Abstract
The Notch signaling pathway is a regulator of self-renewal and differentiation in several tissues and cell types. Notch is a binary cell-fate determinant, and its hyperactivation has been implicated as oncogenic in several cancers including breast cancer and T-cell acute lymphoblastic leukemia (T-ALL). Recently, several studies also unraveled tumor-suppressor roles for Notch signaling in different tissues, including tissues where it was before recognized as an oncogene in specific lineages. Whereas involvement of Notch as an oncogene in several lymphoid malignancies (T-ALL, B-chronic lymphocytic leukemia, splenic marginal zone lymphoma) is well characterized, there is growing evidence involving Notch signaling as a tumor suppressor in myeloid malignancies. It therefore appears that Notch signaling pathway’s oncogenic or tumor-suppressor abilities are highly context dependent. In this review, we summarize and discuss latest advances in the understanding of this dual role in hematopoiesis and the possible consequences for the treatment of hematologic malignancies.
Introduction
Hematopoiesis is a complex process that requires coordination among proliferation, self-renewal, and differentiation of stem and progenitor cells to generate mature cells in the blood. Deregulation or imbalance in these physiological processes can lead to transformation. Notch signaling has been implicated in the regulation of these diverse functions in the hematopoietic system and other tissues. Whereas the importance of Notch1 in lymphocyte development and oncogenic transformation has been well characterized, the relevance of Notch signaling in the specification of other hematopoietic lineages and the hematopoietic stem cell (HSC) has long been a matter of debate. Recent studies of the Notch pathway, including work in our laboratory, have challenged the view that the function of Notch is limited only to promoting thymic T-cell differentiation and the development of marginal zone B cells. Furthermore, there is growing evidence that components of the same oncogenic pathway in lymphocytes may have a growth-suppressive function in myeloid cells, as previously shown in epithelial or head and neck squamous cell carcinomas (SCCs).1-3 Some of the controversy may reflect functional redundancy between receptors, differences between ligands, or the inherent cellular and developmental context-dependent nature of Notch signaling. In this review, we address known and novel roles for Notch signaling in hematopoiesis and its oncogenic and tumor-suppressor functions.
Overview of the Notch signaling pathway
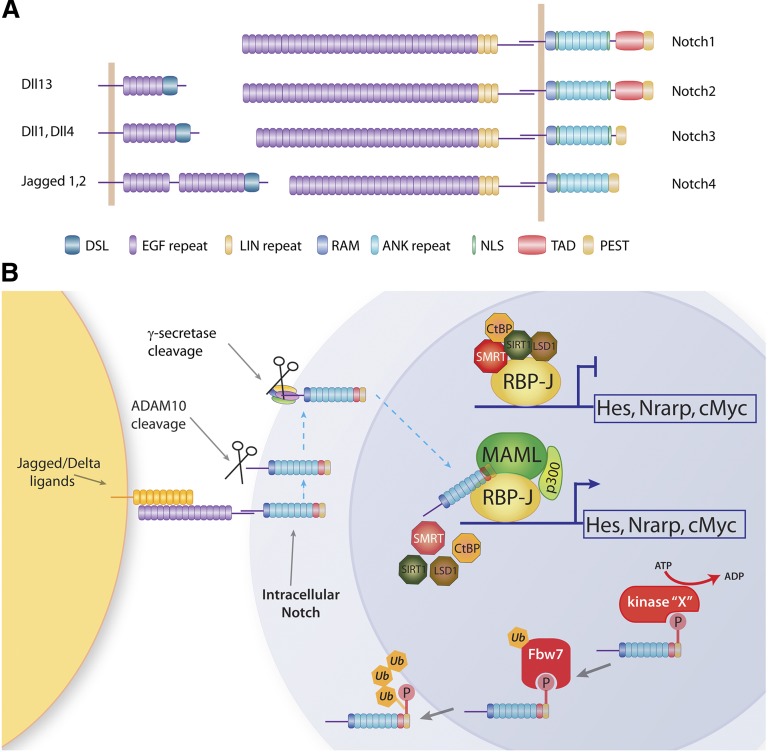
The Notch receptor was first described by Thomas Hunt Morgan in 1917 with the observation of a Drosophila strain with notched wings,4 yet it was not until decades later that Spyros Artavanis-Tsakonas and Michael Young cloned the Notch receptor and attributed the wing-notching phenotype to gene haploinsufficiency.5,6 It is now clear that Notch signaling plays an essential role in many processes during metazoan development and in adult tissues, including fundamental processes such as the determination of cell fates, proliferation, and apoptosis. Although Notch receptors are highly conserved between species, mammals possess 4 distinct Notch receptors in contrast to Drosophila and nematodes that express 1 (Notch) or 2 (LIN-12, GLP-1) receptors, respectively. Notch receptors are single-pass type I transmembrane receptors synthesized as a single precursor that is cleaved during transport in the Golgi by a furin-like convertase (S1 cleavage) and exist as a noncovalently linked heterodimer at the cell surface.7 Notch1 and Notch2 each have 36 epidermal growth factor (EGF)-like repeats, whereas Notch3 has 34 and Notch4 contains 29.8 These differences may be significant, because EGF-like repeats are fucosylated on specific serine and threonine residues by O-fucosyltransferase, and this is required for efficient binding of receptors to ligands.9-11 These O-fucose moieties can be elongated by the addition of N-acetylglucosamine by the Fringe family of 1,3 N-acetylglucosaminyltransferases,12 and these modifications regulate the affinity of Notch receptors for certain ligands.13,14 The intracellular portions of Notch (ICN) consists of ankyrin repeats, a RBP-Jκ associated molecule (RAM) domain, a transactivation domain (TAD), a nuclear localization signal, and a proline-glutamate-serine-threonine-rich (PEST) domain regulating protein stability (Figure 1A).15-17 Notch3 and Notch4 lack the classical TAD, influencing promoter selectivity at least in vitro.18 Experimental evidence indicates that a high-affinity interaction between ICN and RBP-Jκ may also occur through the RAM domain, but it is the ankyrin repeats that are essential for the formation of the transcriptional activation complex and the recruitment of MAML1 (mastermind).17
Figure 1.
Mammal Notch receptors, ligands, and canonical activation pathway. (A) Notch ligands and receptors. There are 5 Notch ligands in mammals: Jagged1, Jagged2, Delta-like1 (Dll1), Delta-like3 (Dll3), and Delta-like4 (Dll4). All ligands have an extracellular domain called DSL (Delta, Serrate, and Lag-2) involved in receptor binding associated with EGF-like repeats. There are 4 Notch receptors in mammals: Notch1 to Notch4. The extracellular domain of the 4 receptors has 3 negative regulatory LIN repeats but varies in the number of EGF-like repeats. The cytoplasmic portion of the receptor contains a RAM domain, nuclear localization signal (NLS), ankyrin (ANK) repeats, and a PEST domain regulating protein stability. Notch1 and Notch 2 have TADs; however, they are absent in Notch3 and Notch4. (B) Cognate interaction between Notch receptors and Notch ligands (Jagged/Delta) triggers 2 consecutive proteolytic cleavages by the ADAM10 metalloprotease and the γ-secretase complex. This generates ICN, the signaling portion of the receptor, which enters the nucleus and displaces corepressors (SMRT and CtBP1) and recruits the coactivator MAML1 and the acetyltransferase p300. ICN activating complex is short-lived, and ICN gets phosphorylated on its PEST domain and subsequently ubiquitinated by Fbw7 and targeted for degradation by the proteasome. ADP, adenosine diphosphate; ATP, adenosine triphosphate.
Ligands for Notch receptors, which include Delta-like and Jagged (known as Delta and Serrate in invertebrates), are also transmembrane proteins with large EGF-like repeats. In mammals, there are 3 Delta-like ligands (Dll1, Dll3, and Dll4) and 2 Jagged ligands (Jag1 and Jag2). Because both receptor and ligand are membrane bound, signaling between cells is short range and requires cell-cell contact. When membrane-bound receptors interact with cognate ligands on an adjacent cell, 2 consecutive proteolytic cleavages of the Notch receptor are initiated. The first S2 cleavage by ADAM10 metalloprotease generates the substrate for the S3 cleavage by the γ-secretase complex, which is composed of Presenilin, APH1, PEN2, and Nicastrin (NCSTN).19 NCSTN serves as a substrate receptor for the γ-secretase complex by recognizing the amino terminus of Notch that is generated following S2 cleavage20 (Figure 1B).
