Abstract
Precursor cell entry into the T-cell developmental pathway can be divided into two phases by the closure of T-lineage commitment. As cells decide against the last alternative options to the T-cell fate, they turn on the transcription factor Bcl11b and silence expression of a group of multipotent progenitor regulatory factors that include hematopoietic transcription factor PU.1. Functional perturbation tests show that Bcl11b is needed for commitment while PU.1 actively participates in keeping open access to alternative fates, until it is silenced; however, PU.1 as well as Bcl11b contributes positively to T-cell development. Our recent work reviewed here sheds light on the transcriptional regulatory network that determines the timing and irreversibility of Bcl11b activation, the ways that Notch signaling from the thymic microenvironment restricts the action of PU.1 to prevent it from diverting cells to non-T fates, and the target genes that PU.1 still regulates under the influence of Notch signaling to contribute to T-cell generation. We argue that T-cell development depends on the sequential operation of two interlaced, but mutually antagonistic, gene regulatory networks, one initially supporting expansion before commitment and the other imposing a “terminal” differentiation process on committed cells.
Introduction
Stem cells are defined by the combination of self-renewal capability with multipotentiality, i.e. the sustained ability to give rise to multiple different types of descendant cells. A corollary of this definition is that when stem cells generate any particular cell type, the differentiating intermediates need to make two decisions: one to terminate self-renewal, and the other to select the particular developmental program to activate while suppressing all of the other alternatives. Understanding of how these two qualitatively different decisions are intertwined is still incomplete. While a great deal is known about how transcription factors collaborate positively to drive particular sets of target genes, giving cells a specific function, it remains much less clear how cells make a clean and coherent break with the other possible programs that were originally available to their precursors. Even less is known about how cells give up the intrinsic capacity for self-renewal and replace it with a more restricted and often limited kind of cell cycle control. One of the best systems available for shedding light on these questions, however, is T cell development in mice.
The early stages of murine T cell development in which cells actually choose the T-cell fate are unusually well-defined, since the hematopoietic progenitors that will generate T cells first migrate to the thymus before making the fate decision. They thus separate themselves physically from the immature blood precursors generating other hematopoietic cell types, and they do this at a very early stage of differentiation when they are still multipotent. Most of the events involved in sequential exclusion of alternative fates consequently take place within the thymus and can be resolved hierarchically by separating the precursors based on multiparameter flow cytometry. Meanwhile, the early T cell development process itself has become accessible to monitoring and manipulation ex vivo by the development of powerful stromal coculture systems, in which the cells develop with high cloning efficiency and high clonal expansion, and in which development can be started, interrupted for manipulation, and restarted again. The molecular biology of cells at successive stages can thus be correlated prospectively with their developmental potentials, and stage-specific gain and loss of function manipulations that are possible in this system have begun to take the machinery of the commitment process apart.
Stem cell fate determination by Notch signals: process and questions
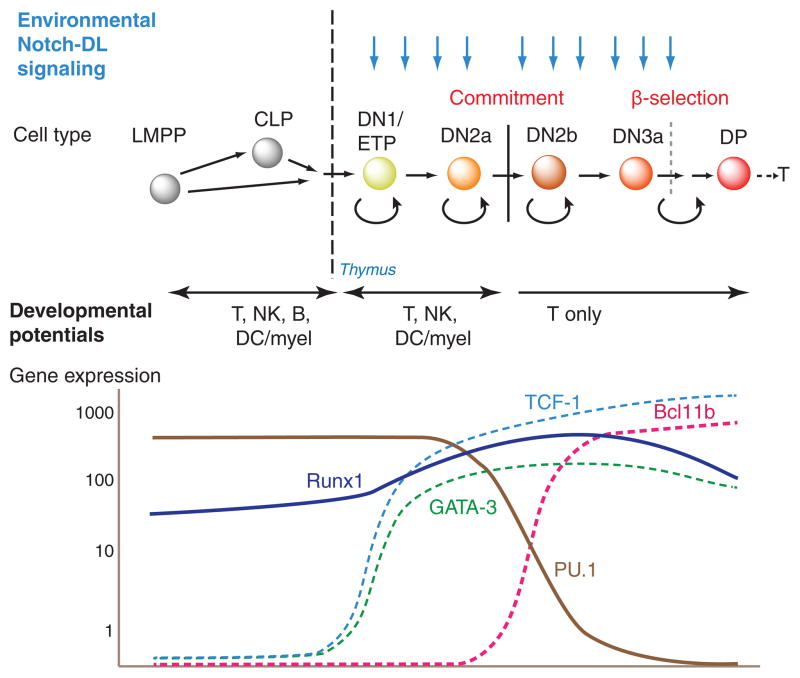
Stem or progenitor cell choice of the T cell fate is completely dependent on environmental signals from Notch-Delta interaction (for recent reviews, see (Radtke et al. 2010; Yuan et al. 2010; Naito et al. 2011; Thompson and Zúñiga-Pflücker 2011)). Progenitor cells cannot develop into T cells unless they express Notch1, and Delta-like 4 (DLL4) is one of the most important features of the thymic microenvironment to drive cells into the T-cell pathway. Notch-Delta interaction is a well-known inductive signal that operates in many developmental systems to trigger a rapid cascade of differentiative consequences. However, what is known about the T-cell program shows that Notch cannot be the only transcriptional input to establish the T-cell fate. The cells need to progress through at least four distinct stages, all under the influence of Notch-Delta signals, with loss of T-cell identity or loss of viability if the signal is interrupted at any stage (Fig. 1, top). The population dynamics along this pathway imply that the progression from stage to stage is slow, with multiple cell cycles under the influence of Notch signals at each stage. Direct Notch target genes are a part of the T-cell differentiation program, as revealed by RNA expression analysis of cells in which Notch signaling is suddenly interrupted by drug treatment or a switch to a Delta-free microenvironment. However, although Notch-dependent T-cell genes are well defined, they are not the same genes at each stage. In fact, some of the strongest Notch target genes are expressed primarily at the beginning of the process, such as Nrarp; others such as Il2ra and HEBalt are activated abruptly in the middle; whereas others such as Ptcra are expressed very preferentially toward the end. Finally, lineage commitment does not occur until after at least two phenotypic stage transitions and numerous cell cycles under the influence of Notch-Delta. The behavior of the cells thus raises three general questions about how the Notch signal drives T cell development. First, why is the lineage commitment so slow, with step times of days (with ~2 cell cycles/day) rather than hours? Second, why are different Notch target genes turned on at each step? Third, what makes the sequence of these individual steps irreversible?
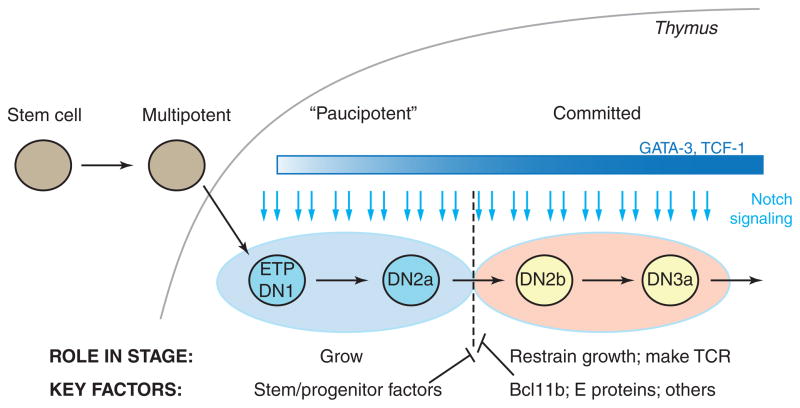
Figure 1.
