysine acetylation sites, as well as multiple sites for caspase cleavage, glycosylation (such as O-GlcNAc modification), methylation, nitrosylation, oxidation, and sumoylation. Changes in cardiac proteasome PTMs, which occur in ischemia and cardiomyopathies, are associated with changes in proteasome activity and proteasome assembly; however several features of this regulation remain to be explored. In this review, we focus on how some of the less common PTMs affect proteasome function and alter cellular protein homeostasis.
Keywords: ubiquitin-proteasome system, acetylation, methylation, ubiquitination, sumoylation, cardiovascular disease
1.1 Introduction
The ubiquitin-proteasome system (UPS) is the main protein degradation system in the heart, degrading up to 90% of the intracellular proteins in some tissues [1]. As much as 30% of newly synthesized proteins are degraded by the proteasome shortly after their synthesis [2]. The UPS is involved in regulating most cellular events in eukaryotes, including cell differentiation, DNA replication and repair, mitosis, transcriptional regulation, and receptor internalization; all of which are important in cardiac biology. The UPS allows cells to readily alter protein expression patterns in response to changing physiological conditions. Moreover, the maintenance of healthy protein turnover by the UPS is critically important in preventing disease through the degradation of oxidized, mutant, denatured, and misfolded proteins [3]. The proteasome is an abundant complex; in liver and kidney cells proteasomes account for approximately 1% of the total cellular protein pool [4]. Cardiac cells contain fewer proteasomes than liver cells, and proteasomes are widely distributed in cytosolic, nuclear, endoplasmic reticular and cytoskeletal compartments [5, 6]. Several defects in protein degradation have been linked to cardiovascular biology and disease [7], including atherosclerosis, familial and idiopathic cardiomyopathies, myocardial ischemia, hypertrophy, reperfusion, and heart failure [8-11]. The mammalian 20S proteasome was first discovered and isolated from human erythrocytes, in which it was termed “cylindrin,” describing the structure of the protein complex [12, 13]. The rapid growth in our understanding and appreciation of the proteasome as a key regulator of virtually all cellular processes has led to the proteasome surfacing as a therapeutic target for combatting many diseases, including cardiovascular diseases (CVDs). However, the anatomical complexity of the proteasome, including heterogeneous subunit assembly, alternate splicing of subunits, and post-translational modifications (PTMs) makes the proteasome a challenging target. While we are beginning to understand how different combinations of subunits affect proteasome activity, the role of alternatively spliced subunits and PTMs are not as well understood. Understanding the role of proteasome PTMs in tuning cellular function is critical, as the relatively long half-lives of mammalian proteasomes (5-8 days) [14, 15] necessitate PTMs as a rapid, immediate means with which to alter proteasome function.
1.2 The Proteasome
The proteasome is comprised of more than 45 subunits. Due to the large number of constituent subunits and independent identifications of proteasome subunits and complexes, the nomenclature presented in the literature has been largely inconsistent. In this review, proteasome subunits are referred to in accordance with the most commonly employed eukaryotic nomenclature published by Baumeister et al., 1998 [16]. The 26S proteasome is composed of the 20S core particle tethered in an ATP-dependent manner to either one or two 19S regulatory complexes [17]. The 20S core particle consists of 28 subunits, positioned as two inner stacked rings of seven unique β subunits, and two outer rings of seven unique α subunits. The 20S core contains three distinct proteolytic activities: caspase-like, trypsin-like and chymotrypsin-like, which are catalyzed by β1, β2 and β5 subunits, respectively [17]. In certain conditions, such as diabetic cardiomyopathy, the β1, β2 and β5 subunits can be replaced with inducible subunits - β1i, β2i and β5i - which possess unique catalytic activities relative to canonical forms. Proteasomes containing β1i, β2i and β5i are referred to as immunoproteasomes. The 19S regulatory complex is larger than the 20S core, consisting of at least 17 subunits. The “base” contains six ATPases (Rpts) and three non-ATPase subunits (Rpn1, 2 and 10), together with the “lid”, which contains eight non-ATPase subunits [18]. The base of the 19S interacts with the outer α ring of the 20S core. The proteasome activator PA28γ (also known as PSME3, 11Sγ, or REGγ) belongs to the 11S family of proteasome activators, which also includes PA28α and PA28β. These proteasome activators bind and activate the 20S proteasome. PA28γ has been shown to promote degradation of important regulatory proteins such as cyclin-dependent kinase inhibitors p21, p16, and p19 and steroid receptor coactivator-3 in an ATP- and ubiquitin-independent manner [19-21]. Several types of proteasomes are known to exist, including 20S, 26S, 11S, intermediate and hybrid-type proteasomes [22]. Electron microscopy has confirmed at least six different proteasome complexes (20S, 19S-20S, 19S-20S-19S, 20S-PA28, PA28-20S-PA28, 19S-20S-PA28) [22]. Two 19S proteasome subunits, Rpn10 and Rpn13, interact with polyubiquitinated substrates. Other accessory proteins such as Rad23, interacts with ubiquitinated proteins and acts as a shuttle for delivering ubiquitin-conjugated proteins to the proteasome.
1.3 Post-Translational Modifications of the Proteasome
Post-translational modifications play critical roles in regulating protein structure and function. The most common PTMs include phosphorylation and ubiquitination. More recently, ubiquitin-like protein (Ubl) modifications (e.g., SUMO, Nedd8, and ISG15) have emerged as vital regulatory mechanisms in directing intracellular processes. These small Ubls covalently attach to their target proteins in a manner similar to ubiquitination. Proteasome subunits are subjected to several PTMs, including N-terminal acetylation, lysine acetylation, methylation, phosphorylation, N-myristoylation, O-linked glycosylation, S-glutathionylation, ubiquitination, proteolysis by caspases, and processing of N-terminal propeptides, the latter being the mechanism by which 20S core particle (CP) catalytic activities are activated. At least 14 different types of PTMs have been identified on proteasome subunits (Table 1). Proteomic studies employing mass spectrometry (MS) have identified the majority of proteasome post-translationally modified sites, including 417 phosphorylation (160 serine, 94 threonine, and 163 tyrosine), over 500 ubiquination, 83 lysine acetylation, 7 O-GlcNAc, 4 sumoylation, 2 methylation, 2 di-methylation, 1 glycosylation, 1 nitrosylation, and 1 myristoylation (Table 1). PTMs identified specifically in cardiac proteasomes are underlined and bolded in Table 1. The biological significance of many PTMs on proteasome subunits is poorly understood. Table 2 shows a summary of the known biological changes that occur when proteasomes are post-translationally modified. Of all the PTMs, the best understood and most investigated is phosphorylation. Since other recent reviews have discussed proteasome phosphorylation in detail [23, 24], this review focuses on the less abundant PTMs.
Table 1.
Post-translational modifications of proteasomes.
| Mo difi cati on |
Proteasome Subunit/Site of Modification | Re fer en ce s |
|---|---|---|
| Lys ine Ac etyl atio n |
α1:55,102,104,116,181;α2:70,92,171; α3:127,160,176,238; α4:157,227;α5:91,203;α7:30,115; α 7:57,65,110,206,230,238;β1:118;β2:237(m);β3:77;β4:68,185,203;β6:77,184,204;β6:201;β1i:5 3,109;Rpt1:116,407,422;Rpt2:258,429;Rpt3:80(m),159(m),238,397,401,418;Rpt4:15,20,68,72 ,206;Rpt6:222;Rpn2:310,401,413,417,849;Rpn3:14;Rpn5:92(m),221,368,448;Rpn6:141,417; Rpn7:117;Rpn8:204,214,293(m),297(m),308,310;Rpn9:298,313;Rpn12:297;rp28:221;PA28α: 35(m),90,232;PA28β:15(m),180;PA28γ:195;PA200:1397; |
[32, 89 - 92 ] |
| N- ter min al Ac etyl atio n |
α1 ;α2;α3;α4;α5;α6;α7;β3;β4;Rpt3;Rpt6;Rpn1 ;Rpn5;Rpn6; | [6, 32 ] |
| Ca spa se cle ava ge |
Aspartic acid residues:
α1:ND;α2:ND;α6:ND;α7:ND;Rpt1 :ND;Rpt2:ND;Rpt5:27;Rpt6:4(m);Rpn2:ND;Rpn10:ND; PA28α:ND; |
[40-42 , 93 ] |
| Di- me thyl atio n |
Rpn2:R199;PA28β:K3; | [90] |
| Gly cos ylat ion |
β3:ND;Rpn11:N241; | [69] |
| HN E |
α1:ND;α2:ND;α3:ND;α4:ND;α5:ND;α6:ND;β3:ND;β4:ND;β6:ND;β1i:ND; | [38, 39
] |
| Me thyl atio n |
PA28γ:K121,212; | [94] |
| N- ter min al my rist oyl atio n |
Rpt2; | [6] |
| Nitr osy lati on |
α2:Y228; | [95] |
| O- Glc NA c |
α1 :S5(m);α4:S130(m);α5:S198(m);α6:S110;β6:S57(m),208(m);Rpt2:ND;Rpt6:T272(m); | [44- 46 ] |
| Oxi dati on |
Rpt3:ND;Rpt5:ND; | [35, 37 , 81 ] |
| Ph osp hor yl- atio n |
Serine