These proteolytic cleavage steps generate ICN, which enters the nucleus to activate the transcription of target genes. Canonical Notch signaling requires the formation of a complex with a DNA-bound transcription factor from the CSL family (RBP-Jκ/CBF-1/KBF2 in mammals). RBP-Jκ binds DNA in a sequence-specific manner and acts as a repressor of transcription in the absence of Notch signaling. Corepressors in this complex include SMRT/NcoR and SHARP (or MINT) and more global repressors such as CtBP1 and SIRT1, which recruit histone deacetylases and the histone demethylase LSD1.21-24 Displacement of corepressors bound to RBP-Jκ by ICN allows the recruitment of coactivators such as mastermind and histone acetyltransferases such as p300 to create a short-lived transcriptional activation complex (Figure 1B). Recent genome-wide chromatin immunoprecipitation assays and sequencing by several groups, including our laboratory, have identified a large number of genes that can be regulated by Notch.25-27 Many of these target genes may be cell-type specific, but there are a few well-characterized transcriptional targets of Notch-RBP-Jκ, including the Hes (hairy enhancer of split) family of transcriptional repressors, the Notch-related ankyrin repeat protein (Nrarp), c-myc, and Deltex.28
Notch signaling is also regulated by ubiquitination. The E3 ligases Neuralized and Mindbomb are responsible for ubiquitination of Notch ligands, which is required for proper trafficking and presentation of active ligands on the cell membrane.29 Notch receptors are also ubiquitinated.30-33 Thus far, only one E3 ligase, Fbw7, an F-box protein that serves as a substrate-recognizing component of the Skp1/Cul1/F-box ubiquitination complex, has been shown to specifically target nuclear Notch for degradation by the proteasome.34 Most of the characterized Fbw7 substrates are potent proto-oncogenes, including c-Myc, cyclin E, and c-Jun.35-37 Mutations that abolish FBW7 function have been identified in primary human tumors and cancer cell lines of various origins, such as breast, ovary, and colon.38-40 Our laboratories identified a “degron” sequence on the –COOH-terminal end of the NOTCH1 PEST domain that is essential for FBW7 binding, ubiquitination, and ICN degradation.41,42 For substrate recognition by FBW7 to occur, the FBW7 degron sequence on the target protein must first be phosphorylated at a core threonine residue.43 CDK8 was shown to phosphorylate and trigger FBW7-dependent degradation of ICN.44 More recent work suggested that another kinase, ILK, could also “prime” NOTCH1 for FBW7-mediated degradation and that GSK3 kinases may also play a role in Notch stability.45,46
Notch signaling in hematopoietic stem and progenitor cells
Experimental evidence indicates that Notch is required for the embryonic development of HSCs,47,48 yet there are differing reports on the role of Notch in adult HSC maintenance and self-renewal (for extensive review, see Bigas et al49). Retroviral transduction of bone marrow progenitors with ICN1 or Hes1 increases the number of HSCs or HSC self-renewal.50-52 It was shown that in the adult bone marrow, there is surface expression of Notch1 and Notch2 in murine hematopoietic stem and multipotent progenitor cells and expression of Jag1 on bone marrow stroma.53 In a more a recent study, the same laboratory demonstrated that human CD34+ cord blood progenitors could be expanded ex vivo in the presence of Dll1 and that this treatment enhanced hematopoietic reconstitution following myeloablative irradiation.54 Additionally, other experimental evidence points toward a role for Notch in HSC, including the presence of Notch ligands on niche stromal cells and detectable expression of receptors as well as activation of Notch/RBP-Jκ transcriptional targets.55,56
Recent studies using mouse models suggest that gain of function of Notch signaling is important at least during the repopulation phase of hematopoietic cells. For example, work from the Rafii group suggested that Notch activation in HSCs by endothelium is important during recovery after myeloablative irradiation.57 Also supporting a specific function for Notch after myeloablation, it was demonstrated that Notch2 controls the tempo of short- and long-term repopulating cells.58 Retroviral transduction of bone marrow progenitors with dominant-negative RBP-Jκ reduced the long-term reconstitution ability of transplanted bone marrow, suggesting again that Notch is important in HSC function.56
In contrast to the consequences of activated Notch signaling on HSC function, loss of Notch signaling through conditional deletion of Notch1 or the ligand Jag1 did not appear to affect adult HSCs, although such findings may be accounted for by compensation by other Notch receptors or ligands.59,60 In contrast to previous experiments with dominant-negative MAML1 (dnMAML1) and dominant-negative RBP-Jκ,56 more recent experiments expressing dnMAML1 by retroviral transduction or from the ROSA26 locus, and most convincingly by deleting Rbp-jκ using Mx1-Cre, indicate that the canonical Notch pathway is not necessary for HSC self-renewal.61 Whether or not Notch is truly dispensable for the maintenance of adult HSCs under homeostatic conditions awaits confirmation using additional models of Notch loss of function.
Notch signaling in megakaryocyte and erythrocyte development
A function for Notch/Dll1 signaling in the megakaryocyte lineage has also been suggested. Initial experiments have shown that plating mouse LSK cells on OP9-Dll1 cells resulted in the appearance of mature megakaryocytes that was not seen in the parental OP9 cells without Notch ligands or in the presence of γ-secretase inhibitors.62 Furthermore, a similar result was obtained when ICN1 or ICN4 was expressed in vitro. In agreement with the later findings, the authors found that megakaryocyte-erythrocyte progenitors (MEPs) expressed higher levels of Notch4 than other Notch receptors. Transplantation of bone marrow transduced with an ICN4 expression vector resulted in an increase in MEPs and a slight increase in bone marrow megakaryocytes, whereas expression of dnMAML1 suppressed MEPs. Based on these findings, the authors suggested that Notch4 is a positive regulator of megakaryopoiesis. However, more recent experiments using human CD34+ progenitor differentiation assays have shown that Dll4 stimulation inhibited the generation of platelets and mature CD41a+CD42b+ megakaryocytes and that this could be rescued by pharmacologic inhibition of Notch signaling using γ-secretase inhibitors.63 Intriguingly, in this system, Notch signaling also resulted in enhanced erythroid cell production. These discrepancies may highlight differential requirement for Notch signaling in mouse and human megakaryopoiesis or implicate different roles of Notch signaling at different stages of megakaryocyte differentiation and maturation. Further studies are needed to precisely decipher the role of Notch signaling in megakaryopoiesis.
Until recently, the function of Notch signaling in erythropoiesis was poorly understood and the existing literature consisted primarily of in vitro differentiation assays showing conflicting results. For example, in murine erythroleukemia cells, Notch1 promotes erythroid differentiation by preventing apoptosis in committed progenitors.64 Knockdown of Notch1 resulted in cell death in both the presence and absence of the erythroid differentiation agent hexamethylene bisacetamide (HMBA). Another study using this cell line confirmed the requirement for Notch during chemically induced differentiation but suggested that Notch regulated cell-cycle proteins.65 In the multipotent FDCP-mix cell line, which can differentiate into myeloid and erythroid cells, expression of ICN1 under the control of tamoxifen worked cooperatively with erythropoietin to accelerate erythroid differentiation.66 In contrast, several other studies claimed that Notch signaling could inhibit erythroid differentiation. Two studies showed that overexpression of ICN1 in the K562 cell line inhibits erythroid differentiation through different mechanisms.67,68 Another study using murine erythroleukemia cell line F5-5 showed that ICN1 overexpression inhibits erythroid differentiation by sustaining GATA2 expression.69 The discrepancies among these studies could be explained by the various chemical treatments used to induce erythroid differentiation, the nature of the cell lines used, or the supraphysiological effect of viral-infection–induced ICN1 over expression.
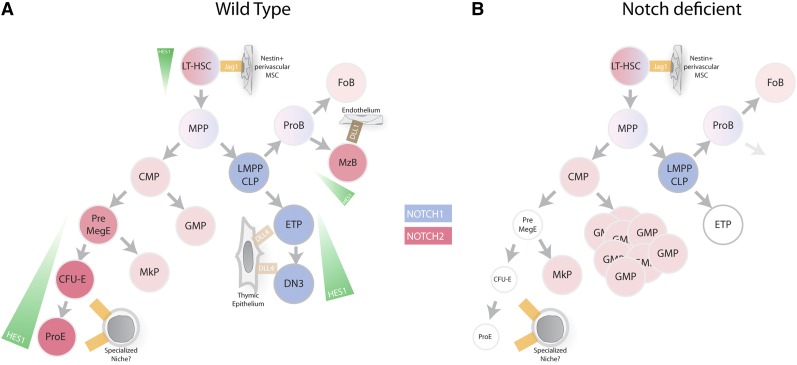
In a recent study, using novel lineage-tracing mouse models for each Notch receptor as well as a reporter strain for Notch activity where green fluorescent protein is knocked in the endogenous Hes1 locus, we have identified that Notch signaling is important for erythroid differentiation. Indeed, we were able to show that early bipotent pre-MEPs as well as downstream erythroid progenitors express high level of Notch2 and show activation of the Notch pathway. Using in vivo gain- and loss-of-function models of Notch2 signaling, we showed that even though it is dispensable at steady state, Notch signaling promotes commitment to the erythroid fate. Our experiments also revealed that Notch signaling is required for recovery following stress-induced ablation of erythroid progenitors, suggesting key roles for Notch signaling in hematopoiesis under conditions of stress70 (Figure 2A).
Figure 2.
Notch signaling in hematopoiesis. (A) Overview of Notch signaling in hematopoietic cells. Blue gradient represents level of Notch1 expression, red gradient represents level of Notch2 expression, and green gradient represents level of Notch activity as determined using Hes1GFP/WT mice. (B) Notch-deficient hematopoiesis. Genetic inhibition of Notch signaling in HSCs leads to accumulation of GMP, reduction of erythrocyte progenitors, and inhibition of marginal zone B-cell differentiation and T-cell differentiation. CFU-E, colony forming unit-erythrocyte; CLP, common lymphoid progenitors; CMP, common myeloid progenitors; DN3, double-negative 3; ETP, early T-cell progenitors; FoB, follicular B cells; GMP, granulocyte/monocyte progenitors; LMPP, lymphoid-primed multipotent progenitors; LT-HSC, long-term hematopoietic stem cells; MegE, megakaryocyte/erythrocyte progenitors; MkP, megakaryocyte progenitors; MPP, multipotent progenitor; MzB, marginal zone B cell; ProB, pro-B cell; ProE, proerythrocyte.
