Abstract
Circulating tumor cells (CTCs) are important targets for study as we strive to better understand, diagnose, and treat cancers. However, CTCs are found in blood at extremely low concentrations; this makes isolation, enrichment, and characterization of CTCs technically challenging. Recently, the development of CTC separation devices has grown rapidly in both academia and industry. Part of this development effort centered on microfluidic platforms, exploiting the advantages of microfluidics to improve CTC separation performance and device integration. In this Focus article, we highlight some of the recent work in microfluidic CTC separation and detection systems and discuss our appraisal of what the field should do next.
Introduction
A cancerous tumor sheds small amounts of tumorous cells into its immediate vasculature; these cells then make their way into the circulatory system, and are thus called circulating tumor cells (CTCs). CTCs have drawn increasing research attention in recent years,1, 2 with many reports indicating their potential value in cancer prognosis, therapy monitoring, and metastasis research.3, 4 The difficulty in using CTCs lies in their extremely low concentrations in blood samples: a normal concentration in a human cancer patient is approximately 1–100 CTCs per mL of blood. This low concentration has hindered research of CTCs in human samples, as the technical difficulties associated with CTC isolation have led to a paucity of analytical tools.
In designing and evaluating CTC isolation systems, it is important to note the three design objectives of an ideal CTC isolation system:
Isolate all of the CTCs in the blood sample (high capture efficiency)
Isolate only the CTCs, with no other cells accidently isolated (high isolation purity)
Perform this isolation quickly (high system throughput)
High capture efficiency is important to obtaining accurate CTC levels and minimizing the required blood sample volume. High capture efficiency also increases the likelihood that captured tumor cells are representative of primary or secondary tumors in solid tissues. High isolation purity is important to the phenotypic and genotypic analyses performed on the CTCs after isolation, as it will reduce analysis variations caused by interference of the normal blood cells. And high system throughput enables the processing of large sample volume in a short time, making the system a viable tool. Numerous engineering efforts have been made in the past decade to develop CTC isolation systems that adhere to these three design objectives.
Many of these efforts have focused on building macro-scale systems for CTC isolation. One such macro-scale system for isolation and counting of CTCs is called Cell Search; Cell Search has been approved by the U.S. Food and Drug Administration (FDA). This semi-automated system works on an immunomagnetic principle, coating magnetic particles with an antibody (anti-EpCAM) and then bonding the antibody to the epithelial adhesion molecules (EpCAM) of CTCs. After magnetic capture of anti-EpCAM labeled CTCs from a blood sample, CTC identification and enumeration are achieved by skilled operators using immunostaining. Although Cell Search has attained FDA approval, the system still has three important drawbacks: it’s only partially automated (rather than fully automated), it has low capture efficiency, and the captured CTCs are no longer viable. In addition to the immunomagnetic approach of Cell Search, CTC isolation has also been achieved in macro-scale systems via the use of size and density gradients. ISET (Isolation by Size of Epithelial Tumor cells) and OncoQuick are able to isolate CTCs from blood cells by their different sizes and density gradients, respectively.5, 6 These systems isolate CTCs without the use of labels and with high throughput, but normally with insufficient isolation purity.
As an alternative to macro-scale CTC isolation systems, researchers have turned to emerging microfluidic technologies to build promising micro-scale CTC isolation systems. Unlike macro-scale platforms, parameters in microfluidic devices can be precisely controlled at the cellular scale (e.g., channel dimensions, flow profile); this precise control facilitates capture efficiency and isolation purity. Furthermore, isolated CTCs can be manipulated to next-stage analysis (e.g., genetic analysis, drug screening) or on-chip cell culturing as part of the cell separation process, speeding up the overall CTC characterization process and eliminating the intermediate procedures required in macro-scale systems. To date, microfluidic-based CTC isolation systems have been studied vigorously, with various cell separation mechanisms devised: magnetic forces, affinity chromatography, size- and/or deformability-based isolation, and dielectrophoresis. In this Focus article, we will summarize the successful work done using these four CTC separation mechanisms, and will discuss our perspective on future work to be done in microfluidic CTC isolation systems.
Magnetic-based CTC separation or detection
Typical magnetic-based cell separation systems use antibody-antigen interactions to bond an antibody-coated magnetic particle to a cell via its surface antigens. Application of a magnetic field then permits separation of the magnetized CTCs from the non-magnetized cells of the ambient blood sample. As described in the introduction section, the FDA has approved a macro-scale immunomagnetic CTC isolation system called Cell Search. Cell Search uses anti-EpCAM antibodies to attach magnetic particles to the EpCAM surface antigens of CTCs. Immunostaining is then used to characterize the CTCs as DAPI positive, CD45 negative, and cytokeratins 8 and 18 positive. Cell Search has been FDA-approved for monitoring CTC levels in patients with metastatic cancer, such as breast cancer, colorectal cancer, and prostate cancer. Clinical investigations with Cell Search indicated that cancer patients with CTC counts equal to or higher than 5 per 7.5 mL of blood, compared to those with counts lower than 5 per 7.5 mL of blood, had a shorter median progression-free survival and shorter overall survival.7 Although Cell Search achieves CTC isolation and has been FDA-approved, the system remains deficient in that it’s only partially automated (rather than fully automated), it has low capture efficiency, and the captured CTCs are no longer viable.
