Abstract
Habitat bioaugmentation and introduction of protective microbiota have been proposed as potential conservation strategies to rescue endangered mammals and amphibians from emerging diseases. For both strategies, insight into the microbiomes of the endangered species and their habitats is essential. Here, we sampled nests of the endangered sea turtle species Eretmochelys imbricata that were infected with the fungal pathogen Fusarium falciforme. Metagenomic analysis of the bacterial communities associated with the shells of the sea turtle eggs revealed approximately 16,664 operational taxonomic units, with Proteobacteria, Actinobacteria, Firmicutes and Bacteroidetes as the most dominant phyla. Subsequent isolation of Actinobacteria from the eggshells led to the identification of several genera (Streptomyces, Amycolaptosis, Micromomospora Plantactinospora and Solwaraspora) that inhibit hyphal growth of the pathogen F. falciforme. These bacterial genera constitute a first set of microbial indicators to evaluate the potential role of microbiota in conservation of endangered sea turtle species.
Introduction
Sea turtles are one of the most endangered groups of animals worldwide with only seven species left [1]. Incidental by-catch, disturbance of nesting beaches, pollution and diseases are major causes of drastic population declines [2]. Among the emerging diseases, the fungal pathogens Fusarium falciforme and F. keratoplasticum are an increasing threat to sea turtle nests, especially to those experiencing environmental stress [3].
Several conservation strategies have been proposed to mitigate the impact of pathogens on endangered species. For example, establishment of ex situ colonies and ‘habitat bioaugmentation and biotherapy’ have been proposed to prevent dispersal of the fungal pathogen Batrachotrichum dendrobatidis in amphibian populations [4]. The latter two strategies encompass the use of protective microbiota, either indigenous or introduced, to limit pathogen infection and spread. These two approaches are adopted in agriculture to control plant diseases [5]–[8]. Also in mammals, the role of gut microbiota in health and disease is now widely studied [9]–[13]. In nature conservation programs, however, these approaches are not common yet. This is due, in part, to a lack of knowledge of the overall diversity of microbiota associated with endangered species and their role, if any, in protecting their hosts against pathogen infection [14].
The structure of microbial communities of different hosts, their genetic diversity, and ecological roles have been studied combining culture-based analysis with polymerase chain reaction (PCR) techniques [15]–[17]. For example, the high-density 16S ribosomal DNA (rDNA) oligonucleotide microarray, referred to as the PhyloChip [18], [19] combined with bacterial isolations has helped identifying key bacterial and archaeal community members in the rhizosphere of plants grown in disease-suppressive soils [20]. In sea turtles, a limited number of culture-based and biochemical studies have allowed describing taxa of bacteria associated with egg failure in several species [21]–[27]. These studies have listed and reported on potentially pathogenic bacteria from unhatched sea turtle eggs. However, full characterization of the microbial community and its effect on hatching of sea turtle eggs has, to our knowledge, never been conducted.
In this study, we investigated the microbial community associated with Fusarium-infected eggs of the critically endangered sea turtle species Eretmochelys imbricate. To that end, we collected eggs from the nesting beach La Playita at Machalilla National Park, Ecuador, in order to survey for bacteria with antifungal activity. For this purpose, PhyloChip analysis was used to identify the bacterial community associated with the turtle eggs. Based on these analyses, targeted isolations of specific bacterial genera were conducted using culture-based techniques followed by in vitro assays to determine the potential antagonistic activity of the selected indigenous microbiota against F. falciforme, the fungal pathogen of sea turtle eggs [3].
Results
Fungal isolation and molecular characterization
A total of 10 fungal isolates were obtained from the eggshells (Figure 1, S1) and initially identified as F. solani based on NCBI BLAST analysis of the ITS nrDNA sequences (Table 1). Phylogenetic analysis of the ITS nrDNA showed that the 10 fungal isolates clustered within the previously described species F. falciforme (Table 1 and Figure S2).
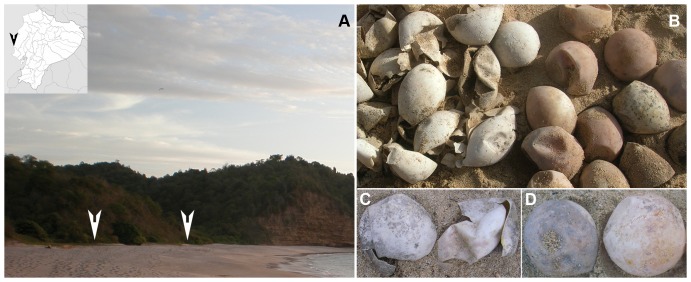
Figure 1. Sea turtle nesting area sampled for this study.
A) Nests of the sea turtle Eretmochelys imbricata in La Playita beach at Machalilla National Park, Ecuador. B) Nest containing hatched and unhatched Fusarium-infected eggs. C) Fusarium-infected hatched eggs. D) Fusarium-infected unhatched eggs.
Table 1. Fusarium falciforme isolates from eggshells of the sea turtle species Eretmochelys imbricata.
| Strain | Source | aGenBank Accession | bMaximum identity |
| 326FUS | Hatched egg | KF179246 | 100% |
| 327FUS | Hatched egg | KF179247 | 99% |
| 328FUS | Hatched egg | KF179248 | 100% |
| 329FUS | Hatched egg | KF179249 | 100% |
| 330FUS | Hatched egg | KF179250 | 100% |
| 331FUS | Hatched egg | KF179251 | 99% |
| 332FUS | Unhatched egg | KF179252 | 100% |
| 333FUS | Unhatched egg | KF179253 | 100% |
| 334FUS | Unhatched egg | KF179254 | 100% |
| 335FUS | Unhatched egg | KF179255 | 100% |
GenBank accession number of the F. falciforme isolates.
BLAST hit corresponds to the NCBI nucleotide database. All the blast hits corresponded with F. solani strains.