Physiological and oncogenic roles for Notch signaling in the lymphoid compartment
Whereas the role of Notch in HSC requires further experimentation, the importance of Notch signaling in lymphocyte development and oncogenic transformation has been thoroughly investigated. It has been well established that Notch1 is essential to direct T-cell lineage commitment at the expense of B-cell development71-73 (Figure 2A). Conditional deletion of Notch1 or Rbp-jκ results in blocked T-cell development and the appearance of ectopic B cells in the thymus.60,71 The oncogenic potential of Notch1 in T-cell leukemia has been demonstrated both experimentally and clinically. In 1991, Ellisen and colleagues identified a t(7;9)(q34;q34.4) translocation in T-cell acute lymphoblastic leukemia (T-ALL) patients that resulted in fusion of the 3′ region of NOTCH1 into the TCRβ locus and consequent overexpression of an active form of Notch1.74 This translocation is rare, found in <1% of T-ALL cases. However, 13 years later, Weng, Ferrando, and colleagues identified activating NOTCH1 mutations in about 56% of T-ALL cases examined, introducing NOTCH1 mutations as the main oncogenic lesion in T-ALL.75 Two major “hot spots” of mutations were characterized: mutations in the HD domain that induce ligand-independent activation, and mutations in the PEST carboxy-terminal domain that increase stability of ICN1.41 Additionally, inactivating mutations were identified in FBW7 and associated with sustained Notch signaling.41,42,76,77 Moreover, human T-cell leukemia can be modeled in mice by overexpression of intracellular Notch1 or Notch3 in hematopoietic progenitors.78-80 Notably, animal models have suggested that NOTCH1 mutations (HD or PEST) are insufficient to induce disease in their own right, even when they are overexpressed, but can collaborate with other oncogenic lesions such as KrasG12D to accelerate T-ALL, suggesting ICN1 dosage is a crucial determinant of Notch oncogenicity.81 We recently generated knockin mice carrying the human NOTCH1 mutant alleles, and our studies also indicated that none of these mutations are sufficient to induce disease (I.A., unpublished observations).
Two recent studies have identified activating NOTCH1 mutations in chronic lymphocytic leukemia (CLL), the most frequent adult leukemia.82,83 Both groups identified similar NOTCH1-activating mutations predicted to impair FBW7-induced Notch1 degradation. It is striking to note that the majority of patients harbor the same frameshift mutation (P2515Rfs*4), in contrast to the more heterogeneous PEST domain mutations that affect T-ALL patients. The reasons for this particular hot spot remain unclear and might involve some highly conserved mechanism. Although the overall frequency of NOTCH1 mutations was not dramatic (from 8.3% to 12.2%), they were primarily found in patients with the more clinically aggressive nonmutated IGV(H) subtype (20.4%) or in Richter syndrome (31.0%) and in chemorefractory CLL (20.8%). These results suggest that although NOTCH1 mutations are not uniformly involved in the molecular pathogenesis of CLL, when present they are associated with poor prognosis and appear to define a distinct high-risk clinical subtype for therapeutic intervention.
Another well-documented role for Notch signaling in the lymphocytic compartment is its role in marginal zone B-cell differentiation in the spleen (Figure 2A). Several studies using conditional deletion of Notch2 and Rbp-Jκ showed lack of marginal zone B cells in the absence of Notch signaling.84,85 Two recent studies identified NOTCH2 mutations in splenic marginal zone lymphoma patients using whole-genome sequencing.86,87 The frequency of mutations ranged from 21% to 25% of cases, making NOTCH2 the most mutated gene in this disease. Most of identified mutations were frameshift or nonsense mutations affecting the PEST domain, suggesting that these were gain-of-function mutations through excision of the Notch2 degron essential for its degradation. However, functional characterization of these mutants needs to be performed.
In contrast to these oncogenic roles in the lymphoid compartment, a study by Zweidler-McKay and colleagues showed that activation of Notch signaling using overexpression of activated forms of the 4 Notch receptors as well as their downstream target, HES1, could induce growth arrest and apoptosis of B-cell acute lymphoblastic leukemia cell lines.88
Physiological and tumor-suppressor role for Notch signaling in the granulocyte/monocyte compartment
The function of Notch signaling in granulocyte/monocyte development is not as well defined as the role of Notch1 in T-cell differentiation or Notch2 in marginal zone B-cell differentiation, and different studies show conflicting results. In vitro and in vivo evidence suggests that Notch suppresses myeloid differentiation.51,52,89-91 Whereas a few studies found that Notch1 activation resulted in accelerated granulocytic differentiation,92,93 their results were challenged by other studies showing that Notch1 activation can inhibit granulocytic differentiation in the same cell line.94,95 However, no obvious effects on myeloid commitment were reported after conditional deletion of Rbp-jκ in mice,71,84 whereas repression of MEP fate and skewing toward common myeloid progenitors and GMPs was observed in a murine dnMamL1-inducible knockin.62
Our laboratory has recently found that conditional Notch loss of function through the deletion of Nicastrin (Ncstn), an essential component of the γ-secretase complex, or compound deletion of Notch1 and Notch2 results in a myeloproliferative syndrome with common features of the human disease chronic myelomonocytic leukemia (CMML).96 CMML is classified as a myelodysplastic/myeloproliferative overlap syndrome, characterized by monocytosis, myeloproliferation, and a high rate of progression to acute myeloid leukemia (AML). This phenotype was characterized by increased monocytes and granulocytes circulating in the blood and invading peripheral tissues and signs of extramedullary hematopoiesis in the spleen and liver. Analysis of myeloerythroid progenitor compartment revealed a skewing toward the GMPs and a reduction of MEPs (Figure 2B). Whole-transcriptome analysis revealed that Notch signaling inhibited a monocytic/granulocytic differentiation program in an early multipotent progenitor. This was at least partially mediated by direct repression of the Pu.1 and Cebpα promoters by Hes1, because Hes1 was able to bind their promoter and enforced expression of Hes1 in hematopoietic stem and progenitor cells pushed differentiation of these cells toward an erythrocytic/megakaryocytic fate at the expense of a granulocytic/monocytic fate. Direct repression of Cebpα by Hes1 was further confirmed by a recent study from Bhandoola’s group.97 Additionally, enforced activation of Notch signaling using a conditional knockin of ICN1 was able to revert Ncstn deletion effects and inhibited commitment of myeloerythroid progenitors to the GMP fate. Finally, sequencing of Notch pathway genes revealed that around 12% of CMML patients harbored inactivating mutations in NCSTN, mastermind-like 1 (MAML1), APH1A, and NOTCH2. These mutations were unique to CMML and were not found in other myeloproliferative disorders such as polycythemia vera and myelofibrosis. These studies suggested that Notch signaling acts to prevent uncontrolled proliferation and transformation of myeloid cells during hematopoietic development.
In agreement with our findings in Ncstn-deficient mice, loss of Notch signaling through conditional deletion of FX results in progenitor cell–autonomous myeloproliferation.90 The mouse protein FX (the homolog of human guanosine diphosphate l-fucose synthase) is required for de novo synthesis of guanosine diphosphate fucose. As previously mentioned, receptor fucosylation is necessary for ligand binding. FX-null animals are deficient in Notch signaling, but this can be rescued through dietary supplementation with fucose. Furthermore, the conditional deletion of the protein O-fucosyltransferase 1 (Pofut1) in hematopoietic cells using Mx1-Cre also resulted in a myeloid hyperplasia.98 As in the FX−/− model, Pofut1 deficiency displayed a Notch loss-of-function phenotype, because Notch signaling requires O-fucosylation of EGF repeats on ligands and receptors. Interestingly, disruption of Notch signaling by conditional deletion of Mindbomb-1 (Mib1) also resulted in a myeloproliferative syndrome.99 Transplantation of wild-type donor cells into lethally irradiated Mib1-deficient recipients also resulted in a myeloproliferative phenotype. Another recent study using conditional deletion of the metalloprotease Adam10, driven by Mx1-cre, led to a similar myeloproliferative disease. Using reciprocal bone marrow transplantation of Adam10-deficient hematopoietic cells in wild-type recipient mice, or wild-type hematopoietic cells in Adam10-deficient recipients, showed that both situations induced a myeloproliferative disorder. This suggests that Notch signaling is important in the hematopoietic compartment to control myeloid progenitor commitment and the effect may be in part mediated through stromal niche cells in a non–cell-autonomous manner.100
Whereas all these studies clearly showed that Notch signaling is a negative regulator of myeloid progenitor commitment and is involved as a tumor suppressor in myeloproliferative disorder, very few studies implicated Notch signaling in AML. Cytogenetic and molecular studies have shown that AML is a heterogeneous disease in which a variety of cytogenetic and molecular alterations have biological and clinical relevance. These include chromosomal abnormalities, which lead to generation of leukemogenic fusion oncoproteins, including MLL fusions, which are associated with adverse outcome. In addition, somatic mutations in CEBPA, FLT3, IDH1/2, NRAS, KIT, TET2, ASXL1, and other genes have been shown to contribute to leukemogenesis and improve AML risk classification.101 A study by Ferrando’s group identified 1 Notch activating mutation in a M0 AML but failed to identify recurrent activating mutations in an extended cohort of patients.102 Another study found consistent downregulation of NOTCH1 expression in 54 AML patients and 6 AML cell lines compared with normal HSCs, which correlated with downregulation of PU.1, potentially explaining the block in myeloid differentiation.103 Several studies by Nara’s group showed that Notch signaling could affect growth and differentiation of some AML cell lines and primary patient samples.104,105
Two different studies published this year by our group and Zweidler-McKay’s group addressed the role of Notch signaling in AML.106,107 Both studies used whole-genome expression data from AML patients to show that the Notch signaling pathway is silenced, as revealed by absence of expression of known Notch target genes, despite high expression levels of NOTCH2 receptors on their cell surface. We then used a mouse model of AML induced by the MLL-AF9 oncofusion protein in combination with an inducible knockin of either ICN1 or ICN2 to demonstrate that activation of Notch signaling can induce apoptosis of AML blasts and regression of disease in vivo. Interestingly, we observed significant reduction of leukemic-initiating cells (LIC) in this model upon Notch activation, and secondary transplantation of remaining LIC showed highly reduced potential to transplant the disease. This suggested that activation of Notch signaling is able to target AML LICs. Whole-transcriptome expression analysis of freshly isolated LIC coupled with in vitro methylcellulose-based differentiation assays showed that Notch activation induced terminal differentiation of leukemic blasts to macrophages and dendritic cells. Both studies also identified the antiapoptotic gene BCL2 as a downregulated gene upon Notch activation. Zweidler-McKay’s group went further, showing that HES1 was mediating part of these effects, and they suggested that it could directly inhibit BCL2 expression.
