Abstract
Picornavirus Type 1 IRESs comprise five principal domains (dII–dVI). Whereas dV binds eIF4G, a conserved AUG in dVI was suggested to stimulate attachment of 43S ribosomal preinitiation complexes, which then scan to the initiation codon. Initiation on Type 1 IRESs also requires IRES trans-acting factors (ITAFs), and several candidates have been proposed. Here, we report the in vitro reconstitution of initiation on three Type 1 IRESs: poliovirus (PV), enterovirus 71 (EV71), and bovine enterovirus (BEV). All of them require eIF2, eIF3, eIF4A, eIF4G, eIF4B, eIF1A, and a single ITAF, poly(C) binding protein 2 (PCBP2). In each instance, initiation starts with binding of eIF4G/eIF4A. Subsequent recruitment of 43S complexes strictly requires direct interaction of their eIF3 constituent with eIF4G. The following events can differ between IRESs, depending on the stability of dVI. If it is unstructured (BEV), all ribosomes scan through dVI to the initiation codon, requiring eIF1 to bypass its AUG. If it is structured (PV, EV71), most initiation events occur without inspection of dVI, implying that its AUG does not determine ribosomal attachment.
Keywords: eIF4G, IRES, PCBP2, poliovirus, translation initiation
Introduction
During canonical eukaryotic translation initiation, 43S preinitiation complexes (comprising 40S subunits, eIF2/GTP/Met-tRNAiMet, eIF3, eIF1 and eIF1A) attach to the cap-proximal region of mRNA and scan along the 5′ untranslated region (5′ UTR) to the initiation codon, where they form 48S initiation complexes with established condon-anticodon base-pairing (Jackson et al, 2010). Attachment of 43S complexes is promoted by eIF4F, eIF4A and eIF4B. eIF4F consists of three subunits: eIF4E (a cap-binding protein), eIF4A (a DEAD-box RNA helicase), and eIF4G (a scaffold for eIF4E and eIF4A, which also binds to eIF3). eIF4A's helicase activity is stimulated by eIF4G and eIF4B. eIFs 4A, 4B and 4F cooperatively unwind the cap-proximal region of mRNA and are thought to promote binding of 43S complexes via the eIF3-eIF4G interaction. They also assist 43S complexes during scanning, but scanning on highly structured 5′ UTRs additionally requires the DExH-box protein DHX29 (Pisareva et al, 2008; Dhote et al, 2012). The key role in maintaining the fidelity of initiation codon selection belongs to eIF1, which discriminates against initiation at non-AUG codons and AUG triplets that have poor nucleotide context or are too close to the 5′-end of mRNA. Start codon recognition and formation of a 48S complex results in dissociation of eIF1, which triggers eIF5-induced hydrolysis of eIF2-bound GTP and release of Pi (Unbehaun et al, 2004). Subsequent joining of a 60S subunit and dissociation of eIFs are mediated by eIF5B (Pestova et al, 2000).
A number of viral mRNAs are translated following initiation by non-canonical 5′ end-independent mechanisms that are collectively termed ‘internal ribosomal entry’. Viral internal ribosomal entry sites (IRESs) are classified into several major groups, based on common sequence motifs and structure. Although each group uses a different mechanism to recruit 40S subunits to the initiation codon, these mechanisms are all based on non-canonical interactions of the IRES with canonical components of the translation apparatus (Jackson et al, 2010). Thus, initiation on picornavirus Type 1 (e.g. poliovirus and Human rhinovirus type 2 (HRV2)) and Type 2 (e.g. encephalomyocarditis virus (EMCV) and foot-and-mouth disease virus (FMDV)) IRESs relies on their specific interaction with the central eIF4A-binding domain of eIF4G (Pestova et al, 1996b; Lomakin et al, 2000; Pilipenko et al, 2000; de Breyne et al, 2009). This interaction allows these IRESs to bypass the requirement for intact eIF4F and to retain activity in virus-infected cells, when host cell translation is ‘shut-off’ following cleavage by viral proteases of eIF4G into an N-terminal fragment that binds eIF4E and a C-terminal fragment that binds eIF4A and eIF3 (Gradi et al, 1998).
Type 1 and Type 2 IRESs are both approximately 450 nt long and consist of five domains, designated II–VI in Type 1 and H–L in Type 2 IRESs. Sequence and secondary structure is conserved within each group, but similarity between groups is minimal except for a Yn-Xm-AUG motif at the 3′-border of each IRES, in which a Yn pyrimidine tract (n = 8–10 nt) is separated by a spacer (m = 18–20 nt) from an AUG triplet. Consequently, the binding sites for eIF4G in domain V in Type 1 IRESs and in domain J in Type 2 IRESs are not homologous. Moreover, whereas the AUG of the Yn-Xm-AUG motif commonly serves as the initiation codon for the viral polyprotein in Type 2 IRESs, in wt Type 1 IRESs it is sequestered within domain VI (dVI) and appears to be silent or only weakly active (Pestova et al, 1994; Kaminski et al, 2010). This triplet has been proposed to stimulate ribosomal entry onto Type 1 IRES, while translation of the viral polyprotein starts at an AUG that is from approximately 30 nt (in HRV2) to approximately 160 nt (in e.g. poliovirus) downstream.
In vitro reconstitution of initiation on Type 2 IRESs established its factor requirements and the outline of its mechanism (Pestova et al, 1996a,b1996b; Lomakin et al, 2000; Pilipenko et al, 2000; Yu et al, 2011a). Thus, 48S complex formation strictly required eIF2, eIF3, the central domain of eIF4G (eIF4G736–1115, designated as ‘eIF4Gm’) and eIF4A, and was enhanced by eIF4B. The requirement for eIF1 and eIF1A was evident only for the FMDV IRES during initiation on a second, downstream AUG (Andreev et al, 2007). After binding to Type 2 IRESs, the eIF4A/eIF4G complex promotes restructuring of their 3′-borders (Kolupaeva et al, 2003), which was suggested to facilitate direct attachment of 43S complexes to the initiation codon. In addition to canonical eIFs, Type 2 IRESs also require specific IRES trans-acting factors (ITAFs). The common ITAF for Type 2 IRESs is the pyrimidine tract binding protein (PTB), but the FMDV IRES additionally requires ITAF45 (Pilipenko et al, 2000).
Initiation on Type 1 IRESs has not been reconstituted in vitro, and much less is known about its mechanism. It has been established that eIF4G binds specifically to domain V of Type 1 IRESs, that this interaction is important for IRES function (de Breyne et al, 2009), that initiation requires eIF4A (Pause et al, 1994), and that as in the case of Type 2 IRESs, eIF4G and eIF4A induce conformational changes downstream of their binding site (de Breyne et al, 2009). However, the exact factor requirements for initiation on Type 1 IRESs remain unknown, and it is also not clear where exactly 43S complexes attach and how they reach the authentic initiation codon. The minimal set of ITAFs required for initiation on Type 1 IRESs has also not yet been established, and a large number of different proteins have been implicated in this process, including poly(C) binding protein 2 (PCBP2; Blyn et al, 1996, 1997; Gamarnik & Andino, 1997), glycyl-tRNA synthetase (GARS; Andreev et al, 2012), La (Meerovitch et al, 1993), PCBP1 (Choi et al, 2004), PTB (Borman et al, 1993; Hellen et al, 1993), SRp20 (Bedard et al, 2007) and upstream of N-Ras (unr; Hunt et al, 1999; Anderson et al, 2007).
Here we report the first reconstitution in vitro of initiation on three distinct Type 1 IRESs from poliovirus (PV), enterovirus 71 (EV71), and bovine enterovirus (BEV) using individual purified translational components. These studies have identified a common set of canonical initiation factors and ITAFs that is necessary and sufficient for accurate and efficient 48S complex formation on these IRESs, and have established the outline of the mechanism of this process.
Results
Factor requirements for ribosomal recruitment to the PV IRES
To facilitate biochemical analysis of PV IRES-mediated initiation, this process was divided into two stages: initial ribosomal recruitment and subsequent scanning to AUG743. To investigate recruitment, we assayed 48S complex formation at AUG586 located at the 3′-border of the IRES. In wt PV mRNA, AUG586 has a poor nucleotide context, and its optimization results in activation of AUG586 in vivo and in cell-free extracts (Pestova et al, 1994). Therefore for our studies, we employed an mRNA comprising PV nts 1–629 with AUG586 in an optimized context, linked to a PV polymerase coding sequence (PV AUG586-good mRNA; Fig 1A). Notably, context optimization also resulted in partial destabilization of dVI.
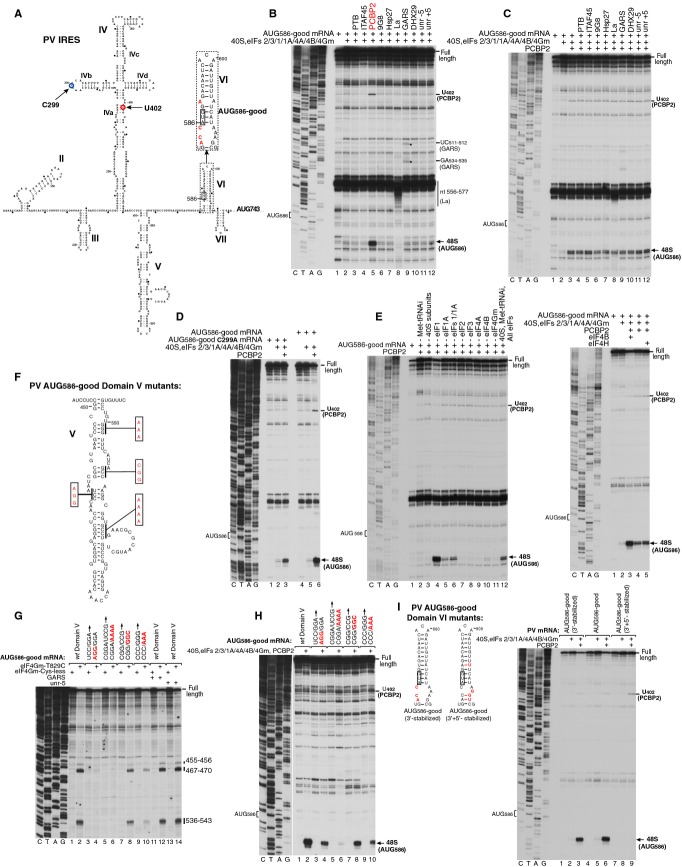
Figure 1.
Factor requirements for initiation at the 3’-border of the PV IRES.
A Model of the PV1M IRES (Genbank V01149). Inset panel shows mutations (red letters) and the structure of dVI in AUG586-good mRNA.
B–E Toeprinting analysis of 48S complex formation at AUG586 on (B, C, E) AUG586-good mRNA, (D) AUG586-good mRNA containing the C299A substitution.
F AUG586-good mRNA containing mutations in domain V.