Bacterial isolation and DNA extraction from sea turtle egg shells
The number of culturable aerobic bacteria, enumerated on 1/10th strength Tryptic Soy Agar (TSA) medium, ranged from 3.1×107 to 8.7×107 Colony Forming Units per area of eggshell (CFU/cm2) from hatched and unhatched turtle eggs respectively. The population density of culturable Actinobacteria, enumerated on semi-selective medium glycerol-arginine agar (GA), ranged from 1.2×104 to 2×105 CFU/cm2 from hatched and unhatched eggs, respectively (Table S1). The Actinobacteria comprised on average, 0.2% of the total aerobic bacteria enumerated on 1/10th TSA (Table S1).
PhyloChip analysis
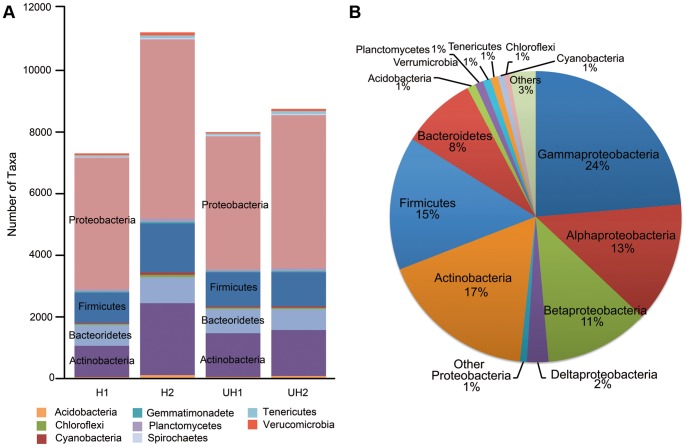
PhyloChip-based metagenomic analysis of the bacterial communities associated with the eggshells revealed the presence of 16,664 operational taxonomic units (OTUs). On average, Proteobacteria (52%), Actinobacteria (17%), Firmicutes (15%) and Bacteroidetes (8%) were detected as the most dominant phyla (Figure 2A, B). No significant differences were detected in overall bacterial phyla composition between hatched and unhatched eggs or between the two nests (Figure 2A). At family level, however, significant (Welsh test, p<0.01; r = 0.75, Anosim) differences in abundance between the two nests were found for the Pseudomonadaceae, which comprised 24% of the Gammaproteobacteria (Figure S3A; Table S2). Furthermore, the Flavobacteriaceae, which comprised 51% of the Bacteroidetes detected, were significantly (r = 1, Anosim) more abundant on shells of hatched eggs than those of unhatched eggs (Figure S3B). Within the Flavobacteriaceae, Chryseobacterium was the second most abundant genus (10%) and C. indologenes and C. gleum were the most represented species (Welch test, p<0.01) in our PhyloChip analysis (Table S3). These two species represented 7 and 5% of the genus Chryseobacterium, respectively.
Figure 2. Composition of the microbial community of shells of Fusarium-infected eggs detected by the PhyloChip analysis.
A) Number of OTUs per phylum detected on hatched (H) and unhatched (UH) eggshells collected from two nests (numbers 1 and 2). Values with >0.25% of occurrence. B) Average distribution of OTUs for all the samples (n = 4).
In vitro activity assay and BOX-PCR based identification of Actinobacteria
The Actinobacteria was the second most abundant bacterial phylum detected on the sea turtle eggshells. Given their well-documented ability to produce an array of antibacterial and antifungal compounds [33]-[35], we isolated Actinobacteria from the eggshells and determined their activity against F. falciforme. Out of a total of 98 randomly selected Actinobacteria isolates from hatched (n = 69) and unhatched eggs (n = 29), thirty-one inhibited hyphal growth of F. falciforme (isolate 331FUS). Among these 31 isolates with antifungal properties, 23 different haplotypes were identified by BOX-PCR fingerprinting. Subsequent 16S rDNA sequencing and phylogenetic analysis indicated that these isolates belong to the genera Streptomyces (16), Amycolaptosis (3), Micromonospora (1), Plantactinospora (4) and Solwaraspora (5) (Table 2). A total of 25 out of 31 of the antagonistic Actinobacteria isolates were obtained from hatched eggs. Out of the 6 isolates obtained from unhatched eggs, 1 corresponded to the genus Planctactinospora and the other five to Streptomyces.
Table 2. 16S rDNA sequence identities of the Actinobacteria, isolated from sea turtle eggshells, that inhibited the hyphal growth of Fusarium falciforme.
| Isolate ACTa | Source | Identity of best BLAST hitb | GenBank accesion | Score | Identity |
| 2 | Hatched egg | Micromonospora sp. | KF179216 | 979 | 99% |
| 121 | Hatched egg | Micromonospora sp. | KF179221 | 1246 | 99% |
| 13 | Hatched egg | Plantactinospora sp. | KF179222 | 1222 | 98% |
| 14 | Hatched egg | Plantactinospora sp. | KF179223 | 1222 | 98% |
| 20 | Hatched egg | Plantactinospora sp. | KF179224 | 1226 | 98% |
| 125 | Unhatched egg | Plantactinospora sp. | KF179225 | 1232 | 99% |
| 1 | Hatched egg | Solwaraspora sp. | KF179226 | 1260 | 100% |
| 16 | Hatched egg | Solwaraspora sp. | KF179227 | 1260 | 100% |
| 19 | Hatched egg | Solwaraspora sp. | KF179228 | 1260 | 100% |
| 23 | Hatched egg | Solwaraspora sp. | KF179229 | 1245 | 100% |
| 108 | Hatched egg | Solwaraspora sp. | KF179230 | 1245 | 100% |
| 145 | Hatched egg | Amylocolaptosis coloradensis | KF179218 | 1134 | 99% |
| 151 | Hatched egg | Amylocolaptosis coloradensis | KF179219 | 1238 | 99% |
| 152 | Hatched egg | Amylocolaptosis coloradensis | KF179220 | 1245 | 99% |
| 147 | Hatched egg | Streptomyces mutabilis | KF179231 | 1265 | 100% |
| 150 | Hatched egg | Streptomyces albogriseolus. | KF179232 | 1250 | 99% |
| 146 | Hatched egg | Streptomyces variabilis | KF179217 | 1065 | 100% |
| 148 | Hatched egg | Streptomyces variabilis. | KF179233 | 1215 | 100% |
| 149 | Hatched egg | Streptomyces variabilis | KF179234 | 1264 | 100% |
| 153 | Hatched egg | Streptomyces variabilis | KF179235 | 1273 | 100% |
| 154 | Hatched egg | Streptomyces variabilis | KF179236 | 1270 | 100% |
| 155 | Hatched egg | Streptomyces variabilis | KF179237 | 1273 | 100% |
| 156 | Hatched egg | Streptomyces variabilis | KF179238 | 1273 | 100% |
| 157 | Hatched egg | Streptomyces variabilis | KF179239 | 1273 | 100% |
| 162 | Hatched egg | Streptomyces variabilis | KF179240 | 1278 | 100% |
| 164 | Unhatched egg | Streptomyces variabilis | KF179241 | 1272 | 100% |
| 166 | Unhatched egg | Streptomyces variabilis | KF179242 | 1269 | 100% |
| 167 | Unhatched egg | Streptomyces variabilis | KF179243 | 1281 | 100% |
| 169 | Unhatched egg | Streptomyces variabilis | KF179244 | 1244 | 100% |
| 170 | Unhatched egg | Streptomyces variabilis | KF179245 | 1277 | 100% |
ACT corresponds to the acronym of the Actinobacterial isolates.
