Abstract
Cellular adaptation to proteotoxic stress at the endoplasmic reticulum (ER) depends on Lys48-linked polyubiquitination by ER-associated ubiquitin ligases (E3s) and subsequent elimination of ubiquitinated retrotranslocation products by the proteasome. The ER-associated E3 gp78 ubiquitinates misfolded proteins by transferring preformed Lys48-linked ubiquitin chains from the cognate E2 Ube2g2 to substrates. Here we demonstrate that Ube2g2 synthesizes linkage specific ubiquitin chains by forming an unprecedented homodimer: The dimerization of Ube2g2, mediated primarily by electrostatic interactions between two Ube2g2s, is also facilitated by the charged ubiquitin molecules. Mutagenesis studies show that Ube2g2 dimerization is required for ER-associated degradation (ERAD). In addition to E2 dimerization, we show that a highly conserved arginine residue in the donor Ube2g2 senses the presence of an aspartate in the acceptor ubiquitin to position only Lys48 of ubiquitin in proximity to the donor E2 active site. These results reveal an unanticipated mode of E2 self-association that allows the E2 to effectively engage two ubiquitins to specifically synthesize Lys48-linked ubiquitin chains.
Keywords: E2 dimerization, ER-associated degradation/ERAD, K48-linked ubiquitin chain, linkage specificity, ubiquitination mechanism
Introduction
Posttranslational modification of proteins with ubiquitin (Ub) regulates many eukaryotic cellular functions. The ubiquitination system in the cell conjugates ubiquitin to substrates either as a single moiety (monoubiquitination) (Ramanathan & Ye, 2011) or a chain containing several ubiquitin molecules (polyubiquitination). Ubiquitin chains with distinct topologies can be generated depending on which of the seven Lys residues in ubiquitin is used to form isopeptide link with the carboxyl terminus of the neighboring ubiquitin (Peng et al, 2003). Among these linkages, Lys48-, Lys11-, and Lys63-linked ubiquitin chains are relatively well characterized. Lys63-linked ubiquitination regulates cellular functions such as endocytosis, nuclear transport, DNA repair, histone remodeling, and kinase activation (Haglund & Dikic, 2005; Li & Ye, 2008; Liu & Chen, 2011), whereas Lys48-and Lys11-linked ubiquitin chains can target substrates to proteasome for degradation (Chau et al, 1989; Jin et al, 2008; Wickliffe et al, 2011b).
The ubiquitination system comprises three types of enzymes, an activating enzyme (E1), a conjugating enzyme (E2), and a ubiquitin ligase (E3). The E1 forms a thioester linkage between its catalytic cysteine and the carboxyl terminal Gly residue of ubiquitin (Schulman & Harper, 2009). An E2 then interacts with the E1 to receive ubiquitin (Schulman & Harper, 2009; Ye & Rape, 2009; Wenzel et al, 2011b; Komander & Rape, 2012). Next, an E3 interacts with the E2 to promote the transfer of ubiquitin to a substrate that is in complex with the E3 (Deshaies & Joazeiro, 2009; Rotin & Kumar, 2009). In higher eukaryotes, there are two E1 enzymes (Schulman & Harper, 2009), approximately 40 E2s, and hundreds of E3 ligases (Deshaies & Joazeiro, 2009; Rotin & Kumar, 2009; Ye & Rape, 2009).
A major task of the ubiquitination system is to assemble ubiquitin chains rapidly on diverse substrates before they dissociate from the E3 enzymes. This is not a trivial issue considering that E2 enzymes generally cannot be recharged with ubiquitin while they are bound to E3 ligases (Eletr et al, 2005; Huang et al, 2005). Hence, for many E3s, the chain building reaction likely involves multiple rounds of E2-E3 interactions that supply the reaction with sufficient ubiquitin moieties. This time-consuming process imposes a challenge for many E3s, particularly those involved in protein quality control in the cell because these enzymes often need to deal with large numbers of misfolded substrates sharing no common degradation signal. Consequently, these E3s at best could only form degenerate, low-affinity interactions with substrates, raising the question of how they can processively assemble ubiquitin chains on the substrates. In addition, certain E3 enzymes carry a high-affinity E2 binding site that could potentially inhibit E2 recharging and thus chain formation. Recent studies suggest several possible solutions to these problems (Deshaies & Joazeiro, 2009; Kleiger et al, 2009). One strategy is to assemble ubiquitin chains on the active site of an E2 enzyme, and then transfer these preassembled ubiquitin chains to substrates (Li et al, 2007). Ubiquitin chain assembly on E2 active site has been reported for several E2s (Haldeman et al, 1997; Cao et al, 2007; Ravid & Hochstrasser, 2007; Bazirgan & Hampton, 2008), but the best characterized example is the ER-associated E2 Ube2g2 (Li et al, 2007, 2009), which acts in conjugation with the RING (Really Interesting New Gene) domain E3 gp78 to ubiquitinate and degrade many misfolded ER proteins (Fang et al, 2001; Song et al, 2005; Christianson et al, 2011). gp78 can rapidly assemble ubiquitin chains on the active site of Ube2g2 and transfer preassembled chains to a substrate (Li et al, 2007). This presumably increases the polyubiquitination efficiency because it eliminates the necessity of forming a stable E3-substrate complex for chain elongation. gp78-mediated polyubiquitination depends on both the RING domain and an additional high-affinity E2 binding motif termed G2BR (Das et al, 2009; Li et al, 2009), and also requires gp78 to form a large oligomer that can nucleate multiple ubiquitin-charged Ube2g2s. However, the precise mechanism of this unusual means of polyubiquitination is still unclear. In particular, Ube2g2 preferentially assembles Lys48-linked ubiquitin chains on its active site (Li et al, 2007), but how this linkage specificity is achieved is unknown.
In this study, we combine biochemical studies with structure-based modeling to dissect the mechanism by which Ube2g2 synthesizes linkage-specific ubiquitin chains on its active site. We show that the Ube2g2 active site-linked ubiquitin chains are formed by a novel Ube2g2 dimer. The E2 dimer interface was determined by structure-based modeling, mutagenesis, and crosslinking experiments, which identify a group of charged residues surrounding Cys48 of the acceptor Ube2g2 as a critical element for E2 dimerization. A Ube2g2 variant bearing the C48A mutation can serve as the ubiquitin donor, but fails to accept ubiquitin to assemble ubiquitin chains at its active site. As a result, the Ube2g2 C48A mutant cannot promote the degradation of a model ERAD substrate in the cell. Importantly, our results also demonstrate that the dimeric Ube2g2 complex can simultaneously recruit two ubiquitin molecules. An electrostatic interaction between the donor E2 and the acceptor ubiquitin is essential for positioning Lys48 of the acceptor ubiquitin in proximity to the active site of the donor E2 for nucleophilic attack. Together, our studies define a unique strategy that allows an E2 to overcome ubiquitin charging barrier imposed by its cognate E3 to form ubiquitin chains with striking efficiency and accuracy.
Results
Define elements critical for active site-associated polyubiquitination
Because Ube2g2/gp78 only assembles Lys48-linked polyubiquitin chains, a ubiquitin mutant bearing lysine to arginine substitution at position 48 (UbK48R) can only be transferred to another ubiquitin containing Lys48, but cannot accept ubiquitin from a donor E2˜ubiquitin thioester complex. Accordingly, we established a single-round ubiquitin turnover assay in which Ube2g2 or its variants were charged with either untagged wild-type (WT) ubiquitin (acceptor) or FLAG-tagged K48R ubiquitin mutant (donor) (Li et al, 2007). The reactions were quenched with N-ethylmaleimide to prevent recharging of Ube2g2. The two charging reactions were combined and incubated in the presence of the purified gp78 cytosolic domain (gp78c). We showed previously that FLAG-UbK48R attached to the active site of the donor Ube2g2 could be transferred to ubiquitin on the active site of another Ube2g2, leading to the formation of Ube2g2-linked hybrid di-ubiquitin consisting of one WT ubiquitin and FLAG–UbK48R (Fig 1A). Using this assay, we first characterized the role of two residues, Tyr83 and His94 in Ube2g2-mediated chain formation. These residues were previously shown to be required for polyubiquitination by Ube2g2, but it is unclear whether they functioned on the donor or acceptor side. When these Ube2g2 mutants were placed at the acceptor position, they could receive ubiquitin from a WT Ube2g2 donor to form di-ubiquitin-linked E2 (Fig 1B, lanes 4–6; 13–15). By contrast, when the same mutants were charged with UbK48R and therefore restricted to serve only as donor, no di-ubiquitin-linked Ube2g2 was formed (Fig 1B, lanes 7–12; 16–21). These results indicate that Tyr83 and His94 act only on the donor E2 side to mediate di-ubiquitin formation. In addition, our data also suggest that active site-linked ubiquitin chains are formed by a unidirectional transferring reaction that occurs in an E2˜Ub dimer complex in which one E2 is dedicated to serve as donor and the other as acceptor. Therefore, Ube2g2 dimerization should play an important role in this reaction.
Figure 1.

- The experimental scheme for the single-round ubiquitin turnover assay. Ube2g2 is charged with either untagged WT ubiquitin (acceptor) or Flag-tagged K48R ubiquitin mutant (donor) by E1 and the reaction is quenched with NEM and EDTA at room temperature for 15 min, and the chase was started after GST-gp78c was added (300 nM).
- Both Y83A and H94A mutations affect the donor function of Ube2g2 as demonstrated by a single-round ubiquitin turnover assay. The samples were analyzed by SDS-PAGE and immunoblotting under a non-reducing condition.
- A Venn diagram showing residues required for either the donor or acceptor functions of Ube2g2. Residues in the acidic loop are colored in red.
- Surface distribution of the essential E2 residues identified from the mutagenesis studies.