G Binding of eIF4G to AUG586-good mRNA containing mutations in domain V as indicated in (F), analyzed by HRC using the eIF4Gm(T829C) mutant. Cleavage sites are indicated on the right.
H, I Toeprinting analysis of 48S complex formation at AUG586 on (H) AUG586-good mRNA containing mutations in domain V and (I) AUG586-good mRNA containing mutations in dVI shown in the left panel.
Data information: For (B–E, H, I), 48S complexes were formed at 100 mM K+. Reaction mixtures contained 40S subunits, Met-tRNAiMet and indicated sets of eIFs and ITAFs. Toeprints caused by binding of GARS, La or PCBP2 and by 48S complexes assembled at AUG586 are shown on the right. The division between sequence lanes and lanes 1–10 in (H) indicates that these two sets of lanes were derived from the same gel, exposed for different lengths of time.
48S complexes were assembled using canonical eIFs (1, 1A, 2, 3, 4A, 4B and 4Gm) supplemented with ITAFs that have been implicated in initiation on Type 1 IRESs (GARS, La, ITAF45, PCBP1, PCBP2, PTB, SRp20 and both unr isoforms), as well as 9G8 (which binds to murine PCBP2; Funke et al, 1996), hsp27 (which binds to eIF4Gm; Cuesta et al, 2000) and DHX29. 48S complex formation was assayed by toeprinting, which involves extension by reverse transcriptase of a primer annealed to the ribosome-bound mRNA. Arrest of cDNA synthesis by the bound 40S subunit yields a ‘toeprint’ at its leading edge, 15–17 nt. downstream of the P site codon. Whereas only trace amounts of 48S complexes assembled at AUG586 in the presence of canonical eIFs alone, 48S complex formation became very efficient when eIFs were supplemented by PCBP2 (Fig 1B) or PCBP1 (supplementary Fig S1). No other ITAF complemented the canonical eIFs or augmented PCBP2's activity (e.g. Fig 1C), even though La and GARS both bound specifically to the IRES, as previously reported (Meerovitch et al, 1993; Andreev et al, 2012). Thus, La's interaction with the IRES led to the appearance of toeprints at nts 556–577 between domains V and VI, whereas binding of GARS resulted in toe-prints at UC511–512 and GA534–535 in domain V (Fig 1B). Consistent with the reported binding of PCBP2 to domain IV (Silvera et al, 1999), inclusion of PCBP2 in reaction mixtures yielded a toe-print at U402 (Figs 1A and B). This toe-print was abrogated by a C299A substitution in subdomain IVb (Fig 1A) reflecting a defect in PCBP2 binding, which correlated with an approximately 60% reduction in 48S complex formation at AUG586 (Fig 1D).
Omission experiments (Fig 1E, left panel) showed that eIFs 2, 3, 4A and 4Gm were essential, whereas eIFs 1A and 4B strongly stimulated 48S complex formation. eIF4B could not be replaced by eIF4H, which is homologous to eIF4B's RRM domain (Fig 1E, right panel). Interestingly, eIF1 substantially reduced 48S complex formation even though AUG586 had good nucleotide context.
To confirm that as for the native AUG743 (de Breyne et al, 2009), initiation on AUG586 also depends on specific interaction of eIF4Gm with domain V, we introduced mutations to disrupt base-pairing in its basal half (Fig 1F) and assayed binding of eIF4G by directed hydroxyl radical cleavage (HRC) using an eIF4Gm mutant containing the single surface-exposed Cys829 (Kolupaeva et al, 2003). In this approach, locally generated hydroxyl radicals cleave mRNA in the vicinity of Fe(II) tethered to a unique cysteine residue on the surface of mRNA-bound protein via the linker 1-(p-bromoacetamidobenzyl)-EDTA (BABE). Reduction in eIF4G binding (Fig 1G) correlated with reduced 48S complex formation at AUG586 (Fig 1H), confirming that the latter depends on eIF4Gm's specific interaction with domain V. Although GARS and unr-5 also interact with domain V of different Type 1 IRESs (Anderson et al, 2007; Andreev et al, 2012; Fig 1B), they did not interfere with binding of eIF4Gm (Fig 1G).
Next, we investigated whether destabilization of dVI in AUG586-good mRNA contributed to the ribosomal accessibility of AUG586, influencing the efficiency of 48S complex formation. Whereas restoration of the secondary structure of the upper part of dVI (A590-U608) did not affect this process, additional restoration of base-pairing in the base by compensatory AAG614–616GGU substitutions abrogated 48S complex formation at AUG586 despite its optimal context (Fig 1I). Destabilization of dVI was therefore required for efficient 48S complex assembly at AUG586 in the in vitro reconstituted system and likely also contributed to its efficient utilization in vivo (Pestova et al, 1994).
48S complex formation on the authentic initiation codon AUG743 of wt poliovirus mRNA
In contrast to AUG586 of AUG586-good mRNA, 48S complexes did not form at AUG743 on wt PV mRNA in similar buffer conditions (Fig 2A, left panel). We therefore considered that initiation on wt mRNA, in which dVI is intact, might additionally require DHX29 to unwind this domain to facilitate accommodation of mRNA in the 40S subunit's mRNA-binding cleft, in a manner analogous to initiation on the Aichivirus IRES, which shares structural features with both Type 1 and Type 2 IRESs, and in which the initiation codon is sequestered in a hairpin at the IRES's 3′-border (Yu et al, 2011b). However, DHX29 did not promote 48S complex formation at AUG743 of wt PV mRNA (Fig 2A, left panel) and did not restore initiation at AUG586-good mRNA with a re-stabilized dVI (Fig 2A, right panel).
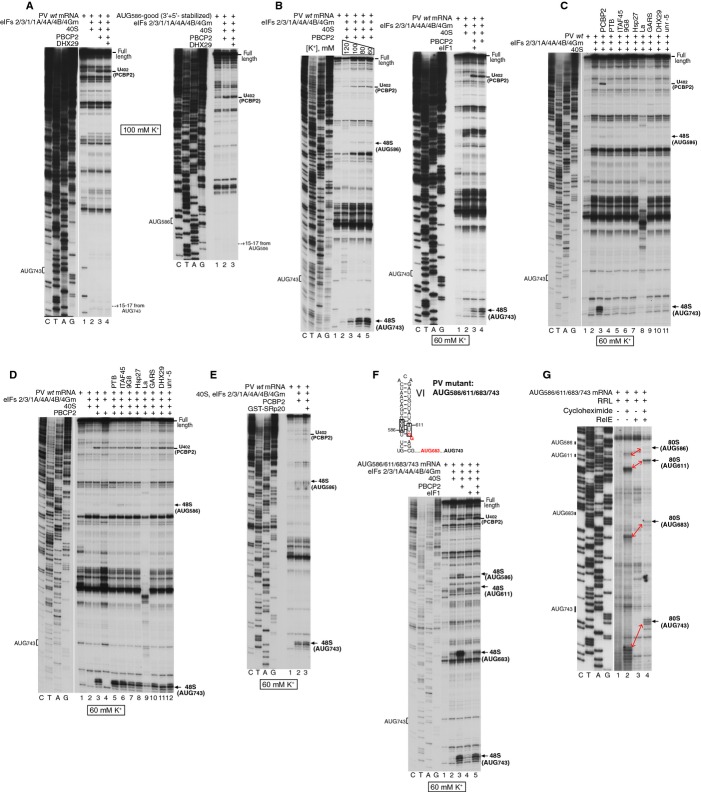
Figure 2.
48S complex formation at the PV initiation codon AUG743.
A–F Toeprinting analysis of 48S complex formation on (A, left panel; B–E) PV wt mRNA, (A, right panel) AUG586 (3′+5′ stabilized) mRNA (Fig 1I), and (F) AUG586/611/683/743 mRNA (F, upper panel). 48S complexes were formed at indicated K+ concentrations. Reaction mixtures contained 40S subunits, Met-tRNAiMet and indicated sets of eIFs and ITAFs. Toeprints caused by 48S complexes assembled at AUG586, AUG611, AUG683 and AUG743 are shown on the right.
G Analysis of 80S ribosomal complexes formed on AUG586/611/683/743 mRNA in RRL in the presence of cycloheximide. After assembly, ribosomal complexes were separated by SDG centrifugation, and fractions corresponding to 80S ribosomes were assayed by toe-printing (lanes 1–2) or RelE cleavage (lanes 3–4). Positions of toe-prints and RelE cleavages corresponding to ribosomal complexes formed at AUG586, AUG611, AUG683 and AUG743 are indicated.
Data information: The divisions between sequence lanes and toe-printing lanes in (A, left panel), (C), (D) and (G) indicate that the two sets of lanes shown in each panel were derived from the same gel, exposed for different lengths of time.
Initiation on Type 1 IRESs has been reported to have a low KCl optimum (Jackson, 1991; Borman et al, 1995), and considerable K+ efflux occurs in poliovirus-infected cells 3 h post-infection (e.g. López-Rivas et al, 1987). This prompted us to investigate 48S complex formation on wt PV mRNA at lower [K+]. Progressive reduction of [K+] from 120 to 60 mM resulted in efficient 48S complex formation at AUG743 in reaction mixtures containing eIFs 2, 3, 1A, 4A, 4B, 4Gm and PCBP2 (Fig 2B, left panel). Interestingly, only trace amounts of 48S complexes formed at AUG586 on wt mRNA despite the absence of eIF1, which discriminates against AUGs in poor nucleotide context (Pestova & Kolupaeva, 2002). These observations suggest that if dVI is intact, ribosomes cannot efficiently inspect the AUG586 codon. Like for initiation at AUG586 (Fig 1E), eIF1 also reduced 48S complex formation at AUG743 on wt PV mRNA (Fig 2B, right panel).
48S complex formation at AUG743 was again dependent on PCBP2 (Fig 2B, right panel), which could not be replaced by other ITAFs (Fig 2C) but was modestly (by ˜20%) augmented by PTB (Fig 2D). PCBP2 binds to SRp20 (Bedard et al, 2007), and since we were unable to obtain it as a soluble recombinant protein, instead used a GST-SRp20 fusion. No influence of GST-SRp20 on PCBP2-dependent 48S complex formation at AUG743 was observed (Fig 2E).
Because our data suggest that efficient ribosomal inspection of wt PV mRNA begins downstream of AUG586, we introduced additional AUGs in the spacer between AUG586 and the native initiation codon (Fig 2F, upper panel) to investigate which could be recognized effectively. The first AUG (AUCAUGG) was placed at nt. 611 in the opposite strand to AUG586, and the second (ACCAUGG) at nt. 683. In the presence of eIFs 2, 3, 1A, 4A, 4B, 4Gm and PCBP2, AUG611 was recognized as weakly as AUG586, whereas AUG683 was selected even more frequently than AUG743 (Fig 2F). This observation suggests that only a few 43S complexes were able to efficiently inspect dVI and the majority began productive inspection somewhere downstream of this domain but before nt. 683. Even though inclusion of eIF1 somewhat reduced 48S complex formation at AUG743, it had much stronger effect on AUG683, indicating that additional negative determinants beyond the immediate context nucleotides might render initiation complexes at AUG683 susceptible to destabilization by eIF1 (Fig 2F, lane 5). As a result, AUG743 became the preferred codon. eIF1 also slightly inhibited 48S complex formation at AUG611 and eliminated it at AUG586. The factor requirements for initiation at AUG611, AUG683 and AUG743 (supplementary Fig S2) were similar to those for initiation at AUG586 in AUG586-good mRNA (Fig 1E).
