Abstract
The primary purpose of this protocol is to prepare genomic DNA libraries that can then be analyzed by massively parallel next-generation sequencing on the Applied Bio-systems SOLiD platform. This protocol can be adapted to next-generation sequencing workflows to ultimately generate up to 1 billion 50 bp sequence tags from the ends of each of the DNA molecules in the library in a single next-generation sequencing run.
1. THEORY
The term next-generation sequencing refers to technologies that have enabled the massively parallel analysis of DNA sequence facilitated through the convergence of advancements in molecular biology, biochemistry and nucleic acid chemistry, electrical and mechanical engineering, and computational biology. Commercialized next-generation sequencing technologies are capable of generating massively parallel sequence data for tens to hundreds of millions of DNA templates simultaneously and generate more than four gigabases of sequence in a single day. These technologies have largely started to replace or significantly augment high-throughput Sanger sequencing for large-scale genomic projects, a trend that is likely to expand as the technologies continue to evolve and advance (Metzker, 2010). These technologies have created significant enthusiasm for the advent of a new era of individualized medicine.
The majority of the existing next-generation sequencing technologies employ massively parallel ‘short-read’ sequencing in which a short sequence (between 25 and 100 bp, depending on the technology and user options) at one or both ends of each of many hundreds of millions of DNA molecules are sequenced simultaneously. These reads are then aligned back to a reference genome assembly to allow measurement of genomic variation such as mutations and single-nucleotide polymorphisms compared to that reference genome assembly. In contrast, classical Sanger sequencing produces long reads of a single DNA molecule at a time, and high-throughput instruments allowing parallel sequencing of hundreds to thousands of sequences in parallel are in routine use. The advantage of the next-generation sequencing platforms is that the cost and throughput of sequencing is significantly lower (estimated to be 100-fold or more cheaper depending on the platform used). The major disadvantage of next-generation sequencing is the inability to generate long reads at this sequence throughput. However, this disadvantage is quickly diminishing due to increasing read-lengths and the use of mate-paired libraries that can circumvent much of the need for long sequence reads.
Because these technologies produce sequence from the ends of each molecule, it is common to fragment genomic DNA to produce DNA libraries with random ends to facilitate whole genome sequence coverage. Such a library, commonly referred to as a fragment library, is the most common type of library sequenced with Next-Generation Sequencing technologies. For each of the existing Next-Generation Sequencing technologies, the protocols used to generate fragment libraries compatible with massively parallel sequencing on that platform are different, but the basic principles are the same. In each case, genomic DNA is randomly fragmented by shearing, sonication, or nebulization. These fragments are modified with adaptor sequences that contain universal amplification and/or sequencing primer sequences. The resulting library of DNA molecules represents the fragment library. In this chapter we describe in detail the procedures used to generate fragment libraries for one of the commercially available next-generation sequencing platforms – the SOLiD platform marketed by Applied Bio-systems. In the accompanying explanatory chapter (see Explanatory Chapter: Next Generation Sequencing), we review concepts and procedures for this platform as well as the other commercially available ones, covering the principles behind preparing the fragment libraries for massively parallel sequencing and the sequencing chemistries.
2. EQUIPMENT
Covaris S2 Sonication System
Microcentrifuge (e.g., Eppendorf model 5417R)
96-well plate PCR thermal cycler (e.g., GeneAmp® PCR System 9700)
NanoDrop® ND-1000 Spectrophotometer
E-Gel iBase and E-Gel Safe Imager (Invitrogen, G6466)
Vortex mixer
PicoFuge® (e.g., Stratagene)
Micropipettors (2, 20, 200, and 1000 μl)
Aerosol barrier pipet tips
Covaris microTUBEs
0.5-ml Eppendorf LoBind centrifuge tubes
1.5-ml Eppendorf LoBind centrifuge tubes
PCR strip tubes
Agilent bioanalyzer 2100 instrument (optional)
3. MATERIALS
Note: Several of the components below should be purchased from New England Biolabs (NEB) and/or from Life Technologies/Applied Bio-systems using the indicated catalog numbers. All oligonucleotide sequences for adaptors and primers can be ordered from commercial vendors such as Integrated DNA Technologies (IDT) or Life Technologies.