Recent research has used magnetic-based methods in microfluidic devices to demonstrate substantial improvements over macro-scale systems like Cell Search. Hoshino et al. achieved magnetic capture of CTCs in a microfluidic channel; the device demonstrated high throughput and high capture efficiency.8 An array of magnets was placed beneath a microfluidic channel, and a blood sample with magnetically-labeled CTCs was passed through the channel (Fig. 1a). The flow profile through the channel was controlled to ensure magnetic attraction forces sufficiently large to immobilize CTCs at the channel bottom. After the entirety of the blood sample had passed through the channel, the magnetically captured CTCs were stained with fluorescent labeled anti-cytokeratin, anti-CD45, and DAPI for final identification. The device was capable of detecting as few as 5 CTCs per mL of blood sample, with a sample throughput of 10 mL/hr. CTC separation was achievable with CTC to blood cell ratios as low as 1:10.9 Average CTC capture efficiencies for COLO205 and SKBR3 cells were 90% and 86%, respectively. Moreover, this device only requires a quarter of the anti-EpCAM coated magnetic particles used by Cell Search. To further demonstrate the capability of this device, clinical relevance with a specific type of cancer needs to be established. In addition, cell viability after magnetic capture is not considered in this study, which limits further analysis of captured CTCs. Nevertheless, the high throughput and capture efficiency achieved in this device demonstrate the potential of employing magnetic CTC separation in microfluidic devices.
Figure 1.
(a) Schematic of a microfluidic device using magnets placed under a main flow channel, along with magnetically-labeled CTCs, to accomplish CTC isolation from a blood sample. (b) Microfluidic device with magnets placed under perpendicularly-oriented side chambers used for collection of magnetically-labeled CTCs; the low fluid shear stresses in the dead-ended side chambers keep the collected CTCs viable. (c) Microfluidic device with a flow-focusing configuration to establish a single-file stream of cells through a microfluidic channel. Device is bonded to a micro-Hall detector array, such that each magnetically-labeled CTC passing over the array induces a Hall voltage and is thus counted. Reproduced from Refs. 8, 9, 10 with permission from RSC and AAAS.
The need for CTC viability after isolation from the ambient blood sample was considered by Ingber and coworkers, who reported a microfluidic magnetic separation device that is capable of capturing and subsequently culturing CTCs.9 The device consists of a main microfluidic channel with multiple chambers attached perpendicularly to the main channel’s sides (Fig. 1b). Magnets were placed under the side chambers to force magnetically-labeled CTCs into the chambers. As a blood sample is injected through the main channel, CTCs are collected in these dead-ended side chambers, where fluid shear stresses acting on the CTCs are minimal. During experiments conducted on the device, 2 to 80 spiked breast cancer cells were isolated from 1 mL of mice blood sample with 90% capture efficiency. While blood was flowed through the main channel at an experimental flow rate of 1.2 mL/hr, the calculated shear stresses experienced by the CTCs collected in the side chambers were less than those experienced in-vivo under physiological conditions. After isolation, the CTCs were cultured for 7 days, demonstrating cell viability.
Magnetism has been used not only as a CTC separation mechanism, but also as a CTC detection mechanism. Issadore et al. utilized the Hall effect to detect and count magnetically-labeled CTCs.10 A 2×4 micro-Hall detector array was fabricated on a substrate, and a microfluidic channel (with a flow-focusing configuration upstream) was bonded on top of the Hall detectors (Fig. 1c). A blood sample is first focused by the flow-focusing configuration to establish a single-file stream of cells. As this stream of cells moves over a Hall detector, each magnetically-labeled CTC induces a Hall voltage and is thus counted. Experiments conducted with the device demonstrated CTC detection levels better than those of the Cell Search system, and no false positives were identified to have occurred. This was accomplished with a high system throughput of 107 cells per minute. Additionally, simultaneous detection of multiple surface markers was accomplished via the use of different types of magnetic nanoparticles. Although the device does not isolate the CTCs, its ability to rapidly and accurately count CTCs—without the use of an optical detection system—could find use in an automated, low cost, and portable CTC analysis tool.
Affinity chromatography CTC separation
CTC separation via affinity chromatography uses the same antibody-antigen interactions used in magnetic-based CTC separation. But while the magnetic-based methods use the interactions to bond magnetic particles to CTCs, affinity chromatography uses them to directly capture the CTCs (thus affinity chromatography does not require the CTC labeling step that is needed in magnetic-based separation, simplifying device application). In affinity chromatography, antibodies are attached to the surfaces of solid structures. The structures are then immersed in a fluid biological sample where the antigens of targeted cells bond with the antibodies, in effect attaching the target cells to the solid structures and separating them from the remainder of the sample.