Because AUG611 was reported to promote efficient initiation in cell-free translation extracts (Hellen et al, 1994), we compared initiation at AUG586, AUG611, AUG683 and AUG743 in RRL. For this, AUG586/611/683/743 mRNA was incubated in the lysate in the presence of cycloheximide, which inhibits elongation after the first round of translocation (Pestova & Hellen, 2003). Assembled complexes were separated by sucrose density gradient (SDG) centrifugation, and fractions corresponding to 80S ribosomal complexes were analyzed by primer extension with or without prior treatment with the bacterial toxin RelE, which cleaves mRNA after the first and second A-site nucleotides (Andreev et al, 2008; Neubauer et al, 2009). Initiation at AUG611 was almost as efficient as at AUG743, whereas substantially fewer complexes formed at AUG683 (Fig 2G). This indicates that the proportion of 43S complexes that were able to inspect dVI in cell-free translation extracts was higher than in the in vitro reconstituted system. Interestingly, in the HRV2 IRES, in which the initiation codon is located at a position equivalent to PV AUG611, a proportion of initiation events could occur without unwinding and inspection of the apical region of dVI (Kaminski et al, 2010), and it is therefore similarly possible that only the basal region of PV dVI was unwound in cell-free extracts to allow ribosomal access to AUG611, while the apex remained base-paired. Initiation at AUG586 was very inefficient, but nonetheless detectable.
Initiation on enterovirus 71 and bovine enterovirus IRESs
To investigate whether initiation on the PV IRES (a member of the species Enterovirus C) is representative of initiation on other Type 1 IRESs, we characterized 48S complex formation on IRESs from EV71 (a member of Enterovirus A) and BEV (a member of Enterovirus E; Zell et al, 1999; Thompson & Sarnow, 2003). EV71 and PV IRESs share approximately 75% sequence identity and have similar structures, including dVI (Fig 3A). However, sequence identity between BEV and these IRESs is below 70%, some structural elements in the BEV IRES are divergent, such as the weakly base-paired dVI (Fig 3A), and it lacks an equivalent of domain VII (dVII).
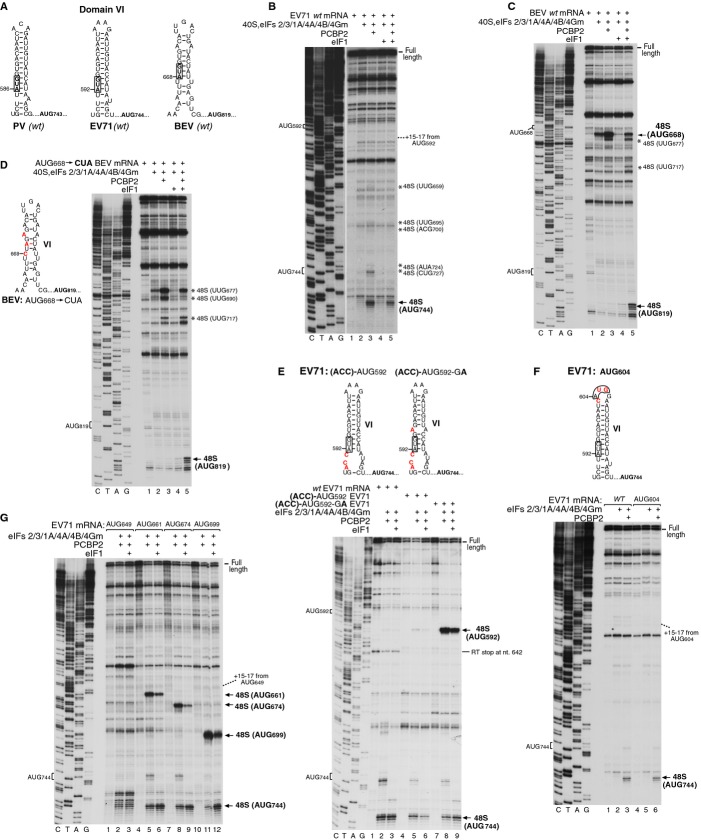
Figure 3.
48S complex formation on EV71 and BEV IRESs.
A Models of dVI in wt PV, EV71 and BEV IRESs.
B–G Toe-printing analysis of 48S complex formation on (B) wt EV71 mRNA, (C) wt BEV mRNA, (D) AUG668→CUA BEV mRNA (left panel), (E) EV71 mRNAs containing destabilizing mutations in dVI (upper panel), (F) EV71 mRNA containing the additional AUG604 (upper panel), and (G) EV71 mRNAs containing additional AUG triplets at positions 649, 661, 674 or 699. 48S complexes were formed at 100 mM K+. Reaction mixtures contained 40S subunits, Met-tRNAiMet and indicated sets of eIFs and ITAFs. Asterisks show toe-prints caused by 48S complexes assembled on near-cognate initiation codons. Toeprints caused by 48S complexes assembled on AUG triplets are indicated by arrows. The division between sequence lanes and lanes 1–5 in (B) indicates that these two sets of lanes were derived from the same gel, exposed for different lengths of time.
48S complex formation was conducted with and without eIF1 to allow complexes to assemble at the poor context AUG codon in dVI at the 3′-border of the IRESs. Although in contrast to the PV IRES, initiation on EV71 and BEV IRESs was relatively efficient at 100 mM K+ (Figs 3B and C), it nevertheless increased when [K+] was reduced to 60 mM (supplementary Fig S3). As for PV, 48S complex formation on these IRESs strongly depended on PCBP2 (Figs 3B and C). However, significant differences were observed in the recognition of the AUG in dVI in the absence of eIF1. Thus, EV71 AUG592 was not recognized, and 48S complexes formed predominantly at the authentic initiation codon AUG744 and to a small extent at intervening near-cognate initiation codons starting with UUG659 (Fig 3B). Although initiation at AUG592 in a cell-free extract was detectable, its level was extremely low (supplementary Fig S4). Notably, utilization of near-cognate codons was very weak, and occurred at a much lower level than during 5′-end-dependent initiation (Pestova & Kolupaeva, 2002). By contrast, on the BEV IRES 48S complexes formed predominantly at AUG668, to a lesser extent at near-cognate codons in dVI and immediately downstream from it (UUG677 and UUG717), but did not form at more distal near-cognate codons or at the authentic initiation codon AUG819 (Fig 3C). Interestingly, 48S complex formation at AUG668 on BEV mRNA was less dependent on PCBP2 than at PV AUG586 on AUG586-good mRNA (Fig 1B).
On the EV71 IRES, eIF1 increased the fidelity of selection of the authentic initiation codon by nearly eliminating initiation at near-cognate codons. However, it also reduced the level of 48S complex formation at the native start codon, as noted above for PV. On BEV mRNA, eIF1 enabled 48S complexes to form at the native AUG819 and reduced their assembly at AUG668 and at downstream near-cognate codons. Inclusion of other ITAFs did not further reduce 48S complex formation at these near-cognate codons (supplementary Fig S5). The fact that eIF1 did not abrogate 48S complex formation at near-cognate codons suggests that if dVI is unstructured so that initiation complexes can assemble at near-cognate codons in or immediately downstream of it, then the IRES stabilizes such complexes, making them less sensitive to eIF1-induced dissociation.
In the absence of eIF1, 48S complexes assembled very efficiently at BEV AUG668, progressively less efficiently at downstream near-cognate codons, but did not form at all at the native initiation codon AUG819, implying that ribosomal entry occurs at or upstream of AUG668, and that in the presence of eIF1, 43S complexes then reach AUG819 by leaky scanning. Interestingly, substitution of AUG668 and the following GUG by non-cognate CUA and GAG triplets, respectively, did not result in 48S complex formation at AUG819 in the absence of eIF1, and 48S complexes assembled only at the upstream near-cognate codons (Fig 3D). Inclusion of eIF1 again promoted 48S complex formation at AUG819 (Fig 3D) at a level that was very similar to that observed on the wt IRES (Fig 3C).
Although AUG592 in dVI of the wt EV71 IRES could not act as an initiation codon, similarly to PV, destabilization of the lower part of the EV71 dVI by optimizing the context of AUG592 (which was manifested by the disappearance of a strong RT stop at nt 642) permitted low-efficiency 48S complex formation at this codon (Fig 3E, lanes 4–6). Additional destabilization of the upper part of dVI by an U596A substitution strongly enhanced recognition of AUG592 (Fig 3E, lanes 7–9). Thus, as for the PV IRES, efficient inspection of dVI of the EV71 IRES required disruption of its secondary structure. Notably, efficient recognition of AUG592 had only a minor inhibitory effect (˜2-fold reduction) on 48S complex formation on the authentic initiation codon AUG744. Similarly, strong enhancement of 48S complex formation at PV dVI AUG586 due to improvement of its nucleotide context resulted in only a minor reduction of initiation at authentic AUG743 (supplementary Fig S6). These data are consistent with the results of previously reported in vitro translation experiments performed using cell-free extracts, where optimization of the nucleotide context of PV AUG586 resulted in an overall increase in the number of initiation events, with strongly enhanced utilization of AUG586 but initiation at AUG743 reduced by only twofold (Pestova et al, 1994). However, the mechanism by which the authentic AUG codon is selected in this case is not clear, and it cannot strictly be excluded that when dVI is destabilized, all 43S complexes inspect dVI and a small proportion reaches the authentic initiation codon by leaky scanning past the dVI AUG.
To localize the site from which 43S complexes could start inspecting the wt EV71 IRES, additional AUGs were introduced at the apex of dVI (AUG604) to minimize disruption of base-pairing, and at several positions (AUG649, AUG661, AUG674 and AUG699) between dVII and the native initiation codon AUG744. As expected, no 48S complexes assembled on AUG604 (Fig 3F). However, AUG649 located 5 nts downstream of dVII was also not recognized (Fig 3G, lanes 1–3), and 48S complex formation started only from AUG661 (Fig 3G, lanes 4–12).
The influence of mutations in domain VI on initiation on PV and EV71 IRESs
Although our data indicate that dVI of PV and EV71 IRESs is not efficiently inspected in the in vitro reconstituted system, the conserved dVI AUG, which is a part of the Yn-Xm-AUG motif, has previously been suggested to form part of the ‘ribosome landing pad’ on Type 1 IRESs, because mutations in this AUG reduced translation of PV, HRV2 and coxsackievirus B1 mRNAs by approximately 70% in a HeLa cell-free translation extract and in RRL supplemented with HeLa cell high-salt S100 cytoplasmic fraction, and to a more variable extent in transfected cells, and this defect was not suppressed by compensatory substitutions restoring the secondary structure of dVI (Iizuka et al, 1991; Meerovitch et al, 1991; Nicholson et al, 1991; Kaminski et al, 2010).
