Abstract
Recent advances strongly suggest that mRNA rather than DNA will be the nucleotide basis for a new class of vaccines and drugs. Therapeutic cancer vaccines against a variety of targets have been developed on this basis and initial clinical experience suggests that preclinical activity can be successfully translated to human application. Likewise, prophylactic vaccines against viral pathogens and allergens have demonstrated their activity in animal models. These successes could be extended preclinically to mRNA protein and gene replacement therapy as well as the induction of pluripotent stem cells by mRNA encoded transcription factors. The production of mRNA-based vaccines and drugs is highly flexible, scalable and cost competitive, and eliminates the requirement of a cold chain. mRNA-based drugs and vaccines offer all the advantages of a nucleotide-based approach at reduced costs and represent a truly disruptive technology that may start a revolution in medicine.
Keywords: mRNA-based vaccines, replicons, RNActive vaccines, therapeutic mRNA
Introduction
The technological advances of molecular biology in recent decades have dramatically changed our understanding of human diseases as well as their diagnosis and treatment. The identification of crucial factors in the pathophysiology of human diseases, made possible by genetic screening, transgenic or knockout animal models to name only a few, also allowed us to develop highly selective, small molecule inhibitors of specific targets that sometimes completely changed the natural course of inexorable diseases, for example, the inhibition of the oncogenic fusion protein bcr-abl in chronic myelogenous leukemia by imatinib, dasatinib, and nilotinib [Panjarian et al. 2013; Quintas-Cardama and Jabbour, 2013]. Our increasing ability to manipulate the genetic code of proteins in question, that is, their DNA, in many instances enabled us to bypass the development of small molecule inhibitors and instead to exploit proteins such as monoclonal antibodies, recombinant naturally occurring or highly engineered proteins for diagnostic and therapeutic purposes to an extent that was almost unimaginable just two decades ago (examples given in the following references) [Bargou et al. 2008; Bendtzen, 2012; Day et al. 2013; Lindsay et al. 2013; Ljung, 2013; Moots and Naisbett-Groet, 2012; Robak, 2012; Sellam et al. 2013; Specenier and Vermorken, 2013]. However, the use of proteins for medical purposes often requires good manufacturing practice (GMP) production processes that are technically and financially very demanding and not very flexible in that each protein requires its own production facility.
It was recognized early on that proteins could principally be expressed either by direct injection of DNA or messenger RNA (mRNA) into target organs [Wolff et al. 1990]. The initial efforts to exploit this effect focused on the development of nucleotide-based vaccines, since the capacity to express proteins on this basis was considered limited. While the first reports on nucleotide-based vaccines showed that vaccines produced on a DNA or mRNA basis had similar activity [Conry et al. 1995; Martinon et al. 1993; Tang et al. 1992], most researchers focused on the development of DNA vaccines in the coming decades. The main reasons for this attitude were a perceived instability of mRNA, difficulties to ensure long-term storage, and importantly, the cost of manufacturing GMP grade material.
Roughly 10 years ago though, Pascolo pointed out that the latter view was erroneous and that the costs of manufacturing mRNA on a large scale would be lower than those to produce DNA [Pascolo, 2004]. Nucleotide vaccines based on mRNA offer the flexibility to encode virtually any protein as antigen in a very short time span, but could be produced with the same production process in the same production facilities. Thus novel vaccines could be made in a very short time with limited financial investments, which is of great importance for pandemic scenarios in infectious diseases and for the possibility of making cancer vaccines against patient-specific cancer-associated or mutated antigens [Kreiter et al. 2012; Petsch et al. 2012]. Moreover, mRNA carries no risk of genomic integration, which might not just be a theoretical risk for DNA. This gives mRNA an inherent safety advantage over DNA-based therapeutics.
This article argues that, contrary to former expectations, it is not DNA that emerges as the nucleotide basis for vaccines, but mRNA might be the ideal basis for the development of new vaccines against infectious pathogens [Petsch et al. 2012] and for cancer vaccines [Kübler et al. 2011; Sebastian et al. 2011, 2012]. With the possibility to do more at reduced costs, mRNA vaccination technology would constitute a truly disruptive technology [Christensen, 1997]. Recent technological advances in understanding the properties of mRNA have widened the conceivable applications of mRNA beyond immunization purposes to the production of proteins in situ [Kariko et al. 2012; Kormann et al. 2011] and to autologous cell therapies based on mRNA-induced pluripotent stem cells [Mandal and Rossi, 2013]. Hence, it may well be that the first drug approved designed on a nucleotide basis will rely on mRNA rather than the seemingly less complicated DNA [Geall et al. 2013; Gilboa, 2012].
DNA vaccines
Vaccines based on nucleotides appeared as the ideal basis to move beyond empirical to rational design of vaccine research and development, and to stimulate the immune system in a directed manner that also generates cellular immunity [Ulmer et al. 2012]. In addition, manufacturing was considered to be relatively simple, inexpensive and scalable with the production process readily adaptable from one vaccine to another or rather from one nucleotide code to another. Though early efforts to develop vaccines based on nucleotides used both DNA [Tang et al. 1992; Ulmer et al. 1993] and RNA [Martinon et al. 1993] for vaccination, the next two decades were largely dominated by research on DNA vaccines, due to the perceived greater ease of use. DNA vaccines were able to induce potent T- and B-cell immune responses in animals against a variety of antigens. This culminated in the development and commercialization of plasmid DNA-based vaccines for animal usage: a melanoma cancer vaccine was approved for dogs [Grosenbaugh et al. 2011], an infectious hematopoietic necrosis virus vaccine for fish and a vaccine for West Nile virus for horses [Davis et al. 2001; Garver et al. 2005; Kurath et al. 2006].