BLAST hit corresponds to the Greengenes database (greengenes.lbl.gov/cgi-bin/nph-blast_interface.cgi).
The data represent the best BLAST hit with 16S rDNA sequences from the GreenGenes database (greengenes.lbl.gov/cgi-bin/nph-blast_interface.cgi).
Based on phylogenetic analysis of the 16S sequences, the antagonistic Streptomyces isolates clustered in three different groups within the 364 Streptomyces OTUs detected by the PhyloChip (Figure S4). The antagonistic isolates classified as S. mutabilis and S. albogriseolus clustered with OTUs classified as the same species detected by the PhyloChip (BS = 74% and BS<50% respectively). The antagonistic S. variabilis isolate clustered with OTUs detected by the PhyloChip classified as S. variabilis and S. aureofaciens (BS = 58%).
Similarly, phylogenetic analysis of the 16S sequences of the antagonistic isolates belonging to Amycolaptosis sp. and Micromonosporaceae, i.e., Micromonospora sp., Plantactinospora sp. and Solwaraspora sp., could be linked with representatives of each of these four Actinobacterial genera detected by PhyloChip analysis (Figure S5A, S5B), The antagonistic isolates identified as Micromonospora sp. clustered with seven OTUs of different species (BS<50%), and those identified as Solwaraspora sp. grouped with one OTU of this genus detected by the PhyloChip (BS<50%). The antagonistic isolate identified as Plantactinospora sp. clustered with one OTU of the species Plantactinospora mayteni (BS = 67%) and the Amycolaptosis coloradensis isolates clustered with representatives of this species detected by the PhyloChip (BS<50%).
Discussion
In this study, we described the microbial community of Fusarium-infected sea turtle eggs from the critically endangered species Eretmochelys imbricata. Due to the extreme difficulties to obtain samples and export permits from authorities for studies on endangered and critically endangered species, only four eggs were allowed to be collected. Hence, the results presented here provide a first ‘glimpse’ into the microflora associated with sea turtle eggs.
The PhyloChip analyses showed that the bacterial community associated with the eggs is mainly represented by the phyla Proteobacteria, Actinobacteria, Firmicutes and Bacteroidetes. In studies on the microbiome of the rhizosphere, members of the Proteobacteria and Firmicutes were described as the most dynamic taxa associated with disease suppression [20]. The potential implication of these bacterial taxa in protection of turtle eggs against Fusarium disease is not yet known.
No significant differences were detected in overall bacterial phyla composition between hatched and unhatched eggs or between the two nests. However, differences in abundance of two representative families of the microbial community of the sea turtle eggs were found. The significant difference in Pseudomonadaceae abundance among nests may reflect the variation in environmental conditions in the nesting area. Honarvar et al [23] demonstrated that bacterial diversity and richness increased with nest density and is higher in the zones closer to vegetation. Pseudomonas species have been previously isolated from cloaca of sea turtle females and eggs [22], [28]. They have been associated with diseases of captive sea turtles although their pathogenicity was not resolved [29]. In soil, the Pseudomonadaceae contribute to natural suppressiveness against several fungal pathogens including Fusarium [20], [30], [31]. For the Flavobacteriaceae, C. indologenes and C. gleum were the most represented OTUs (Welch test, p<0.01) in our PhyloChip analysis (Table S2). Chryseobacterium indologenes has been previously isolated from unhatched eggs of the loggerhead sea turtle Caretta caretta [22] and associated with shell disease of captive freshwater turtles [32]. Conversely, Chryseobacterium sp. strains are also known to exhibit antifungal activity [33]. Hence, the role of Flavobacteriaceae and/or the Pseudomonadaceae in mitigation of Fusarium infections of sea turtle eggs remains unclear. With the combined sample size of 4 eggshells, a first representative analysis of the microbial families that are associated with turtle eggs was performed (Figure 2). However, the differences observed between conditions (nests, hatched, unhatched) on family composition should be interpreted carefully due to the limited sample size per condition (n = 2).
The second most abundant bacterial phylum detected on the sea turtle eggshells was the Actinobacteria. Given their well-documented ability to produce an array of antibacterial and antifungal compounds [34]–[36], we isolated Actinobacteria from the eggshells and determined their activity against F. falciforme. The in vitro activity assays showed that isolated Actinobacteria of the genera Streptomyces, Amycolaptosis, Micromonospora and Plantactinospora are able to inhibit hyphal growth of F. falciforme. Interestingly, most of the antagonistic isolates described in this study were obtained from hatched eggs (Table 2). The majority of the antagonistic isolates belonged to the genus Streptomyces and this genus was the most representative group of the Actinobacteria (Table 2). In plants, Streptomyces species have been implicated in the protection against bacterial [37] and fungal pathogens including Fusarium [38], [39]. Species of the genus Streptomyces and other Actinobacteria with antifungal activity are also well known for their symbiotic associations with insects, protecting these from fungal pathogens [40]. The results of this study suggest that Streptomyces are a component of the bacterial community that reduce infection or proliferation of Fusarium on sea turtle eggs. Whether the Streptomyces, and other antagonistic Actinobacteria species, identified in this study can be used as a bioindicator, or as a component of protective microbiota in the nesting areas, to minimize sea turtle infections by Fusarium or other fungal pathogens remains to be investigated.