- Structure-based sequence alignment of yeast Ubc7p and Ube2g2. Residues critical for the function of Ube2g2 were labeled by asterisks (green, conserved residues; red, non-conserved residues).
- Representative immunoblots showing the activity of the three classes of Ube2g2 mutants using the single-round ubiquitin turnover assay.
Source data are available online for this figure.
We then performed a systematic mutagenesis study in order to identify residues critical for the acceptor function of Ube2g2, which might mediate Ube2g2 dimerization. Based on the published Ube2g2 structure (Arai et al, 2006), we selected 80 surface residues with an average accessible area above 10 Å2 (supplementary Table S1). We mutated each of these residues to alanine. Purified mutant proteins were each analyzed in reciprocal ways using the single-round ubiquitin turnover assay. These studies identified 20 mutants that affected the function of Ube2g2 in different ways (Fig 1C, supplementary Fig S1A–C). As expected, most of these residues are situated in proximity to the catalytic cysteine (Cys89) (Fig 1D and E). Interestingly, most alanine mutations only affected the donor function of Ube2g2 with the exception of C48A and S115A. The C48A mutation preferentially affected the acceptor E2 function whereas the S115A mutant was defective in both donor and acceptor activities (Fig 1C and F).
Ube2g2 forms a functional dimer independent of E3
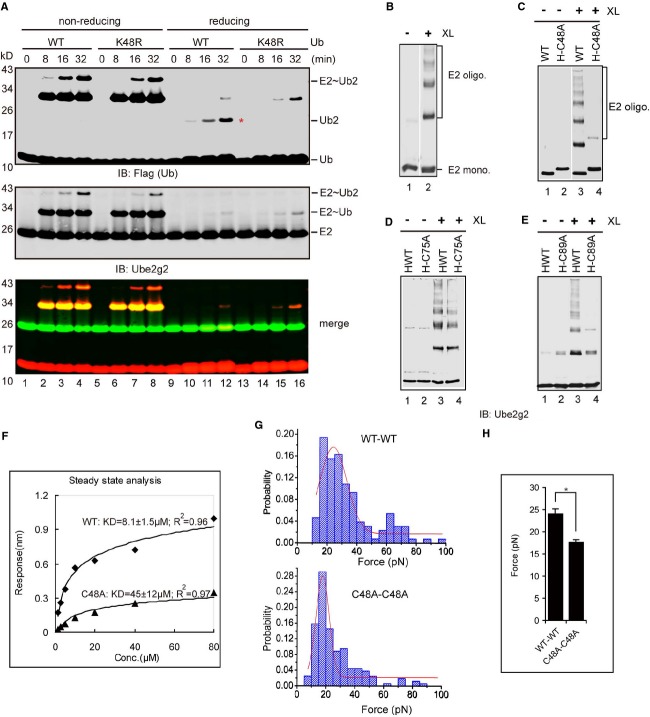
While performing the single-round ubiquitin turnover experiments, we noticed that when both the acceptor Ube2g2˜Ub thioester complex and free ubiquitin were present at similar concentrations, ubiquitin was preferentially transferred to E2 active site-linked ubiquitin rather than to free ubiquitin, forming E2˜Ub-UbK48R instead of free Ub-UbK48R dimer (supplementary Fig S2). This result further confirmed the existence of an interaction between two Ube2g2˜Ub thioesters that establishes the preference of building ubiquitin chains at the Ube2g2 active site. To test whether the interaction between E2˜Ub thioesters was intrinsic to E2 or mediated by the E3 gp78, we performed in vitro ubiquitination experiments in the absence of gp78c. Under this condition, Ube2g2 could form active site-linked di-ubiquitin but no longer ubiquitin chains (Fig 2A, lanes 10–12). Strikingly, the formation of di-ubiquitin was completely dependent on Lys48 of ubiquitin (Fig 2A, lanes 14–16 versus lanes 10–12). Thus, an E3-independent Ube2g2 self-association is sufficient to determine the Lys48 specificity of this E2 enzyme.
Figure 2.
Ube2g2 forms a functional dimer.
A Ube2g2 can form K48-linked di-ubiquitin at the active site in the absence of any E3s. Ube2g2 was incubated with the indicated ubiquitin variants in the absence of E3 for the indicated time periods. Red star indicated di-ubiquitin reduced from the active site. The samples were analyzed by dual color immunoblotting.
B Stabilization of Ube2g2 oligomers by a chemical crosslinker. Where (+) indicated, purified Ube2g2 was treated with the crosslinker BMH (XL). Oligo, oligomer; mono, monomer.
C–E Cys48 is in proximity to Cys89 in the Ube2g2 dimer. WT and (C) His-tagged Ube2g2 C48A (H-C48A), (D) His-tagged Ube2g2 C75A (H-C75A), and (E) His-tagged Ube2g2 C89A (H-C89A) mutant untreated (–) or treated with (+) the crosslinker (XL) BMH were analyzed by immunoblotting.
F Biolayer interferometry analyses of WT and C48A Ube2g2 dimerization. Steady-state analysis was used to calculate the KD of WT and C48A.
G–H Distribution of rupture forces between molecules of WT and C48A mutant were measured by AFM.
Source data are available online for this figure.
We confirmed the dimerization of Ube2g2 using a cysteine-reactive crosslinker Bismaleimidohexane (BMH). After BMH treatment, several high molecular weight products corresponding to Ube2g2 dimer and oligomers were generated, demonstrating that Ube2g2 could indeed interact with each other directly (Fig 2B). The self-association of Ube2g2 was further confirmed by Biolayer Interferometry analysis, which showed that incubation of SA biosensors immobilized with biotinylated Ube2g2 with Ube2g2 elicited a concentration-dependent response, the interactions occurred in a transient fashion with fast association and dissociation rate constants (supplementary Fig S3A).
Cys48 is in proximity to Cys89 in the Ube2g2 dimer
Because the Ube2g2 dimer could be stabilized by a cysteine-reactive homofunctional crosslinker and the C48A mutation affects the acceptor function of Ube2g2, we hypothesized that Cys48 in the acceptor Ube2g2 might be in proximity to another Cys residue in the donor Ube2g2. We mutated the three Cys residues in Ube2g2 to Ala one by one and performed crosslinking experiments with these mutants. High molecular weight crosslinking products were efficiently generated for WT Ube2g2 and the Ube2g2 C75A mutant, but not for the C48A and C89A mutants (Fig 2C–E), suggesting that the crosslinked dimer/oligomers are formed between Cys48 and Cys89 of two neighboring Ube2g2s. Thus, it is likely that in a functional Ube2g2 dimer (see below), Cys48 in one Ube2g2 is in proximity to the catalytic Cys89 of another Ube2g2. This arrangement allows BMH to crosslink multiple Ube2g2 molecules into a higher order oligomer. To see if the C48A mutation affected Ube2g2 dimerization, we used Biolayer Interferometry to measure its dimerization activity. Consistent with our hypothesis, WT Ube2g2 interacted with each other with a KD of around 8 μM, whereas the C48A mutant dimerized with a significantly reduced affinity (Fig 2F, supplementary Fig S3B and C). In addition, atomic force microscopy (AFM) was used to directly measure the rupture force required to dissociate the E2 dimer for the Ube2g2 variants (Fig 2G). The results showed that the most probable rupture force between two WT Ube2g2 molecules was 24.08 ± 1.19 pN, while between two C48A mutants, it was 17.63 ± 0.56 pN (Fig 2H). The change in dimerization affinity and rupture force of the C48A mutant was not due to any folding defects because Circular Dichroism (CD) analyses showed that the secondary structure of the C48A mutant was identical to that of WT Ube2g2 (supplementary Fig S4A). All together, these data suggest that Ube2g2 can form a homo-dimer in which Cys48 of one E2 is in proximity to Cys89 of another E2.
Ube2g2 dimerization plays a role in ER quality control
In order to test whether the C48A mutation inhibited Ube2g2 dimerization in vivo, WT Ube2g2 and the Ube2g2 C48A mutant were each expressed in cells, which were subsequently exposed to a crosslinker. Consistent with the in vitro crosslinking results, WT Ube2g2 formed significantly more oligomers than the C48A mutant, suggesting that the E2 dimer observed in vitro likely occurs in cells (Fig 3A). We next tested whether the dimerization of Ube2g2 is required for Ube2g2-mediated ER quality control. We used a stable TCRα-YFP-expressing cell line to test whether Ube2g2 C48A could still function in ERAD of TCRα, a well-characterized model ERAD substrate whose degradation requires the gp78-Ube2g2 complex for ubiquitination. In agreement with a previous report (Chen et al, 2006), siRNA-mediated knockdown of Ube2g2 increased the half-life of TCRα (Fig 3B). Expression of siRNA resistant WT Ube2g2 but not the C48A mutant at the endogenous level in cells depleted of endogenous Ube2g2 restored TCRα degradation (Fig 3C and D). The correlation between reduced dimerization activity and reduced ERAD activity for the C48A mutant suggests that Ube2g2 dimerization may be important for efficient ubiquitination and substrate degradation in ERAD.
Figure 3.