Outline of early T cell development. Top: succession of early intrathymic developmental stages referred to in the text. LMPP: lymphoid-primed multipotent precursor. CLP: common lymphoid precursor. DN: CD4− CD8− surface TCR−. ETP (Kit-high DN1): Kithigh CD44high CD25− DN. DN2a: Kithigh CD44high CD25+ DN. DN2b: Kitint/+ CD44high CD25+ DN. DN3a: Kitlow CD44low CD25+ DN. DP: CD4+ CD8+ TCRβ+. Blue arrows depict continuous requirement for Notch signaling from ETP through DN3a stage, despite substantial regulatory changes throughout these stages. Curved reflex arrows depict phases of proliferation, either self-renewal or “transit amplifying” type. Lower panel: schematic depicting expression patterns (impressionistic log scale) of three T-lineage transcription factors, GATA-3, TCF-1, and Bcl11b, one pan-hematopoietic transcription factor Runx1/CBFβ, and the multilineage transcription factor PU.1. Other phase 1 transcription factors are regulated similarly to PU.1 (see Fig. 2, Table 1). Figure adapted from (Rothenberg 2012).
An obvious general answer is that Notch signals activate a cascade of transcription factor gene expression changes, which themselves affect the regulatory states in which Notch signals are interpreted in each of the successive stages. In fact, the population of transcription factor genes that is expressed in the cells at the beginning of the process is radically different from the population of factors expressed soon after commitment has occurred. Figure 1 (below) shows some of the most important transcription factor gene expression changes during this process (also see (Rothenberg et al. 2010)). The cells enter the thymus with a legacy of stem and progenitor genes closely shared with those of prethymic common lymphoid precursors and lymphoid-primed multipotent precursors (Kawazu et al. 2007; Tydell et al. 2007; David-Fung et al. 2009; Luc et al. 2012; Zhang et al. 2012; Mingueneau et al. 2013). They initially respond to Notch signals by activating expression of two transcription factors, GATA-3 and TCF-1 (encoded by the Tcf7 gene) within the first major stage, the ETP (or Kit-high DN1) stage (Fig. 1). This then creates a new state that we have referred to as “phase 1” of T-cell development, in which T-cell transcription factors overlap substantially with a population of progenitor factors that are still being expressed (Rothenberg et al. 2010). Notch signaling in the new context turns on additional genes, marking the transition to the DN2a stage, yet even so the cells remain uncommitted. Commitment is only completed as the cells progress from DN2a to DN2b stage, and this event is accompanied by two distinct types of regulatory change. The regulatory gene Bcl11b turns on, followed soon after by Lef1, Ets1, and Ets2; and the stem- and progenitor-associated factors are concomitantly turned off. Transcription factor genes turned off in this period include Sfpi1 (coding for PU.1), Tal1 (coding for SCL), Hhex, and Gfi1b, with Lyl1 turned off one stage later (Yui et al. 2010). This transition now establishes the main outlines of the regulatory state that will persist as the cells stop dividing, undergo TCR gene rearrangement, and embark on their first TCR-dependent selection event, β-selection.
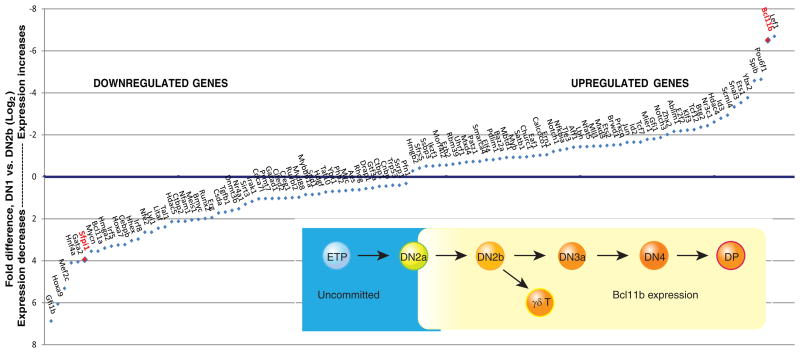
The reciprocal changes in expression of PU.1 and Bcl11b during commitment are particularly dramatic. Figure 2 (main graph) shows that the fold changes in expression of the genes encoding these two factors, between the ETP (Kit+ DN1) and newly committed DN2b stages, are at the extremes for all transcriptional regulatory genes in the genome (Zhang et al. 2012). Bcl11b is one of the two most strongly upregulated, whereas PU.1 (Sfpi1) is one of the six most strongly downregulated. As summarized in the inset of Fig. 2, our work with a fluorescent reporter allele of Bcl11b shows that it in fact turns on in individual cells at the end of the DN2a stage, immediately before the cells make the transition to commitment (H. Y. K., unpublished data)(Rothenberg et al. 2008; Yui et al. 2010; Zhang et al. 2012). PU.1 begins and remains at maximal levels through the DN2a stage, then begins a sharp decline during commitment, although the last residual vestiges of expression are still detectable in DN3a cells (Yui et al. 2010). Both factors are not only regulated in the course of commitment but also functionally implicated in the T-cell lineage commitment mechanism, as described below (Anderson et al. 2002; Dionne et al. 2005; Lefebvre et al. 2005; Franco et al. 2006; Laiosa et al. 2006; Masuda et al. 2007; Ikawa et al. 2010; Li et al. 2010a; Li et al. 2010b). Central questions about the nature and timing of the T-lineage commitment process can therefore be investigated in terms of two discrete problems: why it takes so long for Bcl11b to be turned on, and what PU.1 (and other “phase 1” factors) is doing in the T-lineage progenitor cells until it is turned off.
Figure 2.
Transcription factors that change expression during commitment. Main panel: genes encoding transcriptional regulators that change most in expression between ETP/DN1 stage and newly committed DN2b stage, with fold changes measured by genome-wide RNA-seq analysis (Zhang et al. 2012). Genes annotated as regulatory genes with largest fold decreases in expression or increases in expression (DN2b/DN1 levels <0.5 or >2, FDR<0.05) are arranged on x axis in order of fold change. Values on y axis are log2-transformed ratios of (expression in DN1)/(expression in DN2b). Axis order is set to show downregulated genes below the x axis and upregulated genes above the x axis. PU.1 (Sfpi1) and Bcl11b are highlighted by red labels. Inset: schematic depiction of expression pattern of Bcl11b as defined at the single-cell level by expression of a Bcl11b-IRES-mCitrine fluorescent protein reporter allele described in text (H.Y.K. and E.V.R., unpublished data).
Commitment and the role of Bcl11b
Commitment is a multi-step process that begins even before the cells enter the thymus. The final step of commitment that is coupled with changes in Bcl11b and PU.1 expression is specifically the loss of access to the last two alternative pathways: the natural killer (NK) cell pathway, and the dendritic cell (DC) or myeloid pathway. The cells have lost access to the B-cell developmental option many cell cycles earlier, through a separate mechanism triggered as a result of GATA-3 induction (García-Ojeda et al. 2013; Scripture-Adams et al. 2013). The NK and DC/myeloid pathways are molecularly dissimilar, and most likely are independently controlled. However, what they have in common is that they are both able to be blocked in an ongoing way by environmental Notch signaling, even though they are still available to PU.1-expressing, Bcl11b-negative DN2a cells if Notch signaling is withdrawn (Balciunaite et al. 2005; Masuda et al. 2007; Bell and Bhandoola 2008; Wada et al. 2008; Yui et al. 2010). Only as cells turn on Bcl11b and turn off PU.1, they lose access to both alternatives whether Notch signals are present or not. One feature that makes the transition abrupt is that the cells lose the ability to survive in the absence of Notch signals. They thus become addicted to conditions that block NK, DC and myeloid development alike. The cells remain Notch dependent thereafter until they can express a productive pre-TCR or γδTCR complex.
Bcl11b action is directly required for this step of T-lineage commitment (Ikawa et al. 2010; Li et al. 2010a; Li et al. 2010b). If the Bcl11b gene is deleted, progenitors can still develop in response to Notch-Delta signals all the way to the DN2a stage, but then progression is arrested. The cells turn on the earliest sets of T-cell genes but fail to switch completely to the transcriptional program that characterizes the DN2b/DN3a state (Li et al. 2010a). Importantly, cells that have developed to this stage with ample Notch and cytokine signaling do not die without Bcl11b; instead, they can keep proliferating as DN2a-like cells as long as these signals are provided. If Notch signaling becomes limiting, however, they rapidly tend to switch to an NK cell-like fate (Li et al. 2010a; Li et al. 2010b). Furthermore, cells that have developed without Bcl11b do not give up their access to myeloid fates, as can be seen if a different set of supportive cytokines is provided (Ikawa et al. 2010; Li et al. 2010a). By comparing the effects of Bcl11b deletion from the start of T-cell development with the effects of Bcl11b removal at the DN3a stage, it appears that Bcl11b may play two different roles (L. L., J. A. Zhang, and E.V.R., unpublished results). In one role, it is a direct, continuously required repressor of NK and NKT-like functions (Li et al. 2010b), apparently with the ability to repress the NK-cell regulator Id2 directly (Kastner et al. 2010)(Fig. 3). In the other role, it is an indispensable component required for a hit-and-run process of DN2a to DN2b progression, which excludes myeloid lineage fates, a role that is correlated with the eventual silencing of Sfpi1 and many other phase 1 genes (Ikawa et al. 2010; Li et al. 2010a)(L. L., J. A. Zhang, and E. V. R., unpublished results).