This study provides a first survey of the composition of the bacterial microflora on eggs of endangered sea turtles. Understanding not only the diversity and abundance of bacteria and other microorganisms associated with endangered species, but also the role of these microorganisms in disease suppression may have direct applications for nature conservation programs.
Material and Methods
Ethics Statement
Collection of sea turtle eggshells was done under permissions: 002 RM-DPM-MA and CITES 003/VS. None of the experiments involved sacrificing animals and, therefore, we did not require a specific approval from any institutional animal research ethics committee.
Sample collection
Samples were collected from two selected nests of the sea turtle species Eretmochelys imbricata located in La Playita beach at Machalilla National Park (Ecuador) during the nesting season of 2012 (Figure S1). Four eggs (two hatched and two unhatched) were collected (Figure 1). Immediately after hatching of the eggs (approximately 45 days after the start of the incubation), one hatched and one unhatched egg (containing a nonviable embryo) were collected per nest, all with signs of Fusarium infection [41] (Figure 1). Samples were collected using sterile latex exam gloves and maintained at 4°C in individual bags during 2 days.
Fungal isolation and molecular characterization
To confirm that the turtle eggs were indeed infected by Fusarium species, fragments of the eggshells (1 cm2) were placed on Peptone Dextrose Agar (PDA) and on Malt Agar (Figure S1), both supplemented with rifampicin (100 µg/ml) to prevent bacterial growth, and incubated at 25°C. Pure cultures of the fungal outgrowths were obtained by transferring single hyphal tips to fresh agar media. Pure cultures of the isolates are kept in the culture collection of the Laboratory of Phytopathology at Wageningen University, The Netherlands and the Real Jardín Botánico-CSIC, Spain.
To characterize the fungal isolates, DNA was extracted from mycelium (10 mg) collected from pure cultures. The mycelium was collected in 1.5 ml sterile tubes and 90 µl of NaOH (0.5 M) and two glass beads were added to the suspensions. The suspensions were placed in the Mixer Mill MM400 for 3 min to a frequency of 30 times/s, incubated at room temperature during 2 h and centrifuged at 13,000 rpm for 30 s. The supernatants were diluted 2 and 10 times with 0.1 M Tris-HCl (pH 7.0) for amplification. The primer pairs ITS1/ITS4 were used to amplify the internal transcribed spacer of the nuclear ribosomal DNA (ITS nrDNA). Amplification reactions were performed in 50 µl of reaction that contained 4 µl of DNA sample, 10 µl of 5x Colorless GoTaq Reaction buffer (Promega Co. Ma, US), 2 µl of each primer (10 µM), 2 µl of mix of dNTPs (5 mM), 0.2 µl of 5 U/µl GoTaq DNA polymerase (Promega Co. Ma, US) and 29.8 µl of MiliQ water. The amplification program was: initial denaturalization at 94°C for 5 min; 35 cycles of 94°C for 1 min, 60°C for 1 min and 72°C for 2 min; with a final extension at 72°C for 5 min.
The amplification products were sequenced in both forward and reverse direction (MACROGEN, Amsterdam, The Netherlands). Sequencing results were processed by Sequencher 4.2 (Gene Codes Corporation, Ann Arbor, Michigan, USA) and initially compared with sequences in the National Centre of Biotechnology Information (NCBI) nucleotide databases using BLAST [42]. For precise identification of the Fusarium spp. a phylogenetic analyses was carried out. The generated ITS nrDNA sequences from isolated Fusarium (Table 1), 136 NCBI-GenBank sequences of Fusarium turtle egg isolates (Table S1), and 60 selected sequences of Fusarium spp. from other hosts and environments were included (Table S2). The program Se-Al 2.0a11 Carbon [43] was used for manual alignment of the sequences. Maximum parsimony analysis (MP) [44] was inferred using the heuristic search option in PAUP*v4.0b10. Nonparametric bootstrap support (BS) [45] for each clade was tested based on 10,000 replicates, using the fast-step option. Newly obtained sequences were submitted to GenBank with accession numbers KF179246 through KF179255.
Bacterial isolation and DNA extraction from sea turtle eggshells
For bacterial isolation and DNA extraction, the eggshells (4 cm2) were individually suspended in 10 ml of sterile tap water and vortexed for 2 min. The suspensions were sonicated using an ultrasonic bath (Transsonic 460, Elma) for 2 min and vortexed for an additional 2 min at maximum speed. Each suspension was divided in 1.5 ml aliquots in eppendorf tubes and centrifuged at 13,000 rpm for 30 min. Pellets were resuspended in 100 µl of sterile tap water by vortexing and pipetting and then pooled in a sterile eppendorf tube to a final volume of approximately 700 µl. A 50 µl aliquot of each suspension was mixed with 50 µl of 80% glycerol and these samples were stored in the freezer at −20°C until processed for bacterial isolations. The remaining suspension (approximately 650 µl) was centrifuged at 13,000 rpm during 30 min, supernatants were discarded and pellets were stored at −80°C until processed for DNA extraction. For bacterial isolations, glycerol suspensions were diluted in 10-fold steps up to 10,000 times and, for each dilution, two replicates of 50 µl were plated on 1/10th TSA for total aerobic bacteria and on the semi-selective medium GA supplemented with Nalidixic acid (20 µg/ml) and Trimethoprim (20 µg/ml) for Actinobacteria (Figure S1). Both media were additionally supplemented with Delvocid (100 µg/ml) to prevent fungal growth. TSA plates were incubated at 25°C for 5 days and GA plates were incubated at 30°C for 21 days. Colonies were collected from GA medium. Based on the colony counts, the number of CFU/cm2 was calculated.