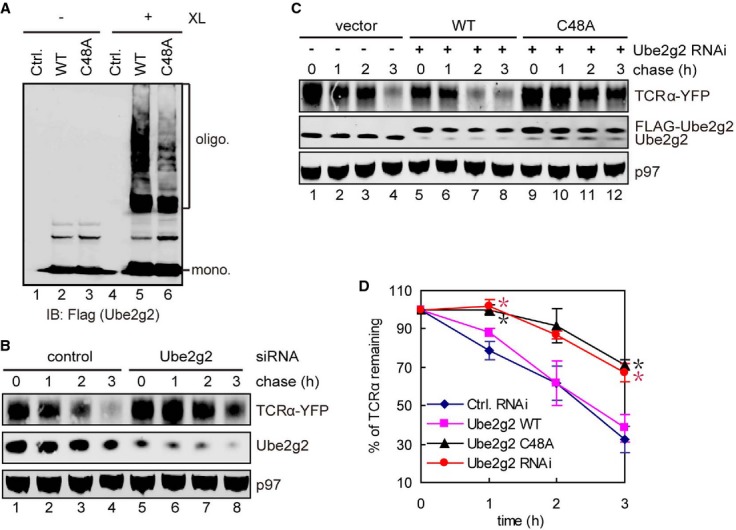
- Dimerization of FLAG-tagged WT and C48A Ube2g2 mutant in cells. Control cells or cells expressing the indicated E2s were treated with BMH (XL). Whole cell extracts were subjected to immunoblotting with FLAG antibodies.
- Knockdown of Ube2g2 stabilizes TCRα. Cells expressing TCRα-YFP were transfected with the indicated siRNA and then treated with cycloheximide. Samples taken at the indicated time points were analyzed by immunoblotting by antibodies against the indicated proteins.
- Ube2g2 C48A is defective in ERAD. TCRα-YFP-expressing cells were transfected with the indicated siRNA together with either a control or vector a rescue plasmid expressing siRNA resistant Ube2g2 WT or C48A. The stability of TCRα was analyzed as in (B).
- Quantification of the experiments shown in (B, C). Error bars, standard error (n = 3) (*P < 0.05).
Source data are available online for this figure.
A structural model of the Ube2g2˜ubiquitin dimer complex
In order to understand how Ube2g2 forms a dimer, we analyzed the surface residues of Ube2g2 (PDB code 3FSH) (Li et al, 2009) around cysteine 48 and catalytic cysteine 89. Many residues around these two cysteines were charged amino acids, suggesting possible electrostatic interactions between two Ube2g2 molecules. In addition, a hydrophobic interaction might be formed between Y152 near Cys48 and Met101 in proximity to Cys89. To test the role of these residues in Ube2g2 dimerization, we mutated these residues, and analyzed the dimerization activity of the resulting mutants. Six charged residues (D46, E50, E129, R148, E149, and K153) and a hydrophobic residue (Y152) around cysteine 48 and six residues (K70, R87, D98, D99, M101 and D139) around cysteine 89 were selected. For charged residues, we substituted them with an amino acid carrying the opposite charge, whereas for hydrophobic residues, we converted them to either Arg or Glu (see below). These substitutions were made individually or in various combinations. We initially used the crosslinking assay to measure the dimerization activities of these mutants. The results revealed that several mutants generated fewer crosslinking products when compared to WT Ube2g2 (Fig 4A and B and data not shown). Among these mutants, two [referred to as M3 (D98R/D99R/M101R) and M4 (E129R/R148E/Y152E), respectively] were selected for further characterization because these mutations reduced Ube2g2 dimerization most significantly (Fig 4B). AFM experiments showed that the most probable rupture forces for the M3 and M4 mutants were significantly reduced comparing with WT Ube2g2 (Fig 4C and supplementary Fig S3D–G). Interestingly, when these two mutants were mixed together, the dimerization of Ube2g2 was restored to almost WT level as indicated by both crosslinking and AFM experiments (Fig 4B, lane 8 versus 5, Fig 4C and supplementary Fig S3G). The introduced mutations did not cause major folding defects in these proteins as revealed by CD spectra analyses (Fig 4D and E). In further support of this notion, the ubiquitin charging efficiency of these two mutants was comparable to that of WT Ube2g2 (Fig 4F). Nonetheless, both mutants were defective in assembly of Ube2g2 active site-linked ubiquitin chains (Fig 4G). In the single-round ubiquitin transfer experiments, both these mutants failed to act as either donor or acceptor (Fig 4H), suggesting that these residues are not only involved in regulating Ube2g2 dimerization, but also have additional functions in chain formation. Accordingly, even though the M3 and M4 mutants could form a hetero-dimer E2 complex, this complex appeared to be inactive as it failed to assemble E2-linked polyubiquitin chains (Fig 4G). Thus, the complementation between the M3 and M4 mutants is limited to their interaction but did not occur functionally. These results strongly suggest that Ube2g2 may use primarily charge-charge interactions to form a dimer (see below).
Figure 4.
Characterization of the Ube2g2 dimer interaction.
A, B Characterization of Ube2g2 dimerization by mutagenesis studies. The indicated Ube2g2 variants were purified and subjected to crosslinking and immunoblotting analyses. Numbers on the gels show the relative amount of the crosslinking products.
C The most probable rupture forces between molecules of Ube2g2 WT, M3 and M4 mutants were measured by AFM. (*P < 0.05).
D, E CD spectroscopy analyses of WT Ube2g2 and the M3 and M4 mutants.
F Charging efficiency was not affected in Ube2g2 M3 and M4 mutants. Ubiquitin charging experiment was carried out at 37°C for 8 min, and the reactions were analyzed by immunoblotting with anti-FLAG (Ub) antibody.
G The Ube2g2 M3 and M4 mutants are defective in assembling active site-linked ubiquitin chains. In vitro ubiquitination was performed using the indicated E2s. The samples were analyzed by immunoblotting under a non-reducing condition.
H M3 and M4 mutations affect both the donor and acceptor function of Ube2g2 as demonstrated by a single-round ubiquitin turnover assay.
Source data are available online for this figure.
We then wished to obtain a structural model of a complex comprising of both the donor and acceptor Ube2g2˜Ub thioesters based on the published E2 and ubiquitin structures and our biochemical results. To this end, we first generated a model of a subcomplex comprising the donor Ube2g2˜Ub and acceptor Ube2g2 using the docking program HADDOCK (de Vries et al, 2010). The donor Ube2g2˜Ub structure was generated by superimposing Ube2g2 (PDB code 3FSH) (Li et al, 2009) to their counterpart in the UbcH5A˜Ub complex (PDB code 4AP4). This donor Ube2g2˜Ub structure was used as molecule 1 for computational docking of the acceptor Ube2g2 [PDB code 3FSH (Li et al, 2009)]. Three distance restraints were specified according to biochemical studies: (i) The carboxyl-terminal carbon atom of Gly76 of the donor ubiquitin should be close to Cys89 (within 2Å) in the donor Ube2g2; (ii) The sulfur atom of Cys48 in the acceptor E2 and Cys89 in donor Ube2g2 were defined as active residues; (iii) E129, R148 and Y152 of acceptor Ube2g2 should be in proximity to D99 and M101 of donor Ube2g2 based on our mutagenesis data (Fig 4). To account for conformational plasticity of the acidic loop region of Ube2g2 and the C-terminal region of ubiquitin, residues 97–115 of donor Ube2g2 and residues 71–76 of donor ubiquitin were defined fully flexible during simulated annealing. The resulting structures (referred to as molecule 2) could be grouped into 10 clusters. Based on the HADDOCK score, binding energy, and buried area, the top-ranked structure in each cluster was chosen for further evaluation (supplementary Fig S5A and B, Table S2). To determine the position of the acceptor ubiquitin in our model, we considered six possibilities according to the reported E2˜ubiquitin structures: UbcH5A˜Ub complex (PDB code 4AP4); Ubc13˜Ub (PDB code 2GMI); Ubc1˜Ub (PDB code 1FXT); UbcH8˜Ub (PDB code 2KJH) and UbcH5b˜Ub (PDB codes 3A33 and 3JVZ) (Hamilton et al, 2001; Eddins et al, 2006; Kamadurai et al, 2009; Serniwka & Shaw, 2009; Sakata et al, 2010; Wenzel et al, 2011a; Plechanovova et al, 2012). We superimposed each of these E2s onto the acceptor Ube2g2 in each of the above mentioned models (supplementary Fig S6A–F). The results showed that the Ubc1˜ubiquitin complex (1FXT) produced no major structural conflict with the model from cluster 2 (supplementary Fig S7A). Moreover, it positioned the acceptor ubiquitin in proximity to Cys89 in the donor E2 for efficient ubiquitin transfer. The whole donor Ube2g2˜Ub/acceptor Ube2g2˜Ub structure was further refined using the HADDOCK to resolve any minor structural conflicts. The final model was further analyzed by docking the E2 binding G2BR domain of gp78 to it, which showed no structural hindrance with the Ube2g2˜Ub thioester dimer complex (supplementary Fig S8A and B).
This procedure generated a model, showing that two Ube2g2 molecules form a V-shaped dimer in which the catalytic cysteine (Cys89) of the donor E2 faces the back side of the acceptor E2 where Cys48 is located. The two residues are separated by 6Å, in line with the spacer arm length of the crosslinker (10Å). On the top side, the two E2 molecules are separated by a 19Å gap, which accommodates the donor and acceptor ubiquitin molecules. The dimer is jointed at the bottom by contacts between the helix 3, sheet 4, and loop 6 of the donor E2 and the helix 4 and loop 3 of the acceptor E2 (supplementary Fig S7A). Consistent with our biochemical studies, the dimer interface is predominantly formed by paired side chain electrostatic interactions, involving Asp99, Asp98, Arg87 and Lys70 from the donor and Arg148, Glu129, Glu50, Asp46 and Glu45 from the acceptor Ube2g2 (supplementary Fig S7B). In addition, Val138 from the donor E2 makes a hydrophobic contact with Tyr152 of the acceptor E2. These interactions bury a total surface area of 1065.9 Å2. Importantly, the interactions allow the formation of a channel between the two E2 protomers. At the bottom of the channel sits the catalytic cysteine of the donor Ube2g2. Intriguingly, the side chain of Lys48 from the acceptor ubiquitin aligns perfectly within the channel and points right towards the catalytic cysteine of the donor E2, immediately suggesting a Ube2g2 dimerization mechanism that engages two ubiquitins in a specific orientation to favor Lys48-linked ubiquitination (supplementary Fig S7B).