residues:α1:17,63,64(m);α2:7,9,77,198(m); α3:13,75(m),81(m),153(m),173(m); α4:167,201; α5 :6,16,43,56, 63,134(m);α6:14,40,54,110,150,177(m),211,230,247(m); α7:13(m),16,97,155(m),158(m),243, 250;β1:12(m),157(m),158,203,204;β2:5,277(m);β3:168,181(m);β4:39(m);β5:139,192(m),204( m),261;β6:48(m),83,167(m),169(m);β7:9,26,30,93(m),206,249;β1i:188;Rpt1:363(r);Rpt2:4,6,9 7(m),297;β2i:229,230;Rpt3:21,28,84,117,346,350;Rpt4:52,201,202,244;Rpt5:9,12(m),345,379 (m);Rpt6:120,136,217,244,256,257,303;Rpn1:16,22(m),147(m),361,363,508;Rpn2:296,315;R pn3:6,45(m),47,105,418,426(m),528(m);Rpn6:14,23,79,81,272,298,320,413,416(m);Rpn7:361 ,366;Rpn8:188,264,268;Rpn9:106,150,323;Rpn10:115,256,266,358,361;Rpn11:34,224;Rpn1 2:54,61,106;S15:2,13,65,121,127,128(m),129;S5b:105(m),193(m),386,389(m),434,466;PA28 α:238(iso2);PA28β:10;PA28γ:24,247,248;PA200:336,339,580,642,727(m),1121,1206,1614,17 46,1753,1758; Threonine residues:α1:73; α2:204(m);α3:33(m),80,97(m),223; α4:31 (m),97(m); α5:36,55,219(m),230(m); α6:42;α7: 186;β1:22(m),23,181(m),190;β2:44,121(m),273(m);β3:36,37,86(m);β5:48(m),158(iso2),262;β 6:38(m),75,78,91,93;β7:27,46,54,208;β1 i(iso2):5;β2i:259,266;Rpt1:63,405,426;Rpt2:53,308,4 34;Rpt3:25,118,316,347;Rpt4:247;Rpt5:9(m),15(m);Rpt6:109,364;Rpn1:9(m),20(m),24,186,49 9;Rpn2:157,270,273,286(m),311,488,585(m);Rpn3:43,393;Rpn5:219;Rpn6:60(m),209,396;Rp n8:186;rp28:207;Rpn9:88,95,131,149;Rpn10:250(m),253(m);Rpn11:266;Rpn12:295;S5b:101( m),210,384(m);PA28γ:23,205(m),207(m);PA200:81,338,572,728(m),729(m),1394; Tyrosine residues:α1:23,96,103,105,107,159,160;α2:6,24,57,76,97,98,101,121 (r),167;α3:143,148,153, 156(m);α4: 106(r),153;α5:8,26,185;α6:128,137,153;α7:8,105,159(m),161,239(r);β1:58(m),59,177;β2:51,1 54;β3:85(m);β3:103,104;β4:3,67,73,134,146,147;β5:99,149,171,220,228,236;β6:8,125,132,13 5,149(m),150,158,215(m);β7:22,75,102,107,186,189,222,223;β1i:217;β1i(iso2):180;β2i:81,15 8;β5i:112,126,162,184,185,234,241,249,269(m);β5i(iso2):5;Rpt1:4,111,174,249,429,432;Rpt2 :25,210,225,439;Rpt3:41,111,112,164,205,417;Rpt4:173,207,328,386;Rpt5:132,135(m);Rpt6: 72,121,148,189;Rpn1:110,158(m),554,906;Rpn2:49,147,494(m),584(m),813,900,950;Rpn3:2 07(r),214(r),264,290,302,392;Rpn5:20,111,137,369,370;Rpn6:72,201,397,415;Rpn7:366;Rpn 8:79(m);S15:41,62,70;Rpn9:156,162,172;Rpn10:326;Rpn11:32,234;Rpn12:315,316;rp28:112, 138;S5b:132,370,457,478;PA28α:249;PA28β:199,239;PA28γ:84,254;PA200:82,213,1489,161 5; |
[46, 78 , 90 , 96 - 132] |
| Su mo ylat ion |
β2:ND;Rpt1:ND;Rpt2:ND;Rpt4:ND;Rpn1:ND;Rpn2:ND;Rpn5:ND,K92;Rpn8:ND;Rpn10:ND;Rp n12:ND;PA28γ:K6,12,14; |
[133
- 135] |
| Ubi quit inat ion |
Lysine
residues:α1:30,45,55,59,71,102,104,116,153,164,171,181,182;α2:39,50,51,53,64,70,92,165, 171,176,227; α3:54,64,67,127,176,180,187,195,199,205,210,231,238,246;α4:28,38(m),52,115,157,174,193 ,204,218,227,234(m);α5:86,91,149,187,192,196,203,209;231;239;α6:30,39,41,50,61,115,189, 208,243,256;α7:29,43,57,65,110,179,183,192,206,222(m),230,238,245;β1:67,183,230;β2:31, 52,72,127,195(m),196(m),225,237,249;β3:15(m),17,41,77,98,192;β4:29,34,37,62,68,162,169, 185,198;β5:91,92,207(m),257;β6:73,77,94,104,146,163(m),164,184,203(m),204,228;β7:147,2 01,240;β1i:53,215;β1i:217;β2i:31,37,68,72,223,236;Rpt1:11,13,17,20,34,46,57,58,66,84,100, 110,116,120,146,181,186,210,222,248,268,316,340,356,402,407,415,422;Rpt2:24,48,69,86,9 1,98,178,217,232,237,258,265,293,326,359,387,396,413,420,423,430;Rpt3:46,62,66,70,80,1 25,174,192,212,217,238,255,273,397,401,409,418;Rpt4:7,20,34,38(m),42,48,72(m),91,168,1 80,197,206,274,298,314,322,333,369(m),383;Rpt5:16,35,53,56,70,79,125,128,144,146,155,1 73,209,211,221,233,245,250,266,278,294,300,327,372;Rpt6:15,27,38,55,88,94,101,125,130, 142,156,162,170,184,196,222,287,290,314,330,346,389,397,402,403;Rpn1:8,27,31,39,41,50, 66,94,105,107,119,141,168,178,189,228,286,292,343,350,397,441,551,712,754,858,860;Rpn 2:20,26,124,136,292,302,310,319,324,354,498(m),543,574(m),720,821 (m),825,827,838,840, 849,853,861,862,865,868,869,890,904,915,934;Rpn3:16,38,54,76,84,89,194,205(m),223,269 , 273,296,321(m),374,404,425,440,455,461,492,503;S5b:143(m),147,416,467(m);Rpn5:52,98, 121,147,153,160,179,212,221,330,400(m),431,448;Rpn6:32,46,59,71,141,175,185,274,304,4 17;Rpn7:13(m),93,107,130,242,349,362,371,372;Rpn8:28,45,46,100,103,113,180,199,204,21 4,279,304;Rpn9:2,31,32,99,105(m),115,122,132,161,174,186,252,298,313(m),321,347,361;R pn10:40,74,81,83,98,122,126,129,133,135,152,262,265(iso4),364(m),365,369;Rpn11:43,94,1 52,154,186,209,215,222,246,253,257,264,273,277;Rpn12:105,111,114(m),120,135,138,141 ( m),218,227,243,281,297,300(m),298,324,337,340(m);S15:91(m),123,198,210(m),211;S5b:14 3(m),147,416,467(m);rp28:23,30,90,153,213,221 ;PA28α:13(m),24,35,36,70(m),155,176,245; PA28β:15,39,108,115,145;PA28γ:14,36,37,86(m),100,110,121,132,195;PA200:56(m),278,91 6,1082(m),1124(m),1294(m),1361(m),1488(m),1459(m),1594(m),1605,1619,1758;PA200:149 9(m); |
[3, 90 , 136- 142] |
Table 2.
Functional roles of post-translational modifications on proteasomes.
| Modification | Proteasome Subunit |
Site of Modification/ Tissue |
Biological Function | Referen -ce |
|---|---|---|---|---|
| Acetylation | PSME3/ PA28γ |
K195/HEK293 | PA28γ acetylation at K195 enhances the monomeric interactions and heptameric formation of the 11S complex. |
[33] |
| HNE modification |
PSMA2/α2 | ND/heart | HNE modification results in decreased proteolytic activities of the 20S proteasome |
[39] |
| PSMA6/α1 | ||||
| PSMA7/α4 | ||||
| O-GlcNAc modification |
PSMC1/Rpt2 | ND/NRK | O-GlcNAc modification of Rpt2 decreases proteasome catalytic activity |
[44] |
| N- myristoylation |
PSMC1/Rpt2 | G2/yeast | N-myristoylation of Rpt2 controls the intracellular localization of the 26S proteasome |
[59] |
| Oxidation | PSMC3/Rpt5 | ND/heart | Oxidation of Rpt3 and Rpt5 decreased chymotrypsin-like activity of the proteasome | [35, 81] |
| PSMC4/Rpt3 | ||||
| Phosphorylat- ion |
PSMA2/α2 | Y121/CWSV1 | Phosphorylation of Y121 is important in nuclear localization of α2. Prevention of Y121 phosphorylation causes growth inhibition and morphologic alterations |
[101] |
| ND/HEK293 | Phosphorylation of α2 by polo-like kinase enhanced chymotrypsin-like activity |
[110] | ||
| PSMA3/α7 | S250/COS-7 | Phosphorylation of Rpt6 plays an important role in stabilizing the association between 19S regulatory complexes and 20S proteasomes |
[143] | |
| Histidine residue/Molt-4 |
Autophosphorylation of histidine residue(s) is important for nucleoside diphosphate kinase-like activity in proteasome |
[144] | ||
| PSMA4/α3 | ND/HEK293 | Phosphorylation of α3 by polo-like kinase enhanced chymotrypsin-like activity |
[145] | |
| PSMA7/α4 | Y153/HEK293 | c-Abl and Arg tyrosine kinases associate with and phosphorylate Y153 of PSMA7 (α4) resulting in decreased proteasome activity and impaired cell cycle regulation |
[111] | |
| PSMB1/β6 | Histidine residue/Molt-4 |
Autophosphorylation of histidine residue(s) is important for nucleoside diphosphate kinase-like activity in proteasome |
[144] | |
| PSMC3/Rpt5 | ND/HELA | Apoptosis signal-regulating kinase 1 (ASK1) phosphorylates Rpt5 resulting in decreased 26S proteasome activity |
[146] | |
| PSMC5/Rpt6 | S120/MDA468 | Serine120 of Rpt6 is phosphorylated by PKA resulting in significant enhancement of proteasome proteolytic activity |
[129] | |
| ND/heart | Phosphorylation of Rpt6 by a tightly associated kinase enhances the association between 19S regulatory complexes and 20S proteasomes. Dephosphorylation of 26S proteasome results in disassembly of 26S into 20S and 19S |
[147] | ||
| PSME/11S | ND/ reticulocyte |
Dephosphorylation abolished 11S ability to activate chymotrypsin-like activity of the proteasome |
[148] | |
| Poly-ADP Ribosylation |
ND | ND/K562 |
In vitro poly-ADP ribosylation activates 20S proteasome activity |
[64] |
| Ubiquitination | PSMD4/ Rpn10 |
K84,84,99,268 /yeast |
Monoubiquitination at four Lys residues lowers the ability of Rpn10 to interact with ubiquitin conjugates |
[67] |
1.4 Regulation of Proteasomes by PTMs
Cardiac proteasome PTMs for which some physiological function is known
1.41 Acetylation
N-terminal acetylation occurs in more than 80% of human proteins, is likely to be irreversible, and may function as a degradation signal [25, 26]. In contrast, acetylation of internal lysine residues is a reversible PTM which modulates protein function [27]. Similar to phosphorylation and dephosphorylation, protein acetylation and deacetylation are major intracellular post-translational regulatory mechanisms [27]. Acetylation of histones is a well-established regulator of gene transcription, but more recently, lysine acetylation of non-histone proteins has surfaced as a rapid and reversible PTM that modifies non-nuclear protein function [28]. Lysine acetylation has been shown to affect protein nuclear localization, stability, transcriptional activity, DNA binding, and protein-protein interactions [27]. The functional effects of phosphorylation and acetylation may complement one another, as they tend to reside in different regions of proteins. While phosphorylation of proteins occurs mainly in unstructured regions, acetylation occurs mainly in regions which contain an ordered secondary structure [29-31].
Acetylation of both CPs and regulatory particles (RPs) has been identified, suggesting that this is a relevant mechanism in which proteasomes rapidly modulate proteolytic activities in response to cellular demands. Recently, a study by Wang et al. identified sites of both N-terminal and internal acetylation of cardiac 20S proteasomes [32]. In vitro treatment of human ischemic heart failure and murine ischemic injured myocardium with two distinct HDAC inhibitors augmented trypsin-like proteolytic activity. Purified 20S proteasomes, isolated from animals in which HDAC inhibition had been administered in vivo, exhibited this enhancement of proteolytic activity, thus demonstrating that 20S proteasomes, independent of regulatory particles or associating partners, housed acetylation sites responsive to global HDAC inhibition. The first cardiac proteasome acetylome was then constructed, identifying acetylation of nine N-termini (α1, α2, α3, α4, α5, α6, α7, β3, β4) and seven internal lysine residues (α1-104, α5-203, α6-30, α6-115, β3-77, β6-203, β7-201). Four lysine and four N-terminal sites were novel identifications, and five internal lysine sites were found to be inducible by HDAC inhibition in vivo. These data suggest that 20S proteasomes have endogenous, regulatory sites of lysine acetylation, which augment proteolytic capacity in response to HDAC inhibition in vivo. Importantly, global HDAC inhibition is in the clinical spotlight as a potential treatment for several diseases, including cardiac, and thus may present an efficacious therapeutic strategy for alleviating proteasome functional insufficiency occurring in certain cardiac diseases.