However, the development of DNA vaccines for humans has not been similarly successful thus far. While a number of clinical trials demonstrated the principle ability of DNA vaccines to induce cellular T- and B-cell responses in humans, the strength of these immune responses was rather lower than that achieved by more conventional approaches. The reasons for this are not quite clear, but might be related to the necessity of DNA vaccines to cross at least two cell membranes, the nuclear membrane in addition to the plasma membrane to achieve antigen expression. Extensive efforts have therefore been directed at various techniques to facilitate the physical delivery of DNA that include sophisticated electroporation (EP) techniques, needle-free approaches, such as particle bombardment and high-pressure delivery, and dermal patches, but also activation of the immune system by encoded immunostimulatory molecules (genetic adjuvants) [Sardesai and Weiner, 2011; Ulmer et al. 2012]. Recent studies with new EP devices reignited hope for DNA vaccines. A therapeutic human papillomavirus (HPV) 16/18 candidate vaccine, VGX-3100, induced robust immune responses to antigens from high-risk HPV serotypes after in vivo EP [Bagarazzi et al. 2012]. It was argued that these contribute to elimination of HPV-infected cells and subsequent regression of the dysplastic process. Malaria DNA vaccine candidates were recently generated from synthetic sequences of four antigens from Plasmodium falciparium (circumsporozoite protein, liver stage antigen 1, thrombospondin-related anonymous protein, and cell-traversal protein for ookinetes and sporozoites), administered in vivo and studied preclinically for their immunogenicity in mice and nonhuman primates (NHPs). Humoral and cellular responses comprising interleukin (IL)-2, interferon γ (IFNγ) and tumor necrosis factor α (TNFα) producing CD4+ and CD8+ T cells were detected against all antigens encoded in mice [Ferraro et al. 2013]. The cellular responses were also found in NHPs, and importantly, antigen-specific CD8+ Granzyme B+ T cells could be detected. An alternative approach by Lu and colleagues showed that priming with DNA vaccines followed by protein boost vaccination regimes results in remarkable immune responses [Vaine et al. 2008; Wang et al. 2012], but this approach gives up some of the advantages of a DNA vaccination only. Against this background of advances in the DNA vaccine field, the recent failure of Allovectin-7 [Bedikian et al. 2010], a bicistronic plasmid encoding human leukocyte antigen (HLA)-B7 and β2 microglobulin formulated with a cationic lipid system, to meet its primary endpoint to achieve durable responses in advanced melanoma in a phase III trial came as a disappointment, but detailed results of the study have not yet been published (http://www.vical.com/). Yet, a phase II trial with TransVax, a therapeutic DNA vaccine targeting cytomegalovirus glycoprotein B and phosphoprotein 65 formulated with poloxamer CRL1005 and benzalkonium chloride, yielded encouraging results [Kharfan-Dabaja et al. 2012]. Cytomegalovirus-seropositive patients after allogeneic hemopoietic stem-cell transplantation showed a reduction in the rate of clinically significant viremia requiring cytomegalovirus-specific antiviral therapy which, however, did not reach statistical significance. A longer follow up revealed a significant reduction of occurrence and recurrence of cytomegalovirus viremia and improved the time to event for viremia episodes compared with placebo. The DNA vaccine was well tolerated and is presently being studied in a phase III trial (http://www.vical.com/). Hence, there clearly is a silver lining on the horizon for DNA vaccines, but despite these recent progresses it remains to be seen whether DNA vaccines can now achieve the breakthrough not seen in the past two decades of research on DNA vaccines.
In addition to the challenges mentioned above, DNA vaccines carry the principle risk of genomic integration as exemplified by the generation of stably transfected cell lines which results from random genomic integration of the transfected DNA which is afterwards selected for. This risk of genomic integration is generally considered to be very small. However, hundreds of millions of doses of a successful prophylactic vaccine against infectious pathogens might be administered to healthy recipients over years. Under these conditions, even a very rare event might become a potentially serious safety problem, particularly in view of the vaccination fatigue in western countries [Finnegan, 2012]. Another issue is the duration of antigen expression which can last for several months. Prolonged antigen expression per se may not necessarily correlate with good immune responses and may even be detrimental to the intended immune effect and lead to exhaustion of T cells [Han et al. 2010; Shin and Wherry, 2007; Wherry et al. 2003]. A different explanation for the ‘underperformance’ of DNA vaccines in humans may be a weaker than presumed built in adjuvanticity of these vaccines. The importance of cytoplasmic DNA sensors for the induction of DNA-dependent immune responses has been increasingly recognized recently [Aoshi et al. 2011; Desmet and Ishii, 2012; Marichal et al. 2011]. It is conceivable that without the assistance of sophisticated delivery methods, not enough DNA might end up in the cytoplasm or that species differences exist in the sensitivity of these sensors to stimulation by DNA. Both effects might impair the efficacy of DNA vaccines in humans.
The promise of mRNA-based vaccines
The potential of mRNA to be used for protein expression was first demonstrated by Wolff and colleagues with the successful expression of a variety of proteins after direct injection of their mRNA into the muscle of mice [Wolff et al. 1990]. This was followed by the first report of a successful mRNA vaccine that demonstrated the induction of anti-influenza cytotoxic T lymphocytes in vivo by immunizing mice with liposomes containing mRNA encoding the influenza virus nucleoprotein [Martinon et al. 1993]. However, liposomal protection of mRNA is not essential as was demonstrated by the repeated injection of an unprotected mRNA coding for carcinoembryonic antigen (CEA) directly into the muscles of mice [Conry et al. 1995]. Mice immunized with unprotected CEA encoding mRNA developed an antibody response to CEA upon challenge with a CEA-expressing tumor cell line, while control mice not receiving mRNA injections did not develop antibodies to CEA. Yet, the immune response required not just administration of the mRNA, but also expression of the encoded antigen by the tumor cell line used for challenge. Five years later, a major leap forward was made by the demonstration that substantial humoral and cellular immune responses could be directly induced by the injection of antigens encoded as naked (unprotected) or protamine-protected mRNA into the ear pinna of mice [Hoerr et al. 2000]. The induced immune response was antigen specific and importantly also functional as indicated by the capacity of induced T cells to lyse cells expressing the antigen. These initial experiments established that mRNA could principally serve as the basis for gene therapy or gene replacement and for the induction of protective cellular or humoral immunity. It was suggested that this approach might be particularly useful for induction of an immune response against a proto-oncogene product or growth factor likely to elicit malignant transformation upon prolonged expression [Conry et al. 1995].
The challenges posed by production
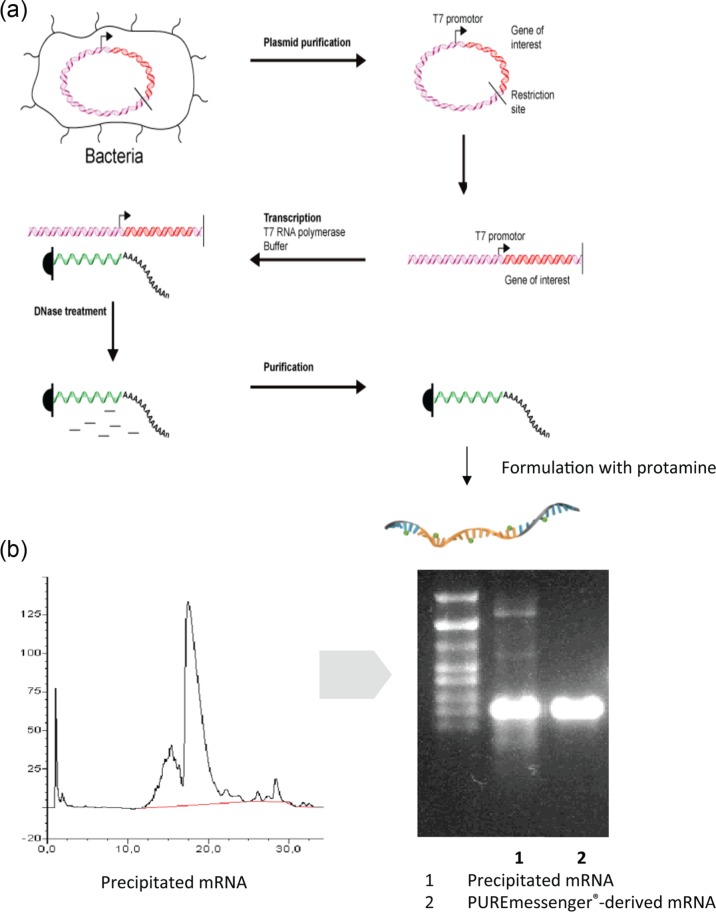
Common laboratory experiences have taught most researchers that RNA is a highly unstable molecule and difficult to work with. This view is heavily influenced by the ubiquitous presence of RNases that can indeed rapidly degrade RNA and thus destroy experiments when appropriate precautions are neglected. However, when looking at its chemical characteristics RNA is in fact a very stable molecule under physiological conditions. At CureVac, the team of F. von der Mülbe demonstrated that it can easily be produced in a process involving the transcription of target RNA by RNA polymerases from a linearized plasmid DNA template, followed by enzymatic destruction of the DNA template by DNases and purification of the resulting mRNA by precipitation and chromatographic methods according to size (Figure 1; detailed description given by Ketterer and colleagues and Pascolo) [Ketterer et al. 2008; Pascolo, 2004, 2006]. This process results in highly pure RNA products and works very well for standard mRNA sizes of a few kilobases, but has also been used successfully to produce mRNAs of sizes up to 15 kb.