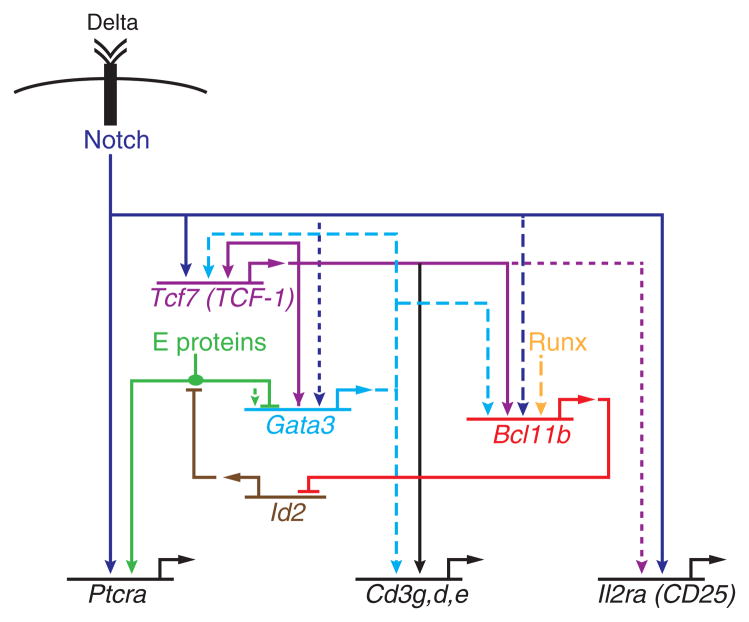
Figure 3.
Feedforward motifs in a gene regulatory network model for T-cell specification. Schematic shows a partial gene regulatory network contributing to the activation and impact of Bcl11b expression. Solid lines: experimental evidence for regulatory impact and direct binding. Dashed lines: evidence for direct binding, possible but not yet demonstrated function at those sites. Dotted lines: evidence for functional effect.
Gene network requirements for commitment probed through Bcl11b regulation
Commitment thus depends on fulfilling the regulatory requirements for inducing Bcl11b. This is a process that involves active removal of epigenetic silencing marks, since the Bcl11b gene begins with strong CpG methylation at multiple regulatory sites (Ji et al. 2010) and histone H3K27me3 modifications persisting all the way into the DN2a stage (Zhang et al. 2012; Li et al. 2013). Both are removed as the cells turn the gene on. One required input is Notch. Bcl11b expression is activated only in cells that have been exposed to Notch signaling, and there are clear binding sites for Notch/RBPJ around its promoter and 5′ introns (Li et al. 2010b). However, Notch/RBPJ is not the only transcription factor controlling its induction. Once induced, Bcl11b expression persists even when Notch signals are removed (Del Real and Rothenberg 2013)(H.Y.K., M.A.Y., and E.V.R., unpublished data). Starting from purified DN2a cells that have not yet turned on Bcl11b expression, we have tested the effects of Notch ligand density on Bcl11b activation in individual cells with a Bcl11b-IRES-fluorescent protein reporter. Quantitative analysis of this system shows that the intensity of Notch-Delta signals affects the frequency of cells turning on Bcl11b much more than it affects the level of Bcl11b expression per cell (H.Y.K., M.A.Y., and E.V.R., unpublished data). Thus, Notch signals may be mostly permissive; other factors must drive the magnitude of actual transcriptional activity. This conclusion actually agrees well with the long period of Notch signaling required before the Bcl11b gene is turned on. Additional factors that are genetically implicated through gain or loss of function experiments include Runx/CBFβ (Guo et al. 2008), GATA-3 (García-Ojeda et al. 2013), and TCF-1 (Weber et al. 2011).
Validating the roles of these inputs depends on identification of concrete protein-DNA interactions needed for regulation. The promoter-proximal region of Bcl11b is not sufficient to drive T-lineage specific expression of a reporter in transfected cells: its expression is similar in pro-T and myeloid cell lines (Li et al. 2013). To look for additional regulatory elements, therefore, we have recently used a strategy based on tracking changes in the status of histone marks during the stages when Bcl11b is first turned on, scanning a region on the order of ~2Mb around the Bcl11b gene. In a gene desert over 800 kb downstream of Bcl11b, we located a region which shifted from repressive to active histone marks across the same developmental interval in which this occurred at the Bcl11b promoter, and we found evidence by chromatin conformation capture experiments that this marked region loops to the Bcl11b promoter/first intron region in a T-lineage specific way (Li et al. 2013). This region has T-lineage cis-regulatory activity in a variety of assays, cooperating specifically with sequences near the Bcl11b promoter and first intron. In a stable transfection system where even a Bcl11b locus bacterial artificial chromosome construct spanning tens of kilobases upstream and downstream of the gene fails to give expression, addition of the far-distal cis-regulatory element is sufficient to confer T-lineage specific expression. This provides a system in which specific transcription factor-target site interactions can be tested for direct causal roles in Bcl11b regulation. In fact, Runx1 and TCF-1 not only bind to activation-marked sites in the first intron and upstream of the Bcl11b promoter, but also in this far-downstream cis-regulatory element. In the far-distal element, both a TCF-1/Runx binding site cluster and a second pair of TCF-1 binding sites are functionally important (Li et al. 2013). GATA-3 may also exert its effects through this element, which it binds weakly, or through additional distal cis-regulatory elements which remain to be characterized. The evidence thus provides good support for a feed-forward network circuit in which not only Notch signals, but also products of the Notch-induced factors TCF-1 (Tcf7) and probably GATA-3 as well, together play essential roles in opening up Bcl11b for expression (Fig. 3).
Phase 1 regulatory genes and the need for repression
The elaborate machinery that seems to be needed to turn on Bcl11b begs the question of what the cells are doing through all the cell cycles they traverse before they undergo commitment. The best estimate we have been able to calculate, using the in vivo repopulation data reported by Petrie and coworkers (Porritt et al. 2003), is that at least ten cell cycles elapse between the time of thymus-settling precursor entry into the thymus and the commitment step when cells turn on Bcl11b (Manesso et al. 2013). In the meantime, the uncommitted ETP and DN2a cells, as purified ex vivo or from in vitro differentiation cultures, are consistently found to express a large set of regulatory genes associated with non-T alternative lineage differentiation, stem cell self-renewal, and/or T-cell leukemogenesis (Table 1)(Tabrizifard et al. 2004; Kawazu et al. 2007; Tydell et al. 2007; David-Fung et al. 2009; McCormack et al. 2010; Belyaev et al. 2012; Zhang et al. 2012; Mingueneau et al. 2013). Any concern that this gene expression pattern might just reflect the difficulty of purifying these early cells away from alternative-lineage contaminants is greatly reduced by finding that the expression of these genes can indeed be stable in cells with early T-lineage markers. Bcl11b-knockout cells, even after repeated sorting for DN2a phenotype and propagation for weeks in culture, continue to express these genes. In cells with normal Bcl11b function, analysis of developing T-lineage cells with dual fluorescent reporters for PU.1 and Bcl11b shows that the same cells that turn on Bcl11b are also initially expressing high levels of PU.1 (H.Y.K., unpublished results). Thus until Bcl11b can be activated, the expression of these stem/progenitor genes demarcates a sustainable “phase 1” part of the T-cell development program.
TABLE 1.