A recent study in HEK293 cells demonstrated that the acetylation of the regulatory PA28γ by CREB binding protein (CBP) at K195 was shown to enhance monomeric interactions and heptameric formation of the 11S complex [33]. Preventing acetylation of PA28γ at K195 via site-directed mutagenesis reduced proteasomal competency for degrading certain proteasome substrates, including p21 and the hepatitis C virus core protein. Functional analysis in HEK293 cells suggested that acetylation of PA28γ plays an important role in the regulation of cell proliferation and cell cycle progression. This acetylation of PA28γ could be reversed by sirtuin 1. Preventing acetylation of PA28γ resulted in faster degradation relative to acetylated PA28γ [33]. These results suggest that proteasome acetylation affects proteasome assembly, activity and half-life and is likely to be important in cardiovascular diseases. Interestingly, N-α-acetyltransferase 10 protein (Naa10p) was recently found to physically associate with the proteasome activator PA28β, and with PA28α in a PA28β-dependent manner resulting in suppressed chymotrypsin-like proteasome activity [34]. However, the acetyltransferase activity of Naa10p is not required for its effect on chymotrypsin-like proteasome activity.
1.42 Carbonylation
A major regulator of cardiac signaling processes is oxidation of amino acid side chains via the addition of carbonyl groups. A study by Divald et al. demonstrated that the 19S subunit, Rpt5, was carbonylated in response to ischemia/reperfusion injury, and that ischemic preconditioning (IPC) could prevent oxidation of Rpt5. This observation was correlated with a 50% decrease in proteasomal chymotrypsin-like activity following I/R injury, which was reduced to 25% when an IPC stimulus was administered [35]. Interestingly, the beneficial effects of IPC on Rpt5 carbonylation could be prevented by proteasome inhibition prior to IPC. As Rpt5 is known to play important roles in the structural integrity of the 19S particle and the 26S proteasome (19S-20S) [36], these studies suggest that IPC may protect the UPS by reducing oxidative damage to 19S subunits, which strengthens both the tethering of the 19S lid to the base and the association of 19S with 20S [35].
Proteomic analysis of SH-SY5Y cells after exposure to endogenous reactive oxygen species (ROS) induction identified the 19S proteasome regulatory subunit, Rpt3, as a major intracellular target of carbonylation [37]. Rpt3 oxidation decreased its ATPase activities as well as the ability of the 26S proteasome to degrade substrates. Rpt3 may be a major molecular target of ROS under conditions of electrophile-induced oxidative stress, and the oxidative modification of Rpt3 may be functionally associated with altered recognition and degradation of proteasomal substrates in cells [37].
1.43 4-Hydroxy-Nonenalyation (4-HNE) Modification
Although the precise mechanisms involved in ischemia/reperfusion-induced inactivation of proteasome activity remain elusive, the modification of proteasome subunits by 4-HNE has been implicated in this process [38]. In vivo coronary occlusion/reperfusion in a rat model induced substantial declines in all three proteasome proteolytic activities. This reduced proteasome activity was associated with the selective modification of specific proteasome subunits (α1, α2, and α4) by 4-HNE [39]. Interestingly, while the occlusion/reperfusion-induced declines in trypsin-like activity were largely preserved in proteasomes purified from rat hearts, the loss in chymotrypsin-like and caspase-like activities were not present in the purified proteasomes. These latter results suggest that decreases in proteasome activity in coronary occlusion/reperfusion is likely due to a combination of direct oxidative modification of the enzyme as well as inhibition of the proteasome by intracellular inhibitory proteins. Incubation of purified cardiac 20S proteasomes with 4-HNE resulted in proteasome inactivation and modification of α1, α2, α4, α5, α6, and β6 subunits [38]. Importantly, oxidative stress appears to cause selective rather than global inhibition of proteasome activity, suggesting that a targeted pool of accumulating proteasomal substrate proteins may perpetuate the damage induced by oxidation.
1.44 Phosphorylation
The proteasome is highly regulated by phosphorylation with over 300 phosphorylation sites identified in mammalian proteasomes (Table 1). Twenty-nine phosphorylation sites have currently been identified on cardiac proteasomes (Table 1). A comprehensive review of the phosphorylation sites on proteasomes has recently been published [23]. Phosphorylation of the proteasome affects its proteolytic activity, assembly and localization (Table 2).
Proteasome PTMs for which some physiological functions are known (but not yet demonstrated in cardiac tissue)
1.45 Caspase Cleavage
Caspase-3 has been shown to specifically cleave three 19S subunits of the proteasome (Rpt5, Rpn2, and Rpn10) [40]. Intriguingly, seven proteasome subunits (α1, α2, α6, α7, Rpt1, Rpt2 and PA28α) were found to be degraded by caspase-7 in caspase-3-deficient MCF-7 cells during apoptosis in vitro and in vivo. This caspase-7 mediated cleavage of proteasome subunits resulted in reduced proteasome activity [41]. Cleavage of Rpt4 by both caspase-3 and -7 was detected in mouse macrophage lysates [42]. These findings suggest that caspase facilitates the execution of apoptosis through downregulation of the 26S proteasome, which in turn regulates the turnover of proapoptotic proteins.
1.46 O-linked N-acetylglucosamine (O-GlcNAc) Modification
Subunits of both 20S and 19S have been shown to be O-GlcNAcylated [43, 44]. More recently, O-GlcNAcylation was shown to directly modulate proteasome activity. OGlcNAcylation of the 19S ATPase subunit Rpt2 inhibits proteasome function, thereby serving as a potential mechanism for controlling intracellular amino acid levels in response to metabolic changes (e.g., nutrient overload and starvation) [43, 44]. Other O-GlcNAc modified proteasome subunits (α1 S5, α4 S130, α5 S198, α6 S110, β6 S57, S208, and Rpt6 T272) were discovered in proteomic analysis [45, 46].
1.47 S-Glutathionylation
Denatured or oxidized proteins can be degraded by free 20S proteasomes via ubiquitin- and ATP-independent processes. It was recently demonstrated that S-glutathionylation of specific Cys residues on 20S subunits (mainly α5) modulated the gating mechanism and activity of yeast 20S proteasomes. S-glutathionylation potentiated 20S gate opening, thereby enhancing degradation of oxidized and partially unfolded proteins [47].
1.48 Methylglyoxal Modification
In diabetic animals, hyperglycemia has been shown to increase intracellular levels of the dicarbonyl methylglyoxal (MGO) which modifies proteins and alters their functions [48]. MGO-derived advanced glycation end product (AGE) modification of renal proteasome subunits showed reduced chymotrypsin-like proteasome activity and increased MGO modification of the 20S β2 subunit in three unique diabetic mouse models [49]. In vitro incubation of purified 20S proteasomes with MGO resulted in arginine modifications on several 20S proteasome subunits (β2 R37, R85; β4 R224, R231; β5 R123, R128) and decreased chymotrypsin-like proteasome activity [49]. As hyperglycemia also increases O-GlcNAc modifications [50], and the aforementioned O-GlcNAc modification of Rpt2 decreases proteasome function, it is conceivable that methylglyoxal and O-GlcNAc could synergistically suppress proteasome activity in diabetic patients.
1.49 Methylation
Lysine and arginine methylation affect intracellular protein localization, protein–protein interactions, and cell signaling [51, 52]. The proteasome subunit α6 in the archaeon Haloferax volcanii was sub-stoichiometrically methylated at five unique sites (D20, E27, E62, E112, and E161) [53]. Alterations in hepatic proteasome subunit methylation following ethanol consumption have been associated with decreased proteasomal chymotrypsin-like activity [54]. The application of the methyl group donor, betaine, to promote methylation alleviated ethanol-elicited proteasome suppression, and tubercidin, a specific methylation inhibitor, suppressed 20S proteasome activity [54]. These results suggest that diminished methylation directly suppresses proteasome function, providing a novel mechanism for regulation of 20S proteasome activity that is independent of ethanol-induced oxidative stress [54]. In the same study, a 25 kDa 20S proteasome subunit was detected as a target of methylation. We have also shown that isolated 20S proteasomes from mouse cardiac tissue are methylated [55]. An important technical consideration for methylation studies is to ensure that carefully controlled methods are used, since in vitro methylation of proteins may occur during sample preparation from the use of methanol [56]. A proteomic screening of human 20S proteasomes exposed to methanol-induced in vitro methylation identified 17 methylation sites from eleven of the fourteen subunits (no methylation was detected in α2, α7 and β2).
1.50 N-Myristoylation
Protein N-myristoylation is an irreversible PTM which covalently links a 14 carbon myristate to the N-terminal glycine of target proteins. N-myristoylation targets modified proteins for cellular membranes [57, 58], and also modulates protein-protein interactions [58]. One proteasome subunit, Rpt2, is N-myristoylated. In normal yeast cells, proteasomes are mainly localized to the nucleus, however non-N-myristoylated mutants of Rpt2 re-localize proteasomes from the nucleus to the cytoplasm, where they formed aggregates [59]. N-myristoylation of Rpt2 therefore controls the intracellular localization of the 26S proteasome in yeast.
1.51 Tyrosine Nitration
Endothelial dysfunction, possibly due to oxidative stress, is an early event in cardiovascular disorders. The activation of 26S proteasomes by peroxynitrite (ONOO2) was recently found to be a common pathway for endothelial dysfunction in mouse models of diabetes (streptozotocin (STZ)-induced type I diabetic mice), hypertension (angiotensin-infused hypertensive mice), and dyslipidemia (high fat-diet-fed LDL receptor knockout (LDLr2/2) mice) [60]. This elevated 26S chymotrypsin-like proteasome activity was associated with tyrosine nitration of the 19S complex, and may be due to increased 26S proteasome assembly [60]. Interestingly, the nitrated proteasome degraded key proteins such as thioredoxin, an enzyme important in cellular homeostasis, at a faster rate than the non-nitrated proteasome.
1.52 Nitrosylation
S-nitrosylation, the covalent incorporation of a nitric oxide (NO) moiety into cysteine thiols, is ubiquitous in biology [61]. S-nitrosylation acts as redox-switch by modulating cysteine thiols in response to intracellular changes in oxidation. NO from S-nitroso-N-acetylepenicillamine (SNAP) inhibits all three 26S proteasome proteolytic activities in a time- and concentration-dependent manner in rat aorta vascular smooth muscle cells (VSMC) [62]. Caspase-like activity was inhibited to the greatest degree (>77%). Nitrosylation also caused changes in proteasome intracellular localization. Exposure of purified 20S proteasomes to S-nitrosoglutathione resulted in the S-nitrosylation of 10 cysteines by NO [62]. A study employing a quantitative switch assay coupled with LC-MS/MS identified 220 S-nitrosylated cysteines on 179 proteins in human pulmonary arterial endothelial cell lysates treated with S-nitrosoglutathione [63]. S-nitrosylated cysteines were identified at 13 sites on 10 proteasome subunits (Rpt1 C389, Rpt4 C170, C347, Rpt5, C387, C396, Rpn2 C806, Rpn6 C222, Rpn9 C114, α1 C154, C161, α7, C42, β3 C19, S15 C81) [63]. S-Nitrosylation may be a mechanism by which NO can exert a reversible effect on proteasome function. A delicate balance of intracellular redox state exists in cells, mainly due to the production of ROS/RNS and the antioxidant systems that remove them. The concentration of ROS/RNS is relatively low under normal homeostatic redox balance. However, increased ROS/RNS production could lead to alterations in the redox balance, resulting in oxidative/nitrosative stress. Oxidative stress may directly link changes in 26S proteasome activity to endothelial dysfunction (prevalent in most types of CVD) as well as ischemia/reperfusion injury.