Figure 1.
Production and thermal stability of messenger RNA (mRNA). (a) Basic structure of CureVac’s good manufacturing practice (GMP) production process of mRNA. All individual steps of the process are performed under GMP conditions. The production of a large number of vaccines in parallel is possible under these conditions. The process can be completed in a few weeks, including more than 39 quality controls demanded for GMP production. Importantly, the process is highly scalable and would allow production of a given vaccine either in one large facility or in several small ones. Costs would be a fraction of those required by a vaccine production site today and can be easily adapted to the production of a new vaccine within days (b) CureVac has developed a scalable, proprietary mRNA purification process (PUREmessenger). Impurities are removed by a chromatography procedure that results in notably purer mRNA than obtainable by standard methods. The highly purified mRNA has also enhanced expression capacity.
Thermal stability of vaccines can pose a major logistical problem for vaccines, particularly in countries where the infrastructure makes it difficult to maintain the cold chain. The problem could be solved, however, by demonstrating that the mRNA-based vaccines can be lyophilized and that the lyophilized vaccines retain their full biological activity [Petsch et al. 2012]. This could also be demonstrated for lyophilized mRNA-based vaccines exposed to thermal stress under International Conference on Harmonisation conditions at temperatures of 25 and 40°C for periods of several years or months, respectively. Even under extreme stress conditions at a temperature of 60°C, stability could be shown for several months. Ongoing experiments suggest that these periods can still be substantially extended (F. von der Mülbe, personal communication).
The production process avoids the use of problematic starting materials such as animal-derived key components, and results in high batch-to-batch reproducibility. Furthermore, the same production process can be used for many different vaccines. This platform characteristic of the production process avoids many costly steps caused by the requirement to fulfill regulatory demands for product-specific validations. The process can be easily adapted to GMP conditions which allowed us to build the first GMP production facility for the production of clinical material. The total production process is highly flexible and scalable, meaning that the process can be easily changed to the production of a new vaccine within a few days and that the process can be scaled to the production of millions of vaccine doses either in one big facility or in several smaller ones. A sensitivity analysis performed with the help of several production experts revealed that vaccines produced on an industrial scale in such a process could be produced at costs that would be competitive in commodity markets, such as the one for influenza vaccines [Petsch et al. 2012]. In fact, contrary to common beliefs, mRNA-based immunotherapy appears to be no more costly than other strategies, such as protein, peptide, DNA, cell or recombinant pathogen based strategies, and might actually be less expensive [Pascolo, 2004].
Ex vivo transfection of dendritic cells with mRNA
Practically any cell type can be transfected with mRNA, including dendritic cells (DCs) [Breckpot et al. 2004]. DCs and other professional and semi-professional antigen-presenting cells (APCs) are critically important for initiating effective immune responses priming CD4 and CD8 T cells against mRNA-encoded antigens. Due to the flexibility of mRNA to encode any antigen and the perceived problems to produce mRNA at a large scale under GMP conditions, efforts to exploit this flexibility for the industrial development of vaccination strategies focused on the ex vivo transfection of APCs early on [Boczkowski et al. 1996; Gilboa and Vieweg, 2004; Sullenger and Gilboa, 2002]. These included the transfection of mRNA-encoded chimeric antigens targeting the antigens to endosomes to enhance major histocompatibility complex (MHC) class II presentation, the combination of various mRNA-encoded antigens and the use of APCs other than DCs for stimulation of the immune system [Bonehill et al. 2003; Breckpot et al. 2003; Gilboa, 2007; O’Neill et al. 2004; Okada et al. 2005; Van den Bosch et al. 2006]. However, a failed trial comparing ex vivo generated, peptide-loaded DC immunotherapy to standard chemotherapy in patients with advanced melanoma was a major setback for DC-based vaccination in general [Schadendorf et al. 2006]. Yet it was argued that DC activation would remain a cornerstone of a successful immunotherapy and novel approaches were fostered (detailed analysis given by Gilboa and Van Lint and colleagues) [Gilboa, 2007; Van Lint et al. 2013].
One of these was developed by Thielemann and colleagues. They developed a cocktail of three different immunostimulatory molecules to systematically alter the activation status of DCs [Bonehill et al. 2008]. The immunostimulatory cocktail consisted of a constitutively active toll-like receptor 4 (TLR4) variant (caTLR4), CD40L for activation of T cells by binding to CD40 and CD70 that provides a survival and proliferation signal to T cells by binding to CD27 and was coadministered with the antigen of interest linked to an MHC class II sorting signal. DCs matured with this so-called ‘TriMix’ mRNA were able to induce specific T cells against the encoded antigen more than 200-fold more effectively than DCs generated with the classical stimulatory cytokine cocktail consisting of IL-1β, TNFα, IL-6 and prostaglandin E2 [Bonehill et al. 2008]. Recent publications suggested that a certain threshold antigen dose is required for the induction of a productive immune response involving antigen-specific T cells [Henrickson et al. 2008], but also that uptake of mRNA and subsequent antigen expression occurs only in immature and not matured DCs [Diken et al. 2011]. A successful DC maturation with the TriMix cocktail would therefore not necessarily have been expected to result in good immunogenicity due to a potentially impaired mRNA uptake by matured DCs. However, this would not pose a logical problem, if the way in which DCs are stimulated and matured has a decisive impact on whether nucleotide-encoded antigens are expressed or not and consequently whether or not immune responses can be generated successfully. Hence, not all activators of the immune system lead to maturation of DCs in a manner detrimental to use in combination with mRNA-expressed antigens [Van Lint et al. 2012].
The first trial in patients with advanced melanoma was performed with a TriMix-DC therapy encoding the melanoma-associated antigens (MAAs) MAGE-A3, MAGE-C2, tyrosinase or gp100, all linked to a HLA class II sorting signal [Van Nuffel et al. 2012b; Wilgenhof et al. 2011b]. MAGE-A3 and -C2 were linked at the N terminus to the signal peptide of lamp1 to ensure transport to the endoplasmic reticulum. Functional CD8+ and CD4+ T cells were elicited, recognizing epitopes derived from encoded antigens that were presented by several HLA types. Importantly, neoepitopes created by the fusion process were also recognized by CD8+ and CD4+ T cells. This proved that DCs generated ex vivo with TriMix mRNA can induce effector CD8+ and CD4+ T cells from the naive T-cell repertoire of patients with melanoma [Van Nuffel et al. 2012b].