Biochemical validation of the Ube2g2 dimer model
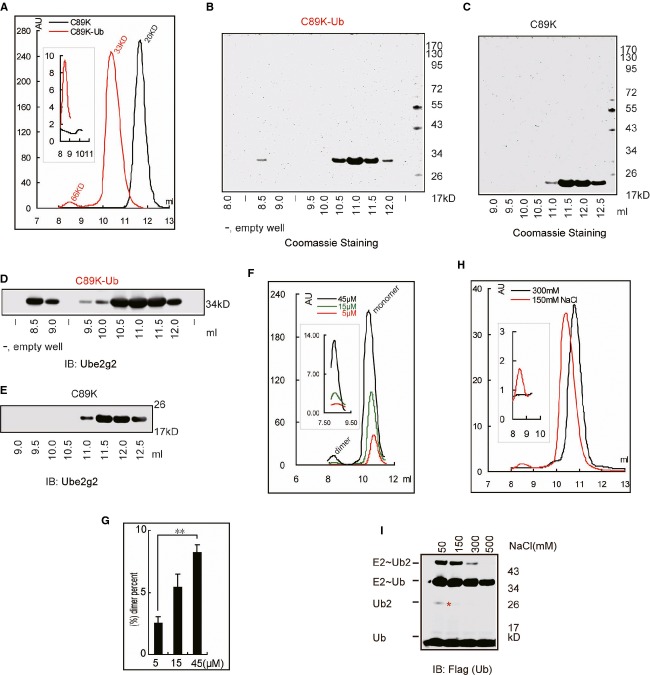
The Ube2g2˜Ub dimer model also reveals physical contacts between the E2s and ubiquitin, suggesting that ubiquitin charged to the Ube2g2 active site may help stabilize the dimer. To test this possibility, we created a C89K Ube2g2 mutant, which could be stably loaded with ubiquitin at the active site at high pH by E1 (Plechanovova et al, 2012). After purification by Ni-NTA affinity chromatography, the sample was applied to a size exclusion column to further purify Ube2g2. A previous study showed that a small fraction of uncharged Ube2g2 can exist as a dimer, but the monomeric peak, once isolated, remained predominantly as a monomer upon re-analysis by size exclusion chromatography (Li et al, 2007) (data not shown). Likewise, when we mock-treated the Ube2g2 C89K mutant with the same condition, but without ATP, and then purified the uncharged Ube2g2 monomer species, it stayed as a monomer as judged by size exclusion chromatography (Fig 5A). However, once charged with ubiquitin, a fraction of the purified Ube2g2˜Ub monomer appeared to form a peak corresponding to a dimer form (Fig 5A). The identity of the putative Ube2g2 C89K-Ub dimer was confirmed by both Coomassie blue staining (Fig 5B and C) and immunoblotting (Fig 5D and E). More Ube2g2 C89K-Ub dimer was formed when its concentration was increased, suggesting that the dimer was likely in equilibrium with the monomer (Fig 5F and G). These results demonstrate that ubiquitin charged on Ube2g2 indeed facilitates its dimerization.
Figure 5.
Electrostatic interaction(s) between two Ube2g2˜Ub molecules.
A Ube2g2 and Ube2g2∼Ub proteins (300 μl 30 μM) were analyzed by size-exclusion chromatography with Superdex 75 10/300GL. The inlet shows an enlarged view of the dimer peak.
B, C Protein fractions from the size-exclusion chromatography experiment were analyzed by SDS-PAGE and Coomassie blue staining. Note that the protein fractions containing Ube2g2 monomer were two-fold diluted before SDS-PAGE analysis.
D, E Samples in (B) and (C) were analyzed by Western blotting with anti-Ube2g2 antibody (Note: The protein fractions containing Ube2g2 monomer were 10-fold diluted before SDS-PAGE analysis.
F, G The equilibrium state of monomer/dimer form of Ube2g2˜Ub was analyzed by size-exclusion chromatography (F) with Superdex 75 10/300 GL using 200 μl 5, 15 and 45 μM Ube2g2˜Ub and the dimer form Ube2g2˜Ub was quantified (G), the percentage of dimer form Ube2g2˜Ub was calculated as: 100 x. Area under dimer/(dimer + monomer) peak (**P < 0.01).
H Size-exclusion chromatography analysis of purified Ube2g2-Ub (150 μl 15 μM) proteins at different concentrations of salt. The inlet shows an enlarged view of the dimer peak. A small shift of the monomeric peak under the high salt condition was seen, which might result from a minor conformational change due to high salt.
I Effect of salt on Ube2g2-catalyzed di-ubiquitin formation in the absence of E3. Ube2g2 was incubated with E1, FLAG-tagged ubiquitin and ATP in the presence of the indicated concentrations of salt for 30 min. The asterisk indicates di-ubiquitin reduced from the active site.
Source data are available online for this figure.
To validate whether Ube2g2˜Ub dimerization is mediated by electrostatic interactions, we analyzed the oligomerization status of Ube2g2 C89K-Ub complex in the presence of a high concentration of salt. The dimeric peak was not detected under the high salt condition, suggesting that the interactions between the two Ube2g2-Ub molecules are sensitive to salt (Fig 5H). Moreover, Ube2g2-catalyzed di-ubiquitin formation could be inhibited by increased concentrations of salt (Fig 5I). Together, these results further confirm that the functional interactions between two Ube2g2 molecules are electrostatic in nature and can be bolstered by the charged ubiquitin molecules.
An ubiquitin sensing residue in Ube2g2 determines Lys48 specificity
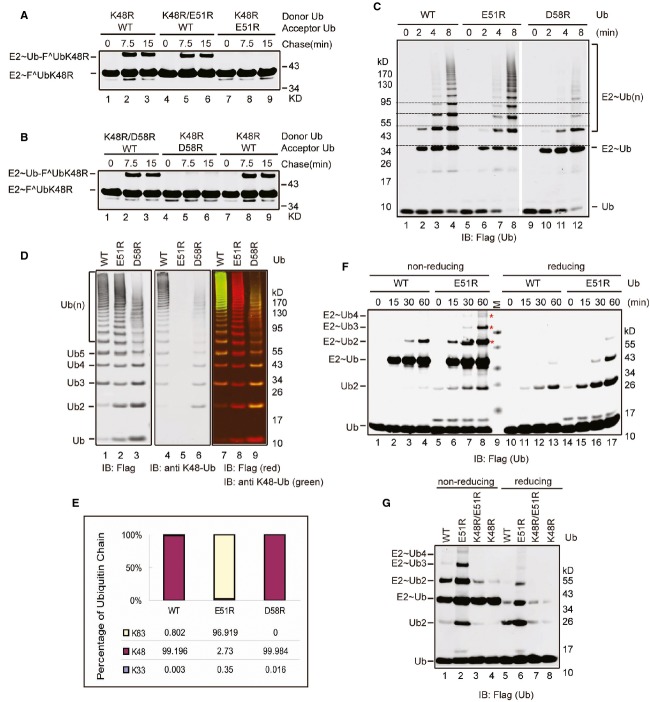
A careful examination of our model suggested that a conserved Arg109 in donor Ube2g2 is located within ion pairing distance with two acidic residues (Glu51 and Asp58) in the acceptor ubiquitin (Fig 6A). Electrostatic interaction between these residues might help orient the acceptor ubiquitin so that only Lys48 can attack the donor E2˜Ub thioester. To test this hypothesis, we first generated an Ube2g2 mutant bearing the R109E mutation. The mutation did not affect global folding of Ube2g2 as demonstrated by CD spectroscopy (supplementary Fig S4B). As anticipated, the charging efficiency of Ube2g2 R109E is similar to that of WT Ube2g2 (supplementary Fig S9). Consistent with our model, this mutant could not be transferred to a ubiquitin conjugated to the acceptor Ube2g2 active site, although it could still accept ubiquitin from a donor Ube2g2 (Fig 6B). In addition, this mutant could not form active site-linked ubiquitin chains on Ube2g2 (Fig 6C), and it also failed to assemble polyubiquitin chains on the gp78 substrate Herp in vitro (Fig 6D). To test whether the Ube2g2 R109E mutant was functional in cells, we expressed siRNA resistant WT Ube2g2 and the R109E mutant at the endogenous level in cells depleted of endogenous Ube2g2. Under this condition, WT Ube2g2 but not the R109E mutant restored the degradation of TCRα (Fig 6E and F). All together, these results suggest that Arginine 109 is essential for the function of Ube2g2 both in vitro and in cells.
Figure 6.
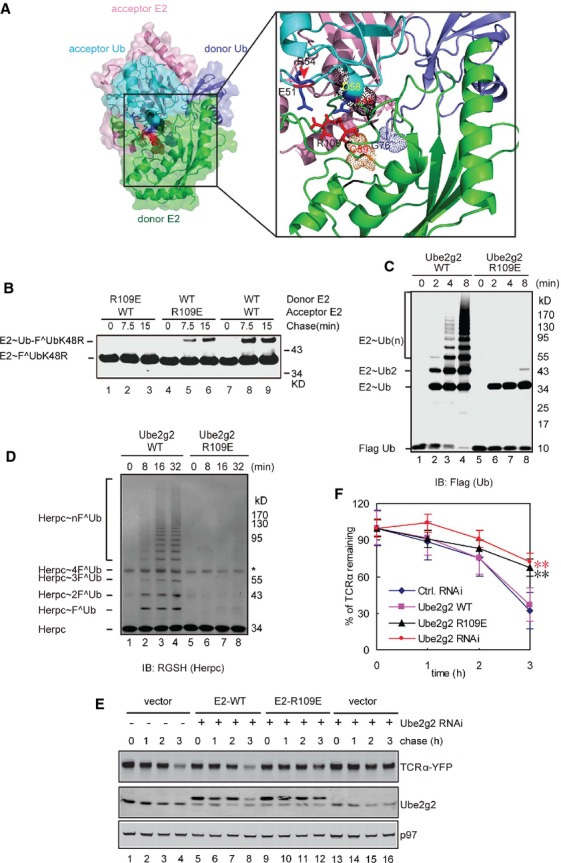
- An interaction between donor Ube2g2 and acceptor ubiquitin as revealed by a structural model. The box shows an enlarged view of the interaction interface. Note that R109 in donor Ube2g2 is in proximity to both E51 and D58 in the acceptor ubiquitin. R109 is also close to the active site of the donor Ube2g2, thus may couple ubiquitin sensing to E2 catalysis.