1.53 Poly-ADP Ribosylation
Treatment of K562 human myelogenous leukemia cells with hydrogen peroxide resulted in a rapid up-regulation of nuclear 20S proteasome activity, which was dependent on poly-ADP ribosylation of the proteasome [64]. The poly-ADP ribosylated nuclear 20S proteasome was found to be more efficient at removing oxidatively damaged histones, suggesting that this mechanism may be important in the nuclear repair system.
1.54 Ubiquitination
It is not surprising that the number of known ubiquinated proteins continues to rise at an exponential rate as more investigators examine the UPS and better techniques become available for isolation of ubiquinated proteins. It is now appreciated that ubiquitination is heterogeneous, as it presents in several different forms and linkages. Ubiquinated proteins are most commonly identified by the presence of a 114.1 Da diglycine (GG) tag on lysine residues or an LRGG-tag (383.2 Da) on internal lysine residues following trypsin digestion. Typical ubiquitinated protein isolation strategies have involved immunoprecipitation with an anti-ubiquitin antibody [65, 66]. The subunits of the proteasome itself are degraded by the UPS, therefore it is not surprising that ubiquitination sites on most proteasome subunits have been detected by liquid chromatography-tandem mass spectrometry (LC-MS/MS). Table 1 lists the known ubiquitination sites identified on proteasome subunits. Since many of these ubiquinated sites likely target subunits for UPS degradation, they will not be discussed further.
It has recently been shown that monoubiquitination of Rpn10 in yeast inhibits the ubiquitin-interacting motif (UIM), thereby reducing the capacity of Rpn10 to interact with polyubiquitinated conjugates [67]. Rpn10 (S5a) is the main polyubiquitin-binding protein that is part of the 26S proteasome. This subunit also exists as free Rpn10 in the cytosol. Rpn10 contains two UIMs, which facilitate polyubiquitin binding. The monoubiquitination of Rpn10 at K71, K84, K99 and K268 was found to be controlled by a NEDD4 ubiquitin-protein ligase member, Rsp5, and the deubiquinating enzyme, Ubp2. The monoubiquitination of Rpn10 was decreased by heat shock, cold shock and cadmium, suggesting that monoubiquitination of Rpn10 may act as a stress sensor that regulates the recruitment of substrates to the proteasome [67]. This is especially interesting since Rpn10 was found to exist in at least two isoforms in murine cardiac proteasomes [6]. The difference between these isoforms is the presence of three residues, GER, at amino acids 255-257 of murine proteasomes. It is possible that these isoforms may have different affinities for polyubiquitinated substrates, however the roles of these isoforms are unknown. Interestingly, Rpn10 itself can be ubiquitinated by several ligases including MuRF1, Siah2, Parkin, APC, and SCFβTRCP1, CHIP, E6AP and Nedd4 [68]. The ubiquitination of Rpn10 by many ubiquitin ligases is unusual, since ligases typically are highly specific enzymes that ubiquitinate a relatively small number of proteins. The ubiquitinated Rpn10 is rapidly degraded by the proteasome, having a half-life of about 30 minutes in C2C12 myoblasts [68].
Proteasome PTMS for which physiological function are not yet determined
1.55 Glycosylation
In Madin-Darby Canine Kidney (MDCK) cells, glycosylation of the proteasome subunits β3 and Rpn11- as determined by Pro-Q Emerald glycoprotein dye on 2D gels - was enhanced after exposure to calcium oxalate dihydrate (COD) crystals, mimicking kidney stone formation [69]. Although the functional effects of these changes are unknown, aberrant glycosylation has been shown to modulate cardiac ion channel activity and electrical signaling through a cell-specific mechanism [70]. These data suggest that abnormal glycosylation is likely to be important in cardiovascular diseases.
1.56 Neddylation
Nedd8 is an additional small ubiquitin-like protein that has been found conjugated to proteins through a process called neddylation. Neddylation has been shown to be important in cell proliferation and development. Affinity purification of GST-Nedd8 modified and associating proteins in HEK293 cells identified 496 proteins, including Rpt2, Rpt4, Rpt5, Rpn1, Rpn2, Rpn3, and Rpn10 [71]. However, since identified proteins could either be direct targets of neddylation or associating proteins of a neddylated target, unequivocal evidence supporting direct neddylation of proteasome subunits remains to be demonstrated. Although the biological significance of neddylation of most proteins is unknown, the presence of significant in vivo polyneddylation suggests that this PTM is functionally important to the cell. Neddylation mimics ubiquitination, and like ubiquitination, neddylation is a reversible and dynamic process.
1.57 Sumoylation
It has been suggested that sumoylation is involved in regulating the cellular stress response [72, 73]. Proteomic analysis of SUMO-4 substrates in HEK293 cells exposed to serum starvation for 24 hours identified 90 proteins, including proteasomal proteins Rpt1 and Rpt6 [74]. Investigation of protein sumoylation in HEK293 cells exposed to oxidative stress (hydrogen peroxide, alkylating agents, and 4-HNE) identified 54 HA-SUMO-1-associated proteins and 38 HA-SUMO-3-associated proteins [73]. HA-SUMO-1-associated proteasomal proteins included Rpn10, while HA-SUMO-3-associated proteasomal proteins included α6 and Rpn10 in cytosol and Rpt1 in nuclei [73]. Tandem affinity purification (TAP) of SUMO-2 conjugates from human U2OS cells before and after exposure to heat shock identified eight putative proteasome subunits (α3, α5, β4, Rpn1, Rpn2, Rpn6, Rpn10, Rpn12) conjugated to SUMO-2, with SUMO-2 conjugated α5 and β4 levels increasing after heat shock [75]. Defective sumoylation activity has been suggested to contribute to the induction of congenital heart disease [76].
1.6 PTMs of the Proteasome and Cardiovascular Disease
The maintenance of cardiac muscle mass depends on the balance between the rates of protein synthesis and degradation. Although several pathways, including the insulin-like growth factor 1/IRS1/PI3K/Akt pathway, lysosomal/autophagy pathway and the myostatin pathway are involved, the UPS plays a central role in maintaining muscle mass [77]. A significant number of studies suggest that impaired proteasomal function directly contributes to heart disease [8-11]. Excluding phosphorylation, modifications of the proteasome lead to either changes in localization, assembly or decreased proteasome function (Table 2). Hearts from animals and humans with several cardiac diseases show decreased proteasome function. This is seemingly contradictory to the data in several different animal models of cardiovascular disease demonstrating that proteasome inhibitors may be a viable option for treatment of cardiovascular disease [78]. However, the clinically approved proteasome inhibitor, bortezomib, has been associated with cardiotoxicity in cancer patients. Also, cardioprotection has been observed by both inhibition and enhancement of proteasome activity. As postulated by Meiners et al. [79], it is likely that the effect of proteasome inhibition depends on the cell type and on the degree of proteasome inhibition. We would also add that the timing of proteasome inhibition (i.e., with respect to disease progression) likely affects the efficacy of proteasome inhibitors for treating cardiac disorders. We have previously shown that purified cardiac 20S proteasomes are more susceptible to proteasome inhibition (by epoxomicin) than purified liver 20S proteasomes [55]. Differential inhibition of proteasomes may result in a shift in the proteasomal substrate repertoires, thereby altering the degradation specificity of proteasomes.
Myocardial ischemia contributes to the pathophysiology of heart failure. Several independent investigations suggest that ischemic injury leads to increased oxidative stress and oxidation of proteasome subunits [80]. These ischemia-dependent changes in proteasome PTMs are associated with changes in proteasome function. Oxidation of Rpt3 and Rpt5 decreased chymotrypsin-like activity of the proteasome [35, 81]. This is consistent with the increased levels of polyubiquitinated proteins observed in patients with ischemic heart disease [82]. The UPS plays a critical role in the regulation of apoptosis. Significant inhibition of the proteasome is typically associated with increased apoptosis, which is likely due to the fact that many apoptotic regulatory proteins, such as Bax and p53, are degraded by the proteasome [83, 84]. Hence, some proteasomal PTMs that cause a decrease in proteasome assembly or function are likely to induce apoptosis, which if allowed to progress, will ultimately result in cell death.
Another major cause of myocardial ischemia is diabetes mellitus [85]. Diabetes, atherosclerosis and hypertension are associated with increased reactive nitrogen species (RNS), which result in nitration of proteasome subunits and an increase in proteasome activity and assembly (Figure 1). This would also be consistent with results from human patients, which exhibit increased chymotrypsin-like proteasome activity in the myocardium of diabetic ischemic patients compared with non-diabetic ischemic patients [85]. However, methylglyoxal and O-GlcNAc modifications of the proteasome secondary to hyperglycemia reduce proteasome activity [48, 49], suggesting that a complex interplay between a myriad of PTMs likely exists, and collectively, they determine the activity of the proteasome. The increased proteasome activity may account for cardiac atrophy, which is found in a percentage of diabetic patients.
Figure 1.
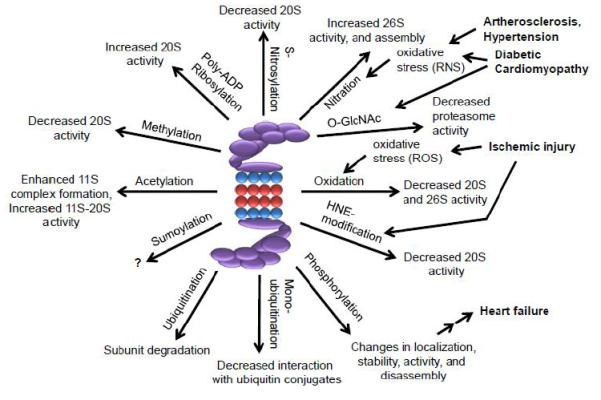
Schematic diagram of the effects of PTMs on proteasome function and their involvement in cardiovascular diseases. The 19S proteasome is shown in purple while the α and β rings of the 20S proteasome are shown in blue and red, respectively. Several cardiovascular diseases are known to alter proteasome PTMs including atherosclerosis, diabetes, hypertension, ischemia/reperfusion and heart failure. Atherosclerosis, diabetes and hypertension, all cause increases in reactive nitrogen species (RNS), resulting in nitration of proteasome subunits, which results in improved proteasome assembly and function. Ischemic injury causes oxidative stress leading to carbonylation of proteasome subunits and impaired proteasome activity. Decreased α7 phosphorylation at residue S250, decreased ATPase activity, and impaired docking of the 19S to the 20S was detected in human end stage heart failure when compared to control hearts [88]. ROS, reactive oxygen species. HNE, hydroxy-nonenalyation.
1.7 Conclusions
Excluding ubiquination sites, more than 515 PTM sites on mammalian 26S proteasomes have been identified (Table 1). Though a remarkable number of PTM sites, it is important to note that the physiological consequences of very few identified sites are currently known. Some investigations employed non-physiological approaches, including overexpression or knockdown, which ultimately affects global PTM levels. Also, while most studies were carried out to determine only one type/group of PTM sites at a particular time, it is becoming evident that significant cross-talk can occur among different PTMs (phosphorylation, acetylation, methylation, ubiquitination, and sumoylation) [86] within the cell, and are likely to occur within the proteasome. To determine the importance of PTM cross-talk, two possible methods offering significant advancement in the field are discussed next. The recent availability of PTM-specific antibodies has provided opportunities for enriching peptides that carry a particular PTM (such as phosphorylation, ubiquitination and acetylation). Purified or semi-purified proteasome samples from control and disease tissue could be digested to form peptides which are then labeled with different iTRAQ (isobaric tags for relative and absolute quantification) labels. Labeled peptides would then be mixed and enriched for phosphopeptides. The unbound peptides could then be enriched for ubiquitinated peptides (using anti-Lys-Gly-Gly antibody) and subsequent unbound peptides enriched for acetylated peptides (using anti-acetylated lysine antibody). All the enriched samples would then be analyzed by LC-MS/MS and will yield significant information about types as well as changes in proteasome PTMs during diseases.