“Phase 1” transcription factors maintained in early T lineage cells
| Characteristic | Genes |
|---|---|
| Expressed until mid ETP stage* | Meis1, Lmo2, Mef2c, Hoxa9 (transcription factors) Flt3 (growth factor receptor) |
| Expressed in two phases, decreasing ETP to DN2a but not silent until after DN3* | Bcl11a, Erg (transcription factors) |
| Expressed in ETP and DN2a, decreasing sharply at DN2b* | Tal1, Gfi1b, Hhex, Sfpi1 (PU.1) |
| Expressed through DN2b stage, then off | Lyl1 Kit (growth factor receptor) |
| Roles in stem cell self-renewal† | Meis1, Tal1, Lyl1, Hhex, Gfi1b, Erg, Lmo2, Kit |
| Roles in lymphomyeloid progenitor self-renewal† | Bcl11a, Lyl1, Kit, Flt3, Hoxa9 |
| T-cell proto-oncogenes† | Tal1, Lyl1, Lmo2, Hhex, Erg, Sfpi1 (if overexpressed), Mef2c |
Expression patterns compiled from RNA-seq, qPCR and microarray data (David-Fung et al. 2009; Yui et al. 2010; Belyaev et al. 2012; Zhang et al. 2012; Mingueneau et al. 2013). The ETP-specific genes are expressed more strongly in samples taken ex vivo from the thymus than from samples of cells differentiating in vitro on OP9-DL stroma.
For functional role groupings, see references in text.
It is obvious that the phase 1 genes must be repressed in normal T-cell development. Aberrant later activity of genes like Tal1, Lyl1, and Lmo2 is a major contributor to T-cell malignancy, while Erg, Hhex, Mef2c, and Sfpi1 deregulation can also promote malignancy (Palomero et al. 2006; Rosenbauer et al. 2006; Zhong et al. 2007; McCormack et al. 2010; Tremblay et al. 2010; Homminga et al. 2011; Thoms et al. 2011). Failure of cells to repress phase 1 genes completely in the DN3 stage, before activating genes associated with β-selection, also appears to underlie a spontaneous high-penetrance T-cell malignancy that we have recently described in NOD background thymocytes (Yui et al. 2013). Moreover, it appears that more than one repressive switch mechanism needs to be used in a coordinated way to ensure normal development. The phase 1 genes vary substantially in the fine-scale timing of their repression and in the degrees to which H3K27me3 repressive marks need to be deposited by Polycomb Repressive Complex 2 to keep them off (Zhang et al. 2012). In the case of PU.1, something is known about the repression machinery that normally silences it, even though PU.1 remains unmarked with H3K27me3. Runx family transcription factors switch from being activators of PU.1 expression to repressors (Huang et al. 2008). At least in part this is because they become engaged at a T-lineage specific silencer element that may quench the activation effects of the major Sfpi1 enhancer (Zarnegar et al. 2010; Zarnegar and Rothenberg 2012). It is tempting to speculate that the Runx-containing repression complex also might include Bcl11b, although this remains to be shown. Other phase 1 genes may use quite different silencing machineries.
Deployment of a phase 1 regulator: the case of PU.1
Is the phase 1 regulatory state actually important for T-cell development, or simply a “problem” that requires ten cell cycles to be resolved? We have investigated this question by focusing on the phase 1 transcription factor PU.1, to determine how it may contribute to T-cell development during the ETP and DN2a stages. This factor poses the question in particularly simple terms. PU.1 can straightforwardly oppose commitment of early T cells, because it is such a powerful driver of genes used in myeloid and dendritic cell fates. It has well-documented genome-wide profiles of target gene binding in different non-T contexts (Ghisletti et al. 2010; Heinz et al. 2010; Pham et al. 2013). Interestingly however, despite its limited role in T-cell precursors, PU.1 can have T-lineage specific targets as well. In ETP, DN2a, and DN2b cells, we find PU.1 binding a distinctive pattern of sites across the genome that differ significantly from the patterns it occupies in macrophage, B, and pre-pro-B cell nuclei. The differences include preferential recruitment to T-lineage enriched sites as well as exclusion from T-lineage closed sites (Zhang et al. 2012). This suggests that PU.1 participates not only in priming the cells for diversion to non-T fates, but also in some functions that are intrinsic to the T-cell program.
Such functions can be suspected because PU.1 is actually required for early T-cell development. When the PU.1 gene is inactivated by Cre deletion in adults, the pipeline for T-cell development is emptied, as well as for the development of obviously PU.1-expressing cell types like macrophages, dendritic cells, and B cells (Dakic et al. 2005; Carotta et al. 2010b). Thus, despite its visibility as a factor that keeps open the door to non-T lineage fates, this is not the most interesting role it can play for the T-cell development program as such. What is it doing for the cells, when it is not threatening them with a shift to a dendritic cell or macrophage identity? We do know that PU.1 impacts on gene expression are highly dependent on the status of Notch signaling within the cells, and Notch signals may help to shape these occupancy profiles. When Notch signals are withdrawn, PU.1 can harshly downregulate numerous T-cell genes and activate a myeloid program, whereas in the presence of Notch signaling, PU.1 has different and more limited effects (Franco et al. 2006; Laiosa et al. 2006; Del Real and Rothenberg 2013). Despite its high levels in ETP stage cells, PU.1 does not block the onset of GATA-3 (or TCF-1) expression in response to Notch signals. Thus, over the next cell cycles if Notch signaling is sustained, PU.1 activity is exerted in a timecourse overlapping with that of the T-cell factors. PU.1 binding occupancy is preferentially correlated with positive transcriptional activity of the linked genes, in early T-lineage cells generally (Zhang et al. 2012). Indeed, it is found associated with promoters and/or histone-marked distal regions of a large fraction of the genes that are expressed during the stages when it is present. At many phase 1 gene loci, the eventual loss of PU.1 binding as Sfpi1 turns off is followed by a deposition of repressive chromatin marks at the previously bound site. Thus, its binding at these sites in Notch-signaled early T cells is at least permissive and perhaps stimulatory for gene expression.
PU.1 targets and the search for phase 1 regulatory functions
A particularly interesting possibility is that the strong, clean pattern of PU.1 binding sites across the genome of early T cells can lead us to previously undiscovered genes that play crucial roles in the poorly understood phase 1 period of T-cell development. Two aspects of phase 1 gene function would be most interesting to determine. First, population dynamics analysis implies that at least half of the total number of cell divisions that postnatal T-cell precursors undergo in the thymus occur in DN1 and DN2a cells before commitment (Petrie and Zuniga-Pflucker 2007; Manesso et al. 2013). Thus, “under the radar” of TCR-dependent proliferation pathways, there is another entire system that builds the T-cell precursor pool before TCR gene rearrangement and before commitment. The genes that support this process are still poorly characterized. Second, there are developmentally regulated timing functions that determine how long phase 1 can continue (David-Fung et al. 2006). In the fetal thymus, phase 1 is a brief transitory period of few cell cycles, with ETP-phenotype cells that have already turned on many DN2-stage genes on an accelerated schedule (Belyaev et al. 2012). In contrast, the adult ETP population shows little if any anticipation of DN2/DN3 gene expression, and in adults the time spent in ETP and DN2 stages is substantially longer with a greatly increased number of cell cycles (Lu et al. 2005). Some of the phase 1 genes may therefore determine the extent of thymic population expansion by controlling the delay of T-lineage commitment.
There is a long step, however, between showing that PU.1 binds to a promoter or non-promoter element of a gene and showing that PU.1 is responsible for the regulation of that gene. PU.1 binds to thousands of sites in these early T cells, at promoters, intronic sites, and intergenic sites, and although many of them are also sites of “accessible” or “active” histone marking, the participation of PU.1 itself at these sites may be functionally irrelevant in many cases. Even though genes linked to PU.1 binding sites are usually being expressed while PU.1 binds, most of them are neither increased nor decreased in expression subsequently when PU.1 levels drop over tenfold. Therefore, additional methods are needed to identify the regulatory functions for which PU.1 is directly responsible.