- The Ube2g2 R109E mutant is defective in the donor function. The donor and acceptor functions of the R109E mutant were tested by the single-round ubiquitin turnover assay. The samples were analyzed by SDS-PAGE and immunoblotting under non-reducing conditions.
- The Ube2g2 R109E mutant cannot assemble active site-linked ubiquitin chains. In vitro ubiquitination was performed using the indicated E2s together with E1, GST-gp78c, FLAG-Ub and ATP. The samples were analyzed by immunoblotting under non-reducing conditions.
- The Ube2g2 R109E mutant failed to assemble polyubiquitin chain on Herpc. In vitro ubiquitination was performed by the indicated E2s as in (C) with the exception that the model substrate Herpc was also included.
- The Ube2g2 R109E mutant is defective in ERAD. TCRα-YFP-expressing cells were transfected with the indicated siRNA together with either a control vector or a rescue plasmid expressing siRNA resistant Ube2g2 or the R109E mutant. Samples taken at the indicated time points were subjected to immunoblotting by antibodies against the indicated proteins.
- Quantification of the experiments shown in (E). Error bars, standard error (n = 4) (**P < 0.01).
Source data are available online for this figure.
We then created ubiquitin mutants bearing either E51R or D58R mutations. Analyses of their CD spectra indicated that the D58R substitution might slightly alter the ubiquitin conformation whereas the UbE51R mutant was well folded (supplementary Fig 10A and B). Single-round ubiquitin turnover assay showed that both these mutants could be transferred to another ubiquitin to form di-ubiquitin, but they failed to accept ubiquitin from other Ube2g2˜Ub molecules (Fig 7A and B). Interestingly, compared to WT ubiquitin, only the ubiquitin D58R mutant had a moderately reduced activity in generating active site-linked ubiquitin chains, whereas the E51R mutant was as effective as WT ubiquitin in synthesizing active site-linked ubiquitin chains (Fig 7C). However, careful evaluation of the ubiquitin chains generated by the E51R mutant uncovered a subtle increase in their mobility on SDS-PAGE gel compared to WT ubiquitin chains produced by Ube2g2 (Fig 7C). This raised the possibility that these chains might be linked by a different lysine residue other than Lys48. This hypothesis was confirmed by the following experiments. First, ubiquitin chains formed by the ubiquitin E51R mutant could not be detected by an antibody specifically recognizing the Lys48 linkage (Fig 7D). Second, mass spectrometry analysis showed that chains made by UbE51R were predominantly linked by Lys63, (Fig 7E). Together, these results suggest a potential ion paring mechanism between Glu51 in ubiquitin and Arg109 in Ube2g2 that determines the Lys48 specificity of this E2 enzyme.
Figure 7.
An ion pairing mechanism determines Lys48 specificity
A, B The UbE51R(A) and UbD58R(B) mutants are defective in accepting ubiquitin from a donor Ube2g2 as revealed by the single-round ubiquitin turnover assay.
C Effect of the UbE51R and UbD58R mutations on polyubiquitin chain assembly on the active site of Ube2g2. In vitro ubiquitination was performed with the indicated E2s. The samples were analyzed by immunoblotting under non-reducing conditions.
D Ubiquitin chains synthesized by UbE51R mutant are not Lys48-linked. Ubiquitination reactions carried out with the indicated ubiquitin variants were analyzed by dual color immunoblotting by anti-FLAG antibody and an antibody specific for Lys48-linked ubiquitin chain.
E Mass spectrometry analysis of the ubiquitin chains synthesized by WT ubiquitin, UbE51R, and UbD58R. Note that only K33-, K48-, and K63-linked ubiquitin chains were identified.
F Ubiquitin E51R mutant partially rescues the ubiquitination defect of Ube2g2 R109E. In vitro ubiquitination experiments were performed with Ube2g2 R109E and the indicated ubiquitin variants.
G Ubiquitin E51R mutant restores Lys48 specificity for Ube2g2 R109E.
Source data are available online for this figure.
To further test the above mentioned ion pairing mechanism, we performed a charge swap experiment to see whether the ubiquitination defect of the Ube2g2 R109E mutant could be rescued by the UbE51R mutant because swapping the charge on E2 and ubiquitin should restore the communication between these residues. Indeed, compared to WT ubiquitin that only formed a negligible amount of di-ubiquitin on Ube2g2 R109E even after prolonged incubation, UbE51R allowed chains containing up to 4 ubiquitin moieties to form on Ube2g2 R109E, demonstrating a partial rescue (Fig 7F, lane 8 versus lane 4). Importantly, ubiquitin chains formed by the Ube2g2 R109E and UbE51R pair were completely dependent on Lys48 in ubiquitin, suggesting that swapping the charge also restores the Lys48 specificity of Ube2g2 (Fig 7G, lanes 2, 6 versus lanes 3, 7). Moreover, compared to WT ubiquitin, UbE51R also allowed Ube2g2 R109E to form di-ubiquitin more efficiently on its active site in the absence of E3 (supplementary Fig S11, lanes 15, 16 versus lanes 7, 8). Collectively, these results demonstrate that Arg109 in Ube2g2 is a ubiquitin sensing residue that communicates with acceptor ubiquitin by an ion pairing mechanism to determine the lysine specificity of the reaction, and thus further validated our E2 dimerization model. Since UbE51R only partially rescues the defect of Ube2g2 R109E, it is possible that Arg109 may also have additional functions in Ube2g2-mediated ubiquitination (see Discussion).
Discussion
Role of E2 dimerization in linkage specification
Our results demonstrate that a direct interaction between a donor Ube2g2 and an acceptor Ube2g2 molecule is essential for building polyubiquitin chains at the active site of the latter. We characterize the Ube2g2 dimer interface by a series of mutagenesis and chemical crosslinking experiments, which show that Cys48 in the acceptor Ube2g2 is in proximity to Cys89 in the donor Ube2g2. Using this information, we construct a structural model of the Ube2g2˜ubiquitin thioester dimer complex, and validate it by extensive biochemical studies (Figs 7 and supplementary Figs S3 and S11).
To our satisfaction, our model not only explains how active site-linked polyubiquitin chains are formed, but also yields valuable information on how the Lys48-linkage specificity is achieved by Ube2g2. The model suggests that two Ube2g2 molecules bind to each other in a front-to-back orientation with each Ube2g2 protomer carrying an ubiquitin covalently at the active site. The interactions between the two E2s, which are primarily electrostatic in nature, help position Lys48 in the acceptor ubiquitin near the donor Ube2g2 active site for nucleophilic attack. Biolayer Interferometry studies show that the Ube2g2 dimerization occurs with extremely fast on and off rates, resulting in a dynamic interaction with a micromolar dissociation constant. Nonetheless, in vitro ubiquitination experiments demonstrate that this interaction is sufficient to produce Lys48-linked di-ubiquitin even in the absence of any E3s. However, Ube2g2 on its own cannot synthesize longer ubiquitin chains. In order to build polyubiquitin chains, the acceptor E2 needs to sequentially engage multiple donor Ube2g2s in a short period of time. This appears to be facilitated by oligomerization of the E3 gp78. Presumably, simultaneous interaction of a large gp78 oligomer carrying multiple donor Ube2g2 molecules increases the local Ube2g2 concentration, which enhances the interactions between donor and acceptor E2s (Li et al, 2009). Consistent with this model, we previously demonstrated that gp78 oligomerization is required for ubiquitin chain formation by Ube2g2 (Li et al, 2009).
Many E2s can form either homo-or hetero-dimers in the cell (Silver et al, 1992; VanDemark et al, 2001; Varelas et al, 2003; David et al, 2011), but the functional relevance of E2 oligomerization has been unclear for most of these enzymes with the exception of the UBE2N-UEV1A hetero-dimer. The UBE2N-UEV1A heterodimer specifically synthesizes Lys63-linked ubiquitin chain. This E2 complex serves as a platform that recruits two ubiquitin molecules; One non-covalently binds UEV1A and the other is covalently linked to the active site of UBE2N (VanDemark et al, 2001). This unique E2 architecture positions Lys63 in UEV1A-bound ubiquitin near the active site of UBE2N, leading to specific formation of Lys63-linked ubiquitin chain (Eddins et al, 2006). Compared with the UBE2N-UEV1A dimer, the dimer interface of Ube2g2 is completely different. Consequently, the spatial orientation of the two ubiquitins bound to Ube2g2 is distinct from that of the UBE2N-UEV1A-bound ubiquitins, which explains why a different acceptor lysine residue is preferentially positioned in proximity to the active site of the donor E2. Importantly, for Ube2g2, both E2s carry covalently bound ubiquitin. Therefore, the transfer of ubiquitin from one E2 to the other leads to the formation of ubiquitin chains on the active site of the latter. The Ube2g2 C48A mutant defective in dimerization also failed to degrade the model ERAD substrate TCRα, therefore suggesting that dimerization is functionally required in cells. Thus, our findings establish a novel mode of E2 dimerization that is functionally required for synthesizing Lys48-linked ubiquitin chain. Given that E2 dimerization is a widespread phenomenon, we propose that E2-E2 interactions may in general have important functions in polyubiquitination, particularly in linkage specification. The distinct forms of E2 dimers may allow E2 enzymes to acquire functional diversity in ubiquitination.