SOLiD Library P1 adaptor plus strand (5′-CCACTACGCCTCCGCTTTCCTCTCTATGGGCAGTCGGTGAT-3′)
SOLiD Library P1 adaptor minus strand (5′-ATCACCGACTGCCCATAGAGAGGAAAGCGGAGGCGTAGTGGTT-3′)
SOLiD Library P2 adaptor plus strand (5′-AGAGAATGAGGAACCCGGGGCAGTT-3′)
SOLiD Library P2 adaptor minus strand (5′-CTGCCCCGGGTTCCTCATTCTCT-3′)
SOLiD Library PCR primer 1 (5′-CCACTACGCCTCCGCTTTCCTCTCTATG-3′)
SOLiD Library PCR primer 2 (5′-CTGCCCCGGGTTCCTCATTCT-3′)
NEBNext End Repair Reaction Buffer Mix (10×) (NEB, E6062A or E6062AA, or as part of E6060S/L, or see recipe below)
NEBNext End Repair Enzyme Mix (NEB, E6061A or E6061AA, or as part of E6060S/L)
SOLiD Library Column Purification Kit (Applied Biosystems, 4443744)
NEBNext Quick Ligation Reaction Buffer (NEB, E6064A or E6064AA, or as part of E6060S/L, or see recipe below)
T4 DNA Ligase (NEB, E6063A or E6063AA or as part of E6060S/L)
LongAmp Taq 2× Master Mix (NEB, E6065A or E6065AA or as part of E6060S/L)
E-Gels compatible with E-Gel iBase system
Tris base
Hydrochloric acid (HCl)
EDTA
Nuclease-free water
Isopropyl alcohol
Ethylene glycol
50-bp DNA ladder (e.g., Invitrogen 10416-014)
3.1. Solutions & buffers
Step 1 1× Low TE Buffer
| Component | Final concentration | Stock | Amount |
|---|---|---|---|
| Tris–HCl (pH 8.0) | 10 mM | 1 M | 10 ml |
| EDTA (pH 8.0) | 0.1 mM | 0.5 M | 200 μl |
Add water to 1 l
Step 2 10× End Repair Reaction Buffer Mix
| Component | Final concentration | Stock | Amount |
|---|---|---|---|
| Tris–HCl (pH 7.5) | 500 mM | 1 M | 2.5 ml |
| MgCl2 | 100 mM | 1 M | 500 μl |
| Dithiothreitol | 100 mM | 1 M | 500 μl |
| ATP | 10 mM | 100 mM | 500 μl |
| dATP | 4 mM | 100 mM | 200 μl |
| dCTP | 4 mM | 100 mM | 200 μl |
| dGTP | 4 mM | 100 mM | 200 μl |
| dTTP | 4 mM | 100 mM | 200 μl |
Add water to 5 ml
Note: This buffer can be purchased from New England Biolabs as part of the NEBNext Master Mix 3 (cat# E6060S/L) or as an individual buffer (cat# E6062A/AA)
Step 3 and also used in Preparation of Adaptor Oligos 5×NEBNext Quick Ligation Reaction Buffer
| Component | Final concentration | Stock | Amount |
|---|---|---|---|
| Tris–HCl (pH 7.6) | 330 mM | 1 M | 1.65 ml |
| MgCl2 | 50 mM | 1 M | 250 μl |
| Dithiothreitol | 5 mM | 1 M | 25 μl |
| ATP | 5 mM | 100 mM | 250 μl |
| Polyethylene glycol (PEG 6000) | 30% | 100% | 1.5 ml |
Add water to 5 ml
Note: This buffer can be purchased from New England Biolabs as part of the NEBNext Master Mix 3 (cat# E6060S/L) or as an individual buffer (cat# E6062A/AA)
4. PROTOCOL
4.1. Preparation
Isolate relatively high-molecular-weight (>5 kb) genomic DNA to use as starting material. One to five microgram of genomic DNA is needed for this protocol (see Preparation of Genomic DNA from Bacteria, Preparation of genomic DNA from Saccharomyces cerevisiae or Isolation of Genomic DNA from Mammalian Cells).