Classical gain of function approaches have yielded several positively regulated PU.1 targets encoding transcription factors that could play roles within the phase 1 T cell program. These include Bcl11a and Lyl1 as well as the very early ETP genes, Lmo2 and Mef2c (A. C. and E.V.R., unpublished results)(Del Real and Rothenberg 2013). Adding PU.1 to DN2a/2b cells, which have already begun to turn these genes off, consistently boosts their expression back to ETP levels or higher. Flt3, which is an important cytokine receptor for prethymic cells and thymus-settling precursors, also requires PU.1 for its expression (Carotta et al. 2010a) (A. C. and E.V.R., unpublished results). However, in order to show whether all these genes are normally regulated by PU.1 at its natural phase 1 levels of expression, it is necessary to show what happens to them when PU.1 is taken away. Precisely because PU.1 is critical for the survival of these earliest T-cell precursors, it is very difficult to isolate them after deleting PU.1, to evaluate the effects on phase 1-specific genes. Even when PU.1 is deleted conditionally at the ETP stage, the population that survives is highly enriched for cells that have escaped complete deletion, so that effects are weak (A. C., unpublished results). To go beyond the limited candidate-gene approach to identify the most important PU.1 targets, therefore, it has been important to devise a strategy that can be applied synchronously, and with high penetrance, to wipe out PU.1 activity in phase 1 cells quickly enough to monitor the effects before the cells die.
We have found that PU.1 dominant negatives can be used in gain of function experiments as a powerful component of such a strategy (A.C., unpublished results). We have tested several variants of PU.1 dominant negatives, the most powerful of which delete all but the PU.1 Ets family DNA-binding domain (Kueh et al. 2013) or convert a truncated PU.1 Ets domain-only molecule into an obligate repressor by attaching the repression domain from Drosophila Engrailed (A. C. and E.V.R., unpublished results). Dominant-negative PU.1 antagonizes the macrophage gene expression programs driven by endogenous PU.1 in myeloid precursors (Kueh et al. 2013), and these constructs are specific enough to be innocuous in a T-lineage cell line immortalized at the DN3 stage, which uses multiple other Ets family transcription factors but not PU.1 any longer (A. C. and E.V.R., unpublished results). Even so, they have strong effects within 24 hr in normal early T cells which do utilize PU.1. Two aspects of the results from forced expression of PU.1-engrailed are particularly interesting. First, dominant-negative transduction can be coupled with genome-wide transcriptome analysis to identify new PU.1 target genes. Second, the results give some insight into the role of PU.1 as a factor controlling T-lineage developmental progression timing itself.
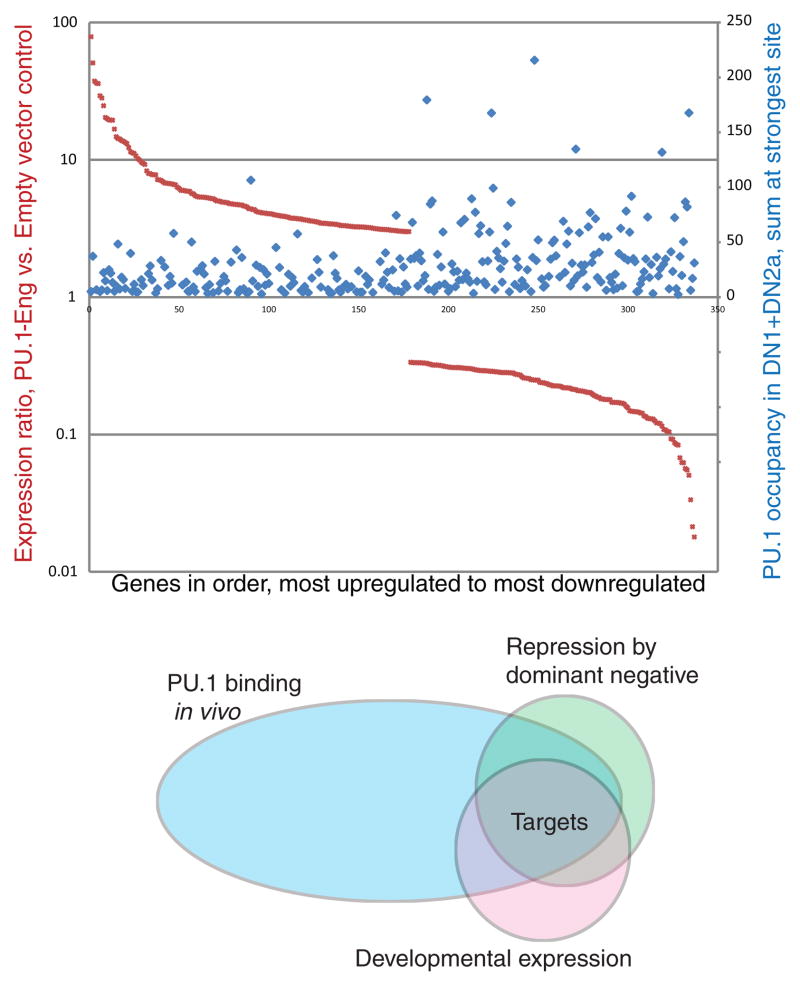
Within 24 hr of transfecting the PU.1 dominant negatives into DN2a/2b T-cell precursors, RNA-seq analysis registers greater than twofold changes in expression of about 1000 genes, about equally divided between increases and decreases in expression. Note that this strategy only predicts the genes repressed by PU.1-engrailed to be direct PU.1 targets; this version of a PU.1 dominant negative should repress genes that natural PU.1 either activates or represses, alike. The genes repressed by PU.1-engrailed are particularly enriched for genes with strong natural PU.1 binding occupancy around them in ETP and DN2a stages, suggesting that the effect depends on direct engagement with high-quality PU.1 sites (Fig. 4, top). The repression is clearly selective for certain binding targets and not others, suggesting that repression only occurs when the dominant negative is recruited to a functionally important cis-regulatory element, not just any PU.1 occupancy site in the genome or even any PU.1 occupancy at a promoter. Thus this search strategy winnows out genes which are highly sensitive to PU.1. Even better selection emerges by filtering PU.1-engrailed repressed genes according to their natural regulation in vivo as PU.1 is turned off (Fig. 4, bottom). Multiple genes we have newly identified as PU.1 targets by these combined criteria have turned out to respond to acute PU.1 deletion too, though these genes include both PU.1-activated and PU.1-repressed genes as well (A. C., S. D., and E.V.R., unpublished results).
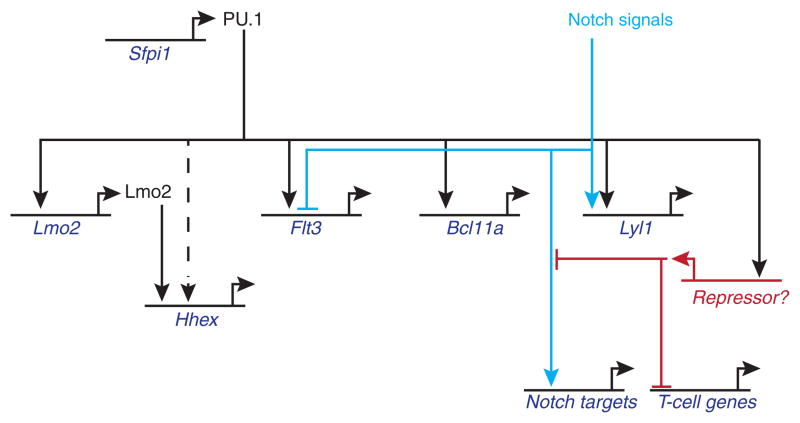
Figure 4.
A dominant negative PU.1-engrailed fusion protein construct represses genes that are enriched for natural PU.1 binding targets in early T cells. Top panel: genome-wide analysis of impact of PU.1-engrailed on expression of target genes, as measured by RNA-seq (dark red symbols, A.C., S. D., and E.V.R., unpublished results), plotted against peak levels of endogenous PU.1 binding associated with the same genes in normal DN1 and DN2a cells (blue symbols)(Zhang et al. 2012). The genes shown were selected for >3x upregulation (left side) or >3x downregulation (right side) by PU.1-engrailed relative to empty vector, 24 hr after transduction, and are ordered on the x axis from most upregulated to most downregulated. Note that the genes repressed by PU.1-engrailed expression (toward the right side) tend to include genes linked to stronger PU.1 binding sites in vivo. In contrast, an equal number of genes are upregulated in cells forced to express PU.1-engrailed, but these do not show any enrichment for strong PU.1 binding, consistent with indirect regulation by PU.1 (A.C., S. D., and E.V.R., unpublished results). Bottom panel: Venn diagram outlining the strategy for combining dominant negative perturbation, mapping of natural PU.1 binding site occupancy, and natural developmental expression patterns to identify new PU.1 targets in an unbiased way.