One of the most striking observations of these studies is that whereas Notch pathway is inactive, AML blasts express high levels of NOTCH2. This led to the exciting hypothesis that Notch signaling could be reactivated in these cells using Notch receptor agonists. Indeed, both studies showed that Notch reactivation was possible in AML cell lines and primary patient cells using recombinant ligands. Activation of Notch using these extracellular stimuli recapitulated previously observed phenotypes with differentiation and apoptosis of the cells. Therefore, highly specific Notch2-activating antibodies or agonists might represent a viable therapeutic strategy for AML patient treatment. Such therapies should not carry a potential risk of induction of T-ALL, because NOTCH2 is not expressed on T cells.
Mechanisms of Notch silencing in AML
The oncogenic and tumor-suppressor functions of the Notch pathway are not only highly cell context dependent but may also be influenced by the genetic landscape within a particular cell type. This is perhaps best exemplified by SCC of the lung: loss-of-function mutations of NOTCH-1 and/or NOTCH-2 have been identified in 12.5% of SCC cases, whereas others have reported Notch pathway activation in the same disease, either through activating mutations of NOTCH-1 (10% of cases) or loss of expression of the negative regulator NUMB (30% of cases).108
Whether the Notch pathway has an oncogenic or tumor-suppressor role in AML is also likely to be dependent on the presence of other genetic lesions. There are, for instance, subgroups of AML patients where activation of the Notch pathway may be important. Notch signaling is integrally involved in the rare subset of patients with acute megakaryocytic leukemia initiated by the OTT-MAL fusion transcript, whereby RBP-Jκ is directly activated by OTT-MAL independently of Notch.109 Also described are 3 AML patients with activating NOTCH-1 mutations clustering together by gene set enrichment analysis in a cohort of 285 AML patients,110 although it is possible these patients would meet European Group of Immunological Markers for Leukemias criteria for mixed-lineage T-ALL/AML or be better classified as early T-cell progenitor T-ALL.111,112 There is also indirect genetic evidence that Notch pathway activation may play an oncogenic role in certain AML subtypes. Tribbles homolog 2 (TRIB2), a direct Notch target, enhances proliferation of myeloid progenitors, and Trib2 transgenic mice develop AML through degradation of Cebpα, a transcription factor that is essential for myeloid differentiation.113 Additionally, transduction of bone marrow cells with AML1-ETO, PML-RARA, or PLZF-RARA fusion constructs has been shown to increase expression of the Notch ligand JAGGED-1, a gene that is also upregulated in some patients with AML.114,115 This is relevant, given recent findings showing upregulation of Notch target genes in both Ctsg-PML-RARA mice and samples from patients with acute promyelocytic leukemia (APML).116 Some APML tumors also displayed sensitivity to Notch pathway inhibitors in vitro, and genetic inhibition of Notch signaling with dnMAML1 reduced APML growth in vivo.
However, these findings are in contrast to non-APML AML, where expression of dnMAML1 in vivo increases leukemic cell proliferation and Notch pathway activation induces apoptosis.107 Furthermore, in the majority of primary AML cases, the expression of Notch target genes is extremely low,106,107,117 which is somewhat surprising given robust expression of many of the Notch ligands and receptors. Interestingly, the Notch coactivator MAML1 is located on chromosome 5q35, in a region commonly retained in 5q MDS but frequently deleted in 5q AML. Out of 5 patients with 5q- AML, where the deletion did not involve MAML1, 3 harbored point mutations of this gene, suggesting inhibition of Notch signaling is involved in disease progression (Figure 3).
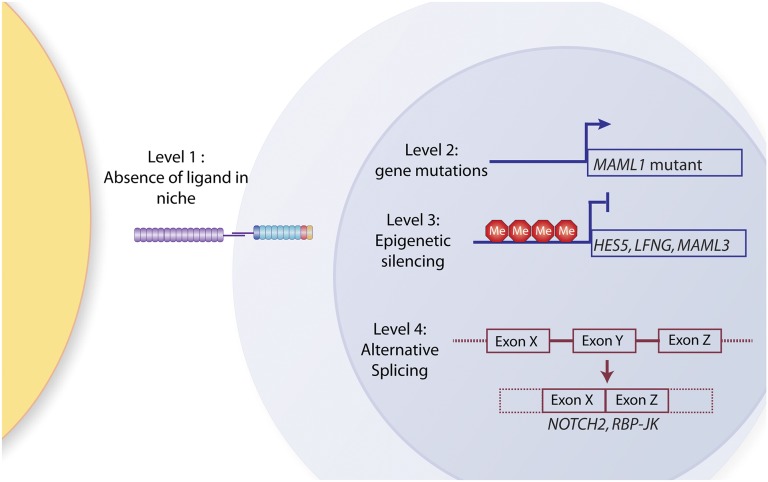
Figure 3.
Mechanisms of Notch signaling silencing in AML. So far, 4 potential levels of silencing of Notch signaling have been identified: (1) AML-initiating cells may reside in niches not expressing Notch ligands; (2) Notch pathway genes might be mutated (so far, only MAML1 mutations have been identified); (3) Notch pathway genes might be epigenetically silenced (for example, HES5, LFNG, and MAML1 have been shown to be hypermethylated in IDH mutant patients; and (4) alternative splicing of Notch pathway genes might lead to expression of inactive isoforms (to date, mis- or alternative-splicing of NOTCH2 and RBP-Jκ have been identified in a large fraction of AML patients).
However, a recent comprehensive genomic analysis of a large AML cohort did not identify loss-of-function mutations in members of the Notch pathway (Cancer Genome Atlas Research Network118), suggesting mechanisms of Notch pathway silencing in AML are distinct from CMML. One possibility is that aberrant methylation patterns in AML disrupt normal Notch signaling events. In support of this, conditional knockin mice with the R132H IDH1 mutation develop splenomegaly and increased numbers of early hematopoietic progenitors and have a DNA methylation pattern similar to those observed in human IDH1- or IDH2-mutant AML, which includes hypermethylation of Notch pathway genes such as LFNG, MAML3, and HES5119 (Figure 3).
Although ablation of Tet2 or Ncstn in mice leads to a slow-onset CMML-like disease,96,120-123 compound Tet2−/− Ncstn−/− knockout mice develop a highly penetrant and aggressive transplantable AML with rapid onset.106 Thus, the Notch pathway plays a clear and distinct tumor-suppressor role in Tet2-mutant AML. Whether expression of mutant IDH1 on a Ncstn-null background cooperates to induce overt AML in mice will be interesting considering mutant IDH results in the production of 2-hydroxyglutarate, which in turn inhibits TET2 function.124-126
One further explanation for the lack of activation of Notch target genes in AML is the presence of alternative splicing of components of the Notch pathway. Alternative splicing involves the differential inclusion or exclusion of exonic and intronic sequences from pre–messenger RNA (mRNA) into the mature mRNA transcript127 and is particularly prevalent in AML.128 Genome-wide analysis using exon arrays identified frequent alternative splicing of NOTCH2 in AML.129 Whereas expression of NOTCH2 splice variants is correlated with downregulation of Notch target gene expression, their functional consequences require further investigation. Analyses of RBP-Jκ mRNA transcripts in AML have revealed the presence of an alternatively spliced transcript lacking a portion of the β-trefoil domain that constitutes the predominant RBP-Jκ isoform in the majority of AML patients (M.R.M and A.T.L., manuscript submitted July 2012). This RBP-Jκ variant is unable to form a complex with NOTCH proteins, cannot bind canonical RBP-Jκ transcriptional sequences, and cannot rescue the distinct vessel phenotype induced by depletion of endogenous csl in a zebrafish model, indicating that the latter is a loss-of-function variant and supporting a tumor-suppressor function for the Notch pathway in the majority of AML patients (Figure 3).