PhyloChip analysis
To identify the bacterial and archaeal communities on the shells of Fusarium–infected sea turtle eggs, metagenomic DNA was isolated from the cell pellets extracted from the hatched and unhatched eggs (Figure S1). The PowerSoil DNA Isolation Kit (MO BIO Laboratories, Inc.) was used for DNA isolation according to the manufacturer's instructions. The DNA concentration was determined by a Nanodrop 1000 Spectrophotometer (Thermo Scientific). The microbial profile for each sample was generated by G3-PhyloChip analysis (Second Genome, CS, USA). All PCR conditions and universal primers used for amplification of 16S rDNA genes of bacteria and archaea were previously described by [18]. Fragmentation of the 16S rDNA amplicons, labelling, hybridization, staining, and scanning of the PhyloChip, as well as data processing to determine absence/presence and HybScores of OTUs was performed according to methods described by [18]. Phyla represented by over 10% of the detected OTUs were analysed in detail. These Phyla were also analysed at the family and genus level. Comparisons of composition between samples were performed using the Bray-Curtis distance with the average as the clustering method. Statistical analyses of the PhyloChip data were performed by Primer-E 6 software (PRIMER-E Ltd., UK).
In vitro activity assay and BOX-PCR based identifications of Actinobacteria
Because Actinobacteria have the ability to produce an array of antibacterial and antifungal compounds [34]–[36], all the bacterial isolates obtained from GA medium were purified and screened for in vitro antagonism against Fusarium isolate 331FUS, which was obtained from the sea turtle eggs in this study. For each bacterial isolate, one 5 mm diameter agar plug from 3-week-old culture plates was inoculated at the periphery of a quadrant of 1/5 strength PDA plates (four plugs per plate in total) and incubated for 4 days at 30°C. After this period, a 5 mm diameter agar plug from a 7-day-old Fusarium plate culture was transferred to the centre of the plate. After an additional 7 days of incubation at 30°C, inhibition of hyphal growth by each of the four bacterial isolates was measured and expressed relative to radial hyphal growth of Fusarium on plates without bacteria.
The genotypic diversity of the bacterial isolates with antagonist activity against Fusarium was assessed by BOX-PCR using the 22-mer BOXA1R oligonucleotide [46], [47]. DNA was extracted from pure cultures using 2 mg of the colonies by microwave treatment as described previously [48]. The suspensions were centrifuged at 13,000 rpm for 30 s and supernatants were used for amplifications. Amplification reactions were performed in 25 µl containing 1 µl of DNA sample, 5 µl of 5x Gitschier buffer [49], 1 µl the BOX1AR primer (10 µm), 1.25 µl of mix of dNTPs (100 mM), 0.4 µl of BSA (10 mg/ml), 2.5 µl of 100% DMSO, 0.4 µl of 5 U/µl GoTaq DNA polymerase (Promega Co. Ma, US) and 13.45 µl of MiliQ water. Amplification was performed following an initial denaturation at 95°C for 2 min; 30 cycles at 94°C for 3 s, 92°C for 30 s, 50°C for 1 min and 65°C for 8 min, with a final extension at 65°C for 8 min [50]. PCR amplification products were detected by electrophoresis in 1% (w/v) agarose gels (5h at 45W). DNA fingerprints were visually compared for similarity; variations in intensity of bands were not taken into account in the analysis. For one isolate of each specific BOX group, the 16S rDNA was amplified with primer pair 8F/1392R [51]. The amplification products were sequenced both forward and reverse (MACROGEN, Amsterdam, The Netherlands). Sequences were processed by Sequencher 4.2 (Gene Codes Corporation, Ann Arbor, Michigan, USA) to obtain the sequence for each isolate. Sequences obtained were compared with those in the NCBI and GreenGenes databases (greengenes.lbl.gov/cgi-bin/nph-blast_interface.cgi). The sequences have been submitted to GenBank with accession numbers KF179216 through KF179245.
The Phylogenetic relationship of the isolated antagonistic Actinobacteria and the Actinobacteria OTUs detected by the PhyloChip was determined per genera. Additional GenBank sequences of Solwarapora sp. (JN633950, JN633958 and JN633962) and Platactinospora sp. (KC336252 and FJ214343) were included in the analysis. The 16S rDNA sequences of Streptomyces ambifaciens (M27245) and Solwaraspora sp. (JN633950) were included as outgroups in the analysis of non-corresponding genera, respectively. The tool MUSCLE available in MEGA5.05 [52] was used to align the 16S rDNA sequences. Maximum parsimony analyses (MP) and BS support where inferred following the methodology explained above.
Supporting Information
Schematic presentation of the metagenomic and classical microbiological approaches and techniques. The scheme represent the approaches used to isolate, identify and characterize the fungal and bacterial community from eggs of the sea turtle species Eretmochelys imbricata nesting at La Playita beach, Machalilla National Park, Ecuador.
(TIF)
Out-group rooted cladogram of the ITS nrDNA region of isolates within the Fusarium solani species complex. One of the most parsimonious trees inferred from the ITS nrDNA sequence data of 136 sea turtle fungal isolates and 60 non-sea turtle fungal isolates. The numbers on the internodes indicate the bootstrap values (BS) of the parsimony analysis. Highlighted isolates correspond to those obtained in this work (n = 10). The arrow indicates the F. falciforme isolate, i.e., 331FUS, used in the dual culture assays to determine the activity of the Actinobacteria.
(TIF)
Cluster analysis (Bray-Curtis) of the microbiome of hatched and unhatched eggs infected by Fusarium falciforme . A) Dendogram of family Pseudomonadaceae (n = 949 OTUs). B) Dendogram of family Flavobacteriaceae (n = 710 OTUs). Abbreviations as in Figure 2.