The sequence of four intradermal vaccinations with this TriMix-DC-MEL therapy followed by IFNα2b treatment was studied in another trial [Wilgenhof et al. 2011b]. Adverse events were mild, comprising grade II injection site reactions in all 29 patients and grade II lethargy and fever in two patients. An antigen-specific response could be determined in 51.7% (12/21) patients after the fourth vaccination. One partial remission and five stable diseases (lasting more than 6 months with regression of metastases) were observed in 17 patients with evaluable disease at baseline [Wilgenhof et al. 2011b]. The clinical course of some patients treated with the autologous RNA DC therapy followed the recently defined immune response related criteria, suggesting that the immunotherapy established a new equilibrium between tumor (growth rate) and cancer immunosurveillance [Dunn et al. 2004; Gulley and Drake, 2011; Madan et al. 2012; Wilgenhof et al. 2011a]. Present efforts strive to augment the antitumor effect by combining the RNA DC therapy with the recently approved checkpoint inhibitor ipilimumab and yielded encouraging results [ClinicalTrials.gov identifier: NCT01302496] [Neyns et al. 2012].
The combined intravenous and intradermal administration of TriMix-DC-MEL therapy was also investigated. A particularly favorable outcome was observed in a patient with stage IVc melanoma and lung, lymph node, bone, and liver metastases whose condition was resistant to darcarbazine. A broad CD8+ and CD4+ T-cell response to the MAA could be measured and the patient developed a sustained objective clinical response [Van Nuffel et al. 2012a]. The complete phase Ib trial in 15 patients with pretreated advanced melanoma resulted in two complete and two partial remissions. The objective responders remained progression free for 24+, 28+, 33+, and 34+ months respectively [Wilgenhof et al. 2013]. An immunoanalysis of the clinical trials performed with TriMix-MEL suggested that functional specific CD8+ T cells distribute both to the skin and peripheral blood of patients. However, in some patients the majority of epitopes were only recognized by CD8+ T cells derived from either skin biopsies or peripheral blood, indicating that some compartmentalization might also be significant in the immune response induced by TriMix-DC-MEL therapy [Benteyn et al. 2013; Van Nuffel et al. 2012b].
An important step forward was achieved in a recent analysis that compared direct intranodal injection of TriMix mRNA targeting trp2, WT1 or ovalbumin to the injection of DCs stimulated ex vivo with TriMix mRNA [Van Lint et al. 2012]. Injection into the lymph nodes created a proinflammatory environment with the expression of MHC class II molecules, inflammatory cytokines and chemokines such as IL-6, IL-15, IFNγ, monocyte chemotactic protein 1 and Interferon-inducible protein 10, as well as granzyme B leading to the induction of antigen-specific CD4+ and CD8+ T cells. Contrary to maturation of DCs with the classical cytokine cocktail, maturation induced by TriMix did not hamper uptake of mRNA by CD11c+ DCs. Direct intranodal vaccination with TriMix mRNA was as effective as TriMix-DC therapy in the induction of cytolytic T lymphocytes and therapeutic responses in a variety of different mouse models [Van Lint et al. 2012]. Since clinical benefits have already been reported after TriMix-DC therapy (see above), one may expect that intranodal administration of a TriMix-based mRNA cocktail would match the efficacy of therapies based on autologous DCs manipulated ex vivo with mRNA. This would make this approach particularly interesting and a newly founded biotech, eTheRNA (Prof K. Thielemans, Free University of Brussels, Brussels, Belgium), strives to build on these results.
DC-based vaccination approaches have also been suggested as a way to achieve functional cure of human immunodeficiency virus (HIV) when traditional treatment regimens have first largely reduced viral load [Vanham and Van Gulck, 2012]. The principle feasibility of this idea has already been demonstrated in humans [Van Gulck et al. 2012]. Experiments with mRNA-based vaccines against HIV target antigens encapsulated by lipoplexes prepared from cationic lipids demonstrated that such lipoplex/mRNA vaccines could be used for subcutaneous vaccination [Pollard et al. 2013]. This procedure is reminiscent of the approach already used in the first report on successful vaccination with an mRNA-based vaccine [Martinon et al. 1993]. The TriMix approach described in more detail above demonstrates that recent technological advances have allowed us to transfer knowledge gained by encouraging results with DC vaccinations in the past to more direct administration of novel mRNA vaccines, which could make the benefits of DC vaccination available to a much wider patient population.
Intranodal applications of mRNA-based vaccines
In the logic of this analysis, Sahin and colleagues opted to engineer mRNA molecules that had characteristics allowing direct administration in vivo to circumvent the demands of DC vaccination. The translational efficacy of RNA-encoded antigens was optimized by modifications of the length and structure of the poly(A) tail as well as the 3’ untranslated region (UTR) between open reading frame (ORF) and poly(A)-tail [Holtkamp et al. 2006]. A length of 120 nucleotides of the poly(A) tail that had to end unmasked, that is, not followed by irrelevant nucleotides stemming from the cloning procedure, was reported to be optimal. Furthermore, a 3’ UTR consisting of two sequential β-globin regions cloned head to tail between the coding region and the poly(A) tail each independently enhanced RNA stability and translation [Holtkamp et al. 2006]. A further improvement of antigen presentation was achieved by adding the MHC class I signal peptide to the N terminus and the MHC class I trafficking signal (MITD) to the C terminus of the antigen [Kreiter et al. 2008]. DCs transfected with RNA encoding such antigen–MITD fusion proteins showed a distinctly higher stimulatory capacity than wild-type controls. Interestingly, not only was the presentation of MHC class I in human and murine DCs improved, but also that of class II epitopes. This was interpreted as a result of the mimicking of the dynamic trafficking of MHC molecules in immature and mature DCs. The role of the secretion signal attached to the fusion proteins was emphasized, which might improve antigen processing by a better concert of protein degradation by endoplasmic reticulum adjacent proteasomes and access to the transporter associated with antigen processing (TAP) molecules. Importantly, the improved antigen presentation led to a polyepitopic expansion of CD4+ and CD8+ T cells, which resulted in distinct CD8+ T-cell specificities and a broad and variable antigen-specific CD4+ repertoire.