In conclusion, similarly as in other tissues, Notch acts as an important cell-fate switch in the hematopoietic system. This binary switch is now well characterized at the level of T cell over B cell, marginal zone B cell over follicular B cell, and erythroid cell over myeloid cell choices. At each of these branch points, Notch signaling is capable of playing either an oncogenic or tumor-suppressor role in the downstream populations if its role is to respectively induce or repress these fates. Additionally, aberrant Notch signaling could also sustain lineage-specific oncogenes or tumor suppressors whose expression would normally be turned off during normal differentiation. Therefore, depending on the context, drugs that inhibit or activate Notch signaling could be particularly relevant. The clinical use of γ-secretase inhibitors is already underway for T-ALL, and the discovery of a tumor-suppressor role for Notch in AML could lead to the development of a novel agonist specifically targeting the Notch2 receptor that might represent a promising strategy for this disease. Furthermore, elucidation of precise downstream mechanisms of Notch-induced AML suppression could open broader therapeutic possibilities.
Acknowledgments
The authors apologize to those whose work was not cited due to space limitations.
I.A. is supported by the National Institutes of Health, Research Project Grant Program of the National Cancer Institute (RO1CA173636, RO1CA133379, RO1CA105129, and RO1CA149655), the Leukemia & Lymphoma Society (Translational Research Program grants), The V Foundation for Cancer Research, the William Lawrence and Blanche Hughes Foundation, and the St. Baldrick’s Foundation for Cancer Research. P.O. was supported by the New York University Medical Scientist Training Program. C.L. was supported by the Helen and Martin Kimmel Center for Stem Cell Research and is currently a Leukemia and Lymphoma Society Fellow. M.R.M. is supported by the Kay Kendall Leukaemia Fund of the United Kingdom, a Team Path To the Cure grant, and the Claudia Adams Barr foundation of the Dana Farber Cancer Institute. A.T.L. is supported by grants from the National Cancer Institute (5P01CA109901, 1R01CA176746, and P30CA06516) and by grants from the William Lawrence and Blanche Hughes Foundation and Alex’s Lemonade Stand. I.A. is a Howard Hughes Medical Institute Early Career Scientist.
Authorship
Contribution: C.L., P.O., I.A., M.R.M., and A.T.L. wrote the review, and C.L. and P.O. designed the figures.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Iannis Aifantis, NYU School of Medicine and Pathology Department, 550 1st Ave, New York, NY 10016; e-mail: iannis.aifantis@nyumc.org; and Camille Lobry, NYU School of Medicine and Pathology Department, 550 1st Ave, New York, NY 10016; e-mail: camille.lobry@nyumc.org.
References
- 1.Nicolas M, Wolfer A, Raj K, et al. Notch1 functions as a tumor suppressor in mouse skin. Nat Genet. 2003;33(3):416–421. doi: 10.1038/ng1099. [DOI] [PubMed] [Google Scholar]
- 2.Agrawal N, Frederick MJ, Pickering CR, et al. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science. 2011;333(6046):1154–1157. doi: 10.1126/science.1206923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stransky N, Egloff AM, Tward AD, et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333(6046):1157–1160. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morgan TH. The theory of the gene. Am Nat. 1917;51:513–514. [Google Scholar]
- 5.Wharton KA, Johansen KM, Xu T, Artavanis-Tsakonas S. Nucleotide sequence from the neurogenic locus notch implies a gene product that shares homology with proteins containing EGF-like repeats. Cell. 1985;43(3 Pt 2):567–581. doi: 10.1016/0092-8674(85)90229-6. [DOI] [PubMed] [Google Scholar]
- 6.Kidd S, Kelley MR, Young MW. Sequence of the notch locus of Drosophila melanogaster: relationship of the encoded protein to mammalian clotting and growth factors. Mol Cell Biol. 1986;6(9):3094–3108. doi: 10.1128/mcb.6.9.3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blaumueller CM, Qi H, Zagouras P, Artavanis-Tsakonas S. Intracellular cleavage of Notch leads to a heterodimeric receptor on the plasma membrane. Cell. 1997;90(2):281–291. doi: 10.1016/s0092-8674(00)80336-0. [DOI] [PubMed] [Google Scholar]
- 8.Radtke F, Raj K. The role of Notch in tumorigenesis: oncogene or tumour suppressor? Nat Rev Cancer. 2003;3(10):756–767. doi: 10.1038/nrc1186. [DOI] [PubMed] [Google Scholar]
- 9.Okajima T, Xu A, Irvine KD. Modulation of notch-ligand binding by protein O-fucosyltransferase 1 and fringe. J Biol Chem. 2003;278(43):42340–42345. doi: 10.1074/jbc.M308687200. [DOI] [PubMed] [Google Scholar]
- 10.Okajima T, Irvine KD. Regulation of notch signaling by o-linked fucose. Cell. 2002;111(6):893–904. doi: 10.1016/s0092-8674(02)01114-5. [DOI] [PubMed] [Google Scholar]
- 11.Rebay I, Fleming RJ, Fehon RG, Cherbas L, Cherbas P, Artavanis-Tsakonas S. Specific EGF repeats of Notch mediate interactions with Delta and Serrate: implications for Notch as a multifunctional receptor. Cell. 1991;67(4):687–699. doi: 10.1016/0092-8674(91)90064-6. [DOI] [PubMed] [Google Scholar]
- 12.Moloney DJ, Panin VM, Johnston SH, et al. Fringe is a glycosyltransferase that modifies Notch. Nature. 2000;406(6794):369–375. doi: 10.1038/35019000. [DOI] [PubMed] [Google Scholar]
- 13.Haines N, Irvine KD. Glycosylation regulates Notch signalling. Nat Rev Mol Cell Biol. 2003;4(10):786–797. doi: 10.1038/nrm1228. [DOI] [PubMed] [Google Scholar]
- 14.Brückner K, Perez L, Clausen H, Cohen S. Glycosyltransferase activity of Fringe modulates Notch-Delta interactions. Nature. 2000;406(6794):411–415. doi: 10.1038/35019075. [DOI] [PubMed] [Google Scholar]
- 15.Radtke F, Wilson A, Mancini SJC, MacDonald HR. Notch regulation of lymphocyte development and function. Nat Immunol. 2004;5(3):247–253. doi: 10.1038/ni1045. [DOI] [PubMed] [Google Scholar]
- 16.Wilson JJ, Kovall RA. Crystal structure of the CSL-Notch-Mastermind ternary complex bound to DNA. Cell. 2006;124(5):985–996. doi: 10.1016/j.cell.2006.01.035. [DOI] [PubMed] [Google Scholar]
- 17.Nam Y, Sliz P, Song L, Aster JC, Blacklow SC. Structural basis for cooperativity in recruitment of MAML coactivators to Notch transcription complexes. Cell. 2006;124(5):973–983. doi: 10.1016/j.cell.2005.12.037. [DOI] [PubMed] [Google Scholar]
- 18.Ong C-T, Cheng H-T, Chang L-W, et al. Target selectivity of vertebrate notch proteins. Collaboration between discrete domains and CSL-binding site architecture determines activation probability. J Biol Chem. 2006;281(8):5106–5119. doi: 10.1074/jbc.M506108200. [DOI] [PubMed] [Google Scholar]
- 19.Bray SJ. Notch signalling: a simple pathway becomes complex. Nat Rev Mol Cell Biol. 2006;7(9):678–689. doi: 10.1038/nrm2009. [DOI] [PubMed] [Google Scholar]
- 20.Shah S, Lee SF, Tabuchi K, et al. Nicastrin functions as a gamma-secretase-substrate receptor. Cell. 2005;122(3):435–447. doi: 10.1016/j.cell.2005.05.022. [DOI] [PubMed] [Google Scholar]
- 21.Mulligan P, Yang F, Di Stefano L, et al. A SIRT1-LSD1 corepressor complex regulates Notch target gene expression and development. Mol Cell. 2011;42(5):689–699. doi: 10.1016/j.molcel.2011.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yatim A, Benne C, Sobhian B, et al. NOTCH1 nuclear interactome reveals key regulators of its transcriptional activity and oncogenic function. Mol Cell. 2012;48(3):445–458. doi: 10.1016/j.molcel.2012.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nagel AC, Krejci A, Tenin G, et al. Hairless-mediated repression of notch target genes requires the combined activity of Groucho and CtBP corepressors. Mol Cell Biol. 2005;25(23):10433–10441. doi: 10.1128/MCB.25.23.10433-10441.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kao HY, Ordentlich P, Koyano-Nakagawa N, et al. A histone deacetylase corepressor complex regulates the Notch signal transduction pathway. Genes Dev. 1998;12(15):2269–2277. doi: 10.1101/gad.12.15.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ntziachristos P, Tsirigos A, Van Vlierberghe P, et al. Genetic inactivation of the polycomb repressive complex 2 in T cell acute lymphoblastic leukemia. Nat Med. 2012;18(2):298–301. doi: 10.1038/nm.2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Palomero T, Lim WK, Odom DT, et al. NOTCH1 directly regulates c-MYC and activates a feed-forward-loop transcriptional network promoting leukemic cell growth. Proc Natl Acad Sci USA. 2006;103(48):18261–18266. doi: 10.1073/pnas.0606108103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hamidi H, Gustafason D, Pellegrini M, Gasson J. Identification of novel targets of CSL-dependent Notch signaling in hematopoiesis. PLoS ONE. 2011;6(5):e20022. doi: 10.1371/journal.pone.0020022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weng AP, Millholland JM, Yashiro-Ohtani Y, et al. c-Myc is an important direct target of Notch1 in T-cell acute lymphoblastic leukemia/lymphoma. Genes Dev. 2006;20(15):2096–2109. doi: 10.1101/gad.1450406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pitsouli C, Delidakis C. The interplay between DSL proteins and ubiquitin ligases in Notch signaling. Development. 2005;132(18):4041–4050. doi: 10.1242/dev.01979. [DOI] [PubMed] [Google Scholar]