The effectiveness of affinity chromatography depends on maximizing the potential for antibody-antigen bonding of the target cells to the capture structures, while still removing the non-target cells. Therefore, sample cells must be frequently brought into contact with the antibody-covered surfaces of the capture structures, and fluid shear stresses must be low enough to permit antibody-antigen bonding while still high enough to remove non-target cells. Microfluidic technologies feature large surface-area-to-volume ratios and allow precise control of fluid shear stress, and therefore are an excellent platform for affinity chromatography. In fact, the implementation of affinity chromatography in microfluidic devices has been demonstrated in rapid point of care diagnosis, such as CD4+ cell counting for HIV monitoring.11
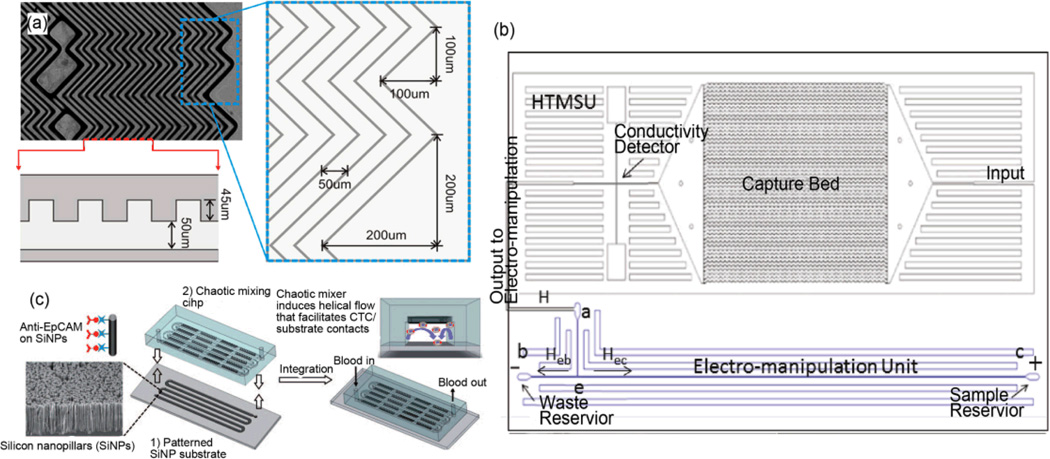
However, microfluidic affinity chromatography is challenging to implement for CTC capture: due to the low concentration of CTCs in blood, a large sample volume (e.g., 1–7.5 mL) must be processed to capture significant amounts of CTCs. For such a large sample volume, contact between the CTCs and the capture structures cannot be ensured by exposing a quiescent sample to the structures. Instead, small amounts of the sample must be continuously flowed past the structures with careful determination of the flow rate. This flow rate determination is necessary to balance the competing needs for high device throughput (fast flow rate) and high capture efficiency (low flow rate, which causes low fluid shear stresses). To increase the probability of CTC capture in microfluidic affinity chromatography, researchers have implemented microstructures within fluid channels. Nagrath et al. fabricated 78,000 anti-EpCAM coated silicon micro-pillars in a microfluidic channel to obtain high capture efficiency while maintaining reasonable system throughput (~1 mL/h).12 In clinical testing, CTCs were identified in 99% of patients’ samples (n=116). Following the work by Nagrath, Stott et al. fabricated a microfluidic channel with a herringbone pattern in the channel ceiling to disrupt the laminar flow profile (Fig. 2a), resulting in a higher potential for CTC bonding13 to the channel surfaces (which are coated with anti-EpCAM antibodies). Compared to the micro-pillar device, the herringbone device has reduced fabrication complexity and improved capture efficiency for spiked PC-3 prostate cancer cells. Further experiments on this device showed CTCs detected in 14 out of 15 prostate cancer patients. After on-chip CTC capture, the herringbone device allows in-situ immunostaining or cell lysis for further characterization. However, feasibility of efficient and viable release of CTCs from this device requires more validation.
Figure 2.
(a) Herringbone pattern in the channel ceiling of a microfluidic device for CTC isolation via affinity chromatography. (b) Schematic of a microfluidic device with integrated systems for CTC capture, enumeration, and electro-manipulation. (c) Image of a substrate whose surface consists of silicon nanopillars coated in anti-EpCAM antibodies, and schematic of the overall microfluidic device featuring a serpentine channel with a herringbone-patterned ceiling to constantly bring sample cells into contact with the substrate for CTC capture. Reproduced from Refs. 13, 15, 16 with permission from NAS, ACS and Wiley.
A straight, flat, short microfluidic channel would have low capture efficiency in an affinity chromatography process. One approach to improving the capture efficiency is the fabrication of microstructures in the flow channel, as described previously. An alternative approach is simply to make the channel long and not straight. Soper and coworkers took this approach, constructing a device with a large number of long, parallel, sinusoidally undulating channels for CTC separation:14 51 parallel channels were fabricated with cross-sectional areas of 35 µm×150 µm (width×depth), and the channel surfaces were coated with anti-EpCAM antibodies (Fig. 2b). With this device, a 1 mL blood sample can be processed within 37 min, with linear velocity optimized at 2 mm/s for maximum CTC capture. Experiments on the device demonstrated that spiked MCF-7 cells were isolated with up to 97% capture efficiency. Thus this approach to affinity chromatography achieved high CTC capture efficiency without the need for microstructures, which are more complex to fabricate than the array of parallel undulating channels. In addition to affinity chromatography for CTC isolation, the device integrates other systems for other analytical purposes. After release of the captured CTCs via treatment of the antibody-antigen bonds with trypsin protease, the stream of CTCs was moved past a conductivity detector to enable automated label-free CTC counting. And after being counted by the conductivity detector, the stream of CTCs was directed to an electrokinetic enrichment unit to move cells to an outlet reservoir for subsequent genetic analysis.15 Although the use of conductive cell counting and electrokinetic enrichment requires more validation for clinical relevance, the device demonstrates the viability of a micro total analysis system (µTAS). By integrating multi-functional units in a single microfluidic device, detection sensitivity can be enhanced while sample loss is reduced.
The aforementioned devices increased the frequency of CTC contact with antibody-coated surfaces by making various design alterations to the device channels. Wang et al. accomplished increased frequency of cell-surface interactions by altering the device substrate. Wang patterned silicon nanopillars on the substrate surface, and coated the pillars with anti-EpCAM antibodies (Fig. 2c).16 A serpentine channel with herringbone patterns along the channel ceiling was bonded to the silicon nanopillar substrate; this setup induced helical flow in the channel in order to repeatedly bring sample cells into contact with the substrate. In experiments testing device performance, at 1 mL/h flow rate, 100 cells/mL MCF-7 cells were captured from buffer solution with 95% efficiency. And in a clinical study, the silicon nanopillar device showed a higher CTC capture efficiency than Cell Search in 17 out of 25 prostate cancer patients’ samples. It is important to note that design alterations made to achieve higher capture efficiency may also induce stronger non-specific cell binding, resulting in lower isolation purity. This trade-off should be understood and balanced based on the emphasis of intended device applications (CTC counting vs. characterization).