As the level and duration of antigen expression are critically important to generate sustained antigen-specific immune responses, modifications of the mRNA cap structure were tried to achieve further improvement [Kuhn et al. 2010]. The m27,2′-OGppspG (β-S-ARCA) phosphorothioate caps enhanced RNA stability and translation efficacy particularly well in immature, but not mature DCs. This is consistent with the already mentioned observation by the same group that the maturation status of DCs has an essential impact on the uptake of mRNA [Diken et al. 2011]. Kreiter and colleagues reported that such modified mRNA led to the priming of antigen-specific CD4+ and CD8+ T cells from naive T cells after repeated intranodal administration, but not after subcutaneous, intradermal or near nodal administration. The mRNA was selectively taken up by resident lymphatic DCs, propagated in an immunostimulatory environment and generated memory T cells in addition to cytolytic effector T cells. The immunological response translated into a good antitumor response in different therapeutic mouse models and increased survival substantially [Kreiter et al. 2010]. The activity of this regimen could be further enhanced by pretreatment of mice by a fusion protein of the extracellular domain of human fms-like tyrosine kinase 3 (FLT3) ligand with the heavy chain constant regions 2 and 3 (CH2–CH3 domain) of human immunoglobulin (Ig)-G4 leading to much increased therapeutic effects of the mRNA. Plasmacytoid DCs that were kept in an immature state by the administration of the FLT3 ligand/IgG4 fusion protein thus allowing increased uptake of the mRNA were essential for the effect [Kreiter et al. 2011]. The approach is currently being tested in a phase I dose escalation trial in advanced melanoma, analyzing the safety and tolerability of intranodal administration of a mRNA-based vaccine targeting tumor-associated antigens [ClinicalTrials.gov identifier: NCT01684241]
The potential of such optimized mRNA-based vaccination procedures to develop much more patient-centered cancer immunotherapies became clear in a recent analysis of the genome of murine B16/F10 melanoma cells. It was found that murine B16/F10 melanoma cells harbor around 962 nonsynonymous mutations, 563 of which are located in expressed genes [Castle et al. 2012]. A detailed analysis of a subgroup of 50 mutations revealed that one-third of these mutations might be immunogenic and that 60% of immune responses are preferentially directed against the mutated sequence rather than the wild-type parent gene. It was suggested that an mRNA-based vaccination platform might be well suited to induce a broad immune response against the mutanome of a patient, opening the way to highly individualized cancer treatment [Diken et al. 2013; Kreiter et al. 2012]. Technology breakthroughs such as deep sequencing have allowed us to clarify the genome and mutations therein of an increasing number of human tumors [Biankin et al. 2012; Killela et al. 2013; Vogelstein et al. 2013]. Despite these advances, as of today it is not yet possible to reliably identify the crucial mutations in an individual patient’s tumor genome (Ultan McDermott, Wellcome Trust Sanger Institute, Cambridge, UK, personal communication). However, in the future hopefully this will be possible, by which time mRNA vaccination technology and also the regulatory requirements might have advanced enough to rapidly generate patient individual cancer vaccines.
Achieving enhanced protein expression with RNA-based replicons
To enhance in vivo expression of foreign proteins encoded by nucleotides, the self-amplifying characteristics of certain RNA viruses, most often members of the α virus family were exploited early on [Johanning et al. 1995; Xiong et al. 1989; Zhou et al. 1994]. The structural genes of such RNA viruses are replaced by the genes of interest while the nonstructural proteins are left intact to ensure replication and protein translation. Liljeström and colleagues described a plasmid-encoded replicon system called DREP (DNA replicon) which was based on self-replicating Semliki Forest virus vectors (replicons) to immunize successfully against lethal challenges with influenza [Berglund et al. 1998, 1999]. Other viruses have been utilized as the basis for such replicons too [Anraku et al. 2002; Dubensky et al. 1996; Hariharan et al. 1998; Kirman et al. 2003]. Once the replicon genes including the gene of interest are read from the plasmid, self-amplification of the replicon leads to powerful protein expression allowing a dose-sparing effect over DNA vaccines. The increased immunogenicity of replicon DNA vaccines in addition to enhanced antigen expression levels may reflect the provision of immunostimulatory ligands by the replicating RNA in the transfected cell that activate pattern recognition receptors such as TLR3, RNA-activated protein kinase (PKR), and melanoma differentiation-associated protein 5 (MDA5) or retinoic acid inducible gene I [(RIG-I) [Diebold et al. 2009; Johansson et al. 2012; Leitner et al. 2000, 2003; Pichlmair et al. 2006; Rehwinkel et al. 2010; Schulz et al. 2005]. In an extension of this work, RNA-based replicons were developed (RREPs) as a safer solution, excluding persistence of the plasmid and a possible genomic integration [Fleeton et al. 2000]. Intramuscular delivery of in vitro transcribed naked RREPs could induce protective immune responses in vivo [Fleeton et al. 2001]. DREPs can also induce potent immune responses after intradermal delivery [Berglund et al. 1998]. This could also be demonstrated for RREPs administered intradermally, but not for naked mRNA administered thus. The immune response elicited by RREPs was comparable to that induced by DREPs, however EP could enhance the effects of intradermally administered RREPs twofold and that of DREPs 12-fold. This might reflect the enhanced entry into the nucleus of DREPs after EP. Since EP for intradermal administration is less cumbersome than after intramuscular administration, this might offer a biosafe alternative for nucleotide-based vaccination in the future.
The efficacy of the administration of naked, not encapsulated RNA replicons can be enhanced by the use of viral replicon delivery systems. While these facilitate delivery to target cells and enhance immunogenicity, they encounter the principle problem of vector immunity [Fleeton et al. 2001]. Apparently, though, this might be overcome by increased dosing of the virus. High titers of neutralizing antibodies and elevated T-regulatory-cell levels after repeated administration of an α virus vector expressing the tumor antigen CEA and encapsulated in virus-like replicon particles (VRPs) to patients with metastatic cancer could be overcome to induce clinically relevant CEA-specific T-cell and antibody responses by increased dosing [Morse et al. 2010]. However, in a recently reported phase I dose escalation trial of VRPs expressing prostate-specific membrane antigen (PSMA) in patients with prostate cancer, weak PSMA-specific enzyme-linked immunosorbent assay signals were detected only in the lower of two dose cohorts. Cellular immune responses could not be found in either of the two dose cohorts. Neutralizing antibodies were found in both cohorts which might indicate that dosing was either suboptimal or that vector immunity annihilated an antigen-specific immune response [Slovin et al. 2013].
Alternatives to VRPs have recently been found in clinically suitable delivery systems for siRNA. A self-amplifying α virus derived mRNA vector system termed SAM (self-amplifying messenger RNA) was developed to encode the F protein of respiratory syncytial virus and encapsulated with a synthetic lipid nanoparticle [Geall et al. 2012]. The authors demonstrated that at very low mRNA doses, very high titers of IgG1 antibodies against the F protein as well as IFNγ-producing CD4+ and CD8+ T cells could be induced in mice after intramuscular administration. The results could be replicated in immunogenicity studies in cotton rats in which the nonviral delivery methods provided protection levels comparable to those achieved with vectors that embedded the self-amplifying RNA in VRPs [Geall et al. 2012]. The authors also showed that lipid nanoparticle encapsulated SAM vaccines elicited functional immune responses against antigens from HIV [Geall et al. 2012]. The SAM system allows rapid production of a functional vaccine against novel viral threats within a very short time interval after the gene sequence of the relevant target is published, as demonstrated by the completion of an immunogenic vaccine against influenza strain H7N9 in a matter of days to very few weeks [Hekele et al. 2013]. Apparently, though, the best immunogenicity is achieved when an interval of at least 8 weeks is used between different vaccinations with the SAM technology. Nevertheless, this approach might represent a viable way to use RNA-based replicons for widespread vaccination against a multitude of infectious diseases [Geall et al. 2013].