- 30.Gupta-Rossi N, Six E, LeBail O, et al. Monoubiquitination and endocytosis direct gamma-secretase cleavage of activated Notch receptor. J Cell Biol. 2004;166(1):73–83. doi: 10.1083/jcb.200310098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gupta-Rossi N, Le Bail O, Gonen H, et al. Functional interaction between SEL-10, an F-box protein, and the nuclear form of activated Notch1 receptor. J Biol Chem. 2001;276(37):34371–34378. doi: 10.1074/jbc.M101343200. [DOI] [PubMed] [Google Scholar]
- 32.Oberg C, Li J, Pauley A, Wolf E, Gurney M, Lendahl U. The Notch intracellular domain is ubiquitinated and negatively regulated by the mammalian Sel-10 homolog. J Biol Chem. 2001;276(38):35847–35853. doi: 10.1074/jbc.M103992200. [DOI] [PubMed] [Google Scholar]
- 33.Wu G, Lyapina S, Das I, et al. SEL-10 is an inhibitor of notch signaling that targets notch for ubiquitin-mediated protein degradation. Mol Cell Biol. 2001;21(21):7403–7415. doi: 10.1128/MCB.21.21.7403-7415.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakayama KI, Nakayama K. Ubiquitin ligases: cell-cycle control and cancer. Nat Rev Cancer. 2006;6(5):369–381. doi: 10.1038/nrc1881. [DOI] [PubMed] [Google Scholar]
- 35.Welcker M, Orian A, Jin J, et al. The Fbw7 tumor suppressor regulates glycogen synthase kinase 3 phosphorylation-dependent c-Myc protein degradation. Proc Natl Acad Sci USA. 2004;101(24):9085–9090. doi: 10.1073/pnas.0402770101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Welcker M, Singer J, Loeb KR, et al. Multisite phosphorylation by Cdk2 and GSK3 controls cyclin E degradation. Mol Cell. 2003;12(2):381–392. doi: 10.1016/s1097-2765(03)00287-9. [DOI] [PubMed] [Google Scholar]
- 37.Wei W, Jin J, Schlisio S, Harper JW, Kaelin WG., Jr The v-Jun point mutation allows c-Jun to escape GSK3-dependent recognition and destruction by the Fbw7 ubiquitin ligase. Cancer Cell. 2005;8(1):25–33. doi: 10.1016/j.ccr.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 38.Kemp Z, Rowan A, Chambers W, et al. CDC4 mutations occur in a subset of colorectal cancers but are not predicted to cause loss of function and are not associated with chromosomal instability. Cancer Res. 2005;65(24):11361–11366. doi: 10.1158/0008-5472.CAN-05-2565. [DOI] [PubMed] [Google Scholar]
- 39.Kwak EL, Moberg KH, Wahrer DC, et al. Infrequent mutations of Archipelago (hAGO, hCDC4, Fbw7) in primary ovarian cancer. Gynecol Oncol. 2005;98(1):124–128. doi: 10.1016/j.ygyno.2005.04.007. [DOI] [PubMed] [Google Scholar]
- 40.Strohmaier H, Spruck CH, Kaiser P, Won KA, Sangfelt O, Reed SI. Human F-box protein hCdc4 targets cyclin E for proteolysis and is mutated in a breast cancer cell line. Nature. 2001;413(6853):316–322. doi: 10.1038/35095076. [DOI] [PubMed] [Google Scholar]
- 41.Thompson BJ, Buonamici S, Sulis ML, et al. The SCFFBW7 ubiquitin ligase complex as a tumor suppressor in T cell leukemia. J Exp Med. 2007;204(8):1825–1835. doi: 10.1084/jem.20070872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.O’Neil J, Grim J, Strack P, et al. FBW7 mutations in leukemic cells mediate NOTCH pathway activation and resistance to gamma-secretase inhibitors. J Exp Med. 2007;204(8):1813–1824. doi: 10.1084/jem.20070876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ye X, Nalepa G, Welcker M, et al. Recognition of phosphodegron motifs in human cyclin E by the SCF(Fbw7) ubiquitin ligase. J Biol Chem. 2004;279(48):50110–50119. doi: 10.1074/jbc.M409226200. [DOI] [PubMed] [Google Scholar]
- 44.Fryer CJ, White JB, Jones KA. Mastermind recruits CycC:CDK8 to phosphorylate the Notch ICD and coordinate activation with turnover. Mol Cell. 2004;16(4):509–520. doi: 10.1016/j.molcel.2004.10.014. [DOI] [PubMed] [Google Scholar]
- 45.Saint Just Ribeiro M, Hansson ML, Lindberg MJ, Popko-Scibor AE, Wallberg AE. GSK3beta is a negative regulator of the transcriptional coactivator MAML1. Nucleic Acids Res. 2009;37(20):6691–6700. doi: 10.1093/nar/gkp724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mo JS, Kim MY, Han SO, et al. Integrin-linked kinase controls Notch1 signaling by down-regulation of protein stability through Fbw7 ubiquitin ligase. Mol Cell Biol. 2007;27(15):5565-74. PMCID: 1952089. [DOI] [PMC free article] [PubMed]
- 47.Kumano K, Chiba S, Kunisato A, et al. Notch1 but not Notch2 is essential for generating hematopoietic stem cells from endothelial cells. Immunity. 2003;18(5):699–711. doi: 10.1016/s1074-7613(03)00117-1. [DOI] [PubMed] [Google Scholar]
- 48.Dzierzak E, Speck NA. Of lineage and legacy: the development of mammalian hematopoietic stem cells. Nat Immunol. 2008;9(2):129–136. doi: 10.1038/ni1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bigas A, Espinosa L. Hematopoietic stem cells: to be or Notch to be. Blood. 2012;119(14):3226–3235. doi: 10.1182/blood-2011-10-355826. [DOI] [PubMed] [Google Scholar]
- 50.Kunisato A, Chiba S, Nakagami-Yamaguchi E, et al. HES-1 preserves purified hematopoietic stem cells ex vivo and accumulates side population cells in vivo. Blood. 2003;101(5):1777–1783. doi: 10.1182/blood-2002-07-2051. [DOI] [PubMed] [Google Scholar]
- 51.Stier S, Cheng T, Dombkowski D, Carlesso N, Scadden DT. Notch1 activation increases hematopoietic stem cell self-renewal in vivo and favors lymphoid over myeloid lineage outcome. Blood. 2002;99(7):2369–2378. doi: 10.1182/blood.v99.7.2369. [DOI] [PubMed] [Google Scholar]
- 52.Carlesso N, Aster JC, Sklar J, Scadden DT. Notch1-induced delay of human hematopoietic progenitor cell differentiation is associated with altered cell cycle kinetics. Blood. 1999;93(3):838–848. [PubMed] [Google Scholar]
- 53.Varnum-Finney B, Purton LE, Yu M, et al. The Notch ligand, Jagged-1, influences the development of primitive hematopoietic precursor cells. Blood. 1998;91(11):4084–4091. [PubMed] [Google Scholar]
- 54.Delaney C, Heimfeld S, Brashem-Stein C, Voorhies H, Manger RL, Bernstein ID. Notch-mediated expansion of human cord blood progenitor cells capable of rapid myeloid reconstitution. Nat Med. 2010;16(2):232–236. doi: 10.1038/nm.2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Calvi LM, Adams GB, Weibrecht KW, et al. Osteoblastic cells regulate the haematopoietic stem cell niche. Nature. 2003;425(6960):841–846. doi: 10.1038/nature02040. [DOI] [PubMed] [Google Scholar]
- 56.Duncan AW, Rattis FM, DiMascio LN, et al. Integration of Notch and Wnt signaling in hematopoietic stem cell maintenance. Nat Immunol. 2005;6(3):314–322. doi: 10.1038/ni1164. [DOI] [PubMed] [Google Scholar]
- 57.Butler JM, Nolan DJ, Vertes EL, et al. Endothelial cells are essential for the self-renewal and repopulation of Notch-dependent hematopoietic stem cells. Cell Stem Cell. 2010;6(3):251–264. doi: 10.1016/j.stem.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Varnum-Finney B, Halasz LM, Sun M, Gridley T, Radtke F, Bernstein ID. Notch2 governs the rate of generation of mouse long- and short-term repopulating stem cells. J Clin Invest. 2011;121(3):1207–1216. doi: 10.1172/JCI43868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mancini SJC, Mantei N, Dumortier A, Suter U, MacDonald HR, Radtke F. Jagged1-dependent Notch signaling is dispensable for hematopoietic stem cell self-renewal and differentiation. Blood. 2005;105(6):2340–2342. doi: 10.1182/blood-2004-08-3207. [DOI] [PubMed] [Google Scholar]
- 60.Radtke F, Wilson A, Stark G, et al. Deficient T cell fate specification in mice with an induced inactivation of Notch1. Immunity. 1999;10(5):547–558. doi: 10.1016/s1074-7613(00)80054-0. [DOI] [PubMed] [Google Scholar]