Additional recent developments in targeted mass spectrometry-based technologies have enabled the sensitive and specific detection and quantification of peptide ions for a global assessment of several PTMs simultaneously. This technology, entitled multiple reaction monitoring (MRM) or selected reaction monitoring (SRM), exploits the specific mass shift occurring on the parent peptide and the daughter peptide fragment (collectively termed a “transition”) that is post-translationally modified [87]. Transitions are selectively targeted and detected by the mass spectrometer, excluding all non-qualifying masses, which increases the sensitivity of detection. Although PTMs that have challenging chemical properties for positive ion mode mass spectrometry (e.g., negatively charged phosphorylation) may require additional enrichment strategies, this technology far exceeds others available for quantifying multiple PTMs simultaneously within a sample and within a technical replicate of the mass spectrometer. This technology will undoubtedly provide an unprecedented understanding of the dynamic cross-talk among all PTMs, and how specific sites on proteins are targeted by PTMs in response to a physiologic stimulus.
The vast array of PTMs occurring on proteasomes suggests that multiple fine-tuning mechanisms regulate proteasome activity. This is most likely important for rapidly modulating proteasome activity in accordance with dynamic intracellular environments. Reduced proteasome activity occurs when the proteasome is modified by non-phosphorylation PTMs (Table 2). This is likely to be physiologically important, since it is unknown to what extent proteasome inhibition determines cardioprotection or cardiotoxicity. This is complicated by the proteasome having three types of proteolytic activities. Given that the chymotrypsin-like activity constitutes the greatest portion of the proteasomal proteolytic activity, an important question is whether cardiotoxicity is caused by inhibition of at least two proteasome proteolytic activities, or if significant inhibition of the chymotrypsin-like activity of the proteasome is sufficient for cardiotoxicity. Significant research is needed to establish how the cardiac proteasome system differs from the proteasome system in other organs, especially those associated with higher incidences of tumors (such as colon cells). When drugs targeting the UPS are used to treat heart diseases, the effects on other organs need to be determined. The effects of different types of proteasome inhibitors (reversible or irreversible), different concentrations (affecting the extent of inhibition), and the length of inhibition (when and how long), are all important in determining the role of proteasome inhibition in cardioprotection and cardiotoxicity.
Once we have a thorough understanding of the role of sumoylation, neddylation, and possibly other ubiquitin-like modifications on proteasome subunits as well as the role of these modifications in cardiac tissue, it is likely that inhibitors specifically targeting the enzymatic activities regulating sumoylation or neddylation may be novel strategies to regulate proteasome activity. Overall, the proteasome is a highly regulated complex that requires intensive research to properly understand the roles of many PTMs in modulating proteasome function. Understanding the role of PTMs in proteasomes is needed to help decipher the proteasome involvement and contribution to most diseases, including cardiovascular diseases.
Highlights.
Cardiac and non-cardiac proteasomes are highly regulated by post-translational modifications.
The proteasome is modulated by less commonly occurring post-translational modifications including poly-ADP ribosylation and nitrosylation.
Several sites on the proteasome are regulated by multiple different post-translational modifications.
Post-translational modifications of the proteasome as occur during cardiovascular diseases are associated with changes in proteasome activity and proteasome assembly.
1.8 Acknowledgements
This work was supported by National Institutes of Health (NIH) Grants HL096819 (A. Gomes) and HL098954 (P. Ping), and a NRSA Grant F32-HL-099029 (S. Scruggs).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
1.9 Conflicts of interest
Nothing declared.
References
- 1.Goll DE, Neti G, Mares SW, Thompson VF. Myofibrillar protein turnover: the proteasome and the calpains. Journal of animal science. 2008;86:E19–35. doi: 10.2527/jas.2007-0395. [DOI] [PubMed] [Google Scholar]
- 2.Schubert U, Anton LC, Gibbs J, Norbury CC, Yewdell JW, Bennink JR. Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature. 2000;404:770–4. doi: 10.1038/35008096. [DOI] [PubMed] [Google Scholar]
- 3.Xu G, Paige JS, Jaffrey SR. Global analysis of lysine ubiquitination by ubiquitin remnant immunoaffinity profiling. Nature biotechnology. 2010;28:868–73. doi: 10.1038/nbt.1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peters JM. Proteasomes: protein degradation machines of the cell. Trends in biochemical sciences. 1994;19:377–82. doi: 10.1016/0968-0004(94)90115-5. [DOI] [PubMed] [Google Scholar]
- 5.Brooks P, Fuertes G, Murray RZ, Bose S, Knecht E, Rechsteiner MC, et al. Subcellular localization of proteasomes and their regulatory complexes in mammalian cells. The Biochemical journal. 2000;346:155–61. Pt 1. [PMC free article] [PubMed] [Google Scholar]
- 6.Gomes AV, Zong C, Edmondson RD, Li X, Stefani E, Zhang J, et al. Mapping the murine cardiac 26S proteasome complexes. Circulation research. 2006;99:362–71. doi: 10.1161/01.RES.0000237386.98506.f7. [DOI] [PubMed] [Google Scholar]
- 7.Pagan J, Seto T, Pagano M, Cittadini A. Role of the ubiquitin proteasome system in the heart. Circulation research. 2013;112:1046–58. doi: 10.1161/CIRCRESAHA.112.300521. [DOI] [PubMed] [Google Scholar]
- 8.Wang X, Li J, Zheng H, Su H, Powell SR. Proteasome functional insufficiency in cardiac pathogenesis. American journal of physiology Heart and circulatory physiology. 2011;301:H2207–19. doi: 10.1152/ajpheart.00714.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Willis MS, Townley-Tilson WH, Kang EY, Homeister JW, Patterson C. Sent to destroy: the ubiquitin proteasome system regulates cell signaling and protein quality control in cardiovascular development and disease. Circulation research. 2010;106:463–78. doi: 10.1161/CIRCRESAHA.109.208801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Su H, Wang X. The ubiquitin-proteasome system in cardiac proteinopathy: a quality control perspective. Cardiovascular research. 2010;85:253–62. doi: 10.1093/cvr/cvp287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Day SM. The ubiquitin proteasome system in human cardiomyopathies and heart failure. American journal of physiology Heart and circulatory physiology. 2013;304:H1283–93. doi: 10.1152/ajpheart.00249.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harris JR. Release of a macromolecular protein component from human erythrocyte ghosts. Biochimica et biophysica acta. 1968;150:534–7. doi: 10.1016/0005-2736(68)90157-0. [DOI] [PubMed] [Google Scholar]
- 13.Harris JR. The isolation and purification of a macromolecular protein component from the human erythrocyte ghost. Biochimica et biophysica acta. 1969;188:31–42. doi: 10.1016/0005-2795(69)90042-7. [DOI] [PubMed] [Google Scholar]
- 14.Cuervo AM, Palmer A, Rivett AJ, Knecht E. Degradation of proteasomes by lysosomes in rat liver. European journal of biochemistry / FEBS. 1995;227:792–800. doi: 10.1111/j.1432-1033.1995.tb20203.x. [DOI] [PubMed] [Google Scholar]
- 15.Hendil KB. The 19 S multicatalytic "prosome" proteinase is a constitutive enzyme in HeLa cells. Biochemistry international. 1988;17:471–7. [PubMed] [Google Scholar]
- 16.Baumeister W, Walz J, Zuhl F, Seemuller E. The proteasome: paradigm of a self-compartmentalizing protease. Cell. 1998;92:367–80. doi: 10.1016/s0092-8674(00)80929-0. [DOI] [PubMed] [Google Scholar]
- 17.Bochtler M, Ditzel L, Groll M, Hartmann C, Huber R. The proteasome. Annual review of biophysics and biomolecular structure. 1999;28:295–317. doi: 10.1146/annurev.biophys.28.1.295. [DOI] [PubMed] [Google Scholar]
- 18.Ehlinger A, Walters KJ. Structural Insights into Proteasome Activation by the 19S Regulatory Particle. Biochemistry. 2013 doi: 10.1021/bi400417a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen X, Barton LF, Chi Y, Clurman BE, Roberts JM. Ubiquitin-independent degradation of cell-cycle inhibitors by the REGgamma proteasome. Molecular cell. 2007;26:843–52. doi: 10.1016/j.molcel.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li X, Amazit L, Long W, Lonard DM, Monaco JJ, O'Malley BW. Ubiquitin- and ATP-independent proteolytic turnover of p21 by the REGgamma-proteasome pathway. Molecular cell. 2007;26:831–42. doi: 10.1016/j.molcel.2007.05.028. [DOI] [PubMed] [Google Scholar]
- 21.Li X, Lonard DM, Jung SY, Malovannaya A, Feng Q, Qin J, et al. The SRC-3/AIB1 coactivator is degraded in a ubiquitin- and ATP-independent manner by the REGgamma proteasome. Cell. 2006;124:381–92. doi: 10.1016/j.cell.2005.11.037. [DOI] [PubMed] [Google Scholar]
- 22.Cascio P, Call M, Petre BM, Walz T, Goldberg AL. Properties of the hybrid form of the 26S proteasome containing both 19S and PA28 complexes. The EMBO journal. 2002;21:2636–45. doi: 10.1093/emboj/21.11.2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scruggs SB, Zong NC, Wang D, Stefani E, Ping P. Post-translational modification of cardiac proteasomes: functional delineation enabled by proteomics. American journal of physiology Heart and circulatory physiology. 2012;303:H9–18. doi: 10.1152/ajpheart.00189.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsukamoto O, Minamino T, Kitakaze M. Functional alterations of cardiac proteasomes under physiological and pathological conditions. Cardiovascular research. 2010;85:339–46. doi: 10.1093/cvr/cvp282. [DOI] [PubMed] [Google Scholar]
- 25.Mogk A, Bukau B. Cell biology. When the beginning marks the end. Science. 2010;327:966–7. doi: 10.1126/science.1187274. [DOI] [PubMed] [Google Scholar]
- 26.Hwang CS, Shemorry A, Varshavsky A. N-terminal acetylation of cellular proteins creates specific degradation signals. Science. 2010;327:973–7. doi: 10.1126/science.1183147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Glozak MA, Sengupta N, Zhang X, Seto E. Acetylation and deacetylation of non-histone proteins. Gene. 2005;363:15–23. doi: 10.1016/j.gene.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 28.Scott I. Regulation of cellular homoeostasis by reversible lysine acetylation. Essays in biochemistry. 2012;52:13–22. doi: 10.1042/bse0520013. [DOI] [PubMed] [Google Scholar]