The other issue that the dominant negative strategy has allowed us to tackle is the way PU.1 intersects with the control of T-cell developmental progression. We discovered that in fact PU.1 quantitatively damps Notch responses in a dose-dependent way as long as it is present in the cells, even though Notch1 itself is expressed and still capable of turning on its own response targets (Fig. 5). In effect, PU.1 sets a threshold for the level of Notch-Delta interaction that is required to elicit a given response (Del Real and Rothenberg 2013). Obtaining a sufficiently strong Notch response is important in turn to keep PU.1 from acting as a net repressor of multiple additional T-cell genes, many of which are not otherwise Notch-dependent. For example, GATA-3 levels can normally remain high in the absence of Notch signals, but PU.1 can repress GATA-3 in the absence of Notch signaling whereas it cannot do this while Notch signaling is sustained (Del Real and Rothenberg 2013). The Notch—PU.1 competition is a dynamic balance that shifts the advantage to the PU.1-dominated or T-cell differentiation programs with changing conditions. If PU.1 is deleted from cells that are progressing from ETP to DN2a stage, in fact the loss now appears to speed up their developmental progression. Up to this point, then, is PU.1 acting as a direct repressor of T-cell differentiation genes?
Figure 5.
A partial gene regulatory network driven by PU.1 in early T-lineage precursors. The figure summarizes representative linkages from the studies described in the text (Franco et al. 2006; Carotta et al. 2010a; Del Real and Rothenberg 2013)(A.C., S.D., S. L. Nutt, S. Carotta, and E.V.R., unpublished results). Note that combinatorial effects from PU.1 and Notch on certain targets can split the expression patterns seen in vivo (e.g. Lyl1 vs. Flt3). Additional phase 1 genes that can be activated by PU.1 in these cells include Mef2c, Meis1, and many myeloid differentiation genes; however, other genes with “phase 1” expression patterns appear to be direct negative, not positive, regulatory targets of PU.1 (not shown; A.C., S.D., S. L. Nutt, S. Carotta, and E.V.R., unpublished results). The effects depicted are those that survive Notch effects on PU.1 activity itself (Franco et al. 2006; Del Real and Rothenberg 2013). Under these conditions at least, PU.1 appears to damp expression of “T-cell genes” such as Tcf7, Ets1, Zfpm1 through an indirect mechanism, based on results with the obligate repressor dominant negative, suggesting the involvement of a PU.1-activated “Repressor X” function(s) (A.C. & E.V.R., unpublished).
The obligate repressor strategy reveals that the mechanism is almost certainly indirect. Rather than repress the T cell genes and Notch response genes, the obligate repressor form of PU.1 instead superactivates them. It can accelerate the onset of expression of DN2-DN3 stage genes even back to the ETP stage. Thus, despite viability problems that eventually overtake them, the cells in which direct PU.1 target genes are repressed begin to develop faster. Thus, PU.1 not only controls expression of a substantial battery of phase 1 regulators and viability genes; it also activates a molecular brake against premature T-cell differentiation, a brake which depends on PU.1 and is ultimately removed (Fig. 5).
Conclusion
The evidence reviewed here shows that precursors entering the thymus must traverse two distinct gene regulatory networks in order to become committed to the T-cell pathway (Fig. 6). The functions one considers integral to mature T cells are conferred upon them in the second network state, the commitment and post-commitment phases. Access to this phase is controlled at least in part by a mechanism that depends on Bcl11b. However, before this stage the cells must operate a completely different gene network, one in which PU.1 plays a role, and one which actively defers the onset of the committed state. We know that PU.1’s role in this network includes both viability functions and two kinds of focused antagonism against the T-cell commitment program – one via damping of Notch signals and the other via another indirect repressive mechanism. Importantly, the impacts of the PU.1 dominant negative or acute PU.1 deletion imply that the indirect repressive mechanism is an important timing controller in normal circumstances (Fig. 6). Thus, the phase 1 genes maintain a stable deferral of T-cell differentiation until the cumulative feed-forward effects of Notch (cf. Fig. 3) finally induce Bcl11b and other regulators that radically shift the balance to extinguish phase 1 gene expression (Fig. 6). This picture of T cell development, transitioning between two mutually opposed network states, suggests why it is so difficult to use single “master regulator” candidate genes to amplify T-cell generation. It explains the significance of the continuity with stem-cell regulatory states. Finally, it suggests that important controllers of the scale and homeostatic setpoints for T-cell population generation may not necessarily be the genes we associate with T-cell differentiation, but rather genes that play their roles during the phase 1 stages, and then allow themselves to be silenced. This picture could therefore have clinical application as well, but only with clear recognition of how effective the cells must be in shutting down the first network when the time comes to shift to the second.
Figure 6.
Two sequential gene regulatory networks compete to control the timing of commitment in T-cell precursors. Summary of discussion in text.
Acknowledgments
We thank our colleagues Michael Elowitz (Caltech), Eric Davidson (Caltech), Carsten Peterson (Lund University), Vijay Chickarmane (Caltech) and Erica Manesso (Lund University) for advice and stimulating theoretical discussions; Marei Dose and Fotini Gounari (University of Chicago) for sharing important ChIP-seq datasets; Irwin Bernstein (Fred Hutchinson Cancer Research Center) for advice and donating DL1-Fc; Stephen Nutt (Walter & Eliza Hall Institute), Mark Leid (Oregon State Health Sciences Center), and Pentao Liu (Cambridge University) for valuable transgenic mouse strains; and Shirley Pease and the Caltech Genetically Modified Mouse Service for generation of new fluorescent reporter mouse strains, all of whom contributed to the experiments reviewed here. We are also indebted to Rochelle Diamond for outstanding laboratory management; Robert Butler, Ni Feng, Maria Lerica Gutierrez Quiloan, and Edward Hernandez for technical support; Diana Perez, Janice Grimm, and Josh Verceles for help with cell sorting; and Ingrid Soto for mouse care. Our work was made possible by support from NIH grants to E.V.R. (RC2 CA148278, R33 HL089123, R01 CA090233, R01 AI083514, and R01 AI095943) and to M.A.Y. (R01 AI064590), a Cancer Research Institute-Irvington Fellowship to H.Y.K., the Albert Billings Ruddock Professorship to E.V.R., the Al Sherman Foundation, and the Louis A. Garfinkle Memorial Laboratory Fund.