- 61.Maillard I, Koch U, Dumortier A, et al. Canonical notch signaling is dispensable for the maintenance of adult hematopoietic stem cells. Cell Stem Cell. 2008;2(4):356–366. doi: 10.1016/j.stem.2008.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mercher T, Cornejo MG, Sears C, et al. Notch signaling specifies megakaryocyte development from hematopoietic stem cells. Cell Stem Cell. 2008;3(3):314–326. doi: 10.1016/j.stem.2008.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Poirault-Chassac S, Six E, Catelain C, et al. Notch/Delta4 signaling inhibits human megakaryocytic terminal differentiation. Blood. 2010;116(25):5670–5678. doi: 10.1182/blood-2010-05-285957. [DOI] [PubMed] [Google Scholar]
- 64.Shelly LL, Fuchs C, Miele L. Notch-1 inhibits apoptosis in murine erythroleukemia cells and is necessary for differentiation induced by hybrid polar compounds. J Cell Biochem. 1999;73(2):164–175. doi: 10.1002/(sici)1097-4644(19990501)73:2<164::aid-jcb3>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 65.Jang MS, Miao H, Carlesso N, et al. Notch-1 regulates cell death independently of differentiation in murine erythroleukemia cells through multiple apoptosis and cell cycle pathways. J Cell Physiol. 2004;199(3):418–433. doi: 10.1002/jcp.10467. [DOI] [PubMed] [Google Scholar]
- 66.Henning K, Schroeder T, Schwanbeck R, Rieber N, Bresnick EH, Just U. mNotch1 signaling and erythropoietin cooperate in erythroid differentiation of multipotent progenitor cells and upregulate beta-globin. Exp Hematol. 2007;35(9):1321–1332. doi: 10.1016/j.exphem.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 67.Ishiko E, Matsumura I, Ezoe S, et al. Notch signals inhibit the development of erythroid/megakaryocytic cells by suppressing GATA-1 activity through the induction of HES1. J Biol Chem. 2005;280(6):4929–4939. doi: 10.1074/jbc.M406788200. [DOI] [PubMed] [Google Scholar]
- 68.Lam LT, Ronchini C, Norton J, Capobianco AJ, Bresnick EH. Suppression of erythroid but not megakaryocytic differentiation of human K562 erythroleukemic cells by notch-1. J Biol Chem. 2000;275(26):19676–19684. doi: 10.1074/jbc.M002866200. [DOI] [PubMed] [Google Scholar]
- 69.Kumano K, Chiba S, Shimizu K, et al. Notch1 inhibits differentiation of hematopoietic cells by sustaining GATA-2 expression. Blood. 2001;98(12):3283–3289. doi: 10.1182/blood.v98.12.3283. [DOI] [PubMed] [Google Scholar]
- 70.Oh P, Lobry C, Gao J, et al. In vivo mapping of notch pathway activity in normal and stress hematopoiesis. Cell Stem Cell. 2013;13(2):190–204. doi: 10.1016/j.stem.2013.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Han H, Tanigaki K, Yamamoto N, et al. Inducible gene knockout of transcription factor recombination signal binding protein-J reveals its essential role in T versus B lineage decision. Int Immunol. 2002;14(6):637–645. doi: 10.1093/intimm/dxf030. [DOI] [PubMed] [Google Scholar]
- 72.Pui JC, Allman D, Xu L, et al. Notch1 expression in early lymphopoiesis influences B versus T lineage determination. Immunity. 1999;11(3):299–308. doi: 10.1016/s1074-7613(00)80105-3. [DOI] [PubMed] [Google Scholar]
- 73.Sambandam A, Maillard I, Zediak VP, et al. Notch signaling controls the generation and differentiation of early T lineage progenitors. Nat Immunol. 2005;6(7):663–670. doi: 10.1038/ni1216. [DOI] [PubMed] [Google Scholar]
- 74.Ellisen LW, Bird J, West DC, et al. TAN-1, the human homolog of the Drosophila notch gene, is broken by chromosomal translocations in T lymphoblastic neoplasms. Cell. 1991;66(4):649–661. doi: 10.1016/0092-8674(91)90111-b. [DOI] [PubMed] [Google Scholar]
- 75.Weng AP, Ferrando AA, Lee W, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306(5694):269–271. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- 76.Malyukova A, Dohda T, von der Lehr N, et al. The tumor suppressor gene hCDC4 is frequently mutated in human T-cell acute lymphoblastic leukemia with functional consequences for Notch signaling. Cancer Res. 2007;67(12):5611–5616. doi: 10.1158/0008-5472.CAN-06-4381. [DOI] [PubMed] [Google Scholar]
- 77.Maser RS, Choudhury B, Campbell PJ, et al. Chromosomally unstable mouse tumours have genomic alterations similar to diverse human cancers. Nature. 2007;447(7147):966–971. doi: 10.1038/nature05886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Izon DJ, Punt JA, Xu L, et al. Notch1 regulates maturation of CD4+ and CD8+ thymocytes by modulating TCR signal strength. Immunity. 2001;14(3):253–264. doi: 10.1016/s1074-7613(01)00107-8. [DOI] [PubMed] [Google Scholar]
- 79.Aster JC, Xu L, Karnell FG, Patriub V, Pui JC, Pear WS. Essential roles for ankyrin repeat and transactivation domains in induction of T-cell leukemia by notch1. Mol Cell Biol. 2000;20(20):7505–7515. doi: 10.1128/mcb.20.20.7505-7515.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pear WS, Aster JC, Scott ML, et al. Exclusive development of T cell neoplasms in mice transplanted with bone marrow expressing activated Notch alleles. J Exp Med. 1996;183(5):2283–2291. doi: 10.1084/jem.183.5.2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chiang MY, Xu L, Shestova O, et al. Leukemia-associated NOTCH1 alleles are weak tumor initiators but accelerate K-ras-initiated leukemia. J Clin Invest. 2008;118(9):3181–3194. doi: 10.1172/JCI35090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Fabbri G, Rasi S, Rossi D, et al. Analysis of the chronic lymphocytic leukemia coding genome: role of NOTCH1 mutational activation. J Exp Med. 2011;208(7):1389–1401. doi: 10.1084/jem.20110921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Puente XS, Pinyol M, Quesada V, et al. Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature. 2011;475(7354):101–105. doi: 10.1038/nature10113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tanigaki K, Han H, Yamamoto N, et al. Notch-RBP-J signaling is involved in cell fate determination of marginal zone B cells. Nat Immunol. 2002;3(5):443–450. doi: 10.1038/ni793. [DOI] [PubMed] [Google Scholar]
- 85.Saito T, Chiba S, Ichikawa M, et al. Notch2 is preferentially expressed in mature B cells and indispensable for marginal zone B lineage development. Immunity. 2003;18(5):675–685. doi: 10.1016/s1074-7613(03)00111-0. [DOI] [PubMed] [Google Scholar]
- 86.Rossi D, Trifonov V, Fangazio M, et al. The coding genome of splenic marginal zone lymphoma: activation of NOTCH2 and other pathways regulating marginal zone development. J Exp Med. 2012;209(9):1537–1551. doi: 10.1084/jem.20120904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kiel MJ, Velusamy T, Betz BL, et al. Whole-genome sequencing identifies recurrent somatic NOTCH2 mutations in splenic marginal zone lymphoma. J Exp Med. 2012;209(9):1553–1565. doi: 10.1084/jem.20120910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zweidler-McKay PA, He Y, Xu L, et al. Notch signaling is a potent inducer of growth arrest and apoptosis in a wide range of B-cell malignancies. Blood. 2005;106(12):3898–3906. doi: 10.1182/blood-2005-01-0355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.de Pooter RF, Schmitt TM, de la Pompa JL, Fujiwara Y, Orkin SH, Zúñiga-Pflücker JC. Notch signaling requires GATA-2 to inhibit myelopoiesis from embryonic stem cells and primary hemopoietic progenitors. J Immunol. 2006;176(9):5267–5275. doi: 10.4049/jimmunol.176.9.5267. [DOI] [PubMed] [Google Scholar]
- 90.Zhou L, Li LW, Yan Q, et al. Notch-dependent control of myelopoiesis is regulated by fucosylation. Blood. 2008;112(2):308–319. doi: 10.1182/blood-2007-11-115204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kawamata S, Du C, Li K, Lavau C. Overexpression of the Notch target genes Hes in vivo induces lymphoid and myeloid alterations. Oncogene. 2002;21(24):3855–3863. doi: 10.1038/sj.onc.1205487. [DOI] [PubMed] [Google Scholar]
- 92.Schroeder T, Just U. Notch signalling via RBP-J promotes myeloid differentiation. EMBO J. 2000;19(11):2558–2568. doi: 10.1093/emboj/19.11.2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Schroeder T, Kohlhof H, Rieber N, Just U. Notch signaling induces multilineage myeloid differentiation and up-regulates PU.1 expression. J Immunol. 2003;170(11):5538–5548. doi: 10.4049/jimmunol.170.11.5538. [DOI] [PubMed] [Google Scholar]
- 94.Milner LA, Bigas A, Kopan R, Brashem-Stein C, Bernstein ID, Martin DI. Inhibition of granulocytic differentiation by mNotch1. Proc Natl Acad Sci USA. 1996;93(23):13014–13019. doi: 10.1073/pnas.93.23.13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Li L, Milner LA, Deng Y, et al. The human homolog of rat Jagged1 expressed by marrow stroma inhibits differentiation of 32D cells through interaction with Notch1. Immunity. 1998;8(1):43–55. doi: 10.1016/s1074-7613(00)80457-4. [DOI] [PubMed] [Google Scholar]