To obtain further insights into this matter, we assayed the activity of PV, EV71 and BEV IRESs containing mutations in dVI (Figs 4A,B and 3D) in cell-free translation extracts and in the in vitro reconstituted system. First, PV AUG586 and EV71 AUG592 were replaced by AAG triplets (the mutation in dVI AUG that had the strongest effect on PV translation in a HeLa cell-free extract; Meerovitch et al, 1991), with or without compensatory mutations in the 3′-half of dVI (Figs 4A and B). Second, dVI was destabilized by substitutions in the 3′-half of the stem (Figs 4A and B). Third, these destabilizing mutations were combined with PV AUG586→AAG or EV71 AUG592→AAG substitutions (Figs 4A and B). Fourth, the EV71 AUG592→CUA mutation was combined with an adjacent GUG595→GAG substitution (with and without compensatory mutations in the 3′-half of dVI), in case the near-cognate GUG could functionally replace AUG592 when it is mutated (Fig 4B). Finally, BEV AUG668 and the following GUG were substituted by non-cognate CUA and GAG triplets, respectively (Fig 3D).
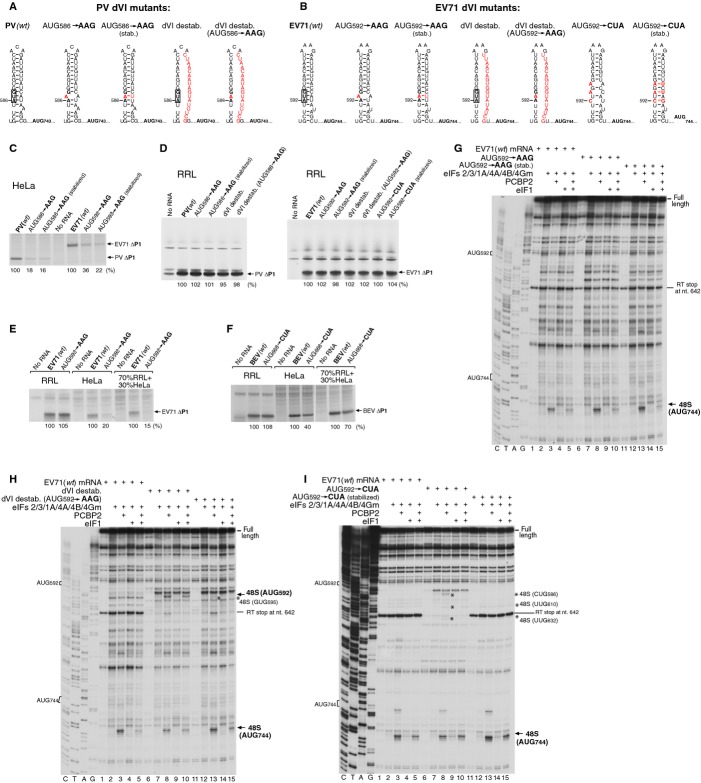
Figure 4.
Influence of mutations in domain VI on 48S complex formation on PV and EV71 IRESs.
A, B Mutations (red) introduced into dVI of (A) PV and (B) EV71 IRESs.
C–F Translation of mRNAs containing mutations in dVI of PV, EV71 (A and B) and BEV (Fig 3D) IRESs in RRL, HeLa cell extract, and RRL supplemented with HeLa cell extract (30%, v/v), as indicated. Translation products were quantified relative to those of wt mRNAs, which were defined as 100%.
G–I Toe-printing analysis of 48S complex formation on EV71 mRNAs containing mutations in dVI (B). 48S complexes were formed at 100 mM K+. Reaction mixtures contained 40S subunits, Met-tRNAiMet and indicated sets of eIFs and PCBP2. Asterisks show toe-prints caused by 48S complexes assembled on near-cognate initiation codons. Toeprints caused by 48S complexes assembled on AUG triplets are indicated by arrows.
Translation was assayed in RRL (Promega Corp., Madison, WI, USA), in Hela cell-free extract (Takara Bio Inc., Otsu, Shiga, Japan), and in a 70:30% (v/v) mixture of the two. Consistent with the previous reports, AUG586→AAG and AUG592→AAG substitutions (with or without compensatory mutations) strongly reduced translational activity of PV and EV71 IRESs in the HeLa extract (Fig 4C). In contrast, no major differences between the levels of translation of wt and any of the mutant PV and EV71 mRNAs were observed in RRL (Fig 4D). However, sensitivity to mutations in dVI AUG was restored by supplementation of RRL with only 30% (v/v) of HeLa extract (Fig 4E). Qualitatively similar results were obtained in the case of BEV AUG668→CUA mutant, although mutations in BEV dVI had less effect on the IRES function compared to PV and EV71 IRESs (Fig 4F). Taking into account the similar levels of activity of the wt IRESs in RRL compared to the HeLa extract (Figs 4E and F), restoration of sensitivity to dVI AUG mutations by addition of only 30% HeLa extract would be more consistent with the presence in a HeLa extract of an inhibitory activity that specifically targets mutant IRESs, rather than a stimulatory activity that enhances translation of wt mRNAs.
In the in vitro reconstituted system, the AUG→AAG substitution in EV71 dVI did not affect the efficiency of 48S complex formation at AUG744, and the minor weakening of the RT stop at nt. 642 showed that it only slightly destabilized dVI (Fig 4G, lanes 6–10). The additional compensatory mutation strengthened dVI (evidenced by a slight increase in the intensity of the RT stop at nt. 642), but similarly did not affect 48S complex formation (Fig 4G, lanes 11–15).
Destabilization of dVI due to mutations in its 3′-half resulted in significant reduction in 48S complex formation at AUG744 in the absence of eIF1, because 43S complexes gained access to dVI, and a substantial proportion of them now stopped at AUG592 (Fig 4H; compare lanes 3 and 8). Inclusion of eIF1 reduced 48S complex formation at AUG592 (Fig 4H, lanes 8, 10), so that the level of initiation at AUG744 was again similar to that on wt mRNA (Fig 4H, lanes 5, 10). However, residual 48S complex formation at AUG592 nevertheless remained high, so that the proportion of initiation events occurring at AUG592 in the in vitro reconstituted system was substantially greater than on the wt mRNA in RRL (supplementary Fig S4). This suggests that in the majority of initiation events in RRL, dVI of the wt mRNA was likely not unwound. Combination of the destabilizing mutations in dVI with the AUG→AAG substitution restored initiation at AUG744 to the level seen in the wt mRNA both in the presence and in the absence of eIF1 (Fig 4H, lanes 3, 5, 13, 15), and only a very low level of initiation at the near-cognate GUG595 occurred in the absence of eIF1 (Fig 4H, lane 13).
The AUG592GUG595→ CUA592GAG595 substitution destabilized dVI and led to a significant level of selection of near-cognate codons in the absence of eIF1, reducing initiation at AUG744 (Fig 4I, compare lanes 3, 8). Inclusion of eIF1 reduced 48S complex formation at these near-cognate codons, so that initiation at AUG744 reached a level similar to that on the wt mRNA (Fig 4I, compare lanes 5, 10). Compensatory substitutions that re-established base-pairing in dVI abrogated 48S complex formation on near-cognate codons so that the level of initiation at AUG744 became similar to that on the wt mRNA irrespective of the presence or absence of eIF1 (Fig 4I, lanes 11–15).
In conclusion, mutations in the conserved AUG triplet in dVI of PV and EV71 IRESs did not affect translation in RRL, or 48S complex formation at the authentic initiation codon in the in vitro reconstituted system, irrespective of whether dVI had been destabilized or not. Moreover, destabilization of dVI, which increased its accessibility for 43S complexes, did not increase the efficiency of initiation at the authentic AUG744 of the EV71 IRES. To the contrary, in the absence of eIF1, destabilization of dVI resulted in efficient utilization of the AUG or near-cognate codons in this domain, which reduced 48S complex formation at AUG744 compared to the wt mRNA. Importantly, in the presence of eIF1, none of the mutations tested here had any significant effect on the level of 48S complex formation at the authentic initiation codon. Taken together with the results of in vitro translation of dVI AUG mutants in different cell-free systems, these data suggest that the dVI AUG does not determine the basic mechanism of initiation on Type 1 IRESs, and instead might function as a regulatory element in a cell-specific manner.
The interaction of PCBP2 with Type 1 IRESs
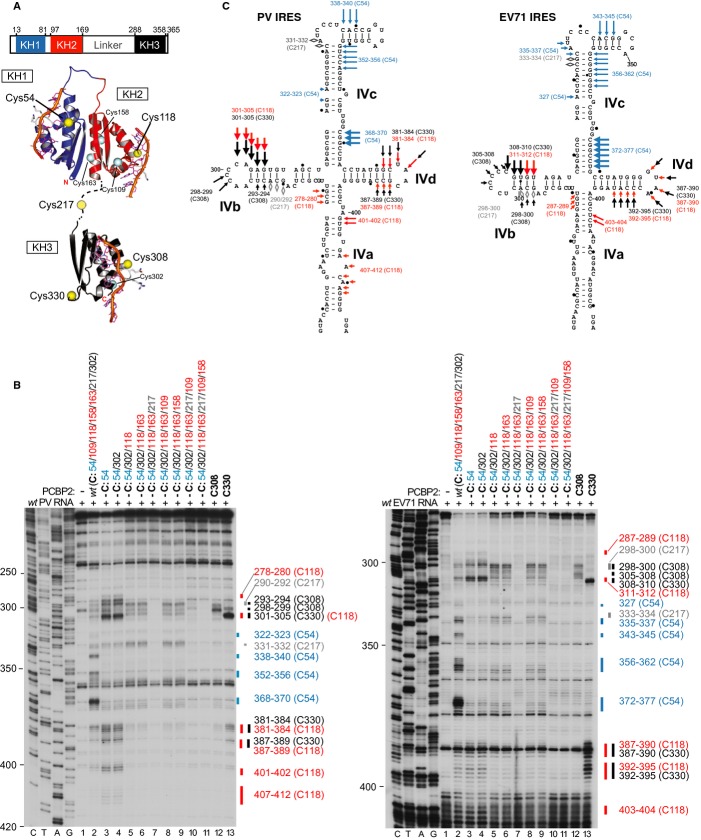
Our data indicate that PCBP2 is a common ITAF that is necessary and sufficient for 48S complex formation on Type 1 IRESs. It comprises three hnRNP K homology (KH) domains: KH1 and KH2 adopt a back-to-back configuration, whereas the C-terminal KH3 is separated from KH1/KH2 by a 119 aa-long linker and is more mobile (Fig 5A; Du et al, 2008). Each KH domain accommodates four bases in the nucleic acid binding cleft formed by α-helix 1, α-helix 2 and an invariant GXXG connecting motif on one side and β-strand 2 and a variable loop on the other (Sidiqi et al, 2005; Du et al, 2008). PCBP2 binds to domain IV of PV and CVB3 IRESs (Blyn et al, 1996; Silvera et al, 1999; Sean et al, 2009). Binding likely involves cooperative interactions of individual KH domains with distinct IRES sites, because it can be impaired by substitutions in all three KH domains. To determine the spatial arrangement of PCBP2's KH domains on EV71 and PV IRESs, we employed the HRC technique.
