Abstract
Initiation is often assumed to be rate-limiting for protein synthesis, but the presence of rare codons nonetheless can influence protein levels. In this issue of The EMBO Journal, Chu et al report that when rare codons are positioned near the start of the coding region, ‘liberation’ of the initiation codon for loading of the next 40S subunit may be rate-limiting for initiation and therefore overall protein synthesis. The sequential nature of translation results in an interdependence in ribosome association either by de novo initiation or recycling. Thus, a more general view emerges where both elongation and initiation can contribute to protein expression.
→See related article http://dx.doi.org/10.1002/embj.201385651
The field of protein synthesis has largely been indoctrinated in the concept that the first committed step in the pathway, ‘initiation’, is rate-limiting. Indeed, there are more than 30 polypeptides associated with initiation, and some 40+ phosphates deposited on different initiation factors. The dominant regulation by the eIF2α kinases and the phosphorylation status of 4E-BP has been well characterized. The former limits the amount of available ternary complex (eIF2•GTP•Met-tRNAi) and the latter restricts the level of active eIF4F. Now, the paper by Chu et al provides support for a more general perspective on the mechanistic coupling between initiation and elongation.
A simplified kinetic model helps to understand the relationship between ribosome association and elongation rates on protein synthesis. Translation can be considered to be a series of sequential reactions in which initiation proceeds with rate constant kinit and is followed by ‘liberation’ of the initiation site at a rate constant klib (Cleland, 1975). The rate constant kinit encompasses ribosome binding and can depend on the concentrations of mRNA and free 40S subunits, and therefore will be the point of regulation under conditions where their concentrations are rate-limiting. The rate constant klib represents movement of the ribosome a sufficient distance from the start codon to allow binding of an additional 40S subunit with rate constant kinit. The rate constant klib is dictated by the individual rate constants for elongation of each codon within the distance that must be cleared to allow another round of initiation.
It follows that since initiation, clearance and elongation are sequential that their relative magnitudes will dictate which will be rate-limiting and therefore the point of regulation for protein expression (Ray, 1983). This can be seen in Fig 1 which shows the magnitude of the overall net rate constant for initiation (kobs) as a function of the intrinsic rate constants for ribosome binding (kinit) and for liberation of the initiation site (klib). When liberation is fast relative to the range of kinit values, the overall observed rate constant for initiation is highly sensitive to kinit. In contrast, when liberation is slow there is little effect of the change in magnitude of kinit. The converse is true and under conditions where kinit is fast relative to klib, changes in klib will affect the net rate constant for initiation. Differences in klib can arise from intrinsic differences in rare codons between mRNAs or from engineering in rare codons as done in the paper by Chu et al. Also, it follows that if the net rate constant for subsequent steps in the translation of the mRNA are fast relative to the rate constant for clearance, then the clearance step(s) will overall be rate limiting.
Figure 1.
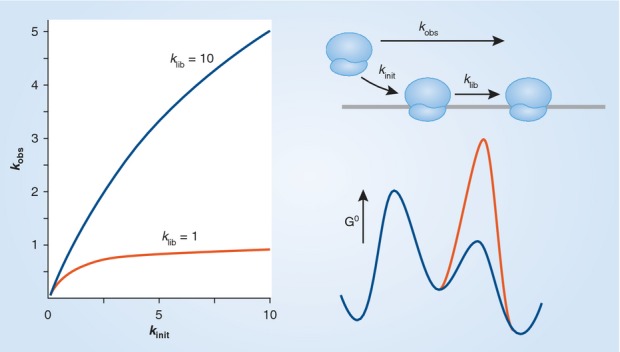
In this simple example the net rate constant for initiation is kobs = kinit klib / (kinit + klib). On the left is an example of the cases in which klib is fast relative to kinit (blue line, klib = 10) and in which klib is slow relative to kinit (red line, klib = 1). These rate constants are pseudo-first order and have units of reciprocal time. On the right is a Gibbs free energy profile of the extreme cases in which kinit is rate limiting (blue line) or klib (red line) is rate limiting.
It follows that all pathways for initiation will be sensitive to clearance, particularly recycling. The documented interaction of the 5′ end of the mRNA (via eIF4F) with the 3′ end of the mRNA (via the poly(A) binding protein) (Wells et al, 1998) provided the explanation for the enhanced translational efficiency of mRNAs that contained an m7G cap and a poly(A) tail. Upon termination, 40S subunits would be recruited more efficiently to the 5′ end of the mRNA than would occur if a free 40S subunit in solution was to be recruited. Consistent with this interpretation was the previous report that formation of full-sized polysomes took considerably longer than just the ribosome transit time for the coding region of the mRNA (Nelson & Winkler, 1987). Here, the explanation was that the more efficient utilization of 40S subunits at termination for initiation tended to outcompete free 40S subunits from binding to the mRNA and thereby, delaying the time required for maximal polysome formation.