Size- and/or deformability-based CTC separation
In many cell separation applications, cells are often separated from an ambient sample by means of their differing physical properties, such as stiffness, size, and density. Such physical property-based separation systems offer the advantages of label-free sorting, high system throughput, and low cost.17, 18 Previous investigations have found that many types of CTCs are significantly larger than other cells contained in peripheral blood.5 This size discrepancy has been harnessed for CTC separation in macro-scale systems; these systems use a porous membrane to retain large CTCs, which are then identified through immunostaining. However, this method suffers from poor capture efficiency and isolation purity, and the CTCs are susceptible to damage from associated large mechanical stresses. Recent studies also revealed that several types of cancer cells or metastatic cancer cells are much more deformable than their benign counterparts.19–22 So far, there have only been a handful of studies trying to separate CTCs based on deformability differentials.
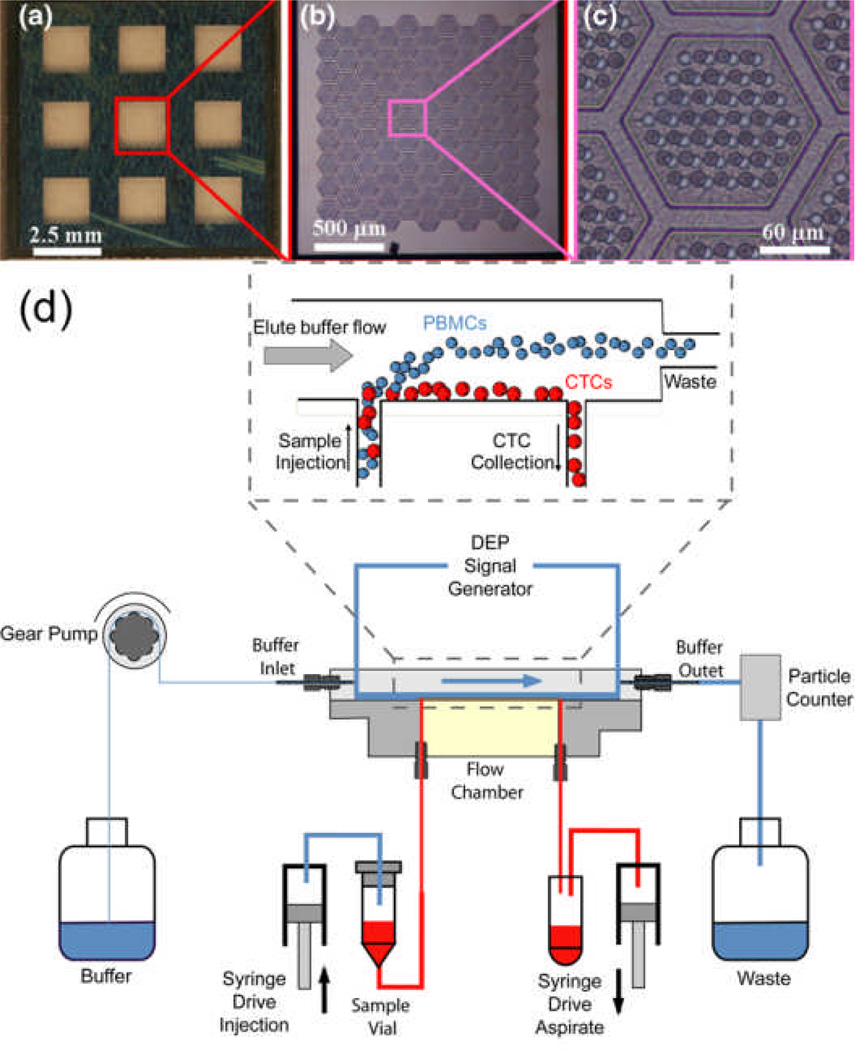
Zheng et al. reported improved size-based CTC separation performance by use of micro-fabricated 3-D microfilters.23 These microfilters are more uniform in size and density (Figs. 3a-c) than macro-scale filters. Experiments with these microfilters demonstrated capture efficiency of 86% for MCF-7 cells with a concentration of 350 cells/mL. Moreover, the system had an impressively high throughput, filtering 1 mL of blood sample within several minutes. Zheng also adjusted the 3-D porous membrane structure of the microfilters to minimize mechanical stress experienced by the cells during the filtration process. As a result, cell viability was high, and isolated CTCs were maintained viable for 2 weeks. This device offers a rapid and convenient CTC isolation method when the CTCs are known to be significantly larger than the ambient blood cells. Another approach successfully utilized cell-size dependent microscale vortices as the cell sorting mechanism for microfluidic CTC separation.24 Deformability-based, as well as both size- and deformability-based, CTC enrichment methods also began to emerge recently. 25, 26
Figure 3.
(a-c) Images of a 3-D microfilter, used for size-based mechanical separation of CTCs, at different magnifications. (d) Schematic of a dielectrophoretic CTC separation device, with detail provided for the flow chamber where an electrode sheet along the chamber floor generates an AC voltage to spatially separate CTCs from PBMCs via a DEP force. Reproduced from Refs. 23, 27 with permission from AIP and Springer.
Dielectrophoretic CTC separation
When dielectric particles are subjected to a non-uniform electric field, they experience a dielectrophoretic (DEP) force. Because the magnitude and direction of the DEP force acting on a particle is dependent in part on the particle’s polarizability, different particle types will experience different DEP forces. Thus DEP can be exploited as a label-free cell separation mechanism.
The DEP force exerted on a cell depends not only on the electrical properties of the cell, but also on the magnitude and frequency (in the case of AC) of the applied electric field. Within certain frequency ranges, a cell may move in the direction of increasing electric field (positive DEP), whereas in other frequency ranges it might move in the direction of decreasing electric field (negative DEP). Gupta et al. designed a microfluidic CTC-isolation device that exploits the varying directionality of the DEP force acting on cancer and blood cells in a suitable electric field.27 A flexible polyimide film sheet was electroplated with interdigitated copper and gold electrodes and used to form the floor of a microfluidic flow chamber. An eluate buffer was introduced at the upstream end of the flow chamber, and a blood sample was introduced along the chamber floor immediately upstream of the flow chamber. By applying an AC voltage across the electrodes in the 45–85 kHz range, a positive DEP force was applied to the cancer cells to drive them towards the flow chamber floor, and a negative DEP force was applied to the sample blood cells to drive them away from the flow chamber floor. Thus at the downstream end of the flow chamber, the cancer cells could be collected via an outlet located at the chamber floor (Fig. 3d).