Figure 5.
- Schematic representation of PCBP2 showing the positions of the three KH domains (upper panel). Ribbon diagrams of PCBP2 KH1-KH2 and KH3 domains (PDB: 2JZX and 2P2R) with the linker between them represented by a dashed line (lower panel). Spheres indicate native (C54, C109, C118, C158, C163, C207, C302) and introduced (C308, C330) cysteines. Cysteines that induced cleavage are colored yellow.
- Primer extension analysis after HRC of PV (left) and EV71 (right) IRESs from Fe(II)-tethered PCBP2.
- Models of the secondary structures of apical subdomains IVa-IVd of PV and EV71 IRESs, marked to show sites of HRC from PCBP2. Arrow size is proportional to cleavage intensity. Sites of HRC (B, C) are colored to match domains in (A).
PCBP2 contains seven naturally occurring cysteines distributed between its three domains (Fig 5A). They were progressively eliminated, and new Cys residues (aa 308 and 330) were introduced into KH3. The resulting mutants containing various subsets of native cysteines or individual newly introduced Cys residues were active in 48S complex formation on the PV IRES (supplementary Fig S7) and were therefore all used in HRC experiments. Sites of cleavage were assigned to individual cysteines by correlating the loss of cleavage at different locations with the elimination of native cysteines and the appearance of additional cleavages with the introduction of novel cysteines (Fig 5B). Cleavages were observed from 5 positions: Cys54 in KH1, Cys118 in KH2, Cys217 in the linker, and Cys308 and Cys330 in KH3. The cleavage patterns in PV and EV71 IRESs were very similar (Fig 5C). Cleavage from KH1 was restricted to subdomain IVc, with strong cuts at the base and medium at the apex, consistent with the reported interaction of KH1 with subdomain IVc of the CVB3 IRES (Zell et al, 2008). Surprisingly, cleavage from KH2 and KH3 overlapped, with strong cleavage from Cys118 and Cys330 at the apex of subdomain IVb and medium-intensity cleavage in subdomain IVd. The strong cleavage from Cys330 in subdomain IVb correlates with the observed direct binding of KH3 to subdomain IVb of the CVB3 IRES (Zell et al, 2008) and with the observation that the C299A substitution impaired IRES function and binding of PCBP2 (Fig 1D). The overlapping pattern of cleavage from Cys118 and Cys330 in subdomains IVb and IVd indicates that they are close to each other, and that although KH3 has a high degree of mobility in solution (Du et al, 2008), it is in close proximity to KH2 when PCBP2 is bound to the IRES. Some additional weak cleavage from KH2 was observed in subdomain IVa. The appearance of two widely separated weak cleavage sites in IVb and IVc from C217, located midway in the linker, may reflect the mobility of the latter.
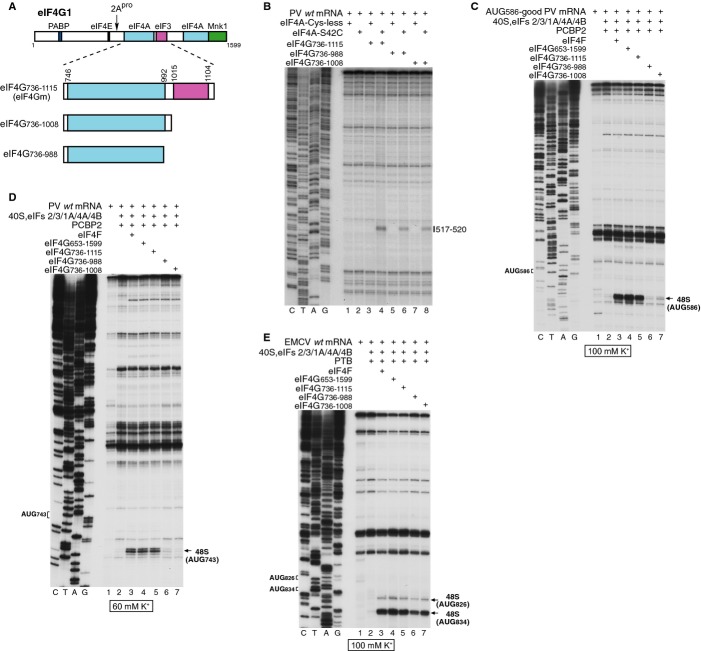
Recruitment of eIF3 by eIF4G is an essential step in 48S complex formation on Type 1 IRESs
We next investigated whether direct eIF4G/eIF3 interaction is essential for initiation on Type 1 IRESs. eIF4Gm truncation mutants lacking the C-terminal eIF3-binding domain (Fig 6A) were able to bind to PV domain V and to recruit eIF4A to the previously reported specific location (de Breyne et al, 2009) as determined by the HRC technique using eIF4A containing the single surface-exposed Cys42 (Fig 6B), albeit with a somewhat reduced efficiency. However, deletion of the eIF3-binding domain abrogated eIF4G's ability to promote 48S complex formation on PV AUG586-good and wt mRNAs (Figs 6C and D), whereas as reported previously (Lomakin et al, 2000), 48S complex formation on the Type 2 EMCV IRES remained robust (Fig 6E, lanes 4–7). Therefore in contrast to Type 2 IRESs, initiation on Type 1 IRESs requires that eIF4G directly recruits eIF3, most likely as a component of the 43S complex.
Figure 6.
Direct interaction of eIF4G with eIF3 is required for recruitment of 43S complexes to the PV IRES.
A Schematic representation of full-length and truncated eIF4G1, showing sites of factor binding and of cleavage by 2Apro.
B Recruitment of eIF4A to the PV IRES by eIF4G mutants, analyzed by HRC using the eIF4A(S42C) mutant. Cleavage sites are indicated on the right.
C–E Toeprint analysis of 48S complex formation on (C) AUG586-good PV, (D) wt PV and (E) EMCV mRNAs, in reaction mixtures that contained 40S subunits, Met-tRNAiMet, eIFs, ITAFs and eIF4F, eIF4G653–1599, eIF4G736–1115, eIF4G736–1008 or eIF4G736–988, as indicated. 48S complexes were formed at indicated K+ concentrations. Toeprints caused by 48S complexes assembled at EMCV AUG826 and AUG834, PV AUG586 and PV AUG743 are indicated on the right.
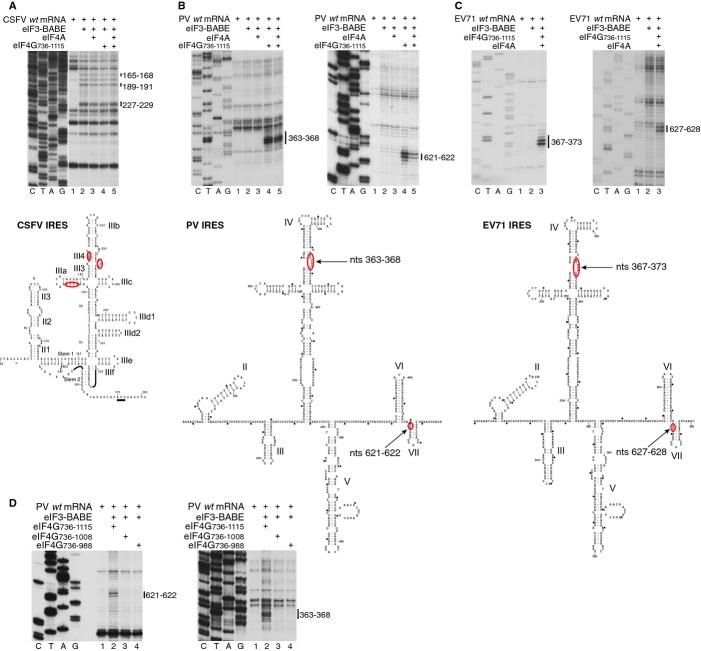
To investigate which regions of Type 1 IRESs might be positioned close to eIF3, we employed HRC using Fe(II)-BABE-derivatized native eIF3 that contains approximately 80 cysteines, some of which might be surface-exposed. To validate this approach, we first mapped cleavage from eIF3 on the classical swine fever virus (CSFV) IRES, which interacts specifically with eIF3 (Pestova et al, 1998). Fe(II)-BABE-derivatized eIF3 induced cleavage in the apical IIIa and IIIb subdomains (Fig 7A), overlapping eIF3's footprint (Sizova et al, 1998). As expected, cleavage did not depend on eIF4G and/or eIF4A (Fig 7A). In similar reactions, specific cleavage was induced in an eIF4Gm-dependent manner at PV nts 363–368 and 621–622 (Fig 7B) and at EV71 nts 367–373 and 627–628 (Fig 7C), which mapped to the apical cruciform region of domain IV and to the junction of dVI and dVII (Figs 7B and C). No cleavage was observed in the absence of eIF4Gm (Figs 7B and C), and deletion of the eIF3-binding domain from eIF4Gm abrogated its ability to promote eIF3-mediated cleavage (Fig 7D). In contrast, no specific cleavage from eIF3 was observed on the Type 2 EMCV IRES in the presence or absence of eIF4Gm (TS and CH, unpublished data).
Figure 7.
Position of eIF3 on Type 1 IRESs assayed by directed hydroxyl radical cleavage.
A–D Primer extension analysis of HRC of (A) CSFV, (B, D) PV and (C) EV71 IRESs from Fe(II)-tethered native eIF3 in the presence of eIF4G736–1115, eIF4G736–1008, eIF4G736–988 and eIF4A, as indicated. Cleavage sites are shown on the right and are mapped onto structural models of CSFV, PV and EV71 IRESs (A–C, lower panels). The division between sequence lanes and lanes 1–5 in (A) indicates that these two sets of lanes were derived from the same gel, exposed for different lengths of time.
Importantly, eIF3 in turn also stabilized eIF4Gm's interaction with Type 1 IRESs. Thus, eIF3 strongly enhanced specific cleavage induced by Fe(II)-BABE-derivatized eIF4Gm(T829C) in domain V of EV71 and PV IRESs (Figs 8A and B). By contrast, it did not increase the intensity of eIF4Gm(T829C)-induced cleavage of the EMCV IRES (Fig 8C). Instead, interaction of eIF4Gm with Type 2 IRESs is strongly enhanced by eIF4A (Lomakin et al, 2000; Kolupaeva et al, 2003), which had only a very minor effect on interaction of eIF4Gm with Type 1 IRESs (Fig 8A; de Breyne et al, 2009). There are therefore significant differences in binding of eIF4G and in the importance of its direct interaction with eIF3 for initiation on Type 1 and on Type 2 IRESs.