References
- Anderson MK, Weiss AH, Hernandez-Hoyos G, Dionne CJ, Rothenberg EV. Constitutive expression of PU.1 in fetal hematopoietic progenitors blocks T cell development at the pro-T cell stage. Immunity. 2002;16:285–296. doi: 10.1016/s1074-7613(02)00277-7. [DOI] [PubMed] [Google Scholar]
- Balciunaite G, Ceredig R, Rolink AG. The earliest subpopulation of mouse thymocytes contains potent T, significant macrophage, and natural killer cell but no B-lymphocyte potential. Blood. 2005;105:1930–1936. doi: 10.1182/blood-2004-08-3087. [DOI] [PubMed] [Google Scholar]
- Bell JJ, Bhandoola A. The earliest thymic progenitors for T cells possess myeloid lineage potential. Nature. 2008;452:764–767. doi: 10.1038/nature06840. [DOI] [PubMed] [Google Scholar]
- Belyaev NN, Biro J, Athanasakis D, Fernandez-Reyes D, Potocnik AJ. Global transcriptional analysis of primitive thymocytes reveals accelerated dynamics of T cell specification in fetal stages. Immunogenetics. 2012;64:591–604. doi: 10.1007/s00251-012-0620-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carotta S, Dakic A, D’Amico A, Pang SH, Greig KT, Nutt SL, Wu L. The transcription factor PU.1 controls dendritic cell development and Flt3 cytokine receptor expression in a dose-dependent manner. Immunity. 2010a;32:628–641. doi: 10.1016/j.immuni.2010.05.005. [DOI] [PubMed] [Google Scholar]
- Carotta S, Wu L, Nutt SL. Surprising new roles for PU.1 in the adaptive immune response. Immunol Rev. 2010b;238:63–75. doi: 10.1111/j.1600-065X.2010.00955.x. [DOI] [PubMed] [Google Scholar]
- Dakic A, Metcalf D, Di Rago L, Mifsud S, Wu L, Nutt SL. PU.1 regulates the commitment of adult hematopoietic progenitors and restricts granulopoiesis. J Exp Med. 2005;201:1487–1502. doi: 10.1084/jem.20050075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David-Fung ES, Butler R, Buzi G, Yui MA, Diamond RA, Anderson MK, Rowen L, Rothenberg EV. Transcription factor expression dynamics of early T-lymphocyte specification and commitment. Dev Biol. 2009;325:444–467. doi: 10.1016/j.ydbio.2008.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David-Fung ES, Yui MA, Morales M, Wang H, Taghon T, Diamond RA, Rothenberg EV. Progression of regulatory gene expression states in fetal and adult pro-T-cell development. Immunol Rev. 2006;209:212–236. doi: 10.1111/j.0105-2896.2006.00355.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Real MM, Rothenberg EV. Architecture of a lymphomyeloid developmental switch controlled by PU.1, Notch and Gata3. Development. 2013;140:1207–1219. doi: 10.1242/dev.088559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dionne CJ, Tse KY, Weiss AH, Franco CB, Wiest DL, Anderson MK, Rothenberg EV. Subversion of T lineage commitment by PU.1 in a clonal cell line system. Dev Biol. 2005;280:448–466. doi: 10.1016/j.ydbio.2005.01.027. [DOI] [PubMed] [Google Scholar]
- Franco CB, Scripture-Adams DD, Proekt I, Taghon T, Weiss AH, Yui MA, Adams SL, Diamond RA, Rothenberg EV. Notch/Delta signaling constrains reengineering of pro-T cells by PU.1. Proc Natl Acad Sci U S A. 2006;103:11993–11998. doi: 10.1073/pnas.0601188103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Ojeda ME, Klein Wolterink RG, Lemaître F, Richard-Le Goff O, Hasan M, Hendriks RW, Cumano A, Di Santo JP. GATA-3 promotes T cell specification by repressing B cell potential in pro-T cells. Blood. 2013;121:1749–1759. doi: 10.1182/blood-2012-06-440065. [DOI] [PubMed] [Google Scholar]
- Ghisletti S, Barozzi I, Mietton F, Polletti S, De Santa F, Venturini E, Gregory L, Lonie L, Chew A, Wei CL, et al. Identification and characterization of enhancers controlling the inflammatory gene expression program in macrophages. Immunity. 2010;32:317–328. doi: 10.1016/j.immuni.2010.02.008. [DOI] [PubMed] [Google Scholar]
- Guo Y, Maillard I, Chakraborti S, Rothenberg EV, Speck NA. Core binding factors are necessary for natural killer cell development, and cooperate with Notch signaling during T cell specification. Blood. 2008;112:480–492. doi: 10.1182/blood-2007-10-120261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homminga I, Pieters R, Langerak AW, de Rooi JJ, Stubbs A, Verstegen M, Vuerhard M, Buijs-Gladdines J, Kooi C, Klous P, et al. Integrated transcript and genome analyses reveal NKX2-1 and MEF2C as potential oncogenes in T cell acute lymphoblastic leukemia. Cancer Cell. 2011;19:484–497. doi: 10.1016/j.ccr.2011.02.008. [DOI] [PubMed] [Google Scholar]
- Huang G, Zhang P, Hirai H, Elf S, Yan X, Chen Z, Koschmieder S, Okuno Y, Dayaram T, Growney JD, et al. PU.1 is a major downstream target of AML1 (RUNX1) in adult mouse hematopoiesis. Nat Genet. 2008;40:51–60. doi: 10.1038/ng.2007.7. [DOI] [PubMed] [Google Scholar]
- Ikawa T, Hirose S, Masuda K, Kakugawa K, Satoh R, Shibano-Satoh A, Kominami R, Katsura Y, Kawamoto H. An essential developmental checkpoint for production of the T cell lineage. Science. 2010;329:93–96. doi: 10.1126/science.1188995. [DOI] [PubMed] [Google Scholar]
- Ji H, Ehrlich LI, Seita J, Murakami P, Doi A, Lindau P, Lee H, Aryee MJ, Irizarry RA, Kim K, et al. Comprehensive methylome map of lineage commitment from haematopoietic progenitors. Nature. 2010;467:338–342. doi: 10.1038/nature09367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastner P, Chan S, Vogel WK, Zhang LJ, Topark-Ngarm A, Golonzhka O, Jost B, Le Gras S, Gross MK, Leid M. Bcl11b represses a mature T-cell gene expression program in immature CD4+CD8+ thymocytes. Eur J Immunol. 2010;40:2143–2154. doi: 10.1002/eji.200940258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawazu M, Yamamoto G, Yoshimi M, Yamamoto K, Asai T, Ichikawa M, Seo S, Nakagawa M, Chiba S, Kurokawa M, et al. Expression profiling of immature thymocytes revealed a novel homeobox gene that regulates double-negative thymocyte development. J Immunol. 2007;179:5335–5345. doi: 10.4049/jimmunol.179.8.5335. [DOI] [PubMed] [Google Scholar]
- Kueh HY, Champhekhar A, Nutt SL, Elowitz MB, Rothenberg EV. Positive feedback between PU.1 and the cell cycle controls myeloid differentiation. Science. 2013;341:670–673. doi: 10.1126/science.1240831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laiosa CV, Stadtfeld M, Xie H, de Andres-Aguayo L, Graf T. Reprogramming of committed T cell progenitors to macrophages and dendritic cells by C/EBPα and PU.1 transcription factors. Immunity. 2006;25:731–744. doi: 10.1016/j.immuni.2006.09.011. [DOI] [PubMed] [Google Scholar]
- Lefebvre JM, Haks MC, Carleton MO, Rhodes M, Sinnathamby G, Simon MC, Eisenlohr LC, Garrett-Sinha LA, Wiest DL. Enforced expression of Spi-B reverses T lineage commitment and blocks β-selection. J Immunol. 2005;174:6184–6194. doi: 10.4049/jimmunol.174.10.6184. [DOI] [PubMed] [Google Scholar]
- Li L, Leid M, Rothenberg EV. An early T cell lineage commitment checkpoint dependent on the transcription factor Bcl11b. Science. 2010a;329:89–93. doi: 10.1126/science.1188989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Zhang JA, Dose M, Kueh HY, Mosadeghi R, Gounari F, Rothenberg EV. A far downstream enhancer for murine Bcl11b controls its T-cell specific expression. Blood. 2013;122:902–911. doi: 10.1182/blood-2012-08-447839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Burke S, Wang J, Chen X, Ortiz M, Lee SC, Lu D, Campos L, Goulding D, Ng BL, et al. Reprogramming of T cells to natural killer-like cells upon Bcl11b deletion. Science. 2010b;329:85–89. doi: 10.1126/science.1188063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu M, Tayu R, Ikawa T, Masuda K, Matsumoto I, Mugishima H, Kawamoto H, Katsura Y. The earliest thymic progenitors in adults are restricted to T, NK, and dendritic cell lineage and have a potential to form more diverse TCRβ chains than fetal progenitors. J Immunol. 2005;175:5848–5856. doi: 10.4049/jimmunol.175.9.5848. [DOI] [PubMed] [Google Scholar]