In device characterization, experiments conducted with peripheral blood mononuclear cells (PBMCs) in buffer solution and spiked with SKOV3 or MDA-MB-231 cancer cells showed capture efficiency of around 70% with a throughput of 1 mL/h. CTC viability after isolation was greater than 97%, and the isolated cells were cultured for one week with a normal proliferation rate. This DEP-based CTC isolation is currently under clinical investigation. Although the system requires no labeling of the cancer cells, it does require that a blood sample be initially processed to isolate all PBMCs (which include CTCs) which are then mixed with a buffer solution; this PBMC/buffer solution is then run through the flow chamber and subjected to DEP separation.
Combination of multiple CTC separation mechanisms
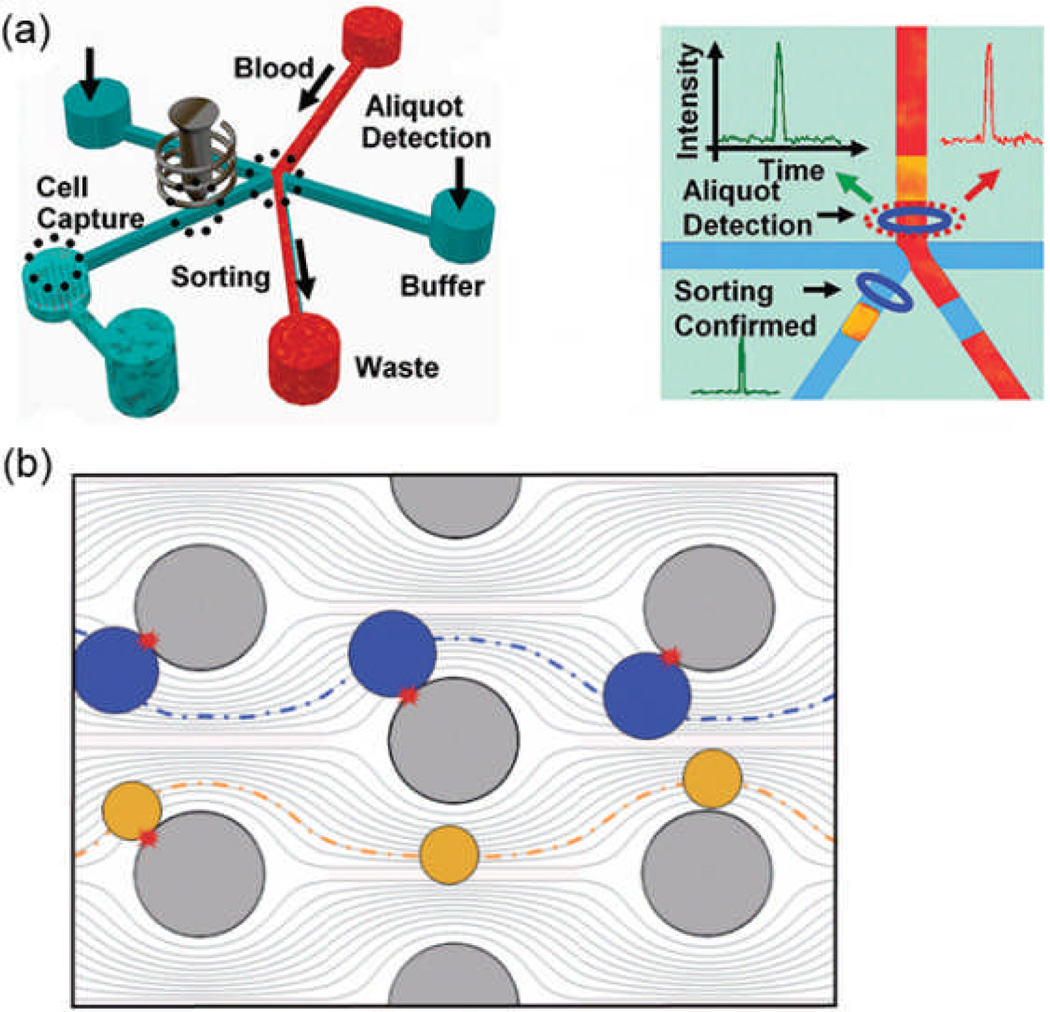
Magnetic-based, affinity chromatography, size/deformability-based, and dielectrophoretic CTC separation approaches each have their own advantages and limitations. Some researchers have combined techniques from multiple separation approaches in an effort to achieve improved separation performance. For example, Chiu and coworkers combined fluorescence-activated cell sorting (FACS) with size-based filtration to enable CTC isolation with high throughput and high capture efficiency.28 Conventional FACS is normally inadequate for CTC applications due to its low throughput and sorting efficiency (individual cells are analyzed in a single-file format). To adapt FACS for CTC isolation, Chiu developed a process called ensemble-decision aliquot ranking (eDAR). As in FACS, the target cells for separation are first labeled with fluorescent tags (anti-EpCAM was one of the antibodies used in experiments carried out with eDAR). However, where FACS analyzes cells one at a time, eDAR divides the sample into volume aliquots, each containing thousands of cells, and then detects the presence of fluorescent CTCs in each aliquot. The blood sample flows continuously within a microfluidic channel through an interrogation region. If no fluorescence is detected as the fluid passes through the interrogation region, the fluid continues down one side of a branched path. If fluorescence is detected, a control solenoid is activated to temporarily release its mechanical pressure on the other side of the branched path, briefly diverting fluid flow down this alternate path. This alternate path leads to a CTC capture chamber, where a filter mechanically separates the CTCs from the blood sample based on their cell size (Fig. 4a). Thus the aliquots are virtual aliquots, in that their volume is defined by a combination of laser illumination volume, bin time of the fluorescence detection system, and switching time of a control solenoid.