Figure 8.
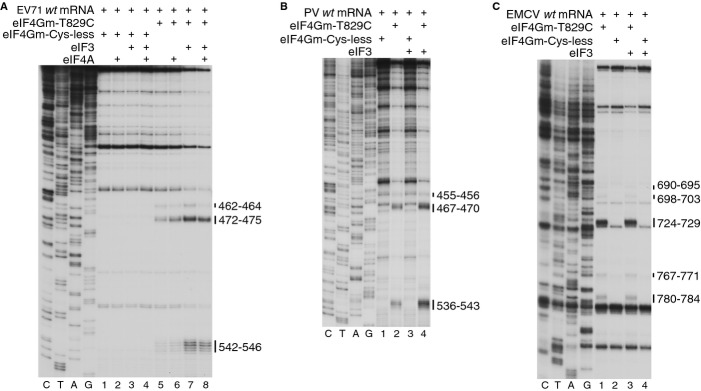
Stimulation by eIF3 of eIF4G's interaction with Type 1 IRESs.
A–C Primer extension analysis of directed hydroxyl radical cleavage of (A) EV71, (B) PV and (C) EMCV IRESs from Fe(II)-tethered eIF4Gm(T829C) in the presence of eIF3 and/or eIF4A as indicated. Cleavage sites are shown on the right.
Discussion
We have determined the minimal set of factors that promote 48S complex formation on three distinct Type 1 IRESs (PV, EV71 and BEV). In each instance, the process required eIF2, eIF3, eIF4A and the central domain of eIF4G, and was strongly stimulated by eIF1A and eIF4B. Initiation on the BEV IRES also depended on eIF1, whereas on PV and EV71 IRESs, eIF1 increased the fidelity of initiation at the authentic start site, even though this was accompanied by an overall reduction in the level of 48S complex formation. All three IRESs also required a single common ITAF, PCBP2. This finding is consistent with earlier data concerning PV IRES function obtained by PCBP2 depletion in cell-free translation extracts (e.g. Blyn et al, 1997; Gamarnik & Andino, 1997). In contrast to some reports (Blyn et al, 1997) but consistent with others (Choi et al, 2004), PCBP2 could be substituted by PCBP1. Other reported potential ITAFs (e.g. GARS, La, ITAF45, PTB, 9G8, SRp20 or unr) were not able to replace PCBP2, and only PTB slightly enhanced PCBP2-dependent 48S complex formation on the authentic initiation codon.
A fundamental point of similarity between the mechanisms of initiation on Type 1 and Type 2 IRESs is that both are based on specific interaction with eIF4G, even though its binding sites on the two types of IRESs are not homologous. A key function of eIF4G in both mechanisms is to recruit eIF4A, which in turn leads to the induction of conformational changes around the 3′-border of the IRES that were suggested to facilitate recruitment of a 43S complex (Kolupaeva et al, 2003; de Breyne et al, 2009). However, a significant difference between initiation on these two IRES classes is that whereas the eIF3-binding domain of eIF4G is not strictly required for initiation on Type 2 IRESs (Lomakin et al, 2000), it is essential for initiation on Type 1 IRESs. In addition to its essential role in recruiting eIF3, this eIF3-binding domain might also engage in interactions with the IRES that stabilize binding of eIF4G/eIF4A, because although eIF4G lacking this domain was able to bind to Type 1 IRESs and promote recruitment of eIF4A, its activity in this process was somewhat reduced. Nevertheless, this observation supports the hypothesis that interaction of eIF4G with eIF3 is an important determinant of the recruitment of 43S complexes to mRNA (Lamphear et al, 1995). eIF4G-mediated recruitment of eIF3 to Type 1 IRESs brings it into close proximity to elements at the apex of domain IV and at the base of dVI, which suggests the possibility of direct interaction between the IRES and eIF3 that might facilitate recruitment of 43S complexes. This potential eIF3-IRES interaction might also be responsible for the observed stimulation by eIF3 of binding of eIF4G to Type 1 IRESs.
Whereas PTB is the common ITAF for Type 2 IRESs (e.g. Pestova et al, 1996a; Pilipenko et al, 2000), all Type 1 IRESs that have been tested to date depend on PCBP2. Both proteins contain multiple RNA-binding domains, including one (in PCBP2; Sidiqi et al, 2005; Du et al, 2008) or two (in PTB; Oberstrass et al, 2005) that are connected by flexible linkers and two that have a fixed orientation to each other and could therefore stabilize RNA in a specific conformation by binding to two separate elements (Lamichhane et al, 2010). However, whereas PTB contacts Type 2 and Aichivirus IRESs at multiple, widely dispersed sites (Kafasla et al, 2009; Yu et al, 2011b), PCBP2 binds to a restricted region at the apex of domain IV of EV71 and PV IRESs (Gamarnik & Andino, 2000; Sean et al, 2009: this study), with the strongest interactions involving binding of KH1 and KH3 domains to subdomains IVc and IVb, respectively (this study; Zell et al, 2008). KH2 induced hydroxyl radical cleavage in subdomains IVb and IVd, which overlapped with cleavage from KH3. This observation suggests that subdomains IVb and IVd are close to each other so that binding of PCBP2 might stabilize the apex of domain IV in a specific compact conformation, and that the flexibly linked KH3 is in close proximity to KH2 when PCBP2 is bound to the IRES. The apex of domain IV is one of the most conserved elements in Type 1 IRESs (e.g. Jackson & Kaminski, 1995), suggestive of its functional importance, and interestingly, sites of strong cleavage from KH1 at the base of subdomain IVc are directly adjacent to sites of cleavage from eIF3. Consequently, PCBP2 could potentially promote recruitment of 43S complexes by direct interaction with eIF3 or even the 40S subunit, or by maintaining the active conformation of the apex of domain IV, which in turn might interact with components of the 43S complex. Low-level stimulation of PCBP2-dependent 48S complex formation by PTB is likely a consequence of its specific binding to the base of domain V, which promotes a slight reorientation of eIF4Gm (Kafasla et al, 2010).
Despite many common characteristics that define the mechanism of initiation on different Type 1 IRESs, there is one important difference, which concerns ribosomal inspection of dVI and utilization as an initiation codon of the AUG triplet of the Yn-Xm-AUG motif, which is sequestered in dVI at the 3′-border of the IRES. In all cases, this AUG is in poor nucleotide context, but even in the absence of eIF1, it is barely recognized in PV and EV71 IRESs, whereas in the BEV IRES, it is used almost exclusively. The extent to which dVI AUGs are used generally correlates with the stability of this domain: it is mostly unstructured in the BEV IRES, and destabilization of dVI in PV and EV71 IRESs results in dVI AUG activation. Unexpectedly, DHX29, which promotes unwinding of a 3′-terminal hairpin in the Aichivirus and related IRESs so that the initiation codon sequestered in it becomes accessible to 43S complexes (Yu et al, 2011b; Sweeney et al, 2012), did not stimulate 48S complex formation at dVI AUGs of Type 1 IRESs in a similar manner.
Inspection of BEV dVI by 43S complexes suggests that 48S complex formation at the BEV authentic initiation codon most likely occurred by leaky scanning past the dVI AUG, and as a result, requires eIF1. In contrast, on PV and EV71 IRESs, 43S complexes could not efficiently recognize naturally occurring or newly introduced AUGs in dVI, as long as its secondary structure was maintained, and in the case of the EV71 IRES, recognition began only from AUG661 (16 nts downstream of dVII), whereas AUG649 (5 nts downstream of dVII) was not selected. Consistently, since 43S complexes did not encounter the AUG in dVI, initiation on PV and EV71 IRESs did not strictly depend on eIF1. In RRL, very poor utilization of EV71 dVI AUG (supplementary Fig S4) could not be explained entirely by the destabilizing influence of eIF1, and would also be more consistent with infrequent inspection of dVI. Moreover, extension of dVII into a very stable hairpin (ΔG = −58 kcal/mol) reduced, but did not abrogate initiation at the authentic initiation codon of the PV IRES in various cell-free extracts (Hellen et al, 1994). Taken together, these data indicate that unwinding and inspection of dVI, and likely also dVII, is not mandatory for initiation on Type 1 IRESs. This raises the questions of which regions of PV and EV71 IRESs first enter the mRNA-binding cleft of the 40S subunit, and how ribosomes reach the authentic initiation codons. Two scenarios can be envisioned: (i) 43S complexes attach downstream of dVII and then scan to the authentic initiation codon, and (ii) they attach upstream of dVI and then scan downstream without unwinding domains VI–VII. Although the observation that an AUG located 5 nts downstream of dVII in the EV71 IRES is not selected as an initiation codon is more consistent with the first scenario, in which steric hindrance would prevent 43S complexes from attaching too close to dVII, the second scenario is also feasible, based on the ability of 43S complexes to bypass stable stems during scanning (Abaeva et al, 2011).
Interestingly, whereas our data indicate that domains VI in the PV and EV71 IRESs are not efficiently inspected in the in vitro reconstituted system and in RRL, the dVI AUG has been previously hypothesized to form part of the ‘ribosome landing pad’, which is sensed by incoming 43S complexes but not used as an initiation codon (Iizuka et al, 1991; Meerovitch et al, 1991; Nicholson et al, 1991; Pilipenko et al, 1992; Kaminski et al, 2010). Although consistent with the previous reports, we found that mutations in the dVI AUG substantially reduced the activity of PV and EV71 IRESs in a HeLa cell-free translation extract, they did not affect the activity of these IRESs in RRL or in the in vitro reconstituted system. This difference could not be explained by potentially more efficient unwinding of dVI in a HeLa extract, because the lack of response to dVI AUG mutations in RRL and in the in vitro reconstituted system did not depend on whether dVI remained intact or was destabilized. Moreover, exactly the same lack of effect on initiation efficiency in RRL and in the in vitro reconstituted system was noted for analogous mutations in the BEV IRES, on which ribosomal complexes do inspect dVI. Taken together with the previously reported data that mutations in the Yn element of the Yn-Xm-AUG motif impaired PV IRES function in a HeLa cell-free extract but not in RRL or a Krebs-2 cell-free extract (Pestova et al, 1991), our current results indicate that the entire Yn-Xm-AUG motif might function as a regulatory element in a cell-specific manner. Our finding that sensitivity to dVI AUG mutations became apparent in RRL on addition of 30% (v/v) HeLa cell-free extract suggests the presence in a HeLa extract of inhibitory, rather than stimulatory, activity that specifically targets mutant IRESs. An interesting precedent to such a hypothetical mechanism is the DRBP76:NF45 heterodimer that binds to domains V/VI of the Type 1 rhinovirus IRES and represses its activity in a cell type-specific manner (Merrill & Gromeier, 2006). However, further investigation of this phenomenon was outside the scope of the present investigation. An interesting related possibility is that such hypothetical factor(s) could also allow regulatable expression of the ORF initiating at the AUG in dVI. Although the spacer between dVI and the authentic initiation codon is commonly described as hypervariable (e.g. Jackson & Kaminski, 1995), the open reading frame (ORF) initiating at the 3′ border of the IRES encodes a 56–76 amino acid-long polypeptide in most members of the species Enterovirus A, B, C, E, F and G that overlaps the authentic initiation codon and is conserved between members of a species and between subsets of species (supplementary Fig S8). The retention of dVI in the majority of enteroviruses hints at a conserved function, which could thus be to regulate the relative levels of expression of the ORF initiating at the AUG in dVI and of the viral polyprotein.