An interesting, often overlooked field for the application of mRNA-based vaccines is allergy. The incidence of allergy has risen dramatically in recent years and now affects up to 25% of the population in western countries, probably a consequence of a more urban rather than the traditional rural lifestyle that leads to a reduced microbial exposure early in life and reduced expression of genes typical of activation of T helper (Th)-1 lymphocytes [Weiss et al. 2012]. The allergic immune response is consequently characterized by a preponderance of allergen specific Th2 cells which secrete the ‘Th2’-type cytokines IL-4, IL-5, and IL-13. These govern alterations in the B-cell switch that lead to production of allergen-specific IgE antibodies that bind to FcϵR receptors on mast cells and basophilic granulocytes on contact with the allergen [Gould and Sutton, 2008]. IL-5 is key to the induction and propagation of eosinophilic granulocytes that migrate from the mucosal surface where initial contact with allergens occurs in tissues deeper in the airways in a process called the ‘allergic march’, leading to severe allergic pathophysiologies such as asthma [Takatsu and Nakajima, 2008]. In a series of papers, Thalhamer and colleagues established that allergen-specific Th1 polarization induced by DNA vaccines encoding a variety of different allergens could prevent allergy [Bauer et al. 2006; Scheiblhofer et al. 2006a, 2006b; Weiss et al. 2006]. While promising, antiallergy DNA-based vaccines would encounter the same safety considerations as prophylactic vaccines against infectious diseases. However, the same group was able to demonstrate that RNA-replicons were as effective as DNA vaccines in preventing allergies in a series of experiments testing up to 29 different allergens [Roesler et al. 2009]. While RNA replicons appeared initially superior to naked RNA compared on a weight basis, a fivefold dose increase of the naked mRNA annihilated the difference. Thus, mRNA-based vaccines appear as a promising path to prevent allergies and ultimately might even find their way into therapy of allergies [Weiss et al. 2012]. An mRNA-based vaccine would be attractive to encode engineered hypoallergic allergens [Thalhamer et al. 2010; Wallner et al. 2011] and might also profit from recently developed laser microporation devices for painless delivery of prophylactic and therapeutic vaccines to the skin [Scheiblhofer et al. 2013].
The use of nonencapsulated mRNA-based vaccines: the RNActive approach
The approaches described above attempted to increase the effectiveness of mRNA-based vaccines either by improving DC maturation (coadministration of costimulatory molecules, class I or II targeting) or increasing the expression capacity (replicon-based approach). A fundamentally different path was followed by the so-called RNActive technology: the expression capacity of mRNA-encoding proteins was greatly enhanced by sequence modifications of the mRNA and immunogenicity of mRNAs created by complexation with protamine.
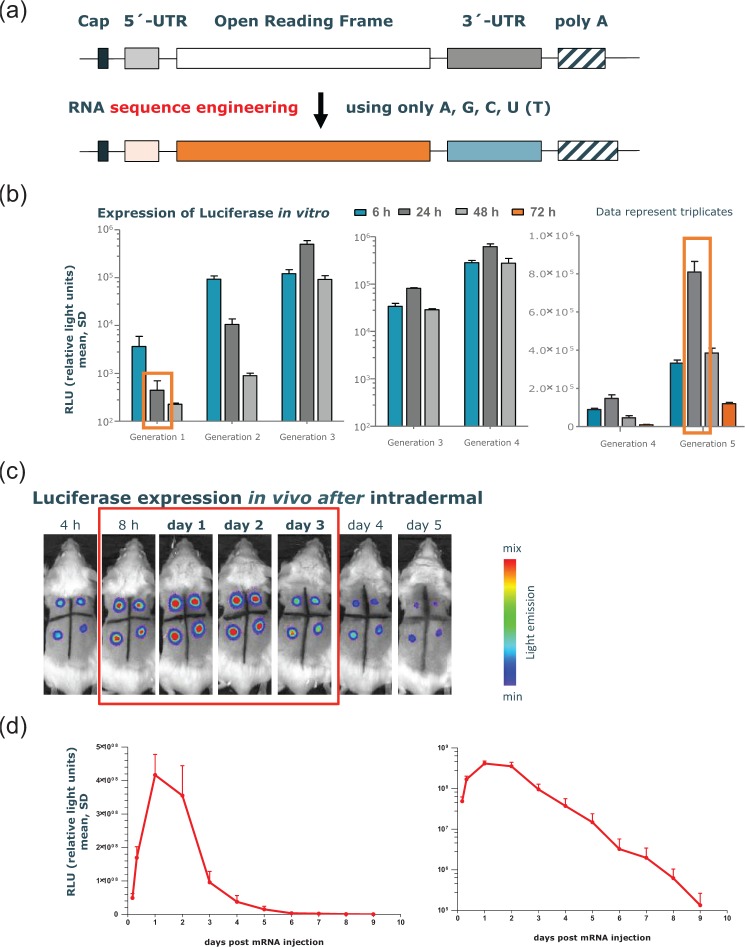
The minimal mRNA structure is characterized by a protein-encoding ORF flanked by two essential elements at the 5’ and 3’ end: the ‘cap’, a 7-methyl-guanosine residue bound to the 5’ end of the RNA via a 5’–5’ triphosphate bond, and the poly(A) tail at the 3’ end [Banerjee, 1980; Wickens, 1990]. Additionally, UTRs at the 5’ (between cap and ORF) and the 3’ end [between ORF and poly(A)-tail] of the ORF affect protein expression [Schlake et al. 2012]. This minimal structure is sequence engineered by use only of the naturally occurring nucleotides A, G, C and U (T) without affecting the primary amino acid sequence encoded by the ORF. The optimization procedure includes experimental procedures for the identification of novel 5’ and 3’ UTRs with favorable impact on mRNA translation and stability, an algorithm optimizing the sequence of the ORF as well as the use of a poly(A)-tail with defined length. Together with ultra-high purification of the mRNA, the expression of luciferase, commonly used as a reporter gene due to its short half life (~2 h), could be increased by four to five orders of magnitude in various test systems (Figure 2). The expression kinetics of encoded proteins were also changed dramatically: peak expression now occurs after 24–48 h with the expression after 72 h matching that of the early time points. Thus the protein expression kinetics of CureVac’s enhanced RNA molecules have started to mimic that of proteins after an influenza virus infection [Julkunen et al. 2001].
Figure 2.
Effects on protein expression by sequence engineering of the principle messenger RNA (mRNA) structure. (a) The classical structure of an mRNA molecule consists of a cap region, followed by an (optional) 5’ untranslated region (UTR), the open reading frame (ORF), an (optional) 3’ UTR, and the poly(A)-tail. Sequence engineering of each subunit of an mRNA molecule with only the naturally occurring nucleotides A, G, C, U that do not affect the primary amino acid sequence encoded by the ORF are the basis of optimized mRNA molecules used in RNActive vaccines. (b) Effect of different generations of sequence-engineered mRNAs (e.g. generated by optimization of the nucleotide content of the ORF or incorporation of different 3’ or 5’ UTRs or combinations thereof) encoding PpLuc produced over the last few years on in vitro expression of luciferase. The mRNA generations encoding Firefly luciferase were electroporated into HeLa cells (generation 1–4) or transfected into human dermal fibroblasts by lipofection (generation 4 and 5) and compared for their in vitro expression of luciferase. The luciferase level was determined at 6, 24 and 48 h, or 72 h post transfection. The dynamic range of the assay does not allow us to compare all mRNA molecules in one experiment. (c) Firefly luciferase encoding mRNA, optimized for translation and stability, was injected intradermally into a BALB/c mouse (four injection sites). The luciferase expression was visualized in the living animal by optical imaging at various time points after mRNA injection and showed maximal protein levels 24–48 h after mRNA injection. (d) Quantitative expression of luciferase over time until 9 days after mRNA injection. Results are shown on a linear scale (left-hand panel) or on a semi-logarithmic scale (right-hand panel). The figure is adapted from Schlake and colleagues [Schlake et al. 2012], details therein.