Prepare double-stranded P1 and P2 adaptors at 50 μM stock concentration.
Mix equal volumes of 125 μM P1 adaptor plus and minus strand oligos. Add 1 part of 5×NEBNext Quick Ligation Reaction Buffer to 4 parts of the plus and minus strand oligo mixture. This will give a final concentration of 50 μM of each oligo and 1× Quick Ligation Buffer. Hybridize oligos by running the following program in a thermal cycler:
| Temperature (°C) | Time (min) |
|---|---|
| 95 | 5 |
| 72 | 5 |
| 60 | 5 |
| 50 | 3 |
| 40 | 3 |
| 30 | 3 |
| 20 | 3 |
| 10 | 3 |
| 4 | Hold |
Store the hybridized oligos at −20 °C until ready for use.
4.2. Duration
| Preparation | 1–5 h |
|---|---|
| Protocol | 1–2 days |
4.3. Caution
Consult your institute’s BioSafety Officer for proper handling of potentially infectious DNA samples and of potentially harmful reagents.
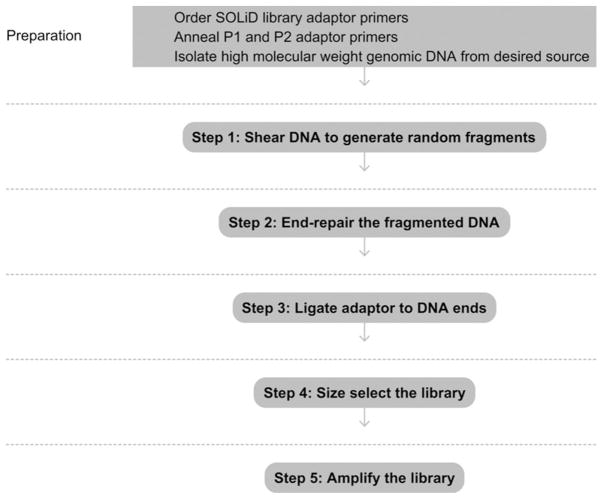
See Fig. 15.1 for the flowchart of the complete protocol.
Figure 15.1.
Flowchart of the complete protocol, including preparation.
5. STEP 1 SHEAR THE DNA TO GENERATE RANDOM FRAGMENTS
5.1. Overview
Fragment the input genomic DNA to make small DNA molecules with a modal size of ~100–200 bp with random ends. This is done by sonication in a Covaris S2 System.
5.2. Duration
30 min
-
1.1
Dilute the desired amount of DNA to 100 μl in 1× Low TE Buffer in a 1.5-ml LoBind centrifuge tube.
-
1.2
Slowly transfer the DNA to a microTUBE loaded into the Covaris S2 through the presplit septa, being careful not to introduce any bubbles into the bottom of the tube.
-
1.3
Shear the DNA using the following Covaris S2 System conditions:
Number of cycles: 6
Bath temperature: 5 °C
Bath temperature limit: 30 °C
Mode: frequency sweeping
Water quality testing function: off
Duty cycle: 10%
Intensity: 5
Cycles/burst: 100
Time: 60 s
-
1.4
Insert a pipette tip through the presplit septa, and slowly transfer the sheared DNA from the microTUBE to a new 1.5-ml LoBind centrifuge tube.
5.3. Caution
Follow Covaris S2 System instructions for proper setup, operation, and maintenance of the instrument.
See Fig. 15.2 for the flowchart of Step 1.
Figure 15.2.