Materials and Methods
Plasmid construction; purification of ribosomal subunits, initiation factors, methionyl tRNA synthetase, recombinant ITAFs; preparation of mRNAs; and aminoacylation of tRNAs are described in the Supplemental Material, which also contains detailed protocols for all experimental procedures.
In vitro translation
EV71, PV and BEV mRNAs were translated in RRL (Promega Corp.), HeLa cell-free extract (Takara Bio Inc.) or their mixture in the presence of [35S]methionine (Figs 4C–F). Translation products were resolved by SDS-PAGE.
Toe-printing analysis of initiation complex formation
48S complexes were assembled on wt and mutant PV, EV71, BEV and EMCV mRNAs in the presence of 40S subunits, initiation factors and ITAFs as indicated in Figs 1(B–E, H, I), 2(A–F), 3(B–G), 4(G–I), 6(C–E), supplementary S1, S2 and S3, and were analyzed by primer extension using AMV RT and 32P-labeled primers (Pisarev et al, 2007).
Analysis of ribosomal complex formation on PV and EV71 mRNAs in RRL
80S ribosomes were allowed to assemble on 32P-5′-end-labeled PV and EV71 mRNAs (Figs 2G and supplementary S4) by incubation in RRL supplemented with cycloheximide, and were then purified by centrifugation through 10–30% SDGs. Aliquots from ribosomal fractions were then analyzed by primer extension using AMV RT and 32P-labeled primers (Pisarev et al, 2007) or incubated with RelE and analyzed in parallel by primer extension using AMV RT.
Directed hydroxyl radical cleavage
Purified, Fe(II)-BABE-derivatized eIF4Gm-T829C, eIF4A-S42C, PCBP2 mutants and native eIF3 were incubated with wt or mutant PV, EV71 or CSFV IRES-containing mRNAs in the presence of eIFs and ITAFs as indicated in Figs 1(G), 5(B), 6(B), 7 and 8. Cleavage reactions were initiated with 0.05% H2O2 and 5 mM ascorbic acid, and quenched using thiourea (Kolupaeva et al, 2003). Sites of hydroxyl radical cleavage were determined by primer extension using AMV RT and 32P-labeled primers.
Acknowledgments
We thank T. Heise, R. Jackson, B. Lelj, P. Schimmel, B. Semler and J. Smiley for plasmids, and C. Neubauer and V. Ramakrishnan for RelE. This work was supported by NIH grant AI51340 to C.U.T.H.
Author contributions
TS, TP and CH conceived and designed experiments.TS and IA performed the experiments. TS, IA, TP and CH analyzed the data. TP and CH wrote the paper. TS and IA commented on the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supplementary information for this article is available online: http://emboj.embopress.org
References
- Abaeva IS, Marintchev A, Pisareva VP, Hellen CU, Pestova TV. Bypassing of stems versus linear base-by-base inspection of mammalian mRNAs during ribosomal scanning. EMBO J. 2011;30:115–129. doi: 10.1038/emboj.2010.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson EC, Hunt SL, Jackson RJ. Internal initiation of translation from the human rhinovirus-2 internal ribosome entry site requires the binding of Unr to two distinct sites on the 5′ untranslated region. J Gen Virol. 2007;88:3043–3052. doi: 10.1099/vir.0.82463-0. [DOI] [PubMed] [Google Scholar]
- Andreev DE, Fernandez-Miragall O, Ramajo J, Dmitriev SE, Terenin IM, Martinez-Salas E, Shatsky IN. Differential factor requirement to assemble translation initiation complexes at the alternative start codons of foot-and-mouth disease virus RNA. RNA. 2007;13:1366–1374. doi: 10.1261/rna.469707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreev D, Hauryliuk V, Terenin I, Dmitriev S, Ehrenberg M, Shatsky I. The bacterial toxin RelE induces specific mRNA cleavage in the A site of the eukaryote ribosome. RNA. 2008;14:233–239. doi: 10.1261/rna.693208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreev DE, Hirnet J, Terenin IM, Dmitriev SE, Niepmann M, Shatsky IN. Glycyl-tRNA synthetase specifically binds to the poliovirus IRES to activate translation initiation. Nucleic Acids Res. 2012;40:5602–5614. doi: 10.1093/nar/gks182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedard KM, Daijogo S, Semler BL. A nucleo-cytoplasmic SR protein functions in viral IRES-mediated translation initiation. EMBO J. 2007;26:459–467. doi: 10.1038/sj.emboj.7601494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blyn LB, Swiderek KM, Richards O, Stahl DC, Semler BL, Ehrenfeld E. Poly(rC) binding protein 2 binds to stem-loop IV of the poliovirus RNA 5′ noncoding region: identification by automated liquid chromatography-tandem mass spectrometry. Proc Natl Acad Sci USA. 1996;93:11115–11120. doi: 10.1073/pnas.93.20.11115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blyn LB, Towner JS, Semler BL, Ehrenfeld E. Requirement of poly(rC) binding protein 2 for translation of poliovirus RNA. J Virol. 1997;71:6243–6246. doi: 10.1128/jvi.71.8.6243-6246.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borman A, Howell MT, Patton JG, Jackson RJ. The involvement of a spliceosome component in internal initiation of human rhinovirus RNA translation. J Gen Virol. 1993;74:1775–1788. doi: 10.1099/0022-1317-74-9-1775. [DOI] [PubMed] [Google Scholar]
- Borman AM, Bailly JL, Girard M, Kean KM. Picornavirus internal ribosome entry segments: comparison of translation efficiency and the requirements for optimal internal initiation of translation in vitro. Nucleic Acids Res. 1995;23:3656–3663. doi: 10.1093/nar/23.18.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Breyne S, Yu Y, Unbehaun A, Pestova TV, Hellen CUT. Direct functional interaction of initiation factor eIF4G with type 1 internal ribosomal entry sites. Proc Natl Acad Sci USA. 2009;106:9197–9202. doi: 10.1073/pnas.0900153106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K, Kim JH, Li X, Paek KY, Ha SH, Ryu SH, Wimmer E, Jang SK. Identification of cellular proteins enhancing activities of internal ribosomal entry sites by competition with oligodeoxynucleotides. Nucleic Acids Res. 2004;32:1308–1317. doi: 10.1093/nar/gkh300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuesta R, Laroia G, Schneider RJ. Chaperone hsp27 inhibits translation during heat shock by binding eIF4G and facilitating dissociation of cap-initiation complexes. Genes Dev. 2000;14:1460–1470. [PMC free article] [PubMed] [Google Scholar]
- Dhote V, Sweeney TR, Kim N, Hellen CU, Pestova TV. Roles of individual domains in the function of DHX29, an essential factor required for translation of structured mammalian mRNAs. Proc Natl Acad Sci USA. 2012;109:E3150–E3159. doi: 10.1073/pnas.1208014109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z, Fenn S, Tjhen R, James TL. Structure of a construct of a human poly(C)-binding protein containing the first and second KH domains reveals insights into its regulatory mechanisms. J Biol Chem. 2008;283:28757–28766. doi: 10.1074/jbc.M803046200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funke B, Zuleger B, Benavente R, Schuster T, Goller M, Stévenin J, Horak I. The mouse poly(C)-binding protein exists in multiple isoforms and interacts with several RNA-binding proteins. Nucleic Acids Res. 1996;24:3821–3828. doi: 10.1093/nar/24.19.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamarnik AV, Andino R. Two functional complexes formed by KH domain containing proteins with the 5′ noncoding region of poliovirus RNA. RNA. 1997;3:882–892. [PMC free article] [PubMed] [Google Scholar]
- Gamarnik AV, Andino R. Interactions of viral protein 3CD and poly(rC) binding protein with the 5′ untranslated region of the poliovirus genome. J Virol. 2000;74:2219–2226. doi: 10.1128/jvi.74.5.2219-2226.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gradi A, Svitkin YV, Imataka H, Sonenberg N. Proteolysis of human eukaryotic translation initiation factor eIF4GII, but not eIF4GI, coincides with the shutoff of host protein synthesis after poliovirus infection. Proc Natl Acad Sci USA. 1998;95:11089–11094. doi: 10.1073/pnas.95.19.11089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellen CU, Pestova TV, Wimmer E. Effect of mutations downstream of the internal ribosome entry site on initiation of poliovirus protein synthesis. J Virol. 1994;68:6312–6322. doi: 10.1128/jvi.68.10.6312-6322.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellen CU, Witherell GW, Schmid M, Shin SH, Pestova TV, Gil A, Wimmer E. A cytoplasmic 57-kDa protein that is required for translation of picornavirus RNA by internal ribosomal entry is identical to the nuclear pyrimidine tract-binding protein. Proc Natl Acad Sci USA. 1993;90:7642–7646. doi: 10.1073/pnas.90.16.7642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt SL, Hsuan JJ, Totty N, Jackson RJ. unr, a cellular cytoplasmic RNA-binding protein with five cold-shock domains, is required for internal initiation of translation of human rhinovirus RNA. Genes Dev. 1999;13:437–448. doi: 10.1101/gad.13.4.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iizuka N, Yonekawa H, Nomoto A. Nucleotide sequences important for translation initiation of enterovirus RNA. J Virol. 1991;65:4867–4873. doi: 10.1128/jvi.65.9.4867-4873.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson RJ. Potassium salts influence the fidelity of mRNA translation initiation in rabbit reticulocyte lysates: unique features of encephalomyocarditis virus RNA translation. Biochim Biophys Acta. 1991;1088:345–358. doi: 10.1016/0167-4781(91)90124-5. [DOI] [PubMed] [Google Scholar]
- Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol. 2010;11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson RJ, Kaminski A. Internal initiation of translation in eukaryotes: the picornavirus paradigm and beyond. RNA. 1995;1:985–1000. [PMC free article] [PubMed] [Google Scholar]
- Kafasla P, Morgner N, Pöyry TA, Curry S, Robinson CV, Jackson RJ. Polypyrimidine tract binding protein stabilizes the encephalomyocarditis virus IRES structure via binding multiple sites in a unique orientation. Mol Cell. 2009;34:556–568. doi: 10.1016/j.molcel.2009.04.015. [DOI] [PubMed] [Google Scholar]