‘Self adjuvanticity’ was bestowed on this sequence-engineered, expression-enhanced mRNA by complexation with protamine. The mRNA/protamine complexes form larger particles than uncomplexed mRNA (~250–350 nm versus ~50 nm) and activate the immune system in a process involving the endosome resident TLR7 [Fotin-Mleczek et al. 2011; Kallen et al. 2013; Scheel et al. 2005]. In vitro experiments revealed that the uncomplexed mRNA is taken up by an adenosine triphosphate dependent process and could subsequently be detected in cytoplasm and lysosomes [Lorenz et al. 2011]. The final formulation of an RNActive vaccine is obtained by mixing the immunostimulating mRNA/protamine complexes with the antigen-expressing ‘naked’ sequence-engineered mRNA. An optimal ratio between the two components could be established that ensures both good antigen expression and good immunostimulation after intradermal administration [Fotin-Mleczek et al. 2011].
The two-component, self-adjuvanted RNActive vaccines generated by this technology elicit strong and balanced immune responses: Th1 and Th2, humoral and cellular as well as effector and memory responses are induced. Highly effective vaccines could be engineered on this basis that induce powerful CD4+ and CD8+ T-cell responses and work well in different tumor models after repeated, frequent administration [Fotin-Mleczek et al. 2011, 2012] as well as vaccines against infectious diseases such as influenza that induce protective, long-lived humoral immune responses and only require administration as prime, boost [Petsch et al. 2012]. Importantly, in species varying from mice over ferrets to large pigs, RNActive vaccines were effective after the easy intradermal administration and did not require the more complicated intranodal administration [Kreiter et al. 2010, 2011; Van Lint et al. 2012]. In fact, intradermal administration of RNActive vaccines was as immunogenic as intranodal administration of a conventional mRNA vaccine [Kallen et al. 2013].
This difference between RNActive vaccines and other mRNA-based vaccines may result from the enhanced protein expression capacity of RNActive vaccines and a favorable way of activating the immune system. Activation of TLR7/8 proved to be a critical component of a new, largely improved cocktail for DC maturation that led to strong Th1 responses [Spranger et al. 2010, 2012]. Moreover, newly developed small molecule TLR7/8 agonists were shown to colocalize to an MCH class II containing compartment of human plasmacytoid DCs [Iavarone et al. 2011; Russo et al. 2011]. The production of the type I interferons IFNα and IFNβ by these cells appears to be instrumental for generation of strong immune responses, particularly Th1 and memory responses, that are required to reject tumors [Desmet and Ishii, 2012; Diamond et al. 2011]. Activation of TLR7/8 might therefore be a particularly useful way to activate the immune system not just to combat cancer, but also for successful vaccination strategies against chronic infections [Bernstein et al. 2012; Mbow et al. 2010; Walsh et al. 2012]. Furthermore, the expression of different TLRs is cell type specific, meaning that different APCs react specifically to activators such as pathogen-associated molecular patterns or endogenous damage associated molecular patterns (extensively reviewed by Desmet and Ishii) [Desmet and Ishii, 2012], For example, TLR7 expression occurs mostly in plasmacytoid DCs and B cells in humans, while TLR8 is expressed in monocytes, macrophages, and conventional DCs, but not plasmacyotid DCs. Since DC subsets appear to have different preferred localizations that can also be affected by the inflammatory status of a patient [Hartmann et al. 2003, 2006; Naik et al. 2006; Wollenberg et al. 2002], the route of administration might also affect the vaccination efficiency. Translation of experimental results from animal models to humans thus also requires consideration of route of administration used and an analysis of the specific challenges or advantages posed by it.
Clinical experiences with mRNA-based vaccines
Clinical trials that used direct injection of mRNA instead of in vitro treatment of DCs and the subsequent administration of these modified cells to patients were first initiated around 10 years ago. The first trial included 15 patients with melanoma stage III or IV [Weide et al. 2008]. A cDNA library was prepared from the metastatic lesions of patients and transcribed into an mRNA library. The autologous mRNA library was then administered to patients with granulocyte macrophage colony-stimulating factor (GM-CSF) as adjuvant. The repeated injection of the mRNA library vaccine was feasible and safe. The immunoanalysis, hampered by the technological difficulties to measure immune responses against not strictly defined targets, suggested that humoral responses could be induced in four patients and that there might have been transient increases in the frequencies of CD4+ and CD8+ T cells. An objective response could not be observed, but two patients had a mixed response and a favorable clinical course was observed in five patients.
In the next trial, protamine-stabilized mRNAs coding for Melan-A, tyrosinase, gp100, Mage-A1, Mage-A3 and survivin were injected intradermally into 21 patients with metastatic melanoma with GM-CSF as adjuvant [Weide et al. 2009]. In 10 patients keyhole limpet hemocyanin (KLH) was added to the vaccine. No adverse event greater than grade II was observed. Two of four immunologically evaluable patients showed an antigen-specific T-cell response. Interestingly, a decrease of Foxp3+/CD4+ T-regulatory cells was observed in patients also receiving KLH, whereas myeloid suppressor cells (CD11b+HLA-DRlo monocytes) were reduced in the patients not receiving KLH. One of seven patients with measurable disease showed a partial response of lung metastases after 12 vaccinations. A histopathologically proven bone metastasis detected in this patient 16 months after the start of vaccination was surgically removed and this patient remained relapse free.
Thirty patients with stage IV renal cell cancer were treated with naked mRNA coding for the tumor-associated antigens mucin 1 (MUC1), CEA, human epidermal growth factor receptor 2 (Her-2/neu), telomerase, survivin, and melanoma-associated antigen 1 (MAGE-A1) with GM-CSF as adjuvant [Rittig et al. 2011]. The first 14 patients received a less intensive induction schedule than the consecutive 16 patients. Both cohorts received monthly vaccinations afterwards and in both groups the vaccinations were well tolerated. Immunologically evaluable material was available for 17 patients, of which 12 showed an immune response. Six patients in the first and nine in the second cohort had stable disease lasting longer than 3 months; one patient in the first cohort had a confirmed partial response with shrinkage of cervical and mediastinal lymph nodes. One patient in the second cohort who required abdominal paracentesis every other day had a decline of the paracentesis frequency in line with a decline of the tumor marker CA-125 and regression of abdominal tumor sites. Ultimately he remained free of paracentesis for over 3 months.
The first clinical trials with self-adjuvanted RNActive® vaccines (CureVac GmbH, Tübingen, Germany) were performed in patients with advanced castrate-resistant prostate carcinoma and stage IIIB/IV non-small cell lung cancer (NSCLC) that was at least stable after first-line platinum-based chemotherapy or chemoradiation. Both studies suggested that the application of RNActive vaccines to humans was safe. The four tumor associated antigens prostate specific antigen, prostate stem-cell antigen, prostate-specific membrane antigen, and six transmembrane epithelial antigen of the prostate 1 were selected for the first-in-man phase I/IIa study in patients with prostate cancer and designated CV9103 [Kübler et al. 2011]. The NSCLC antigen cocktail CV9201 consisted of five tumor-associated antigens: MAGE-C1, MAGE-C2, NY-ESO-1, survivin, and 5T4 [Sebastian et al. 2011, 2012].