Flowchart of Step 1.
6. STEP 2 END-REPAIR THE FRAGMENTED DNA
6.1. Overview
DNA fragments generated by sonication in Step 1 will be end polished to produce a library of DNA fragments with blunt, 5′-phosphorylated ends that are ready for ligation. The end polishing is accomplished by using the T4 DNA polymerase, which can fill in 5′ overhangs via its polymerase activity and recess 3′ overhangs via its 3′→5′ exonuclease activity. The phosphorylation of 5′ ends is accomplished by T4 polynucleotide kinase.
6.2. Duration
~1 h
-
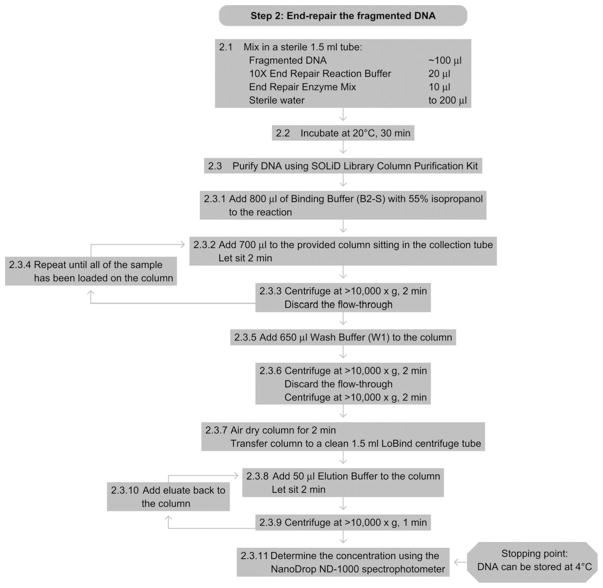
2.1Mix the following components in a sterile LoBind centrifuge tube:
Fragmented DNA ~100 μl (recovered from sonicator) 10× End Repair Reaction Buffer Mix 20 μl End Repair Enzyme Mix 10 μl Sterile water to 200 μl -
2.2
Incubate at 20 °C for 30 min.
-
2.3
Purify DNA using SOLiD Library Column Purification Kit:
-
2.3.1
Add 4 volumes (in this case add 800 μl since the reaction is 200 μl) of Binding Buffer (B2-S) with 55% isopropanol to the reaction from above.
-
2.3.2
Add 700 μl of this mixture to the provided column sitting in the collection tube and let column stand for 2 min.
-
2.3.3
Centrifuge the column at >10 000×g for 1 min and discard the flow-through.
-
2.3.4
Repeat Steps 2.3.2 and 2.3.3 until all of the sample has been loaded onto the column.
-
2.3.5
Add 650 μl of Wash Buffer (W1) to the column.
-
2.3.6
Centrifuge the column at >10 000×g for 2 min and discard flow through. Repeat centrifugation again without adding any additional W1 buffer to remove the residual Wash Buffer.
-
2.3.7
Air-dry the column for 2 min and transfer the column from the collection tube to a clean 1.5-ml LoBind centrifuge tube.
-
2.3.8
Add 50 μl of Elution Buffer (E1) to the column and let stand for 2 min.
-
2.3.9
Centrifuge the column at >10 000×g for 1 min.
-
2.3.10
Add the eluate from Step 2.3.9 back to the column, and let stand for 2 min. Centrifuge the column at >10 000×g for 1 min.
-
2.3.11
Calculate the final concentration using the NanoDrop ND-1000 spectrophotometer according to manufacturer’s instructions (alternatively see Explanatory Chapter: Nucleic Acid Concentration Determination).
-
2.3.1
6.3. Tip
Read the kit manual before first use to make sure all buffers are properly prepared and to get familiarized with the kit components and procedures.
6.4. Tip
The best yields are achieved when 5 μg or less of genomic DNA were initially used. If more than this amount was used in the above steps, it is best to use multiple columns.