Figure 4.
(a) Schematic of device setup for ensemble-decision aliquot ranking CTC sorting, and a drawing of virtual volume aliquots being separated from the blood sample to form actual volume aliquots. (b) Schematic of geometrically-enhanced differential immunocapture obstacles distorting the streamlines of a flowing fluid, leading to increased cell-surface collisions for the larger cells (the targeted CTCs) and fewer cell-surface collisions for the smaller cells (other cells in the sample). Reproduced from Refs. 28, 29 with permission from Wiley and RSC.
Due to the scarcity of CTCs in a blood sample, testing for CTC fluorescence in an aliquot containing thousands of cells, and then filtering only those aliquots that contain CTCs, serves to greatly improve system throughput. In eDAR, the virtual aliquot volume was set at 2 nL, which was found to optimize both detection sensitivity and system throughput (3 mL/h). Capture efficiency was determined to be 93% for both MCF-7 and SKBr-3 cells with concentrations as low as 5 cells/mL. Also, by implementing multiple light sources and detectors, this system allows multi-color sorting with either “AND” or “OR” logic, resulting in flexible sorting options. It should be noted that the cost of this system will be higher than other microfluidic CTC isolation devices due to the use of laser sources, optics, and multiple avalanche photodiode detectors.
In affinity chromatography, increasing the probability of sample cell collision with the antibody-coated capture surfaces is crucial to achieving high capture efficiencies and high system throughput. However, increasing this probability of cell-surface collisions for all sample cells (i.e., both target and non-target cells) will increase the probability of non-specific binding, reducing isolation purity. Kirby and coworkers attempted to resolve this tradeoff between capture efficiency and isolation purity by preferentially increasing the probability of cell-surface collisions for target cells, rather than all cells. To do this, Kirby introduced an array of staggered circular or octagonal posts to an affinity chromatography CTC capture device.29 The posts acted as obstacles to fluid flow, and were coated with antibodies in order to serve as capture surfaces. The positioning of the posts created flow streamlines that brought larger cells into contact with post surfaces more frequently than smaller cells (Fig. 4b); thus Kirby called the approach geometrically-enhanced differential immunocapture (GEDI). In GEDI device experiments, a capture efficiency of 85% and isolation purity of 68% were achieved using a flow rate under 1 mL/h for a blood sample with spiked prostate cancer cells (150–220 cells/mL). Patients’ samples were also tested with a J591 antibody-coated GEDI microfluidic device. CTCs were found in 90% of total samples (n=20). After capture, prostate cancer cells were tested for taxanes treatment; microtubule bundling was detected using immunofluorescence staining.30
Perspectives
In the previous sections, we have examined work done in microfluidic CTC separation or detection devices. These devices function based on four general mechanisms: magnetic forces acting on magnetically-labeled CTCs, affinity chromatography targeting some surface molecule of CTCs, size/deformability difference between CTCs and other blood sample cells, and dielectrophoresis exploiting the differing polarizabilities of CTCs and other blood sample cells. We have also examined devices that combine aspects of multiple separation mechanisms. Whatever the functional mechanisms, all of these devices have sought to meet the design objectives of high capture efficiency, high isolation purity, and high system throughput.
It is essential to recognize that different cancer types—and even different cancer sub-types—will produce CTCs with different characteristics. Current research indicates that no single biological or physical standard can be used to define all types of CTCs.31 Efforts on device development should begin taking this into account. For example, accumulated knowledge of CTC physical properties (e.g., size, stiffness, density) for different cancer types should be codified. This would allow a size-based CTC separation device to be optimized for a particular cancer type; it would also allow that device to incorporate separation methods using other physical parameters (e.g., stiffness and density) to better differentiate between CTCs and ambient blood cells.25, 32
As another example of how device development efforts should account for different cancer types producing CTCs with different characteristics, consider the antibody-antigen bonds used in magnetic labeling, fluorescence labeling, and affinity chromatography. The cell surface molecule EpCAM is widely used to target CTCs, but certain types of cancer are EpCAM-negative. Not only does EpCAM expression differ among cancer types, it can also vary significantly over the progression of a single cancer type. Clearly, EpCAM is not an ideal option for all device applications. Depending on the application, other antibodies—or mixtures of antibodies—should be used to appropriately target CTCs. Another option for certain types of cancer is the use of aptamers,33 generated via SELEX (Systematic Evolution of Ligands by Exponential Enrichment). Recent study showed that 3D DNA network consisted of repetitive aptamer domains resulted in higher cell capture efficiency under high flow rate in cell affinity chromatography.34 This result is promising for improving the performance of current CTC-capture devices. Future CTC-isolation devices and operating procedures should be made compatible with multiple antibodies or aptamers, thus improving device applicability.
A final point regarding different cancer types producing CTCs with different characteristics: as the catalogue of CTC separation devices grows, researchers need to begin performing comparative studies across cancer types. By analyzing blood from a single patient in multiple separation devices—or, alternately, using a single device to analyze samples from multiple patients, each with a different cancer type—we can begin establishing type-specific performance profiles for CTC separation devices. This will enable clinicians to select the most suitable system for each patient.35
In addition to considering the variability of CTCs, researchers should continue the trend of creating integrated microfluidic systems that not only separate CTCs, but perform subsequent cell analyses and/or cultures as well. Such cell analyses will produce detailed CTC characterizations that should reveal information about cancer progress and metastasis (hopefully, correlation between CTCs and early cancer diagnosis can be established). In-situ genetic or proteomic profiling for CTCs can be established without sample loss or excessive dilution. And culturing of CTCs will enable drug screenings and detailed metastatic studies.