- 96.Klinakis A, Lobry C, Abdel-Wahab O, et al. A novel tumour-suppressor function for the Notch pathway in myeloid leukaemia. Nature. 2011;473(7346):230–233. doi: 10.1038/nature09999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.De Obaldia ME, Bell JJ, Wang X, et al. T cell development requires constraint of the myeloid regulator C/EBP-α by the Notch target and transcriptional repressor Hes1. Nat Immunol. 2013;14(12):1277–1284. doi: 10.1038/ni.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yao D, Huang Y, Huang X, et al. Protein O-fucosyltransferase 1 (Pofut1) regulates lymphoid and myeloid homeostasis through modulation of Notch receptor ligand interactions. Blood. 2011;117(21):5652–5662. doi: 10.1182/blood-2010-12-326074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kim YW, Koo BK, Jeong HW, et al. Defective Notch activation in microenvironment leads to myeloproliferative disease. Blood. 2008;112(12):4628–4638. doi: 10.1182/blood-2008-03-148999. [DOI] [PubMed] [Google Scholar]
- 100.Yoda M, Kimura T, Tohmonda T, et al. Dual functions of cell-autonomous and non-cell-autonomous ADAM10 activity in granulopoiesis. Blood. 2011;118(26):6939–6942. doi: 10.1182/blood-2011-06-357210. [DOI] [PubMed] [Google Scholar]
- 101.Bacher U, Schnittger S, Haferlach T. Molecular genetics in acute myeloid leukemia. Curr Opin Oncol. 2010;22(6):646–655. doi: 10.1097/CCO.0b013e32833ed806. [DOI] [PubMed] [Google Scholar]
- 102.Palomero T, McKenna K, O-Neil J, et al. Activating mutations in NOTCH1 in acute myeloid leukemia and lineage switch leukemias. Leukemia. 2006;20(11):1963–1966. doi: 10.1038/sj.leu.2404409. [DOI] [PubMed] [Google Scholar]
- 103.Chen PM, Yen CC, Wang WS, et al. Down-regulation of Notch-1 expression decreases PU.1-mediated myeloid differentiation signaling in acute myeloid leukemia. Int J Oncol. 2008;32(6):1335–1341. doi: 10.3892/ijo_32_6_1335. [DOI] [PubMed] [Google Scholar]
- 104.Tohda S, Kogoshi H, Murakami N, Sakano S, Nara N. Diverse effects of the Notch ligands Jagged1 and Delta1 on the growth and differentiation of primary acute myeloblastic leukemia cells. Exp Hematol. 2005;33(5):558–563. doi: 10.1016/j.exphem.2005.01.010. [DOI] [PubMed] [Google Scholar]
- 105.Tohda S, Murata-Ohsawa M, Sakano S, Nara N. Notch ligands, Delta-1 and Delta-4 suppress the self-renewal capacity and long-term growth of two myeloblastic leukemia cell lines. Int J Oncol. 2003;22(5):1073–1079. [PubMed] [Google Scholar]
- 106.Lobry C, Ntziachristos P, Ndiaye-Lobry D, et al. Notch pathway activation targets AML-initiating cell homeostasis and differentiation. J Exp Med. 2013;210(2):301–319. doi: 10.1084/jem.20121484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kannan S, Sutphin RM, Hall MG, et al. Notch activation inhibits AML growth and survival: a potential therapeutic approach. J Exp Med. 2013;210(2):321–337. doi: 10.1084/jem.20121527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Westhoff B, Colaluca IN, D’Ario G, et al. Alterations of the Notch pathway in lung cancer. Proc Natl Acad Sci USA. 2009;106(52):22293–22298. doi: 10.1073/pnas.0907781106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Mercher T, Raffel GD, Moore SA, et al. The OTT-MAL fusion oncogene activates RBPJ-mediated transcription and induces acute megakaryoblastic leukemia in a knockin mouse model. J Clin Invest. 2009;119(4):852–864. doi: 10.1172/JCI35901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wouters BJ, Jordà MA, Keeshan K, et al. Distinct gene expression profiles of acute myeloid/T-lymphoid leukemia with silenced CEBPA and mutations in NOTCH1. Blood. 2007;110(10):3706–3714. doi: 10.1182/blood-2007-02-073486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Coustan-Smith E, Mullighan CG, Onciu M, et al. Early T-cell precursor leukaemia: a subtype of very high-risk acute lymphoblastic leukaemia. Lancet Oncol. 2009;10(2):147–156. doi: 10.1016/S1470-2045(08)70314-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bene MC, Castoldi G, Knapp W, et al. Proposals for the immunological classification of acute leukemias. European Group for the Immunological Characterization of Leukemias (EGIL). Leukemia. 1995;9(10):1783–1786. [PubMed] [Google Scholar]
- 113.Keeshan K, He Y, Wouters BJ, et al. Tribbles homolog 2 inactivates C/EBPalpha and causes acute myelogenous leukemia. Cancer Cell. 2006;10(5):401–411. doi: 10.1016/j.ccr.2006.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Alcalay M, Meani N, Gelmetti V, et al. Acute myeloid leukemia fusion proteins deregulate genes involved in stem cell maintenance and DNA repair. J Clin Invest. 2003;112(11):1751–1761. doi: 10.1172/JCI17595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ross ME, Mahfouz R, Onciu M, et al. Gene expression profiling of pediatric acute myelogenous leukemia. Blood. 2004;104(12):3679–3687. doi: 10.1182/blood-2004-03-1154. [DOI] [PubMed] [Google Scholar]
- 116.Grieselhuber NR, Klco JM, Verdoni AM, et al. Notch signaling in acute promyelocytic leukemia. Leukemia. 2013;27(7):1548–1557. doi: 10.1038/leu.2013.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Bullinger L, Döhner K, Bair E, et al. Use of gene-expression profiling to identify prognostic subclasses in adult acute myeloid leukemia. N Engl J Med. 2004;350(16):1605–1616. doi: 10.1056/NEJMoa031046. [DOI] [PubMed] [Google Scholar]
- 118.Cancer Genome Atlas Research Network. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368(22):2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Sasaki M, Knobbe CB, Munger JC, et al. IDH1(R132H) mutation increases murine haematopoietic progenitors and alters epigenetics. Nature. 2012;488(7413):656–659. doi: 10.1038/nature11323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Quivoron C, Couronné L, Della Valle V, et al. TET2 inactivation results in pleiotropic hematopoietic abnormalities in mouse and is a recurrent event during human lymphomagenesis. Cancer Cell. 2011;20(1):25–38. doi: 10.1016/j.ccr.2011.06.003. [DOI] [PubMed] [Google Scholar]
- 121.Ko M, Bandukwala HS, An J, et al. Ten-Eleven-Translocation 2 (TET2) negatively regulates homeostasis and differentiation of hematopoietic stem cells in mice. Proc Natl Acad Sci USA. 2011;108(35):14566–14571. doi: 10.1073/pnas.1112317108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Li Z, Cai X, Cai CL, et al. Deletion of Tet2 in mice leads to dysregulated hematopoietic stem cells and subsequent development of myeloid malignancies. Blood. 2011;118(17):4509–4518. doi: 10.1182/blood-2010-12-325241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Moran-Crusio K, Reavie L, Shih A, et al. Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell. 2011;20(1):11–24. doi: 10.1016/j.ccr.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Gross S, Cairns RA, Minden MD, et al. Cancer-associated metabolite 2-hydroxyglutarate accumulates in acute myelogenous leukemia with isocitrate dehydrogenase 1 and 2 mutations. J Exp Med. 2010;207(2):339–344. doi: 10.1084/jem.20092506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Xu W, Yang H, Liu Y, et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of α-ketoglutarate-dependent dioxygenases. Cancer Cell. 2011;19(1):17–30. doi: 10.1016/j.ccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Figueroa ME, Abdel-Wahab O, Lu C, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18(6):553–567. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Luco RF, Allo M, Schor IE, Kornblihtt AR, Misteli T. Epigenetics in alternative pre-mRNA splicing. Cell. 2011;144(1):16–26. doi: 10.1016/j.cell.2010.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Adamia S, Haibe-Kains B, Pilarski PM, et al. A genome-wide aberrant RNA splicing in patients with acute myeloid leukemia identifies novel potential disease markers and therapeutic targets. Clin Cancer Res. 2014;20(5):1135–1145. doi: 10.1158/1078-0432.CCR-13-0956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Adamia S, Bar-Natan M, Haibe-Kains B, et al. NOTCH2 and FLT3 gene mis-splicing are common events in patients with acute myeloid leukemia (AML): new potential targets in AML [published online ahead of print February 26, 2014]. Blood. 2014 doi: 10.1182/blood-2013-02-481507. [DOI] [PMC free article] [PubMed] [Google Scholar]