6.5. Tip
This is a stopping point and purified DNA can be stored at 4 °C, or can be taken directly for Adaptor Ligation.
See Fig. 15.3 for the flowchart of Step 2.
Figure 15.3.
Flowchart of Step 2.
7. STEP 3 ADAPTOR LIGATION
7.1. Overview
The primary goal of this step is to ligate adaptors to the DNA library. These adaptors are necessary for subsequent amplification and sequencing of the DNA library on the SOLiD instrument. Ligation of double-stranded DNA adaptors is accomplished by use of T4 DNA ligase. The double-stranded adaptors do not have 5′ phosphates and contain a 5′ overhang on one end to prevent ligation in the incorrect orientation. The lack of 5′ phosphate leads to generation of a nick on one strand of the ligated DNA. This will be nick translated and removed during an upcoming amplification step.
7.2. Duration
30–60 min
-
3.1
Calculate the amount of 50 μM stock P1 and P2 adaptors needed for ligation. The goal is to have a 30:1 ratio of each adaptor to DNA molecules in the library. Assuming a modal size of ~165 bp using the sonication conditions listed above and 660 pg per base pair of DNA, one would have 9.2 pmol μg−1 of DNA. Therefore, for a 30:1 ratio, we would need 5.5 μl of the 50 μM P1 and P2 adaptor stocks per 1 μg of DNA library.
-
3.2Mix the following components in a sterile LoBind centrifuge tube:
End repaired DNA from Step 2 Variable 5× NEBNext Quick Ligation Reaction Buffer 40 μl P1 DNA adaptor (50 μM) Variable P2 DNA adaptor (50 μM) Variable T4 DNA ligase 10 μl Sterile water to 200 μl -
3.3
Incubate at 20 °C for 15 min.
-
3.4
Purify DNA as described in Step 2.3, except that the DNA should be eluted in 65 μl instead of 50 μl of Elution Buffer (E1).
7.3. Tip
This is a stopping point and purified DNA can be stored at 4 °C, or can be taken directly for size selection.
See Fig. 15.4 for the flowchart of Step 3.
Figure 15.4.

Flowchart of Step 3.
8. STEP 4 SIZE SELECTION OF LIBRARY
8.1. Overview
In this step, the adaptor-ligated library will be size-selected to ~200–250 bp size range. By doing this, we remove unligated adaptors and also select the optimal size-range for subsequent emulsion PCR and sequencing. In the procedure described below, we will use the Invitrogen E-Gel system for convenient loading, running, and elution of the size selected DNA.
8.2. Duration
1 h
-
4.1
Remove the combs from the E-Gel and load onto the E-Gel iBase system linked to the E-gel Safe Imager Real-Time Transilluminator according to the manufacturer’s instructions.
-
4.2
Load 20 μl of the ligated, purified DNA from Step 3 into each of three wells in the top row for each sample. When loading, skip the center well, the wells to the right and left of the center well, and the two outer most wells.
-
4.3
Load 10 μl of the 50-bp ladder (0.1 μg μl−1) to the center top well. Add 7 μl of water to fill the well.
-
4.4
Fill empty wells in the top row with 20 μl of nuclease-free water.
-
4.5
Fill the middle row of wells in the gel with 20 μl of nuclease-free water. Add 20 μl of nuclease free water to the center middle row of wells.
-
4.6
Run the gel with iBase program SizeSelect 2% (preprogrammed in iBase) for a total of precisely 11 min and 40 s. Monitor the gel in real-time using the real-time transilluminator. If needed, the wells in the middle row should be filled with nuclease-free water. When the 200 bp band from the ladder is at the bottom of the middle row center well but still within the well, if the run has not stopped already, then stop the run.
-
4.7
Collect the solutions from the wells, which should now contain DNA in the range of 200–250 bp, and pool them according to samples.
-
4.8
Wash each of the middle row collection wells with 25 μl of nuclease-free water and collect the wash solution and add to the retrieved material collected in Step 4.7.