(TIF)
Out-group rooted phylogenetic tree inferred from the 16S rDNA sequence data from isolates of Streptomyces spp. Data includes isolates of Streptomyces spp. (n = 16) with activity against Fusarium falciforme, and those detected by the PhyloChip analysis (n = 364). The numbers at the internodes indicate the bootstrap values (BS) of the parsimony analysis.
(TIF)
Out-group rooted phylogenetic trees inferred from sequence data from isolates of the Amycolaptosis sp. and Micromonosporaceae. Phylogenetic trees were inferred from the 16S rDNA data from isolates from both taxa, with activity against Fusarium falciforme, and those detected by the PhyloChip analysis. A) Phylogenetic tree from the isolates of the Amycolaptosis sp. (n = 3) with activity against F. falciforme, and those detected by the PhyloChip analysis (n = 29). B) Phylogenetic tree from isolates of the Micromonosporaceae (n = 11) with activity against F. falciforme, those detected by the PhyloChip analysis (n = 33), and additional GenBank strains (n = 5). The numbers at the internodes of the phylogenetic trees indicate the bootstrap values (BS) of the parsimony analysis.
(TIF)
Number of bacteria isolated from the shells of hatched and unhatched eggs of the sea turtle species Eretmochelys imbricata on 1/10th TSA agar medium (total aerobic bacteria) and on GA medium (semi-selective for Actinobacteria). Presented are the Colony Forming Units (CFU/cm2) for each of the two media and for each of the two hatch statuses. For each hatch status, a mean value of 2 eggs is given. SD refers to the standard deviation.
(DOCX)
Most abundant microbial communities from Fusarium -infected eggshells of the sea turtle species Eretmochelys imbricata . Data shown represent the most abundant phyla and families detected by the PhyloChip. The families highlighted in grey are most represented (with >10%) per phylum.
(DOCX)
Chryseobacterium species found significantly more abundant on eggshells of hatched than of unhatched eggs of the sea turtle species Eretmochelys imbricata (Welsh test, p<0.01; r = 1, Anosim).
(DOCX)
Acknowledgments
Thanks to Dr. Jesús Muñoz for support. We want to thank Chunxu Song, Yiying Liu and Viviane Cordovéz-da-Cunha for their valuable advices on the isolation and characterization of the bacterial communities.
Funding Statement
This work was supported by grants of Ministerio de Economía y Competitividad, Spain (CGL2012-39357) and by Ms-Ph D program on Biodiversity and Conservation of Tropical Areas, CSIC-UIMP. The Dutch government funded program BE-Basic supported JMSR and MV, JMSR was supported by grant of Consejo Superior de Investigaciones Científicas, CSIC (JAEPre 0901804). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.IUCN (2004) IUCN Red List of Threatened Species. A Global Species Assessment; Baillie JEM, Hilton-Taylor C, Stuart SN, editors. Gland, Switzerland and Cambridge, UK: IUCN.
- 2. Wallace BP, DiMatteo AD, Bolten AB, Chaloupka MY, Hutchinson BJ, et al. (2011) Global conservation priorities for marine turtles. PLoS ONE 6: e24510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sarmiento-Ramírez JM, Abella-Pérez E, Phillott AD, Sim J, Martin MP, et al. (2014) Global distribution of two fungal pathogens threatening endangered sea turtles. PLoS ONE 9: e85853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Woodhams CD, Bosch Jaime, Briggs Cheryl, Cashins Scott, Davis R Leyla, et al. (2011) Mitigating amphibian disease: strategies to maintain wild populations and control chytridiomycosis. Frontiers in Zoology 8: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Berendsen RL, Pieterse CMJ, Bakker PAHM (2012) The rhizosphere microbiome and plant health. Trends in Plant Science 17: 478–486. [DOI] [PubMed] [Google Scholar]
- 6. Yang CH, Crowley DE, Menge JA (2001) 16S rDNA fingerprinting of rhizosphere bacterial communities associated with healthy and Phytophthora infected avocado roots. FEMS Microbiology Ecology 35: 129–136. [DOI] [PubMed] [Google Scholar]
- 7. Bautista-Baños S, Sivakumar D, Bello-Pérez A, Villanueva-Arce R, Hernández-López M (2013) A review of the management alternatives for controlling fungi on papaya fruit during the postharvest supply chain. Crop Protection 49: 8–20. [Google Scholar]
- 8.Gnanamanickam SS (2002) Biological Control of Crop Diseases; Gnanamanickam SS, editor. New York, NY: Marcel Dekker. 449 p. [Google Scholar]
- 9. Sekirov I, Russell SL, Antunes LCM, Finlay BB (2010) Gut microbiota in health and disease. Physiological Reviews 90: 859–904. [DOI] [PubMed] [Google Scholar]
- 10. Nicholson JK, Holmes E, Kinross J, Burcelin R, Gibson G, et al. (2012) Host-gut microbiota metabolic interactions. Science 336: 1262–1267. [DOI] [PubMed] [Google Scholar]
- 11. Xu J, Lazou AI, Prykhodko O, Olsson C, Ahrné S, et al. (2013) Intake of blueberry fermented by Lactobacillus plantarum affects the gut microbiota of L-NAME trated rats. Evidence-Based Complemetary and Alternative Medicine 2013: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Viladomiu M, Hontecillas R, Yuan L, Lu P, Bassaganya-Riera J (2013) Nutritional protective mechanisms against gut inflammation. The Journal of Nutritional Biochemistry 24: 929–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. D'Aversa F, Tortora A, Ianiro G, Ponziani F, Annicchiarico B, et al. (2013) Gut microbiota and metabolic syndrome. Internal and emergency medicine 8: 11–15. [DOI] [PubMed] [Google Scholar]
- 14.Daskin HJ, Alford AR (2012) Content-dependent symbioses and their potential roles in wildlife diseases. Proceedings of the Royal Society B 279.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Avaniss-Aghajani E, Jones K, Chapman D, Brunk C (1994) A molecular technique for identification of bacteria using small subunit ribosomal RNA sequences. Biotechniques 17: 144–149. [PubMed] [Google Scholar]