Interaction between donor E2 and acceptor ubiquitin contributes to Lys48 specificity
In addition to E2 dimerization, our studies also identify an ion pairing mechanism that contributes to the Lys48 specificity of Ube2g2. We show that Arg109, a conserved residue in the 3/10 helix of Ube2g2, can communicate with an aspartate (Asp51) in the acceptor ubiquitin, which helps orient Lys48 of the acceptor ubiquitin near the donor E2 active site. Importantly, this interaction prevents other Lys residues in the acceptor ubiquitin from attacking the donor E2 active site. This activity is particularly important for polyubiquitination considering that structural studies have revealed tremendous flexibility in the linker region between E2 and a covalently bound ubiquitin. Thus, without proper E2-acceptor ubiquitin interactions, the acceptor ubiquitin may adopt a variety of conformations (supplementary Fig S6), allowing a non-specific lysine to attack donor E2. Indeed, when acting with a ubiquitin mutant that has Asp51 converted to a positively charged residue, Ube2g2 preferentially produces Lys63-linked ubiquitin chain. Arg109 may have additional functions because the chain building activity of Ube2g2 R109E is only partially restored by the ubiquitin E51R mutant. Given that Arg109 in Ube2g2 is near the E2 active site, it is conceivable that the interaction between the acceptor ubiquitin and Arg109 in donor E2 may also influence the architecture of the E2 active site. We propose that Arg109 may function as a ubiquitin sensor that couples ubiquitin binding to re-organization of the E2 active site, which enhances both the efficiency and linkage specificity of Ube2g2-mediated polyubiquitination.
Until now, the mechanisms of ubiquitin linkage specification have only been studied for a few E2s. In each case, formation of a given ubiquitin linkage requires specific interactions between donor E2 and acceptor ubiquitin. Taking the yeast chain-forming E2 UBC1 as an example, three residues (Thr84, Gln122 and Ala124) near the UBC1 active site appears to contact a critical Tyr residue near Lys48 in the acceptor ubiquitin, allowing UBC1 to specifically catalyze Lys48-linked ubiquitination (Rodrigo-Brenni et al, 2011). For Lys11-linked ubiquitin chains, electrostatic interactions between the donor E2-ubiquitin complex and acceptor ubiquitin play an essential role. The interactions involve a network of ionic contacts between Lys6, Lys63 of ubiquitin, and Glu131, Glu139 on helix 3 of Ube2S (Wickliffe et al, 2011a). Finally, for Cdc34, an acidic loop near the E2 active site was proposed to interact with the acceptor ubiquitin in a way that allows only Lys48 in the acceptor ubiquitin to attack the donor E2 active site (Petroski & Deshaies, 2005). These results, together with our new findings, demonstrate that E2s can use distinct surface areas to engage acceptor ubiquitin in a variety of modes, resulting in distinct lysine specificity of polyubiquitination.
Materials and Methods
Antibodies, proteins, crosslinking reagents and siRNAs
FLAG and K48-linkage-specific polyubiquitin antibodies were purchased from Abmart (Shanghai, China) and Cell signaling Technology (Danvers, MA, USA), respectively. E1, FLAG-Ubiquitin, and WT ubiquitin were purchased from Boston Biochem (Cambridge, MA, USA). The crosslinking reagents BMH and DTDP were purchased from Pierce (Rockford, IL, USA) and Sigma (St. Louis, MO, USA), respectively. siRNAs to knockdown Ube2g2 were designed and synthesized by GenePharma (Shanghai, China).
Plasmids, strain and cell line
Plasmids pET28-Ube2g2, pGEX-gp78c, pET28-FLAG-UbK48R, and pET3-Ub were described previously (Li et al, 2007). FLAG-Ube2g2 was amplified and cloned into pcDNA3.1 and pRK. pET28-FLAG-UbK48R, pET3-Ub, pET28-Ube2g2 single/double/multiple point(s) alanine substitutions, were generated by site-directed mutagenesis. pET28a-FLAG-Ube2g2, pRK-Flag-Ube2g2 and pcDNA-FLAG-Ube2g2 resistant to siRNA (the sequence CAGTGCAGAGTGTGGAGAA in Ube2g2 cDNA was replaced by CCGTCCAAAGCGTCGAAAA) were generated by site-directed mutagenesis too. All plasmids were amplified in E. coli Top10, and recombinant proteins were expressed in E. coli BL 21 or Rossetta 2. Stable cell line expressing TCRα-YFP (293T-TCRα-YFP) cells were described previously (Soetandyo et al, 2010).
Protein purification
Purification of Ube2g2 and gp78c was described previously (Ye et al, 2003). Purified E2 variants and E3 were further fractionated by size exclusion chromatography on Superdex75 and Superose 6 column, respectively, in 50 mM Tris–HCl, pH 8.0, 150 mM potassium chloride, 5% glycerol, and 2 mM magnesium chloride. His-FLAG-tagged UbK48R was purified under denaturing condition (8M urea) following the instructions provided by Qiagen (Venlo, Netherlands). Purified proteins were refolded in PBS.
In vitro ubiquitination assay
Ubiquitin charging experiment was carried out as follows: E1 (60 nM) and Ube2g2 (E2, 300 nM) were incubated with FLAG-tagged Ub (10 μM) at 37°C in the reaction buffer (25 mM Tris-HCl, pH 7.4, 2 mM magnesium/ATP, 0.2 mM DTT). Samples taken at indicated time points were mixed with the Laemmli buffer in the absence or presence of a reducing reagent. Ubiquitination was conducted using the same condition described above with the addition of the purified E3 gp78c (300 nM).
Single-round ubiquitin turnover assay
To monitor single-round turnover of Ub precharged to Ube2g2 or its variants, E2 variants (300 nM) were incubated with E1 (60 nM), together with either FLAG-UbK48R(as donor) or WT Ub (as acceptor) at 37°C for 15 min in the reaction buffer (25 mM Tris–HCl, pH 7.4, 2 mM magnesium/ATP, 0.1 mM DTT). The reaction was treated with 50 mM EDTA and 10 mM NEM for 15 min at room temperature. Equal volume of donor and acceptor was mixed together with GST-gp78c (300 nM) at 37°C for different times. The reaction was stopped by addition of the Laemmli buffer. Samples were analyzed by immunoblotting with antibodies indicated in the figure legends.
Ube2g2 C89K-Ub dimer synthesis and purification
Recombinant proteins (GST-E1, His-Ube2g2 C89K and His-Flag-Ub K48R) were expressed in E. coli. and purified as described previously (Li et al, 2007). For purification of Ube2g2-Ub with a stable Ube2g2-Ub thioester, the Ube2g2 C89K mutant was generated and purified by. Ni-NTA affinity resin. Before fractionated by size exclusion chromatography on Superdex75, His-Ube2g2 C89K was treated with thrombin overnight at 4°C to remove the His tag.
The isopeptide bond-linked Ube2g2(C89K)-Ub conjugate was prepared as descried previously (Plechanovova et al, 2012), incubating Ube2g2(C89K) (100 μM) with His-Ub K48R (100 μM) and E1 (1 μM) at 35°C for 26 hrs in a buffer containing 3 mM ATP, 5 mM MgCl2, 50 mM Tris pH 10.0, 150 mM NaCl and 0.8 mM TCEP. The E2–Ub conjugate was purified by Ni-affinity chromatography and size exclusion chromatography (Superdex75). The peak fractions corresponding to the monomeric form of the E2-Ub complex are pooled and concentrated before re-analysis by size exclusion chromatography with Superdex 75 10/300 GL of GE (Fairfield, CT, USA).
Circular dichroism spectroscopy
To determine the secondary structure of Ube2g2 and Ub variants, the CD spectra of the proteins were taken at 0.1 mg/ml in 50 mM Tris, using a Chirascan Plus CD spectrometer (Applied Photophysics, Surrey, UK) and a 1 mm quartz sample cell equilibrated at 20°C. Scans between 200 and 260 nm were performed at a scan rate of 50 nm per min.
Protein interaction studies via Biolayer Interferometry
For the affinity study, Octet RED96 (ForteBio, CA, USA), equipped with streptavidin (SA) biosensors, was applied (Bourhis et al, 2010; Volkan et al, 2012). SA biosensors were pre-wetted for 15 min in a buffer containing 10 mM HEPES pH7.4, 150 mM NaCl, 3 mM EDTA, then incubated with 15 μg/ml biotinylated Ube2g2 variants in the same buffer for 300 s. The loading level of Ube2g2 variants reaches to saturation (around 2.4 ± 0.2 nm). To study the interactions of biotinylated Ube2g2 variants on the biosensor with unlabeled Ube2g2, The biosensors were incubated with 200 μl protein samples (1.25–80 μM) or as a negative control with the buffer for 600s and then incubated with a protein free buffer (10 mM HEPES pH7.4, 150 mM NaCl, 3 mM EDTA) for 600s to allow dissociation. Blank binding cycles using buffer only were used to correct the baseline shift during the analyses. Steady state analysis in Data Analysis version 7.0 (Fortebio) was used to calculate the KD. Responses (average from 585 to 595 s) were used in analysis. The Model Equation: Response = Rmax×Conc./(KD + Conc.). R2 is the coefficient of determination estimating the goodness of a curve fit. All tests were repeated in triplicate.