- Kafasla P, Morgner N, Robinson CV, Jackson RJ. Polypyrimidine tract-binding protein stimulates the poliovirus IRES by modulating eIF4G binding. EMBO J. 2010;29:3710–3722. doi: 10.1038/emboj.2010.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminski A, Pöyry TA, Skene PJ, Jackson RJ. Mechanism of initiation site selection promoted by the human rhinovirus 2 internal ribosome entry site. J Virol. 2010;84:6578–6589. doi: 10.1128/JVI.00123-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolupaeva VG, Lomakin IB, Pestova TV, Hellen CU. Eukaryotic initiation factors 4G and 4A mediate conformational changes downstream of the initiation codon of the encephalomyocarditis virus internal ribosomal entry site. Mol Cell Biol. 2003;23:687–698. doi: 10.1128/MCB.23.2.687-698.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamichhane R, Daubner GM, Thomas-Crusells J, Auweter SD, Manatschal C, Austin KS, Valniuk O, Alain FH-T, Rueda D. RNA looping by PTB: evidence using FRET and NMR spectroscopy for a role in splicing repression. Proc Natl Acad Sci USA. 2010;107:4105–4110. doi: 10.1073/pnas.0907072107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamphear BJ, Kirchweger R, Skern T, Rhoads RE. Mapping of functional domains in eukaryotic protein synthesis initiation factor 4G (eIF4G) with picornaviral proteases. Implications for cap-dependent and cap-independent translational initiation. J Biol Chem. 1995;270:21975–21983. doi: 10.1074/jbc.270.37.21975. [DOI] [PubMed] [Google Scholar]
- Lomakin IB, Hellen CU, Pestova TV. Physical association of eukaryotic initiation factor 4G (eIF4G) with eIF4A strongly enhances binding of eIF4G to the internal ribosomal entry site of encephalomyocarditis virus and is required for internal initiation of translation. Mol Cell Biol. 2000;20:6019–6029. doi: 10.1128/mcb.20.16.6019-6029.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Rivas A, Castrillo JL, Carrasco L. Cation content in poliovirus-infected HeLa cells. J Gen Virol. 1987;68:335–342. doi: 10.1099/0022-1317-68-2-335. [DOI] [PubMed] [Google Scholar]
- Meerovitch K, Nicholson R, Sonenberg N. In vitro mutational analysis of cis-acting RNA translational elements within the poliovirus type 2 5′ untranslated region. J Virol. 1991;65:5895–5901. doi: 10.1128/jvi.65.11.5895-5901.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meerovitch K, Svitkin YV, Lee HS, Lejbkowicz F, Kenan DJ, Chan EK, Agol VI, Keene JD, Sonenberg N. La autoantigen enhances and corrects aberrant translation of poliovirus RNA in reticulocyte lysate. J Virol. 1993;67:3798–3807. doi: 10.1128/jvi.67.7.3798-3807.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrill MK, Gromeier M. The double-stranded RNA binding protein 76:NF45 heterodimer inhibits translation initiation at the rhinovirus type 2 internal ribosome entry site. J Virol. 2006;80:6936–6942. doi: 10.1128/JVI.00243-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neubauer C, Gao YG, Andersen KR, Dunham CM, Kelley AC, Hentschel J, Gerdes K, Ramakrishnan V, Brodersen DE. The structural basis for mRNA recognition and cleavage by the ribosome-dependent endonuclease RelE. Cell. 2009;139:1084–1095. doi: 10.1016/j.cell.2009.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson R, Pelletier J, Le SY, Sonenberg N. Structural and functional analysis of the ribosome landing pad of poliovirus type 2: in vivo translation studies. J Virol. 1991;65:5886–5894. doi: 10.1128/jvi.65.11.5886-5894.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberstrass FC, Auweter SD, Erat M, Hargous Y, Henning A, Wenter P, Reymond L, Amir-Ahmady B, Pitsch S, Black DL, Allain FH. Structure of PTB bound to RNA: specific binding and implications for splicing regulation. Science. 2005;309:2054–2057. doi: 10.1126/science.1114066. [DOI] [PubMed] [Google Scholar]
- Pause A, Méthot N, Svitkin Y, Merrick WC, Sonenberg N. Dominant negative mutants of mammalian translation initiation factor eIF-4A define a critical role for eIF-4F in cap-dependent and cap-independent initiation of translation. EMBO J. 1994;13:1205–1215. doi: 10.1002/j.1460-2075.1994.tb06370.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova TV, Hellen CU. Translation elongation after assembly of ribosomes on the Cricket paralysis virus internal ribosomal entry site without initiation factors or initiator tRNA. Genes Dev. 2003;17:181–186. doi: 10.1101/gad.1040803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova TV, Hellen CU, Shatsky IN. Canonical eukaryotic initiation factors determine initiation of translation by internal ribosomal entry. Mol Cell Biol. 1996a;16:6859–6869. doi: 10.1128/mcb.16.12.6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova TV, Hellen CU, Wimmer E. Translation of poliovirus RNA: role of an essential cis-acting oligopyrimidine element within the 5′ nontranslated region and involvement of a cellular 57-kilodalton protein. J Virol. 1991;65:6194–6204. doi: 10.1128/jvi.65.11.6194-6204.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova TV, Hellen CU, Wimmer E. A conserved AUG triplet in the 5′ nontranslated region of poliovirus can function as an initiation codon in vitro and in vivo. Virology. 1994;204:729–737. doi: 10.1006/viro.1994.1588. [DOI] [PubMed] [Google Scholar]
- Pestova TV, Kolupaeva VG. The roles of individual eukaryotic translation initiation factors in ribosomal scanning and initiation codon selection. Genes Dev. 2002;16:2906–2922. doi: 10.1101/gad.1020902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova TV, Lomakin IB, Lee JH, Choi SK, Dever TE, Hellen CU. The joining of ribosomal subunits in eukaryotes requires eIF5B. Nature. 2000;403:332–335. doi: 10.1038/35002118. [DOI] [PubMed] [Google Scholar]
- Pestova TV, Shatsky IN, Fletcher SP, Jackson RJ, Hellen CU. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev. 1998;12:67–83. doi: 10.1101/gad.12.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova TV, Shatsky IN, Hellen CU. Functional dissection of eukaryotic initiation factor 4F: the 4A subunit and the central domain of the 4G subunit are sufficient to mediate internal entry of 43S preinitiation complexes. Mol Cell Biol. 1996b;16:6870–6878. doi: 10.1128/mcb.16.12.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilipenko EV, Gmyl AP, Maslova SV, Svitkin YV, Sinyakov AN, Agol VI. Prokaryotic-like cis elements in the cap-independent internal initiation of translation on picornavirus RNA. Cell. 1992;68:119–131. doi: 10.1016/0092-8674(92)90211-t. [DOI] [PubMed] [Google Scholar]
- Pilipenko EV, Pestova TV, Kolupaeva VG, Khitrina EV, Poperechnaya AN, Agol VI, Hellen CU. A cell cycle-dependent protein serves as a template-specific translation initiation factor. Genes Dev. 2000;14:2028–2045. [PMC free article] [PubMed] [Google Scholar]
- Pisarev AV, Unbehaun A, Hellen CU, Pestova TV. Assembly and analysis of eukaryotic translation initiation complexes. Methods Enzymol. 2007;430:147–177. doi: 10.1016/S0076-6879(07)30007-4. [DOI] [PubMed] [Google Scholar]
- Pisareva VP, Pisarev AV, Komar AA, Hellen CU, Pestova TV. Translation initiation on mammalian mRNAs with structured 5′UTRs requires DExH-box protein DHX29. Cell. 2008;135:1237–1250. doi: 10.1016/j.cell.2008.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sean P, Nguyen JH, Semler BL. Altered interactions between stem-loop IV within the 5′ noncoding region of coxsackievirus RNA and poly(rC) binding protein 2: effects on IRES-mediated translation and viral infectivity. Virology. 2009;389:45–58. doi: 10.1016/j.virol.2009.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidiqi M, Wilce JA, Vivian JP, Porter CJ, Barker A, Leedman PJ, Wilce MC. Structure and RNA binding of the third KH domain of poly(C)-binding protein 1. Nucleic Acids Res. 2005;33:1213–1221. doi: 10.1093/nar/gki265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvera D, Gamarnik AV, Andino R. The N-terminal K homology domain of the poly(rC)-binding protein is a major determinant for binding to the poliovirus 5′-untranslated region and acts as an inhibitor of viral translation. J Biol Chem. 1999;274:38163–38170. doi: 10.1074/jbc.274.53.38163. [DOI] [PubMed] [Google Scholar]
- Sizova DV, Kolupaeva VG, Pestova TV, Shatsky IN, Hellen CU. Specific interaction of eukaryotic translation initiation factor 3 with the 5′ nontranslated regions of hepatitis C virus and classical swine fever virus RNAs. J Virol. 1998;72:4775–4782. doi: 10.1128/jvi.72.6.4775-4782.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney TR, Dhote V, Yu Y, Hellen CU. A distinct class of internal ribosomal entry site in members of the Kobuvirus and proposed Salivirus and Paraturdivirus genera of the Picornaviridae. J Virol. 2012;86:1468–1486. doi: 10.1128/JVI.05862-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson SR, Sarnow P. Enterovirus 71 contains a type I IRES element that functions when eukaryotic initiation factor eIF4G is cleaved. Virology. 2003;315:259–266. doi: 10.1016/s0042-6822(03)00544-0. [DOI] [PubMed] [Google Scholar]
- Unbehaun A, Borukhov SI, Hellen CU, Pestova TV. Release of initiation factors from 48S complexes during ribosomal subunit joining and the link between establishment of codon-anticodon base-pairing and hydrolysis of eIF2-bound GTP. Genes Dev. 2004;18:3078–3093. doi: 10.1101/gad.1255704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Abaeva IS, Marintchev A, Pestova TV, Hellen CU. Common conformational changes induced in type 2 picornavirus IRESs by cognate trans-acting factors. Nucleic Acids Res. 2011a;39:4851–4865. doi: 10.1093/nar/gkr045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Sweeney TR, Kafasla P, Jackson RJ, Pestova TV, Hellen CU. The mechanism of translation initiation on Aichivirus RNA mediated by a novel type of picornavirus IRES. EMBO J. 2011b;30:4423–4436. doi: 10.1038/emboj.2011.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zell R, Ihle Y, Effenberger M, Seitz S, Wutzler P, Görlach M. Interaction of poly(rC)-binding protein 2 domains KH1 and KH3 with coxsackievirus RNA. Biochem Biophys Res Commun. 2008;377:500–503. doi: 10.1016/j.bbrc.2008.09.156. [DOI] [PubMed] [Google Scholar]
- Zell R, Sidigi K, Henke A, Schmidt-Brauns J, Hoey E, Martin S, Stelzner A. Functional features of the bovine enterovirus 5′-non-translated region. J Gen Virol. 1999;80:2299–2309. doi: 10.1099/0022-1317-80-9-2299. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.