8.3. Tip
Alternatively, the user could run the library on a standard 1% agarose gel along with a 50-bp DNA ladder (see Agarose Gel Electrophoresis). After running, the portion of the DNA library corresponding to 200–250 bp could be excised using a sterile blade. This excised gel fragment can be purified with a standard DNA gel extraction procedure such as those available in commercialized kits (e.g., Qiagen Gel Extraction kit). However, the procedure described above will allow more standardized generation of fragment libraries.
See Fig. 15.5 for the flowchart of Step 4.
Figure 15.5.
Flowchart of Step 4.
9. STEP 5 AMPLIFICATION OF THE LIBRARY
9.1. Overview
In this step, the size-selected fragment library is amplified to a small extent to prepare for downstream emulsion PCR and sequencing steps. The initial phase of amplification involves a step for nick translation of the nick remaining at the 5′ ligation junction of the adaptors. This nick will get translated to the end of the adaptor to remove the nick completely. The next steps use standard PCR with primers specific to the P1 and P2 adaptors to amplify the library. The polymerase used for amplification can also carry out the nick translation step, and therefore only one enzyme is needed.
9.2. Duration
~1–2 h
-
5.1Mix the following components in a sterile LoBind centrifuge tube:
DNA from size selection step Variable P1 adaptor primer (50 μM stock) 10 μl P2 adaptor primer (50 μM stock) 10 μl 2× LongAmp Taq Master Mix 250 μl Sterile water to 500 μl -
5.2
Alliquot 125 μl into each of four PCR tubes.
-
5.3Place in a thermal cycler and carry out the following cycling program:
Cycle process Temperature Time Cycles Nick translation 72 °C 20 min 1 Initial denaturation 95 °C 5 min 1 Denaturation 95 °C 15 s 2–10a Annealing 62 °C 15 s Extension 70 °C 1 min Final extension 70 °C 5 min 1 Hold 4 °C 1 1 aAdjust the number of cycles based on starting amount of DNA as follows:- 100 ng to 1 μg: 6–8 cycles
- 1–2 μg: 4–6 cycles
- 2–5 μg: 2–3 cycles
-
5.4
Clean up amplified library according to protocols described in Step 2.3.
-
5.5
Quantitate and quality control the generated library using an Agilent Bioanalyzer 2100 (optional), according to the manufacturer’s recommendations. Samples should then be ready for sequencing workflows including emulsion PCR, bead enrichment, and sequencing. Refer to Applied Biosystems web site or consult a SOLiD Next-Generation Sequencing service facility for protocols on these subsequent steps.
9.3. Tip
These cycling conditions represent starting guidelines. It may be necessary to optimize the number of cycles to avoid overamplification while obtaining enough library material to proceed to downstream emulsion PCR steps. Amplified products should be tested for quality by analysis with an Agilent Bioanalyzer or agarose gel to ensure that the predominant products (>75%) are within the size selected range. Otherwise, it is likely that the product was overamplified producing PCR artifacts that will compromise sequence real estate.
See Fig. 15.6 for the flowchart of Step 5.
Figure 15.6.

Flowchart of Step 5.
Referenced Literature
- Metzker ML. Sequencing technologies – The next generation. Nature Reviews Genetics. 2010;11:31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
Related Literature
- These protocols were adapted from the Applied Biosystems protocols in the Applied Bio-systems SOLiD™ 4 System Library Preparation Guide (www.appliedbiosystems.com), as well as the protocols supplied with the NEBNext DNA Sample Prep Master Mix Set 3 (NEB E6060S/L) kit.
Referenced Protocols in Methods Navigator
- Explanatory Chapter: Next Generation Sequencing
- Preparation of Genomic DNA from Bacteria
- Preparation of genomic DNA from Saccharomyces cerevisiae
- Isolation of Genomic DNA from Mammalian Cells
- Explanatory Chapter: Nucleic Acid Concentration Determination
- Agarose Gel Electrophoresis