A first step in creating such integrated microfluidic systems is combining multiple CTC separation methods on a single device. For example, high-throughput, size/deformability-based separation can be used initially to obtain a small volume of mostly CTCs. This small, mostly pure volume can then be moved to a highly specific stop-flow affinity chromatography module that results in a highly pure CTC population. Once this highly pure CTC population is obtained from the blood sample, various kinds of microfluidic methods (e.g., hydrodynamic flow, pneumatic valve, electric and acoustic manipulation36) can be used to move cells between different analytical modules. Such integrated microfluidic systems hold much promise, and they are realizable right now. Researchers can move beyond demonstration of individual techniques, and begin creating fully functionalized CTC analysis devices. This is not to say that we should cease our search for novel individual techniques. In fact, some new CTC separation approaches have recently shown particular promise (e.g., hydrodynamic separation,37 acoustic sorting38, 39), and deserve further investigation.
Lastly, we should acknowledge that although many microfluidic CTC isolation and detection devices have shown promising results, the macro-scale system Cell Search remains the only FDA-approved instrument for CTC isolation. To gain more traction in the medical industry, current and future microfluidic systems should be validated with more clinically relevant testing. Meanwhile, reproducibility, robustness, cost, and operational complexity of current and future microfluidic systems should be optimized with clinical applications foremost in mind.
Acknowledgements
The authors acknowledge Prof. Subra Suresh for his helpful discussions and insightful suggestions. This research was supported by National Institutes of Health (NIH) Director’s New Innovator Award (1DP2OD007209-01).
References
- 1.Yu M, Stott S, Toner M, Maheswaran S, Haber DA. J. Cell Biol. 2011;192:373–382. doi: 10.1083/jcb.201010021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alunni-Fabbroni M, Sandri MT. Methods. 2010;50:289–297. doi: 10.1016/j.ymeth.2010.01.027. [DOI] [PubMed] [Google Scholar]
- 3.Botteri E, Sandri MT, Bagnardi V, Munzone E, Zorzino L, Rotmensz N, Casadio C, Cassatella MC, Esposito A, Curigliano G, Salvatici M, Verri E, Adamoli L, Goldhirsch A, Nole F. Breast Cancer Res. Treat. 2010;122:211–217. doi: 10.1007/s10549-009-0668-7. [DOI] [PubMed] [Google Scholar]
- 4.Pierga JY, Bidard FC, Mathiot C, Brain E, Delaloge S, Giachetti S, de Cremoux P, Salmon R, Vincent-Salomon A, Marty M. Clin. Cancer Res. 2008;14:7004–7010. doi: 10.1158/1078-0432.CCR-08-0030. [DOI] [PubMed] [Google Scholar]
- 5.Vona G, Sabile A, Louha M, Sitruk V, Romana S, Schutze K, Capron F, Franco D, Pazzagli M, Vekemans M, Lacour B, Brechot C, Paterlini-Brechot P. Am. J. Pathol. 2000;156:57–63. doi: 10.1016/S0002-9440(10)64706-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Muller V, Stahmann N, Riethdorf S, Rau T, Zabel T, Goetz A, Janicke F, Pantel K. Clin. Cancer Res. 2005;11:3678–3685. doi: 10.1158/1078-0432.CCR-04-2469. [DOI] [PubMed] [Google Scholar]
- 7.Cristofanilli M, Budd GT, Ellis MJ, Stopeck A, Matera J, Miller MC, Reuben JM, Doyle GV, Allard WJ, Terstappen L, Hayes DF. N. Eng. J. Med. 2004;351:781–791. doi: 10.1056/NEJMoa040766. [DOI] [PubMed] [Google Scholar]
- 8.Hoshino K, Huang YY, Lane N, Huebschman M, Uhr JW, Frenkel EP, Zhang XJ. Lab Chip. 2011;11:3449–3457. doi: 10.1039/c1lc20270g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kang JH, Krause S, Tobin H, Mammoto A, Kanapathipillai M, Ingber DE. Lab Chip. 2012;12:2175–2181. doi: 10.1039/c2lc40072c. [DOI] [PubMed] [Google Scholar]
- 10.Issadore D, Chung J, Shao HL, Liong M, Ghazani AA, Castro CM, Weissleder R, Lee H. Sci. Transl. Med. 2012;4:141ra192. doi: 10.1126/scitranslmed.3003747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moon S, Gurkan UA, Blander J, Fawzi WW, Aboud S, Mugusi F, Kuritzkes DR, Demirci U. PLoS One. 2011;6:e21409. doi: 10.1371/journal.pone.0021409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nagrath S, Sequist LV, Maheswaran S, Bell DW, Irimia D, Ulkus L, Smith MR, Kwak EL, Digumarthy S, Muzikansky A, Ryan P, Balis UJ, Tompkins RG, Haber DA, Toner M. Nature. 2007;450:1235–1239. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stott SL, Hsu CH, Tsukrov DI, Yu M, Miyamoto DT, Waltman BA, Rothenberg SM, Shah AM, Smas ME, Korir GK, Floyd FP, Gilman AJ, Lord JB, Winokur D, Springer S, Irimia D, Nagrath S, Sequist LV, Lee RJ, Isselbacher KJ, Maheswaran S, Haber DA, Toner M. Proc. Natl. Acad. Sci. U.S.A. 2010;107:18392–18397. doi: 10.1073/pnas.1012539107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Adams AA, Okagbare PI, Feng J, Hupert ML, Patterson D, Gottert J, McCarley RL, Nikitopoulos D, Murphy MC, Soper SA. J. Am. Chem. Soc. 2008;130:8633–8641. doi: 10.1021/ja8015022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dharmasiri U, Njoroge SK, Witek MA, Adebiyi MG, Kamande JW, Hupert ML, Barany F, Soper SA. Anal. Chem. 2011;83:2301–2309. doi: 10.1021/ac103172y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang ST, Liu K, Liu JA, Yu ZTF, Xu XW, Zhao LB, Lee T, Lee EK, Reiss J, Lee YK, Chung LWK, Huang JT, Rettig M, Seligson D, Duraiswamy KN, Shen CKF, Tseng HR. Angew. Chem., Int. Ed. 2011;50:3084–3088. doi: 10.1002/anie.201005853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mao X, Huang TJ. Lab Chip. 2012;12:4006–4009. doi: 10.1039/c2lc90100e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shi J, Huang H, Stratton Z, Huang YP, Huang TJ. Lab Chip. 2009;9:3354–3359. doi: 10.1039/b915113c. [DOI] [PubMed] [Google Scholar]
- 19.Suresh S, Spatz J, Mills JP, Micoulet A, Dao M, Lim CT, Beil M, Seufferlein T. Acta Biomater. 2005;1:15–30. doi: 10.1016/j.actbio.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 20.Suresh S. Acta Biomater. 2007;3:413–438. doi: 10.1016/j.actbio.2007.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Suresh S. Nat. Nanotechnol. 2007;2:748–749. doi: 10.1038/nnano.2007.397. [DOI] [PubMed] [Google Scholar]
- 22.Cross SE, Jin YS, Rao J, Gimzewski JK. Nat. Nanotechnol. 2007;2:780–783. doi: 10.1038/nnano.2007.388. [DOI] [PubMed] [Google Scholar]
- 23.Zheng SY, Lin HK, Lu B, Williams A, Datar R, Cote RJ, Tai YC. Biomed. Microdevices. 2011;13:203–213. doi: 10.1007/s10544-010-9485-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hur SC, Mach AJ, Di Carlo D. Biomicrofluidics. 2011;5 doi: 10.1063/1.3576780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tan SJ, Yobas L, Lee GYH, Ong CN, Lim CT. Biomed. Microdevices. 2009;11:883–892. doi: 10.1007/s10544-009-9305-9. [DOI] [PubMed] [Google Scholar]
- 26.Hur SC, Henderson-MacLennan NK, McCabe ERB, Di Carlo D. Lab Chip. 2011;11:912–920. doi: 10.1039/c0lc00595a. [DOI] [PubMed] [Google Scholar]
- 27.Gupta V, Jafferji I, Garza M, Melnikova VO, Hasegawa DK, Pethig R, Davis DW. Biomicrofluidics. 2012;6:024133. doi: 10.1063/1.4731647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schiro PG, Zhao MX, Kuo JS, Koehler KM, Sabath DE, Chiu DT. Angew. Chem., Int. Ed. 2012;51:4618–4622. doi: 10.1002/anie.201108695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gleghorn JP, Pratt ED, Denning D, Liu H, Bander NH, Tagawa ST, Nanus DM, Giannakakou PA, Kirby BJ. Lab Chip. 2010;10:27–29. doi: 10.1039/b917959c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kirby BJ, Jodari M, Loftus MS, Gakhar G, Pratt ED, Chanel-Vos C, Gleghorn JP, Santana SM, Liu H, Smith JP, Navarro VN, Tagawa ST, Bander NH, Nanus DM, Giannakakou P. PLoS One. 2012;7:e35976. doi: 10.1371/journal.pone.0035976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van de Stolpe A, Pantel K, Sleijfer S, Terstappen LW, den Toonder JMJ. Cancer Res. 2011;71:5955–5960. doi: 10.1158/0008-5472.CAN-11-1254. [DOI] [PubMed] [Google Scholar]
- 32.Park JM, Lee JY, Lee JG, Jeong H, Oh JM, Kim YJ, Park D, Kim MS, Lee HJ, Oh JH, Lee SS, Lee WY, Huh N. Anal. Chem. 2012;84:7400–7407. doi: 10.1021/ac3011704. [DOI] [PubMed] [Google Scholar]
- 33.Sheng WA, Chen T, Katnath R, Xiong XL, Tan WH, Fan ZH. Anal. Chem. 2012;84:4199–4206. doi: 10.1021/ac3005633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao W, Cui CH, Bose S, Guo D, Shen C, Wong WP, Halvorsen K, Farokhzad OC, Teo GSL, Phillips JA, Dorfman DM, Karnik R, Karp JM. Proc. Natl. Acad. Sci. U.S.A. 2012;109:19626–19631. doi: 10.1073/pnas.1211234109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Farace F, Massard C, Vimond N, Drusch F, Jacques N, Billiot F, Laplanche A, Chauchereau A, Lacroix L, Planchard D, Le Moulec S, Andre F, Fizazi K, Soria JC, Vielh P. Brit. J. Cancer. 2011;105:847–853. doi: 10.1038/bjc.2011.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ding XY, Lin SCS, Kiraly B, Yue HJ, Li SX, Chiang IK, Shi JJ, Benkovic SJ, Huang TJ. Proc. Natl. Acad. Sci. U.S.A. 2012;109:11105–11109. doi: 10.1073/pnas.1209288109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sun JS, Li MM, Liu C, Zhang Y, Liu DB, Liu WW, Hu GQ, Jiang XY. Lab Chip. 2012;12:3952–3960. doi: 10.1039/c2lc40679a. [DOI] [PubMed] [Google Scholar]
- 38.Ding XY, Lin SSC, Lapsley MI, Li SX, Guo X, Chan CY, Chiang IK, Wang L, McCoy JP, Huang TJ. Lab Chip. 2012;12:4228–4231. doi: 10.1039/c2lc40751e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Augustsson P, Magnusson C, Nordin M, Lilja H, Laurell T. Anal. Chem. 2012;84:7954–7962. doi: 10.1021/ac301723s. [DOI] [PMC free article] [PubMed] [Google Scholar]