- 29.Malik R, Nigg EA, Korner R. Comparative conservation analysis of the human mitotic phosphoproteome. Bioinformatics. 2008;24:1426–32. doi: 10.1093/bioinformatics/btn197. [DOI] [PubMed] [Google Scholar]
- 30.Gnad F, Ren S, Cox J, Olsen JV, Macek B, Oroshi M, et al. PHOSIDA (phosphorylation site database): management, structural and evolutionary investigation, and prediction of phosphosites. Genome biology. 2007;8:R250. doi: 10.1186/gb-2007-8-11-r250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim SC, Sprung R, Chen Y, Xu Y, Ball H, Pei J, et al. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Molecular cell. 2006;23:607–18. doi: 10.1016/j.molcel.2006.06.026. [DOI] [PubMed] [Google Scholar]
- 32.Wang D, Fang C, Zong NC, Liem DA, Cadeiras M, Scruggs SB, et al. Regulation of Acetylation Restores Proteolytic Function of Diseased Myocardium in Mouse and Human. Molecular & cellular proteomics : MCP. 2013 doi: 10.1074/mcp.M113.028332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu J, Wang Y, Li L, Zhou L, Wei H, Zhou Q, et al. Site-specific Acetylation of the Proteasome Activator REGgamma Directs Its Heptameric Structure and Functions. The Journal of biological chemistry. 2013;288:16567–78. doi: 10.1074/jbc.M112.437129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Min L, Xu H, Wang J, Qu L, Jiang B, Zeng Y, et al. N-alpha-acetyltransferase 10 protein is a negative regulator of 28S proteasome through interaction with PA28beta. FEBS letters. 2013;587:1630–7. doi: 10.1016/j.febslet.2013.04.016. [DOI] [PubMed] [Google Scholar]
- 35.Divald A, Kivity S, Wang P, Hochhauser E, Roberts B, Teichberg S, et al. Myocardial ischemic preconditioning preserves postischemic function of the 26S proteasome through diminished oxidative damage to 19S regulatory particle subunits. Circulation research. 2010;106:1829–38. doi: 10.1161/CIRCRESAHA.110.219485. [DOI] [PubMed] [Google Scholar]
- 36.DeMartino GN. Purification of PA700, the 19S regulatory complex of the 26S proteasome. Methods in enzymology. 2005;398:295–306. doi: 10.1016/S0076-6879(05)98024-5. [DOI] [PubMed] [Google Scholar]
- 37.Ishii T, Sakurai T, Usami H, Uchida K. Oxidative modification of proteasome: identification of an oxidation-sensitive subunit in 26 S proteasome. Biochemistry. 2005;44:13893–901. doi: 10.1021/bi051336u. [DOI] [PubMed] [Google Scholar]
- 38.Farout L, Mary J, Vinh J, Szweda LI, Friguet B. Inactivation of the proteasome by 4-hydroxy-2-nonenal is site specific and dependant on 20S proteasome subtypes. Archives of biochemistry and biophysics. 2006;453:135–42. doi: 10.1016/j.abb.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 39.Bulteau AL, Lundberg KC, Humphries KM, Sadek HA, Szweda PA, Friguet B, et al. Oxidative modification and inactivation of the proteasome during coronary occlusion/reperfusion. The Journal of biological chemistry. 2001;276:30057–63. doi: 10.1074/jbc.M100142200. [DOI] [PubMed] [Google Scholar]
- 40.Sun XM, Butterworth M, MacFarlane M, Dubiel W, Ciechanover A, Cohen GM. Caspase activation inhibits proteasome function during apoptosis. Molecular cell. 2004;14:81–93. doi: 10.1016/s1097-2765(04)00156-x. [DOI] [PubMed] [Google Scholar]
- 41.Jang M, Park BC, Lee AY, Na KS, Kang S, Bae KH, et al. Caspase-7 mediated cleavage of proteasome subunits during apoptosis. Biochemical and biophysical research communications. 2007;363:388–94. doi: 10.1016/j.bbrc.2007.08.183. [DOI] [PubMed] [Google Scholar]
- 42.Demon D, Van Damme P, Vanden Berghe T, Deceuninck A, Van Durme J, Verspurten J, et al. Proteome-wide substrate analysis indicates substrate exclusion as a mechanism to generate caspase-7 versus caspase-3 specificity. Molecular & cellular proteomics : MCP. 2009;8:2700–14. doi: 10.1074/mcp.M900310-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sumegi M, Hunyadi-Gulyas E, Medzihradszky KF, Udvardy A. 26S proteasome subunits are O-linked N-acetylglucosamine-modified in Drosophila melanogaster. Biochemical and biophysical research communications. 2003;312:1284–9. doi: 10.1016/j.bbrc.2003.11.074. [DOI] [PubMed] [Google Scholar]
- 44.Zhang F, Su K, Yang X, Bowe DB, Paterson AJ, Kudlow JE. O-GlcNAc modification is an endogenous inhibitor of the proteasome. Cell. 2003;115:715–25. doi: 10.1016/s0092-8674(03)00974-7. [DOI] [PubMed] [Google Scholar]
- 45.Overath T, Kuckelkorn U, Henklein P, Strehl B, Bonar D, Kloss A, et al. Mapping of O-GlcNAc sites of 20 S proteasome subunits and Hsp90 by a novel biotin-cystamine tag. Molecular & cellular proteomics : MCP. 2012;11:467–77. doi: 10.1074/mcp.M111.015966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Trinidad JC, Barkan DT, Gulledge BF, Thalhammer A, Sali A, Schoepfer R, et al. Global identification and characterization of both O-GlcNAcylation and phosphorylation at the murine synapse. Molecular & cellular proteomics : MCP. 2012;11:215–29. doi: 10.1074/mcp.O112.018366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Silva GM, Netto LE, Simoes V, Santos LF, Gozzo FC, Demasi MA, et al. Redox control of 20S proteasome gating. Antioxidants & redox signaling. 2012;16:1183–94. doi: 10.1089/ars.2011.4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Giardino I, Edelstein D, Brownlee M. Nonenzymatic glycosylation in vitro and in bovine endothelial cells alters basic fibroblast growth factor activity. A model for intracellular glycosylation in diabetes. The Journal of clinical investigation. 1994;94:110–7. doi: 10.1172/JCI117296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Queisser MA, Yao D, Geisler S, Hammes HP, Lochnit G, Schleicher ED, et al. Hyperglycemia impairs proteasome function by methylglyoxal. Diabetes. 2010;59:670–8. doi: 10.2337/db08-1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yao D, Taguchi T, Matsumura T, Pestell R, Edelstein D, Giardino I, et al. High glucose increases angiopoietin-2 transcription in microvascular endothelial cells through methylglyoxal modification of mSin3A. The Journal of biological chemistry. 2007;282:31038–45. doi: 10.1074/jbc.M704703200. [DOI] [PubMed] [Google Scholar]
- 51.Chen C, Nott TJ, Jin J, Pawson T. Deciphering arginine methylation: Tudor tells the tale. Nature reviews Molecular cell biology. 2011;12:629–42. doi: 10.1038/nrm3185. [DOI] [PubMed] [Google Scholar]
- 52.Black JC, Whetstine JR. Tipping the lysine methylation balance in disease. Biopolymers. 2013;99:127–35. doi: 10.1002/bip.22136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Humbard MA, Reuter CJ, Zuobi-Hasona K, Zhou G, Maupin-Furlow JA. Phosphorylation and methylation of proteasomal proteins of the haloarcheon Haloferax volcanii. Archaea. 2010;2010:481725. doi: 10.1155/2010/481725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Osna NA, White RL, Donohue TM, Jr., Beard MR, Tuma DJ, Kharbanda KK. Impaired methylation as a novel mechanism for proteasome suppression in liver cells. Biochemical and biophysical research communications. 2010;391:1291–6. doi: 10.1016/j.bbrc.2009.12.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gomes AV, Young GW, Wang Y, Zong C, Eghbali M, Drews O, et al. Contrasting proteome biology and functional heterogeneity of the 20 S proteasome complexes in mammalian tissues. Molecular & cellular proteomics : MCP. 2009;8:302–15. doi: 10.1074/mcp.M800058-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen G, Liu H, Wang X, Li Z. In vitro methylation by methanol: proteomic screening and prevalence investigation. Analytica chimica acta. 2010;661:67–75. doi: 10.1016/j.aca.2009.12.012. [DOI] [PubMed] [Google Scholar]
- 57.Johnson DR, Bhatnagar RS, Knoll LJ, Gordon JI. Genetic and biochemical studies of protein N-myristoylation. Annual review of biochemistry. 1994;63:869–914. doi: 10.1146/annurev.bi.63.070194.004253. [DOI] [PubMed] [Google Scholar]
- 58.Rudnick DA, McWherter CA, Gokel GW, Gordon JI. MyristoylCoA:protein N-myristoyltransferase. Advances in enzymology and related areas of molecular biology. 1993;67:375–430. doi: 10.1002/9780470123133.ch5. [DOI] [PubMed] [Google Scholar]
- 59.Kimura A, Kato Y, Hirano H. N-myristoylation of the Rpt2 subunit regulates intracellular localization of the yeast 26S proteasome. Biochemistry. 2012;51:8856–66. doi: 10.1021/bi3007862. [DOI] [PubMed] [Google Scholar]
- 60.Xu J, Wang S, Zhang M, Wang Q, Asfa S, Zou MH. Tyrosine nitration of PA700 links proteasome activation to endothelial dysfunction in mouse models with cardiovascular risk factors. PloS one. 2012;7:e29649. doi: 10.1371/journal.pone.0029649. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 61.Seth D, Hausladen A, Wang YJ, Stamler JS. Endogenous protein S-Nitrosylation in E. coli: regulation by OxyR. Science. 2012;336:470–3. doi: 10.1126/science.1215643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kapadia MR, Eng JW, Jiang Q, Stoyanovsky DA, Kibbe MR. Nitric oxide regulates the 26S proteasome in vascular smooth muscle cells. Nitric oxide : biology and chemistry / official journal of the Nitric Oxide Society. 2009;20:279–88. doi: 10.1016/j.niox.2009.02.005. [DOI] [PubMed] [Google Scholar]
- 63.Murray CI, Uhrigshardt H, O'Meally RN, Cole RN, Van Eyk JE. Identification and quantification of S-nitrosylation by cysteine reactive tandem mass tag switch assay. Molecular & cellular proteomics : MCP. 2012;11:M111–013441. doi: 10.1074/mcp.M111.013441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ullrich O, Reinheckel T, Sitte N, Hass R, Grune T, Davies KJ. Poly-ADP ribose polymerase activates nuclear proteasome to degrade oxidatively damaged histones. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:6223–8. doi: 10.1073/pnas.96.11.6223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ventadour S, Jarzaguet M, Wing SS, Chambon C, Combaret L, Bechet D, et al. A new method of purification of proteasome substrates reveals polyubiquitination of 20 S proteasome subunits. The Journal of biological chemistry. 2007;282:5302–9. doi: 10.1074/jbc.M610005200. [DOI] [PubMed] [Google Scholar]
- 66.Vasilescu J, Zweitzig DR, Denis NJ, Smith JC, Ethier M, Haines DS, et al. The proteomic reactor facilitates the analysis of affinity-purified proteins by mass spectrometry: application for identifying ubiquitinated proteins in human cells. Journal of proteome research. 2007;6:298–305. doi: 10.1021/pr060438j. [DOI] [PubMed] [Google Scholar]
- 67.Isasa M, Katz EJ, Kim W, Yugo V, Gonzalez S, Kirkpatrick DS, et al. Monoubiquitination of RPN10 regulates substrate recruitment to the proteasome. Molecular cell. 2010;38:733–45. doi: 10.1016/j.molcel.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Uchiki T, Kim HT, Zhai B, Gygi SP, Johnston JA, O'Bryan JP, et al. The ubiquitin-interacting motif protein, S5a, is ubiquitinated by all types of ubiquitin ligases by a mechanism different from typical substrate recognition. The Journal of biological chemistry. 2009;284:12622–32. doi: 10.1074/jbc.M900556200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chiangjong W, Sinchaikul S, Chen ST, Thongboonkerd V. Calcium oxalate dihydrate crystal induced changes in glycoproteome of distal renal tubular epithelial cells. Molecular bioSystems. 2011;7:1917–25. doi: 10.1039/c1mb05052d. [DOI] [PubMed] [Google Scholar]