A few additional facts germane to understanding rate-limiting steps in initiation are: in most reports of polysome profiles from log-phase growth, the amount of 40S, 60S and 80S ribosome (the latter which may or may not represent new initiations) is roughly 20 to 30% of the total ribosome pool. As a consequence, approximately 80% (or more) of the 40S subunits are in polysomes. Secondly, from estimates of the levels of initiation factor proteins, the most common numbers come out to be roughly 0.3–1.0 mole of factor per mole of ribosome (John Hershey and Christopher Fraser, personal communication). Thus, if 70–80% of the ribosomes are associated with elongation, then there is more than an equal molar amount of initiation factor for newly initiating 40S subunits (note, the initiation factors could still be limiting if the affinity for the 40S subunit was poor). Thus, understanding the individual steps that contribute to kinit is the key to understanding translational control.
The article by Chu et al clearly shows several variations where changes in the elongation rate influence protein expression as manipulated through the use of rare codons. In a similar study by Shah and coworkers (Shah et al, 2013), they developed a whole cell model for the study of protein synthesis in yeast. In this, the authors concluded that ‘protein production in healthy yeast cells is typically limited by the availability of free ribosomes’. Where these two come together is that the availability of free 40S subunits is limited by 40S subunits being released at termination, not by free 40S subunits in solution. It should be noted that the Chu et al article and previous publications implying control at the level of elongation (i.e. Nielsen & McConkey, 1980) were based upon cells in log-phase growth. It still remains that initiation factors can be the dominant control point under conditions of reduced protein synthesis and that the global control through restricted levels of ternary complex or restricted levels of eIF4F activity actually have different consequences (Merrick, 2003, 2010).
Thus, the rate-limiting step in the sequential process of translation is the forward step for which a change in its rate constant produces the largest effect on the overall rate. This fundamental feature is well illustrated by the translational control mechanism revealed by Chu et al that responds to the speed of ribosome movement immediately after the start codon. So, yes Virginia, there is control of initiation by elongation.1 The trick perhaps is to find the experiment to understand the underlying values for klib and kinit.
Acknowledgments
The authors wish to thank Drs. John Hershey and Christopher Fraser for their comments on the relative molar ratio of initiation factors to 40S subunits
Note
In response to 8-year-old Virginia O'Hanlon's question ‘Please tell me the truth, is there a Santa Claus?’, newspaper columnist Francis Church replied ‘Yes Virginia, there is a Santa Claus.’ Church's letter in the New York Sun was originally published in 1897 and was published annually at Christmas time in the Sun for almost 50 years.
Conflict of interest
The authors declare that they have no conflict of interest.
References
- Chu D, Kazana E, Bellanger N, Singh T, Tuite MF, von der Haar T. Translation elongation can control translation initiation on eukaryotic mRNAs. EMBO J. 2014;33:21–34. doi: 10.1002/embj.201385651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleland WW. Partition analysis and the concept of net rate constants as tools in enzyme kinetics. Biochemistry. 1975;14:3220–3224. doi: 10.1021/bi00685a029. [DOI] [PubMed] [Google Scholar]
- Merrick WC. Initiation of protein synthesis in eukaryotes. Biochem Mol Biol Educ. 2003;31:378–385. [Google Scholar]
- Merrick WC. Eukaryotic protein synthesis: still a mystery. J Biol Chem. 2010;285:21197–21201. doi: 10.1074/jbc.R110.111476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson EM, Winkler MM. Regulation of mRNA entry into polysomes Parameters affecting polysome size and the fraction of mRNA in polysomes. J Biol Chem. 1987;262:11501–11506. [PubMed] [Google Scholar]
- Nielsen PJ, McConkey EH. Evidence for control of protein synthesis in HeLa cells via the elongation rate. J Cell Physiol. 1980;104:269–281. doi: 10.1002/jcp.1041040302. [DOI] [PubMed] [Google Scholar]
- Ray WJ., Jr Rate-limiting step: a quantitative definition Application to steady-state enzymatic reactions. Biochemistry. 1983;22:4625–4637. doi: 10.1021/bi00289a003. [DOI] [PubMed] [Google Scholar]
- Shah P, Ding Y, Niemczyk M, Kudla G, Plotkin JB. Rate-limiting steps in yeast protein translation. Cell. 2013;153:1589–1601. doi: 10.1016/j.cell.2013.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells SE, Hillner PE, Vale RD, Sachs AB. Circularization of mRNA by eukaryotic translation initiation factors. Mol Cell. 1998;2:135–140. doi: 10.1016/s1097-2765(00)80122-7. [DOI] [PubMed] [Google Scholar]