- Luc S, Luis TC, Boukarabila H, Macaulay IC, Buza-Vidas N, Bouriez-Jones T, Lutteropp M, Woll PS, Loughran SJ, Mead AJ, et al. The earliest thymic T cell progenitors sustain B cell and myeloid lineage potential. Nat Immunol. 2012;13:412–419. doi: 10.1038/ni.2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manesso E, Chickarmane V, Kueh HY, Rothenberg EV, Peterson C. Computational modelling of T-cell formation kinetics: output regulated by initial proliferation-linked deferral of developmental competence. J R Soc Interface. 2013;10:20120774. doi: 10.1098/rsif.2012.0774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda K, Kakugawa K, Nakayama T, Minato M, Katsura Y, Kawamoto H. T cell lineage determination precedes the initiation of TCRβ rearrangement. J Immunol. 2007;179:3699–3706. doi: 10.4049/jimmunol.179.6.3699. [DOI] [PubMed] [Google Scholar]
- McCormack MP, Young LF, Vasudevan S, de Graaf CA, Codrington R, Rabbitts TH, Jane SM, Curtis DJ. The Lmo2 oncogene initiates leukemia in mice by inducing thymocyte self-renewal. Science. 2010;327:879–883. doi: 10.1126/science.1182378. [DOI] [PubMed] [Google Scholar]
- Mingueneau M, Kreslavsky T, Gray D, Heng T, Cruse R, Ericson J, Bendall S, Spitzer MH, Nolan GP, Kobayashi K, et al. The transcriptional landscape of αβ T cell differentiation. Nat Immunol. 2013;14:619–632. doi: 10.1038/ni.2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito T, Tanaka H, Naoe Y, Taniuchi I. Transcriptional control of T-cell development. International immunology. 2011;23:661–668. doi: 10.1093/intimm/dxr078. [DOI] [PubMed] [Google Scholar]
- Palomero T, Odom DT, O’Neil J, Ferrando AA, Margolin A, Neuberg DS, Winter SS, Larson RS, Li W, Liu XS, et al. Transcriptional regulatory networks downstream of TAL1/SCL in T-cell acute lymphoblastic leukemia. Blood. 2006;108:986–992. doi: 10.1182/blood-2005-08-3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrie HT, Zuniga-Pflucker JC. Zoned out: functional mapping of stromal signaling microenvironments in the thymus. Annu Rev Immunol. 2007;25:649–679. doi: 10.1146/annurev.immunol.23.021704.115715. [DOI] [PubMed] [Google Scholar]
- Pham T-H, Minderjahn J, Schmidl C, Hoffmeister H, Schmidhofer S, Chen W, Langst G, Benner C, Rehli M. Mechanisms of in vivo binding site selection of the hematopoietic master transcription factor PU.1. Nucleic Acids Res. 2013;41:6391–6402. doi: 10.1093/nar/gkt355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porritt HE, Gordon K, Petrie HT. Kinetics of steady-state differentiation and mapping of intrathymic-signaling environments by stem cell transplantation in nonirradiated mice. J Exp Med. 2003;198:957–962. doi: 10.1084/jem.20030837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radtke F, Fasnacht N, MacDonald HR. Notch signaling in the immune system. Immunity. 2010;32:14–27. doi: 10.1016/j.immuni.2010.01.004. [DOI] [PubMed] [Google Scholar]
- Rosenbauer F, Owens BM, Yu L, Tumang JR, Steidl U, Kutok JL, Clayton LK, Wagner K, Scheller M, Iwasaki H, et al. Lymphoid cell growth and transformation are suppressed by a key regulatory element of the gene encoding PU.1. Nat Genet. 2006;38:27–37. doi: 10.1038/ng1679. [DOI] [PubMed] [Google Scholar]
- Rothenberg EV. Transcriptional drivers of the T-cell lineage program. Curr Opin Immunol. 2012;24:132–138. doi: 10.1016/j.coi.2011.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothenberg EV, Moore JE, Yui MA. Launching the T-cell-lineage developmental programme. Nat Rev Immunol. 2008;8:9–21. doi: 10.1038/nri2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothenberg EV, Zhang J, Li L. Multilayered specification of the T-cell lineage fate. Immunol Rev. 2010;238:150–168. doi: 10.1111/j.1600-065X.2010.00964.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scripture-Adams DD, Li L, Arias AM, Elihu KJ, Butler I, RR, Champhekar A, Zhang JA, Rothenberg EV. GATA-3 dose-dependent checkpoints in early T cell commitment. J Immunol. 2013 doi: 10.4049/jimmunol.1301663. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabrizifard S, Olaru A, Plotkin J, Fallahi-Sichani M, Livak F, Petrie HT. Analysis of transcription factor expression during discrete stages of postnatal thymocyte differentiation. J Immunol. 2004;173:1094–1102. doi: 10.4049/jimmunol.173.2.1094. [DOI] [PubMed] [Google Scholar]
- Thompson PK, Zúñiga-Pflücker JC. On becoming a T cell, a convergence of factors kick it up a Notch along the way. Semin Immunol. 2011;23:350–359. doi: 10.1016/j.smim.2011.08.007. [DOI] [PubMed] [Google Scholar]
- Thoms JAI, Birger Y, Foster S, Knezevic K, Kirschenbaum Y, Chandrakanthan V, Jonquieres G, Spensberger D, Wong JW, Oram SH, et al. ERG promotes T-acute lymphoblastic leukemia and is transcriptionally regulated in leukemic cells by a stem cell enhancer. Blood. 2011;117:7079–7089. doi: 10.1182/blood-2010-12-317990. [DOI] [PubMed] [Google Scholar]
- Tremblay M, Tremblay CS, Herblot S, Aplan PD, Hebert J, Perreault C, Hoang T. Modeling T-cell acute lymphoblastic leukemia induced by the SCL and LMO1 oncogenes. Genes Dev. 2010;24:1093–1105. doi: 10.1101/gad.1897910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tydell CC, David-Fung ES, Moore JE, Rowen L, Taghon T, Rothenberg EV. Molecular dissection of prethymic progenitor entry into the T lymphocyte developmental pathway. J Immunol. 2007;179:421–438. doi: 10.4049/jimmunol.179.1.421. [DOI] [PubMed] [Google Scholar]
- Wada H, Masuda K, Satoh R, Kakugawa K, Ikawa T, Katsura Y, Kawamoto H. Adult T-cell progenitors retain myeloid potential. Nature. 2008;452:768–772. doi: 10.1038/nature06839. [DOI] [PubMed] [Google Scholar]
- Weber BN, Chi AW, Chavez A, Yashiro-Ohtani Y, Yang Q, Shestova O, Bhandoola A. A critical role for TCF-1 in T-lineage specification and differentiation. Nature. 2011;476:63–68. doi: 10.1038/nature10279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan JS, Kousis PC, Suliman S, Visan I, Guidos CJ. Functions of notch signaling in the immune system: consensus and controversies. AnnuRevImmunol. 2010;28:343–365. doi: 10.1146/annurev.immunol.021908.132719. [DOI] [PubMed] [Google Scholar]
- Yui MA, Feng N, Rothenberg EV. Fine-scale staging of T cell lineage commitment in adult mouse thymus. J Immunol. 2010;185:284–293. doi: 10.4049/jimmunol.1000679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yui MA, Feng N, Zhang JA, Liaw CY, Rothenberg EV, Longmate JA. Loss of T cell progenitor checkpoint control underlies leukemia initiation in Rag1-deficient nonobese diabetic mice. J Immunol. 2013;190:3276–3288. doi: 10.4049/jimmunol.1202970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarnegar MA, Chen J, Rothenberg EV. Cell-type-specific activation and repression of PU.1 by a complex of discrete, functionally specialized cis-regulatory elements. Mol Cell Biol. 2010;30:4922–4939. doi: 10.1128/MCB.00354-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarnegar MA, Rothenberg EV. Ikaros represses and activates PU.1 cell-type-specifically through the multifunctional Sfpi1 URE and a myeloid specific enhancer. Oncogene. 2012;31:4647–4654. doi: 10.1038/onc.2011.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JA, Mortazavi A, Williams BA, Wold BJ, Rothenberg EV. Dynamic transformations of genome-wide epigenetic marking and transcriptional control establish T cell identity. Cell. 2012;149:467–482. doi: 10.1016/j.cell.2012.01.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Y, Jiang L, Hiai H, Toyokuni S, Yamada Y. Overexpression of a transcription factor LYL1 induces T- and B-cell lymphoma in mice. Oncogene. 2007;26:6937–6947. doi: 10.1038/sj.onc.1210494. [DOI] [PubMed] [Google Scholar]