- 70.Montpetit ML, Stocker PJ, Schwetz TA, Harper JM, Norring SA, Schaffer L, et al. Regulated and aberrant glycosylation modulate cardiac electrical signaling. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:16517–22. doi: 10.1073/pnas.0905414106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jones J, Wu K, Yang Y, Guerrero C, Nillegoda N, Pan ZQ, et al. A targeted proteomic analysis of the ubiquitin-like modifier nedd8 and associated proteins. Journal of proteome research. 2008;7:1274–87. doi: 10.1021/pr700749v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shao R, Zhang FP, Tian F, Anders Friberg P, Wang X, Sjoland H, et al. Increase of SUMO-1 expression in response to hypoxia: direct interaction with HIF-1alpha in adult mouse brain and heart in vivo. FEBS letters. 2004;569:293–300. doi: 10.1016/j.febslet.2004.05.079. [DOI] [PubMed] [Google Scholar]
- 73.Manza LL, Codreanu SG, Stamer SL, Smith DL, Wells KS, Roberts RL, et al. Global shifts in protein sumoylation in response to electrophile and oxidative stress. Chemical research in toxicology. 2004;17:1706–15. doi: 10.1021/tx049767l. [DOI] [PubMed] [Google Scholar]
- 74.Guo D, Han J, Adam BL, Colburn NH, Wang MH, Dong Z, et al. Proteomic analysis of SUMO4 substrates in HEK293 cells under serum starvation-induced stress. Biochemical and biophysical research communications. 2005;337:1308–18. doi: 10.1016/j.bbrc.2005.09.191. [DOI] [PubMed] [Google Scholar]
- 75.Golebiowski F, Matic I, Tatham MH, Cole C, Yin Y, Nakamura A, et al. System-wide changes to SUMO modifications in response to heat shock. Science signaling. 2009;2:ra24. doi: 10.1126/scisignal.2000282. [DOI] [PubMed] [Google Scholar]
- 76.Wang J, Chen L, Wen S, Zhu H, Yu W, Moskowitz IP, et al. Defective sumoylation pathway directs congenital heart disease. Birth defects research Part A, Clinical and molecular teratology. 2011;91:468–76. doi: 10.1002/bdra.20816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Banerjee A, Guttridge DC. Mechanisms for maintaining muscle. Current opinion in supportive and palliative care. 2012;6:451–6. doi: 10.1097/SPC.0b013e328359b681. [DOI] [PubMed] [Google Scholar]
- 78.Moser K, White FM. Phosphoproteomic analysis of rat liver by high capacity IMAC and LC-MS/MS. Journal of proteome research. 2006;5:98–104. doi: 10.1021/pr0503073. [DOI] [PubMed] [Google Scholar]
- 79.Meiners S, Ludwig A, Stangl V, Stangl K. Proteasome inhibitors: poisons and remedies. Medicinal research reviews. 2008;28:309–27. doi: 10.1002/med.20111. [DOI] [PubMed] [Google Scholar]
- 80.Powell SR, Divald A. The ubiquitin-proteasome system in myocardial ischaemia and preconditioning. Cardiovascular research. 2010;85:303–11. doi: 10.1093/cvr/cvp321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Predmore JM, Wang P, Davis F, Bartolone S, Westfall MV, Dyke DB, et al. Ubiquitin proteasome dysfunction in human hypertrophic and dilated cardiomyopathies. Circulation. 2010;121:997–1004. doi: 10.1161/CIRCULATIONAHA.109.904557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Weekes J, Morrison K, Mullen A, Wait R, Barton P, Dunn MJ. Hyperubiquitination of proteins in dilated cardiomyopathy. Proteomics. 2003;3:208–16. doi: 10.1002/pmic.200390029. [DOI] [PubMed] [Google Scholar]
- 83.Yang Y, Li CC, Weissman AM. Regulating the p53 system through ubiquitination. Oncogene. 2004;23:2096–106. doi: 10.1038/sj.onc.1207411. [DOI] [PubMed] [Google Scholar]
- 84.Zhang HG, Wang J, Yang X, Hsu HC, Mountz JD. Regulation of apoptosis proteins in cancer cells by ubiquitin. Oncogene. 2004;23:2009–15. doi: 10.1038/sj.onc.1207373. [DOI] [PubMed] [Google Scholar]
- 85.Marfella R, Di Filippo C, Portoghese M, Siniscalchi M, Martis S, Ferraraccio F, et al. The ubiquitin-proteasome system contributes to the inflammatory injury in ischemic diabetic myocardium: the role of glycemic control. Cardiovascular pathology : the official journal of the Society for Cardiovascular Pathology. 2009;18:332–45. doi: 10.1016/j.carpath.2008.09.008. [DOI] [PubMed] [Google Scholar]
- 86.Yang XJ, Seto E. Lysine acetylation: codified crosstalk with other posttranslational modifications. Molecular cell. 2008;31:449–61. doi: 10.1016/j.molcel.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lam MP, Scruggs SB, Kim TY, Zong C, Lau E, Wang D, et al. An MRM-based workflow for quantifying cardiac mitochondrial protein phosphorylation in murine and human tissue. Journal of proteomics. 2012;75:4602–9. doi: 10.1016/j.jprot.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Day SM, Divald A, Wang P, Davis F, Bartolone S, Jones R, et al. Impaired assembly and post-translational regulation of 26S proteasome in human end-stage heart failure. Circulation Heart failure. 2013;6:544–9. doi: 10.1161/CIRCHEARTFAILURE.112.000119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, et al. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–40. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 90.Technology CS. PhosphoSite Plus. 2013 p. PhosphoSitePlus ( http://www.phosphosite.org) is an open, comprehensive, manually curated and interactive resource for studying experimentally observed post-translational modifications, primarily of human and mouse proteins. .
- 91.Zhao S, Xu W, Jiang W, Yu W, Lin Y, Zhang T, et al. Regulation of cellular metabolism by protein lysine acetylation. Science. 2010;327:1000–4. doi: 10.1126/science.1179689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Simon GM, Cheng J, Gordon JI. Quantitative assessment of the impact of the gut microbiota on lysine epsilon-acetylation of host proteins using gnotobiotic mice. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:11133–8. doi: 10.1073/pnas.1208669109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mahrus S, Trinidad JC, Barkan DT, Sali A, Burlingame AL, Wells JA. Global sequencing of proteolytic cleavage sites in apoptosis by specific labeling of protein N termini. Cell. 2008;134:866–76. doi: 10.1016/j.cell.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Jung SY, Li Y, Wang Y, Chen Y, Zhao Y, Qin J. Complications in the assignment of 14 and 28 Da mass shift detected by mass spectrometry as in vivo methylation from endogenous proteins. Analytical chemistry. 2008;80:1721–9. doi: 10.1021/ac7021025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhan X, Desiderio DM. Nitroproteins from a human pituitary adenoma tissue discovered with a nitrotyrosine affinity column and tandem mass spectrometry. Analytical biochemistry. 2006;354:279–89. doi: 10.1016/j.ab.2006.05.024. [DOI] [PubMed] [Google Scholar]
- 96.Dephoure N, Zhou C, Villen J, Beausoleil SA, Bakalarski CE, Elledge SJ, et al. A quantitative atlas of mitotic phosphorylation. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:10762–7. doi: 10.1073/pnas.0805139105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Olsen JV, Vermeulen M, Santamaria A, Kumar C, Miller ML, Jensen LJ, et al. Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Science signaling. 2010;3:ra3. doi: 10.1126/scisignal.2000475. [DOI] [PubMed] [Google Scholar]
- 98.Mayya V, Lundgren DH, Hwang SI, Rezaul K, Wu L, Eng JK, et al. Quantitative phosphoproteomic analysis of T cell receptor signaling reveals system-wide modulation of protein-protein interactions. Science signaling. 2009;2:ra46. doi: 10.1126/scisignal.2000007. [DOI] [PubMed] [Google Scholar]
- 99.Lee SH, Park Y, Yoon SK, Yoon JB. Osmotic stress inhibits proteasome by p38 MAPK-dependent phosphorylation. The Journal of biological chemistry. 2010;285:41280–9. doi: 10.1074/jbc.M110.182188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wu F, Wang P, Zhang J, Young LC, Lai R, Li L. Studies of phosphoproteomic changes induced by nucleophosmin-anaplastic lymphoma kinase (ALK) highlight deregulation of tumor necrosis factor (TNF)/Fas/TNF-related apoptosis-induced ligand signaling pathway in ALK-positive anaplastic large cell lymphoma. Molecular & cellular proteomics : MCP. 2010;9:1616–32. doi: 10.1074/mcp.M000153-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Benedict CM, Clawson GA. Nuclear multicatalytic proteinase subunit RRC3 is important for growth regulation in hepatocytes. Biochemistry. 1996;35:11612–21. doi: 10.1021/bi960889p. [DOI] [PubMed] [Google Scholar]
- 102.Gu TL, Deng X, Huang F, Tucker M, Crosby K, Rimkunas V, et al. Survey of tyrosine kinase signaling reveals ROS kinase fusions in human cholangiocarcinoma. PloS one. 2011;6:e15640. doi: 10.1371/journal.pone.0015640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lu H, Zong C, Wang Y, Young GW, Deng N, Souda P, et al. Revealing the dynamics of the 20 S proteasome phosphoproteome: a combined CID and electron transfer dissociation approach. Molecular & cellular proteomics : MCP. 2008;7:2073–89. doi: 10.1074/mcp.M800064-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Shiromizu T, Adachi J, Watanabe S, Murakami T, Kuga T, Muraoka S, et al. Identification of Missing Proteins in the neXtProt Database and Unregistered Phosphopeptides in the PhosphoSitePlus Database As Part of the Chromosome-Centric Human Proteome Project. Journal of proteome research. 2013;12:2414–21. doi: 10.1021/pr300825v. [DOI] [PubMed] [Google Scholar]
- 105.Christensen GL, Kelstrup CD, Lyngso C, Sarwar U, Bogebo R, Sheikh SP, et al. Quantitative phosphoproteomics dissection of seven-transmembrane receptor signaling using full and biased agonists. Molecular & cellular proteomics : MCP. 2010;9:1540–53. doi: 10.1074/mcp.M900550-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Rush J, Moritz A, Lee KA, Guo A, Goss VL, Spek EJ, et al. Immunoaffinity profiling of tyrosine phosphorylation in cancer cells. Nature biotechnology. 2005;23:94–101. doi: 10.1038/nbt1046. [DOI] [PubMed] [Google Scholar]
- 107.Sui S, Wang J, Yang B, Song L, Zhang J, Chen M, et al. Phosphoproteome analysis of the human Chang liver cells using SCX and a complementary mass spectrometric strategy. Proteomics. 2008;8:2024–34. doi: 10.1002/pmic.200700896. [DOI] [PubMed] [Google Scholar]
- 108.Wang X, Chen CF, Baker PR, Chen PL, Kaiser P, Huang L. Mass spectrometric characterization of the affinity-purified human 26S proteasome complex. Biochemistry. 2007;46:3553–65. doi: 10.1021/bi061994u. [DOI] [PubMed] [Google Scholar]
- 109.Weber C, Schreiber TB, Daub H. Dual phosphoproteomics and chemical proteomics analysis of erlotinib and gefitinib interference in acute myeloid leukemia cells. Journal of proteomics. 2012;75:1343–56. doi: 10.1016/j.jprot.2011.11.004. [DOI] [PubMed] [Google Scholar]