- 16. Head IM, Saunders JR, Pick RW (1998) Microbial evolution, diversity and ecology: a decade of ribosomal RNA analysis of uncultured microorganisms. Microbial Ecology 35: 1–21. [DOI] [PubMed] [Google Scholar]
- 17. Kim W (2012) Application of metagenomic techniques: understanding the unrevealed human microbiota and explaining the in clinical infectious diseases. Journal of Bacteriology and Virology 42: 262–275. [Google Scholar]
- 18. Hazen TC, Dubinsky EA, DeSantis TZ, Andersen GL, Piceno YM, et al. (2010) Deep-sea oil plume enriches indigenous oil-degrading bacteria. Science 330: 204–208. [DOI] [PubMed] [Google Scholar]
- 19. DeAngelis KM, Brodie EL, DeSantis TZ, Andersen GL, Lindow SE, et al. (2009) Selective progressice response of soil microbial community to wild oats roots. The ISME Journal 3: 168–178. [DOI] [PubMed] [Google Scholar]
- 20.Mendes R, Kruijt M, Bruijn Id, Dekkers E, Voor Mvd, et al.. (2011) Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science 332. [DOI] [PubMed]
- 21. Al-Bahry S, Mahmouda I, Elshafie A, Al-Harthy A, Al-Ghafri S, et al. (2009) Bacterial flora and antibiotic resistance from eggs of green turtles Chelonia mydas: an indication of polluted effluents. Marine Pollution Bulletin 58: 720–725. [DOI] [PubMed] [Google Scholar]
- 22. Craven KS, Awong-Taylor J, Griffiths L, Bass C, Muscarella M (2007) Identification of bacterial isolates from unhatched loggerhead (Caretta caretta) sea turtle eggs in Georgia, USA. Marine Turtle Newsletter 115: 9–11. [Google Scholar]
- 23. Honarvar S, Spotila JR, O'Connor M (2011) Microbial community structure in sand on two olive ridley arribada nesting beaches, Playa La Flor, Nicaragua and Playa Nancite, Costa Rica. Journal of Experimental Marine Biology and Ecology 409: 339–344. [Google Scholar]
- 24.Mo CL, Salas I, Caballero M (1990) Are fungi and bacteria responsible for olive ridley's egg lost? In: Richardson T, Richardson J, Donnelly M, editors. Tenth Annual Workshop on Sea Turtle Biology and Conservation: NOAA Technical Memorandum NMFS-SEFC-278. pp. 249–252.
- 25. Wyneken J, Burke TK, Salmon M, Pedersen DK (1988) Egg failure in natural and relocated sea turtle nests. Journal of Herpetology 22: 88–96. [Google Scholar]
- 26. Awong-Taylor J, Craven KS, Griffiths L, Bass C, Muscarella M (2008) Comparison of biochemical and molecular methods for the identification of bacterial isolates associated with failed loggerhead sea turtle eggs. Journal of Applied Microbiology 104: 1244–1251. [DOI] [PubMed] [Google Scholar]
- 27.Girondot M, Fretey J, Prouteau I, Lescure J (1990) Hatchling success for Dermochelys coriacea in a French Guiana hatchery. NOAA Technical Memorandum NMFS-SEFC- 278.. [Google Scholar]
- 28. Santoro M, Orrego CM, Hernández Gómez G (2006) Flora bacteriana cloacal y nasal de Lepidochelys olivacea (Testudines: Cheloniidae) en el pacífico norte de Costa Rica. Revista de Biología Tropical 54: 43–48. [PubMed] [Google Scholar]
- 29. Glazebrook JS, Campbell RSF, Thomas AT (1993) Studies on an ulcerative stomatitis -obstructive rhinitis-pneumonia disease complex in hatchling and juvenile sea turtles, Chelonia mydas and Caretta caretta . Disease of Aquatic Organisms 16: 133–147. [Google Scholar]
- 30. Haas D, Defago G (2005) Biological control of soil-borne pathogens fy fluorescent Pseudomonas . Nature Reviews Microbiology 3: 307–319. [DOI] [PubMed] [Google Scholar]
- 31. Weller DM, Raaijmakers JM, Gardener BBM, Thomashow LS (2002) Microbial populations responsible for specific soil suppressiveness to plant pathonges Annual Review of Phytopathology. 40: 309–348. [DOI] [PubMed] [Google Scholar]
- 32. Hernandez-Divers SJ, Hensel P, Gladden J, Hernandez-Divers SM, Buhlmann KA, et al. (2009) Investigation of shell disease in map turtles (Grapnemys spp.). Journal of Wildlife disease 45: 637–652. [DOI] [PubMed] [Google Scholar]
- 33. Ming-Ming Y, Liu-Ping X, Qing-Yun X, Jing-Hui Y, Quan X, et al. (2012) Screening potential bacterial biocontrol agents towards Phytophthora capsici in pepper. European Journal of Plant Pathology 134: 811–820. [Google Scholar]
- 34. Goodfellow M, Fiedler H-P (2010) A guide to successful bioprospecting: informed by actinobacterial systematics. Antonie Van Leeuwenhoek 98: 119–142. [DOI] [PubMed] [Google Scholar]
- 35. Nachtigall J, Kulik A, Helaly S, Bull AT, Goodfellow M, et al. (2011) Atacamycins A-C, 22-membered antitumor macrolactones produced by Streptomyces sp Jounal of Antibiotics. 64: 775–780. [DOI] [PubMed] [Google Scholar]
- 36. Tanaka Y, Omura S (1990) Metabolism and Products of Actinomycetes - An Introduction. Actinomycetologica 4: 13–14. [Google Scholar]
- 37. El-Abyad MS, El-Sayed MA, El-Shanshoury AR, El-Sabbagh SM (1993) Towards the biological control of fungal and bacterial diseases of tomato using antagonistic Streptomyces spp. Plant and Soil 149: 185–193. [Google Scholar]
- 38. Köberl M, Ramadan EM, Adam M, Cardinale M, Hallmann J, et al. (2013) Bacillus and Streptomyces were selected as broad-spectrum antagonists against soilborne pathogens from arid areas in Egypt. FEMS Microbiology Letters 342: 168–178. [DOI] [PubMed] [Google Scholar]
- 39. Doumbou CL, Hamby Salove MK, Crawford DL, Beaulieu C (2001) Actinomycetes, promising tools to control plant diseases and to promote plant growth. Phytoprotection 82: 85–102. [Google Scholar]
- 40. Currie CR, Scott JA, Summerbell RC, Malloch D (1999) Fungus-growing ants use antibiotic-producing bacteria to control garden parasites. Nature 398: 701–704. [Google Scholar]
- 41. Sarmiento-Ramírez JM, Abella E, Martín MP, Tellería MT, López-Jurado LF, et al. (2010) Fusarium solani is responsible for mass mortalities in nests of loggerhead sea turtle, Caretta caretta, in Boavista, Cape Verde. FEMS Microbiology Letters 312: 192–200. [DOI] [PubMed] [Google Scholar]
- 42. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local aligment search tool. Journal of Molecular Biology 215: 403–410. [DOI] [PubMed] [Google Scholar]
- 43.Rambaut A (2002) Se-Al Sequence Alignment Editor 2.0a11 ed: University of Oxford.