Atomic force microscopy experiments
The rupture force measurements of various protein pairs are performed with a custom-made AFM. Commercial cantilevers (Bruker AFM Probes, Camarillo, CA, USA) calibrated by the thermal fluctuation method (Hutter & Bechhoefer, 1993). To functionalize the AFM, the cantilever tip and the polystyrene dish surface were incubated with WT Ube2g2 or mutation proteins at 4°C overnight. BSA was used as a substitute for Ube2g2 proteins for negative controls. After six washes with a HEPES-containing buffer (10 mM HEPES, 150 mM NaCl, pH 7.4) the tip and dish surface were blocked with BSA solution (1%BSA in HEPES buffer) at room temperature for 1 h. If the cantilever tip and the polystyrene dish functionalized with different proteins, we use hyphen to connect their names (e.g. ‘A–B’). The protein before hyphen adsorbed on the cantilever tip (e.g. the ‘A’ in ‘A–B’); and the protein after hyphen adsorbed on the polystyrene dish (e.g. the ‘B’ in ‘A–B’). In each event, the cantilever tip was first pressed on the surface for 1 s at force about 10 pN, and then lifted up at the speed of 200 nm/s. The cantilever deflection was collected and analyzed to count the adhesion rate and to calculate the unbinding forces. The adhesion rate for each protein pair was averaged over 200 events. For most experiments, successful adhesion events are < 30% of the total events, suggesting that the observed adhesion events are dominated by single molecule adhesion. Moreover, only single-peak dissociation events were included in force calculation to further ensure that the forces measured were dissociation force of single protein pair. Two hundred total events were used to calculate the adhesion rate for each experimental repetition. The mean and the standard error of mean were analyzed from the corresponding repetitions. The histograms of the rupture forces were fitted with Gaussian distribution  P, probability; f, force; A and w, Gaussian parameters) using ORIGIN software (Microcal Software, Northhampton, MA, USA). The maximum force from the fitting (f0) was taken as the most probable rupture force for each molecular pair. The fitting algorithm can give the standard error for each parameter. The Tukey test was used for post hoc tests of significant differences between groups using ORIGIN software.
P, probability; f, force; A and w, Gaussian parameters) using ORIGIN software (Microcal Software, Northhampton, MA, USA). The maximum force from the fitting (f0) was taken as the most probable rupture force for each molecular pair. The fitting algorithm can give the standard error for each parameter. The Tukey test was used for post hoc tests of significant differences between groups using ORIGIN software.
Quantitative mass spectrometry of ubiquitin linkages
Stable isotope-labeled GG-linked ubiquitin peptides were synthesized, quantified by amino acid analysis, and used for absolute quantification. Labeled peptide standards are added during trypsin digestion of the native proteins, The resulting peptide samples were dissolved in a sample buffer (6% acetic acid, 0.005% heptafluorobutyric acid, 5% acetonitrile, and 0.1% trifluoroacetic acid) and analyzed by LC-MS using an LTQ-Orbitrap mass spectrometer (Thermo Finnigan, San Jose, CA, USA). The MS instrument was operated in selective reaction monitoring (SRM) mode to increase sensitivity as described previously (Xu et al, 2009).
Computational docking
Docking was performed with the HADDOCK Web server (de Vries et al, 2010). All HADDOCK runs were performed with 1000 structures for rigid body docking, five trials for rigid body minimization, sampling of 180 degrees rotated solutions during rigid body EM, 200 structures for semi-flexible refinement, 200 structures for explicit solvent refinement, epsilon constant for the electrostatic energy term equal to 10, solvated docking mode as ‘on’ with water as solvent and clustering parameters, i.e. backbone rmsd cut-off for clustering equal to 10.0 Å, minimum cluster size equal to 4. Structural coordinates for the final model are provided in the Supplementary Information.
Cell culture and transfection
293T-TCRα-YFP cells were maintained at 37°C and 5% CO2 in DMEM supplemented with penicillin, streptomycin, and 10%FBS. To generate cells with FLAG-Ube2g2 variants expression and endogenous Ube2g2 knock down, 293T-TCRα-YFP were co-transfected with siRNA and pcDNA-Flag-Ube2g2 variants. Twenty four hours after transfection, the expression of FLAG-Ube2g2 and knockdown of Ube2g2 was confirmed by western blotting. Cycloheximide experiments were performed as described previously (Soetandyo et al, 2010).
Crosslinking experiments
Chemical crosslinked was performed as follows: For in vitro assay, 5 μM Ube2g2 variants were incubated on ice for 15 min in the presence or absence of 2 μM BMH (or DTDP). For in vivo assay, cell pellets were resuspended in cold NP40 buffer (50 mM Tris pH8.0, 0.5% NP40, 150 mM NaCl, 5 mM MgCl2, EDTA-free protease inhibitor mixture (Roche Applied Science, Mannheim, Germany) and incubated on ice for 15 min. Then, the lysate was cleared by centrifugation and incubated on ice for 30 min in the presence or absence of 10 mM BMH (or DTDP). Crosslinked products were detected by western blotting.
Acknowledgments
We thank Xinhua Lin and Jun Tang for critical reading of the manuscript, Huimin Wang and Jianwu Wang for their assistance in the Biolayer Interferometry study. This work is supported by a National Natural Science Foundation of China (Grant No. 30970603), the Major Basic Research Program (Grant No. 2012CB944404), the Knowledge Innovation Program (Grant No. KSCX2-YW-N-071), the One Hundred Talents Program of Chinese Academy of Sciences, and the intramural research program at NIDDK of the National Institutes of Health, USA.
Author contributions
WL planned and performed most of the experiments and helped to write the manuscript. YS and YZ performed and analysed some biochemical experiments. CL and PW built the E2 dimer model; YL and LZ performed quantitative Mass Spectrometry of ubiquitin linkages, JL and PX analyzed the data. YY and WL supervised the project, designed the experiments and wrote the article.
Conflict of interest
The authors declare that they have no conflict of interest.
Supplementary information for this article is available online: http://emboj.embopress.org
References
- Arai R, Yoshikawa S, Murayama K, Imai Y, Takahashi R, Shirouzu M, Yokoyama S. Structure of human ubiquitin-conjugating enzyme E2 G2 (UBE2G2/UBC7) Acta Crystallogr Sect F Struct Biol Cryst Commun. 2006;62:330–334. doi: 10.1107/S1744309106009006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazirgan OA, Hampton RY. Cue1p is an activator of Ubc7p E2 activity in vitro and in vivo. J Biol Chem. 2008;283:12797–12810. doi: 10.1074/jbc.M801122200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourhis E, Tam C, Franke Y, Bazan JF, Ernst J, Hwang J, Costa M, Cochran AG, Hannoush RN. Reconstitution of a Frizzled8.Wnt3a.LRP6 signaling complex reveals multiple Wnt and Dkk1 binding sites on LRP6. J Biol Chem. 2010;285:9172–9179. doi: 10.1074/jbc.M109.092130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J, Wang J, Qi W, Miao HH, Wang J, Ge L, DeBose-Boyd RA, Tang JJ, Li BL, Song BL. Ufd1 is a cofactor of gp78 and plays a key role in cholesterol metabolism by regulating the stability of HMG-CoA reductase. Cell Metab. 2007;6:115–128. doi: 10.1016/j.cmet.2007.07.002. [DOI] [PubMed] [Google Scholar]
- Chau V, Tobias JW, Bachmair A, Marriott D, Ecker DJ, Gonda DK, Varshavsky A. A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science. 1989;243:1576–1583. doi: 10.1126/science.2538923. [DOI] [PubMed] [Google Scholar]
- Chen B, Mariano J, Tsai YC, Chan AH, Cohen M, Weissman AM. The activity of a human endoplasmic reticulum-associated degradation E3, gp78, requires its Cue domain, RING finger, and an E2-binding site. Proc Natl Acad Sci USA. 2006;103:341–346. doi: 10.1073/pnas.0506618103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson JC, Olzmann JA, Shaler TA, Sowa ME, Bennett EJ, Richter CM, Tyler RE, Greenblatt EJ, Wade Harper J, Kopito RR. Defining human ERAD networks through an integrative mapping strategy. Nat Cell Biol. 2011;14:93–105. doi: 10.1038/ncb2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das R, Mariano J, Tsai YC, Kalathur RC, Kostova Z, Li J, Tarasov SG, McFeeters RL, Altieri AS, Ji X, Byrd RA, Weissman AM. Allosteric activation of E2-RING finger-mediated ubiquitylation by a structurally defined specific E2-binding region of gp78. Mol Cell. 2009;34:674–685. doi: 10.1016/j.molcel.2009.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David Y, Ziv T, Admon A, Navon A. The E2 ubiquitin-conjugating enzymes direct polyubiquitination to preferred lysines. J Biol Chem. 2011;285:8595–8604. doi: 10.1074/jbc.M109.089003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- Eddins MJ, Carlile CM, Gomez KM, Pickart CM, Wolberger C. Mms2-Ubc13 covalently bound to ubiquitin reveals the structural basis of linkage-specific polyubiquitin chain formation. Nat Struct Mol Biol. 2006;13:915–920. doi: 10.1038/nsmb1148. [DOI] [PubMed] [Google Scholar]