- 110.Feng Y, Longo DL, Ferris DK. Polo-like kinase interacts with proteasomes and regulates their activity. Cell growth & differentiation : the molecular biology journal of the American Association for Cancer Research. 2001;12:29–37. [PubMed] [Google Scholar]
- 111.Liu X, Huang W, Li C, Li P, Yuan J, Li X, et al. Interaction between c-Abl and Arg tyrosine kinases and proteasome subunit PSMA7 regulates proteasome degradation. Molecular cell. 2006;22:317–27. doi: 10.1016/j.molcel.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 112.Wisniewski JR, Nagaraj N, Zougman A, Gnad F, Mann M. Brain phosphoproteome obtained by a FASP-based method reveals plasma membrane protein topology. Journal of proteome research. 2010;9:3280–9. doi: 10.1021/pr1002214. [DOI] [PubMed] [Google Scholar]
- 113.Rigbolt KT, Prokhorova TA, Akimov V, Henningsen J, Johansen PT, Kratchmarova I, et al. System-wide temporal characterization of the proteome and phosphoproteome of human embryonic stem cell differentiation. Science signaling. 2011;4:rs3. doi: 10.1126/scisignal.2001570. [DOI] [PubMed] [Google Scholar]
- 114.Moritz A, Li Y, Guo A, Villen J, Wang Y, MacNeill J, et al. Akt-RSK-S6 kinase signaling networks activated by oncogenic receptor tyrosine kinases. Science signaling. 2010;3:ra64. doi: 10.1126/scisignal.2000998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Luo W, Slebos RJ, Hill S, Li M, Brabek J, Amanchy R, et al. Global impact of oncogenic Src on a phosphotyrosine proteome. Journal of proteome research. 2008;7:3447–60. doi: 10.1021/pr800187n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ge F, Xiao CL, Yin XF, Lu CH, Zeng HL, He QY. Phosphoproteomic analysis of primary human multiple myeloma cells. Journal of proteomics. 2010;73:1381–90. doi: 10.1016/j.jprot.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 117.Eang R, Girbal-Neuhauser E, Xu B, Gairin JE. Characterization and differential expression of a newly identified phosphorylated isoform of the human 20S proteasome beta7 subunit in tumor vs. normal cell lines. Fundamental & clinical pharmacology. 2009;23:215–24. doi: 10.1111/j.1472-8206.2009.00665.x. [DOI] [PubMed] [Google Scholar]
- 118.Manes NP, Dong L, Zhou W, Du X, Reghu N, Kool AC, et al. Discovery of mouse spleen signaling responses to anthrax using label-free quantitative phosphoproteomics via mass spectrometry. Molecular & cellular proteomics : MCP. 2011;10:M110–000927. doi: 10.1074/mcp.M110.000927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wilson-Grady JT, Haas W, Gygi SP. Quantitative comparison of the fasted and re-fed mouse liver phosphoproteomes using lower pH reductive dimethylation. Methods. 2013;61:277–86. doi: 10.1016/j.ymeth.2013.03.031. [DOI] [PubMed] [Google Scholar]
- 120.Choudhary C, Olsen JV, Brandts C, Cox J, Reddy PN, Bohmer FD, et al. Mislocalized activation of oncogenic RTKs switches downstream signaling outcomes. Molecular cell. 2009;36:326–39. doi: 10.1016/j.molcel.2009.09.019. [DOI] [PubMed] [Google Scholar]
- 121.Hsu PP, Kang SA, Rameseder J, Zhang Y, Ottina KA, Lim D, et al. The mTOR-regulated phosphoproteome reveals a mechanism of mTORC1-mediated inhibition of growth factor signaling. Science. 2011;332:1317–22. doi: 10.1126/science.1199498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kim BG, Lee JH, Ahn JM, Park SK, Cho JH, Hwang D, et al. 'Two-stage double-technique hybrid (TSDTH)' identification strategy for the analysis of BMP2-induced transdifferentiation of premyoblast C2C12 cells to osteoblast. Journal of proteome research. 2009;8:4441–54. doi: 10.1021/pr900231a. [DOI] [PubMed] [Google Scholar]
- 123.Schreiber TB, Mausbacher N, Keri G, Cox J, Daub H. An integrated phosphoproteomics work flow reveals extensive network regulation in early lysophosphatidic acid signaling. Molecular & cellular proteomics : MCP. 2010;9:1047–62. doi: 10.1074/mcp.M900486-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Wang Z, Udeshi ND, Slawson C, Compton PD, Sakabe K, Cheung WD, et al. Extensive crosstalk between O-GlcNAcylation and phosphorylation regulates cytokinesis. Science signaling. 2010;3:ra2. doi: 10.1126/scisignal.2000526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Herskowitz JH, Seyfried NT, Duong DM, Xia Q, Rees HD, Gearing M, et al. Phosphoproteomic analysis reveals site-specific changes in GFAP and NDRG2 phosphorylation in frontotemporal lobar degeneration. Journal of proteome research. 2010;9:6368–79. doi: 10.1021/pr100666c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Gevaert K, Staes A, Van Damme J, De Groot S, Hugelier K, Demol H, et al. Global phosphoproteome analysis on human HepG2 hepatocytes using reversed-phase diagonal LC. Proteomics. 2005;5:3589–99. doi: 10.1002/pmic.200401217. [DOI] [PubMed] [Google Scholar]
- 127.Huttlin EL, Jedrychowski MP, Elias JE, Goswami T, Rad R, Beausoleil SA, et al. A tissue-specific atlas of mouse protein phosphorylation and expression. Cell. 2010;143:1174–89. doi: 10.1016/j.cell.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Munton RP, Tweedie-Cullen R, Livingstone-Zatchej M, Weinandy F, Waidelich M, Longo D, et al. Qualitative and quantitative analyses of protein phosphorylation in naive and stimulated mouse synaptosomal preparations. Molecular & cellular proteomics : MCP. 2007;6:283–93. doi: 10.1074/mcp.M600046-MCP200. [DOI] [PubMed] [Google Scholar]
- 129.Zhang F, Hu Y, Huang P, Toleman CA, Paterson AJ, Kudlow JE. Proteasome function is regulated by cyclic AMP-dependent protein kinase through phosphorylation of Rpt6. The Journal of biological chemistry. 2007;282:22460–71. doi: 10.1074/jbc.M702439200. [DOI] [PubMed] [Google Scholar]
- 130.Rikova K, Guo A, Zeng Q, Possemato A, Yu J, Haack H, et al. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell. 2007;131:1190–203. doi: 10.1016/j.cell.2007.11.025. [DOI] [PubMed] [Google Scholar]
- 131.Pighi C, Gu TL, Dalai I, Barbi S, Parolini C, Bertolaso A, et al. Phospho-proteomic analysis of mantle cell lymphoma cells suggests a pro-survival role of B-cell receptor signaling. Cell Oncol (Dordr) 2011;34:141–53. doi: 10.1007/s13402-011-0019-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Villen J, Beausoleil SA, Gerber SA, Gygi SP. Large-scale phosphorylation analysis of mouse liver. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:1488–93. doi: 10.1073/pnas.0609836104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Tatham MH, Matic I, Mann M, Hay RT. Comparative proteomic analysis identifies a role for SUMO in protein quality control. Science signaling. 2011;4:rs4. doi: 10.1126/scisignal.2001484. [DOI] [PubMed] [Google Scholar]
- 134.Matic I, Schimmel J, Hendriks IA, van Santen MA, van de Rijke F, van Dam H, et al. Site-specific identification of SUMO-2 targets in cells reveals an inverted SUMOylation motif and a hydrophobic cluster SUMOylation motif. Molecular cell. 2010;39:641–52. doi: 10.1016/j.molcel.2010.07.026. [DOI] [PubMed] [Google Scholar]
- 135.Wu Y, Wang L, Zhou P, Wang G, Zeng Y, Wang Y, et al. Regulation of REGgamma cellular distribution and function by SUMO modification. Cell research. 2011;21:807–16. doi: 10.1038/cr.2011.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kim W, Bennett EJ, Huttlin EL, Guo A, Li J, Possemato A, et al. Systematic and quantitative assessment of the ubiquitin-modified proteome. Molecular cell. 2011;44:325–40. doi: 10.1016/j.molcel.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Wagner SA, Beli P, Weinert BT, Nielsen ML, Cox J, Mann M, et al. A proteome-wide, quantitative survey of in vivo ubiquitylation sites reveals widespread regulatory roles. Molecular & cellular proteomics : MCP. 2011;10:M111–013284. doi: 10.1074/mcp.M111.013284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Danielsen JM, Sylvestersen KB, Bekker-Jensen S, Szklarczyk D, Poulsen JW, Horn H, et al. Mass spectrometric analysis of lysine ubiquitylation reveals promiscuity at site level. Molecular & cellular proteomics : MCP. 2011;10:M110–003590. doi: 10.1074/mcp.M110.003590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Shi Y, Chan DW, Jung SY, Malovannaya A, Wang Y, Qin J. A data set of human endogenous protein ubiquitination sites. Molecular & cellular proteomics : MCP. 2011;10:M110–002089. doi: 10.1074/mcp.M110.002089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Wagner SA, Beli P, Weinert BT, Scholz C, Kelstrup CD, Young C, et al. Proteomic analyses reveal divergent ubiquitylation site patterns in murine tissues. Molecular & cellular proteomics : MCP. 2012;11:1578–85. doi: 10.1074/mcp.M112.017905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Meierhofer D, Wang X, Huang L, Kaiser P. Quantitative analysis of global ubiquitination in HeLa cells by mass spectrometry. Journal of proteome research. 2008;7:4566–76. doi: 10.1021/pr800468j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Denis NJ, Vasilescu J, Lambert JP, Smith JC, Figeys D. Tryptic digestion of ubiquitin standards reveals an improved strategy for identifying ubiquitinated proteins by mass spectrometry. Proteomics. 2007;7:868–74. doi: 10.1002/pmic.200600410. [DOI] [PubMed] [Google Scholar]
- 143.Bose S, Stratford FL, Broadfoot KI, Mason GG, Rivett AJ. Phosphorylation of 20S proteasome alpha subunit C8 (alpha7) stabilizes the 26S proteasome and plays a role in the regulation of proteasome complexes by gamma-interferon. The Biochemical journal. 2004;378:177–84. doi: 10.1042/BJ20031122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Yano M, Mori S, Kido H. Intrinsic nucleoside diphosphate kinase-like activity is a novel function of the 20 S proteasome. The Journal of biological chemistry. 1999;274:34375–82. doi: 10.1074/jbc.274.48.34375. [DOI] [PubMed] [Google Scholar]
- 145.Van Hoof D, Munoz J, Braam SR, Pinkse MW, Linding R, Heck AJ, et al. Phosphorylation dynamics during early differentiation of human embryonic stem cells. Cell stem cell. 2009;5:214–26. doi: 10.1016/j.stem.2009.05.021. [DOI] [PubMed] [Google Scholar]
- 146.Um JW, Im E, Park J, Oh Y, Min B, Lee HJ, et al. ASK1 negatively regulates the 26 S proteasome. The Journal of biological chemistry. 2010;285:36434–46. doi: 10.1074/jbc.M110.133777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Satoh K, Sasajima H, Nyoumura KI, Yokosawa H, Sawada H. Assembly of the 26S proteasome is regulated by phosphorylation of the p45/Rpt6 ATPase subunit. Biochemistry. 2001;40:314–9. doi: 10.1021/bi001815n. [DOI] [PubMed] [Google Scholar]
- 148.Li N, Lerea KM, Etlinger JD. Phosphorylation of the proteasome activator PA28 is required for proteasome activation. Biochemical and biophysical research communications. 1996;225:855–60. doi: 10.1006/bbrc.1996.1263. [DOI] [PubMed] [Google Scholar]