- 44.Swofford DL (2003) PAUP*: Phylogenetic Analysis Using Parsimony (*and other methods). 4.0 ed: Sinauer Associates.
- 45. Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39: 783–791. [DOI] [PubMed] [Google Scholar]
- 46. Versalovic J, Koeuth T, R.Lupski J (1991) Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Research 19: 6823–6831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Martin B, Humbert O, Camara M, Guenzi E, Walker J, et al. (1992) A highly conserved repeated DNA element located in the chromosome of Streptococcus pneumoniae . Nucleic Acids Research 20: 3479–3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tendulkar SR, Gupta A, Chattoo BB (2003) A simple protocol for isolation of fungal DNA. Biotechnology Letters 25: 1941–1944. [DOI] [PubMed] [Google Scholar]
- 49. Kogan SC, Doherty M, Gitschier J (1987) An improved method for prenatal diagnosis of genetic diseases by analysis of amplified DNA sequences. The New England Journal of Medicine 317: 985–990. [DOI] [PubMed] [Google Scholar]
- 50.Rademaker JLW, Louws FJ, De Brujin FJ (1997) Characterization of ecologically important microbes by rep-PCR genomic fingerprint. In: Akkermans ADL, Van Elsas JD, De Brujin JD, editors. Molecular Microbial Ecology Manual. Dordrecht: Kluwer Academic Publishers. pp. 1–26.
- 51. Turner S, Pryer KM, Miao VPW, Palmer JD (1999) Investigating deep phylogenetic relationships among Cyanobacteria and plastids by small rRNA sequence analysis. The Journal of Eukaryotic Microbiology 46: 327–338. [DOI] [PubMed] [Google Scholar]
- 52. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, et al. (2011) MEGA5: Molecular Evolutionary Genetics Analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Schematic presentation of the metagenomic and classical microbiological approaches and techniques. The scheme represent the approaches used to isolate, identify and characterize the fungal and bacterial community from eggs of the sea turtle species Eretmochelys imbricata nesting at La Playita beach, Machalilla National Park, Ecuador.
(TIF)
Out-group rooted cladogram of the ITS nrDNA region of isolates within the Fusarium solani species complex. One of the most parsimonious trees inferred from the ITS nrDNA sequence data of 136 sea turtle fungal isolates and 60 non-sea turtle fungal isolates. The numbers on the internodes indicate the bootstrap values (BS) of the parsimony analysis. Highlighted isolates correspond to those obtained in this work (n = 10). The arrow indicates the F. falciforme isolate, i.e., 331FUS, used in the dual culture assays to determine the activity of the Actinobacteria.
(TIF)
Cluster analysis (Bray-Curtis) of the microbiome of hatched and unhatched eggs infected by Fusarium falciforme . A) Dendogram of family Pseudomonadaceae (n = 949 OTUs). B) Dendogram of family Flavobacteriaceae (n = 710 OTUs). Abbreviations as in Figure 2.
(TIF)
Out-group rooted phylogenetic tree inferred from the 16S rDNA sequence data from isolates of Streptomyces spp. Data includes isolates of Streptomyces spp. (n = 16) with activity against Fusarium falciforme, and those detected by the PhyloChip analysis (n = 364). The numbers at the internodes indicate the bootstrap values (BS) of the parsimony analysis.
(TIF)
Out-group rooted phylogenetic trees inferred from sequence data from isolates of the Amycolaptosis sp. and Micromonosporaceae. Phylogenetic trees were inferred from the 16S rDNA data from isolates from both taxa, with activity against Fusarium falciforme, and those detected by the PhyloChip analysis. A) Phylogenetic tree from the isolates of the Amycolaptosis sp. (n = 3) with activity against F. falciforme, and those detected by the PhyloChip analysis (n = 29). B) Phylogenetic tree from isolates of the Micromonosporaceae (n = 11) with activity against F. falciforme, those detected by the PhyloChip analysis (n = 33), and additional GenBank strains (n = 5). The numbers at the internodes of the phylogenetic trees indicate the bootstrap values (BS) of the parsimony analysis.
(TIF)
Number of bacteria isolated from the shells of hatched and unhatched eggs of the sea turtle species Eretmochelys imbricata on 1/10th TSA agar medium (total aerobic bacteria) and on GA medium (semi-selective for Actinobacteria). Presented are the Colony Forming Units (CFU/cm2) for each of the two media and for each of the two hatch statuses. For each hatch status, a mean value of 2 eggs is given. SD refers to the standard deviation.
(DOCX)
Most abundant microbial communities from Fusarium -infected eggshells of the sea turtle species Eretmochelys imbricata . Data shown represent the most abundant phyla and families detected by the PhyloChip. The families highlighted in grey are most represented (with >10%) per phylum.
(DOCX)
Chryseobacterium species found significantly more abundant on eggshells of hatched than of unhatched eggs of the sea turtle species Eretmochelys imbricata (Welsh test, p<0.01; r = 1, Anosim).
(DOCX)