- Eletr ZM, Huang DT, Duda DM, Schulman BA, Kuhlman B. E2 conjugating enzymes must disengage from their E1 enzymes before E3-dependent ubiquitin and ubiquitin-like transfer. Nat Struct Mol Biol. 2005;12:933–934. doi: 10.1038/nsmb984. [DOI] [PubMed] [Google Scholar]
- Fang S, Ferrone M, Yang C, Jensen JP, Tiwari S, Weissman AM. The tumor autocrine motility factor receptor, gp78, is a ubiquitin protein ligase implicated in degradation from the endoplasmic reticulum. Proc Natl Acad Sci USA. 2001;98:14422–14427. doi: 10.1073/pnas.251401598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haglund K, Dikic I. Ubiquitylation and cell signaling. EMBO J. 2005;24:3353–3359. doi: 10.1038/sj.emboj.7600808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldeman MT, Xia G, Kasperek EM, Pickart CM. Structure and function of ubiquitin conjugating enzyme E2-25K: the tail is a core-dependent activity element. Biochemistry. 1997;36:10526–10537. doi: 10.1021/bi970750u. [DOI] [PubMed] [Google Scholar]
- Hamilton KS, Ellison MJ, Barber KR, Williams RS, Huzil JT, McKenna S, Ptak C, Glover M, Shaw GS. Structure of a conjugating enzyme-ubiquitin thiolester intermediate reveals a novel role for the ubiquitin tail. Structure. 2001;9:897–904. doi: 10.1016/s0969-2126(01)00657-8. [DOI] [PubMed] [Google Scholar]
- Huang DT, Paydar A, Zhuang M, Waddell MB, Holton JM, Schulman BA. Structural basis for recruitment of Ubc12 by an E2 binding domain in NEDD8's E1. Mol Cell. 2005;17:341–350. doi: 10.1016/j.molcel.2004.12.020. [DOI] [PubMed] [Google Scholar]
- Hutter JL, Bechhoefer J. Calibration of atomic-force microscope tips. Rev Sci Instrum. 1993;64:1868–1873. [Google Scholar]
- Jin L, Williamson A, Banerjee S, Philipp I, Rape M. Mechanism of ubiquitin-chain formation by the human anaphase-promoting complex. Cell. 2008;133:653–665. doi: 10.1016/j.cell.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamadurai HB, Souphron J, Scott DC, Duda DM, Miller DJ, Stringer D, Piper RC, Schulman BA. Insights into ubiquitin transfer cascades from a structure of a UbcH5B approximately ubiquitin-HECT(NEDD4L) complex. Mol Cell. 2009;36:1095–1102. doi: 10.1016/j.molcel.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleiger G, Saha A, Lewis S, Kuhlman B, Deshaies RJ. Rapid E2-E3 assembly and disassembly enable processive ubiquitylation of cullin-RING ubiquitin ligase substrates. Cell. 2009;139:957–968. doi: 10.1016/j.cell.2009.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komander D, Rape M. The ubiquitin code. Annu Rev Biochem. 2012;81:203–229. doi: 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]
- Li W, Tu D, Brunger AT, Ye Y. A ubiquitin ligase transfers preformed polyubiquitin chains from a conjugating enzyme to a substrate. Nature. 2007;446:333–337. doi: 10.1038/nature05542. [DOI] [PubMed] [Google Scholar]
- Li W, Tu D, Li L, Wollert T, Ghirlando R, Brunger AT, Ye Y. Mechanistic insights into active site-associated polyubiquitination by the ubiquitin-conjugating enzyme Ube2g2. Proc Natl Acad Sci USA. 2009;106:3722–3727. doi: 10.1073/pnas.0808564106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Ye Y. Polyubiquitin chains: functions, structures, and mechanisms. Cell Mol Life Sci. 2008;65:2397–2406. doi: 10.1007/s00018-008-8090-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Chen ZJ. Expanding role of ubiquitination in NF-kappaB signaling. Cell Res. 2011;21:6–21. doi: 10.1038/cr.2010.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Schwartz D, Elias JE, Thoreen CC, Cheng D, Marsischky G, Roelofs J, Finley D, Gygi SP. A proteomics approach to understanding protein ubiquitination. Nat Biotechnol. 2003;21:921–926. doi: 10.1038/nbt849. [DOI] [PubMed] [Google Scholar]
- Petroski MD, Deshaies RJ. Mechanism of lysine 48-linked ubiquitin-chain synthesis by the cullin-RING ubiquitin-ligase complex SCF-Cdc34. Cell. 2005;123:1107–1120. doi: 10.1016/j.cell.2005.09.033. [DOI] [PubMed] [Google Scholar]
- Plechanovova A, Jaffray EG, Tatham MH, Naismith JH, Hay RT. Structure of a RING E3 ligase and ubiquitin-loaded E2 primed for catalysis. Nature. 2012;489:115–120. doi: 10.1038/nature11376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanathan HN, Ye Y. Cellular strategies for making monoubiquitin signals. Crit Rev Biochem Mol Biol. 2011;47:17–28. doi: 10.3109/10409238.2011.620943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravid T, Hochstrasser M. Autoregulation of an E2 enzyme by ubiquitin-chain assembly on its catalytic residue. Nat Cell Biol. 2007;9:422–427. doi: 10.1038/ncb1558. [DOI] [PubMed] [Google Scholar]
- Rodrigo-Brenni MC, Foster SA, Morgan DO. Catalysis of lysine 48-specific ubiquitin chain assembly by residues in E2 and ubiquitin. Mol Cell. 2011;39:548–559. doi: 10.1016/j.molcel.2010.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotin D, Kumar S. Physiological functions of the HECT family of ubiquitin ligases. Nat Rev Mol Cell Biol. 2009;10:398–409. doi: 10.1038/nrm2690. [DOI] [PubMed] [Google Scholar]
- Sakata E, Satoh T, Yamamoto S, Yamaguchi Y, Yagi-Utsumi M, Kurimoto E, Tanaka K, Wakatsuki S, Kato K. Crystal structure of UbcH5b˜ubiquitin intermediate: insight into the formation of the self-assembled E2˜Ub conjugates. Structure. 2010;18:138–147. doi: 10.1016/j.str.2009.11.007. [DOI] [PubMed] [Google Scholar]
- Schulman BA, Harper JW. Ubiquitin-like protein activation by E1 enzymes: the apex for downstream signalling pathways. Nat Rev Mol Cell Biol. 2009;10:319–331. doi: 10.1038/nrm2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serniwka SA, Shaw GS. The structure of the UbcH8-ubiquitin complex shows a unique ubiquitin interaction site. Biochemistry-Us. 2009;48:12169–12179. doi: 10.1021/bi901686j. [DOI] [PubMed] [Google Scholar]
- Silver ET, Gwozd TJ, Ptak C, Goebl M, Ellison MJ. A chimeric ubiquitin conjugating enzyme that combines the cell cycle properties of CDC34 (UBC3) and the DNA repair properties of RAD6 (UBC2): implications for the structure, function and evolution of the E2s. EMBO J. 1992;11:3091–3098. doi: 10.1002/j.1460-2075.1992.tb05381.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soetandyo N, Wang Q, Ye Y, Li L. Role of intramembrane charged residues in the quality control of unassembled T-cell receptor alpha-chains at the endoplasmic reticulum. J Cell Sci. 2010;123:1031–1038. doi: 10.1242/jcs.059758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song BL, Sever N, DeBose-Boyd RA. Gp78, a membrane-anchored ubiquitin ligase, associates with Insig-1 and couples sterol-regulated ubiquitination to degradation of HMG CoA reductase. Mol Cell. 2005;19:829–840. doi: 10.1016/j.molcel.2005.08.009. [DOI] [PubMed] [Google Scholar]
- VanDemark AP, Hofmann RM, Tsui C, Pickart CM, Wolberger C. Molecular insights into polyubiquitin chain assembly: crystal structure of the Mms2/Ubc13 heterodimer. Cell. 2001;105:711–720. doi: 10.1016/s0092-8674(01)00387-7. [DOI] [PubMed] [Google Scholar]
- Varelas X, Ptak C, Ellison MJ. Cdc34 self-association is facilitated by ubiquitin thiolester formation and is required for its catalytic activity. Mol Cell Biol. 2003;23:5388–5400. doi: 10.1128/MCB.23.15.5388-5400.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkan E, Ford BA, Pinkner JS, Dodson KW, Henderson NS, Thanassi DG, Waksman G, Hultgren SJ. Domain activities of PapC usher reveal the mechanism of action of an Escherichia coli molecular machine. Proc Natl Acad Sci USA. 2012;109:9563–9568. doi: 10.1073/pnas.1207085109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries SJ, van Dijk M, Bonvin AM. The HADDOCK web server for data-driven biomolecular docking. Nat Protoc. 2010;5:883–897. doi: 10.1038/nprot.2010.32. [DOI] [PubMed] [Google Scholar]
- Wenzel DM, Lissounov A, Brzovic PS, Klevit RE. UBCH7 reactivity profile reveals parkin and HHARI to be RING/HECT hybrids. Nature. 2011a;474:105–108. doi: 10.1038/nature09966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenzel DM, Stoll KE, Klevit RE. E2s: structurally economical and functionally replete. Biochem J. 2011b;433:31–42. doi: 10.1042/BJ20100985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickliffe KE, Lorenz S, Wemmer DE, Kuriyan J, Rape M. The mechanism of linkage-specific ubiquitin chain elongation by a single-subunit E2. Cell. 2011a;144:769–781. doi: 10.1016/j.cell.2011.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickliffe KE, Williamson A, Meyer HJ, Kelly A, Rape M. K11-linked ubiquitin chains as novel regulators of cell division. Trends Cell Biol. 2011b;21:656–663. doi: 10.1016/j.tcb.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P, Duong DM, Seyfried NT, Cheng D, Xie Y, Robert J, Rush J, Hochstrasser M, Finley D, Peng J. Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell. 2009;137:133–145. doi: 10.1016/j.cell.2009.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y, Meyer HH, Rapoport TA. Function of the p97-Ufd1-Npl4 complex in retrotranslocation from the ER to the cytosol: dual recognition of nonubiquitinated polypeptide segments and polyubiquitin chains. J Cell Biol. 2003;162:71–84. doi: 10.1083/jcb.200302169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y, Rape M. Building ubiquitin chains: E2 enzymes at work. Nat Rev Mol Cell Biol. 2009;10:755–764. doi: 10.1038/nrm2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.