The phase I/IIa prostate-carcinoma study with CV9103 showed an unexpectedly high level of antigen-specific T cells after five intradermal vaccinations administered over a 6-month period in around 80% of immunologically evaluable patients with prostate carcinoma independent of their HLA background [Kübler et al. 2011]. Others have also advocated mRNA-based vaccination as a method to overcome HLA restriction of patients with tumor [Van Nuffel et al. 2012c]. Antigen-specific T-cell immune responses were detected against all antigens independent of their cellular localization. Less clear results were observed for antigen-unspecific B cells that appeared to be increased and natural killer cells that seemed to show a slightly increased activation. The majority of patients showing an antigen-specific immune response reacted against more than one antigen, a phenomenon recently associated with increased survival in a study of a peptide vaccine in patients with renal cell carcinoma [Walter et al. 2012]. While this study was not designed to assess clinical efficacy, individual patients showed interesting clinical courses that might indicate clinical benefit. The overall survival of vaccinees and its correlation with immune responses is presently being analyzed (manuscript in preparation).
Based on these initial results, a controlled phase IIb study has opened enrolment of patients with castrate-resistant prostate carcinoma with asymptomatic or minimally symptomatic metastasis to ascertain the clinical efficacy of CV9104, a further developed version of CV9103, in a systematic manner. The study will employ more frequent vaccination in the induction phase followed by maintenance vaccinations at prolonged intervals. The primary endpoint of this study of around 180 patients in nine European countries is overall survival. Additionally a number of secondary endpoints will investigate the mechanism of action, associated with improved survival and the impact on subsequent therapies [ClinicalTrials.gov identifier: NCT01817738].
Similarly to the prostate carcinoma study, the number of vaccinations in the phase I/IIa trial of patients with NSCLC was limited to five intradermal immunizations, but the more life-threatening disease of these patients required a more intensive vaccination schedule. Similar to CV9103, the NSCLC cocktail CV9201 showed a favorable safety profile. An antigen-specific humoral and cellular immune response was determined in roughly two-thirds of the treated patients. A significant increase of pregerminal center B cells by a factor of at least two was observed in more than half of the patients and associated with an increase in total CD4+ effector T cells during treatment. Together, more than 80% of the treated patients with NSCLC had a detectable antigen-specific immune response or an increase in germinal center B cells despite their heavy pretreatment with platinum-based chemotherapy (Sebastian, manuscript in preparation). Based on these results, a phase Ib study has been launched in patients with metastatic NSCLC to ascertain an assumed synergistic effect between vaccination and radiation [ClinicalTrials.gov identifier: NCT01915524].
Novel options: gene therapy
Wolff and colleagues demonstrated that mRNA can be used to express foreign proteins in vivo [Wolff et al. 1990]. The potential of self-amplifying RNA replicons to improve this was recognized early on [Johanning et al. 1995; Zhou et al. 1994]. However, while self-amplifying RNA replicons drive up protein expression, the much enhanced protein expression might also lead to immunogenic suicidal cell death, a problem that could be aggravated by activation of proteins sensing double-stranded RNA such as double-stranded PKR (dsPKR) [Berglund et al. 1998; Diebold et al. 2009]. This would probably increase the potency of a vaccine, but would clearly be unwanted for the expression of proteins for nonimmunogenic purposes. Mammalian mRNA, however, not only contains the nucleotides A, G, C, U but also modified nucleosides such as 5-methylcytidine and N6 methyladenosine [Kariko and Weissman, 2007]. These as well as another prominent modified nucleoside, pseudouridine (ψ), reduce activation of the immune system through RNA sensors such as TLR3, TLR7, TLR8, and dsPKR [Kariko and Weissman, 2007]. Bacteria lack such modified nucleosides, but they are abundant in viruses where they are thought to contribute to immunoavoidance of the virus [Kariko et al. 2005, 2008; Koski et al. 2004]. Nucleoside modifications can limit activation of 2’-5’-oligoadenylate synthetase and slow cleavage by RNase L. Activation of PKR could be diminished by incorporation of pseudouridine (ψ) into mRNA which resulted in enhanced protein expression, possibly due to the reduced inhibitory impact of immune stimulation on protein translation [Anderson et al. 2010].
Pseudouridine (ψ) was therefore used to prepare mRNA coding for erythropoietin. Injection of the erythropoietin encoding pseudouridine (ψ) mRNA intraperitoneally could substantially increase reticulocyte counts and hematocrit mice [Kariko et al. 2012]. Neither increased cytokine levels nor newly generated antierythropoietin antibodies levels could be measured in mice after repeated injection of the erythropoietin pseouridine (ψ) mRNA. In a pilot study in macaques, elevated erythropoietin levels were found after a single intraperitoneal administration. An internal analysis at CureVac, however, revealed that the enhanced, sequence-engineered mRNA used in the RNActive vaccines does not cause relevant immune activation when the mRNA complex with protamine is absent (unpublished observation). A mRNA thus prepared coding for erythropoietin led to a clear, biologically relevant increase in reticulocytes in mice upon a single intramuscular injection. Corresponding pseudouridine (ψ)-containing mRNA was not any more effective at increasing reticulocytes (unpublished observation). Other researchers replaced a relatively small proportion of only 25% of uridine and cytidine with 2-thiouridine and 5-methyl cytidine which caused a synergistic decrease of mRNA binding to pattern recognition receptors and reduced the release of immunogenic cytokines [Kormann et al. 2011]. A single intramuscular injection of such a modified erythropoietin mRNA raised the hematocrit from 51.5% to 64.2 % after 28 days. Importantly, the authors could also demonstrate that they could rescue a lethal congenital phenotype of a mouse strain caused by lack of surfactant protein B (SP-B) by aerosolic administration of modified SP-B mRNA. A very recent paper showed that direct intramyocardial injection of a synthetic modified RNA (modRNA) encoding human vascular endothelial growth factor A (VEGF-A) with lipofectamine as vehicle increased functionality and long-term survival in the expansion and directed differentiation of endogenous heart progenitors in a mouse myocardial infarction model [Zangi et al. 2013]. VEGF-A prepared as a nonmodified mRNA was ineffective and VEGF-A prepared as a DNA vector had even detrimental effects in some readouts, possibly due to the persisting expression of VEGF-A. In contrast, VEGF-A modRNA showed a pulse-like cardiac expression profile in vivo with a rapid onset of protein expression after a few hours and a peak after 18 h, which might be ideal to mimic the often transient nature of paracrine signals. Conspicuously, the in vivo expression profile of a luciferase modRNA closely resembled that of luciferase encoded by CureVac’s enhanced engineered mRNA (see above) after direct injection without vehicle into the mouse dermis [Figure 2(c)], while the in vitro expression of VEGF-A modRNA appeared to be shorter than that of luciferase expressed in vitro with the enhanced engineered mRNA [Figure 2(b)] [Zangi et al. 2013]. This may indicate that a sequence-engineered, expression-enhanced mRNA prepared from nonmodified nucleotide might match or even surpass synthetic modified mRNA as a delivery system for proteins.
The use of modified mRNA, as described, pursued commercially by companies such as Ethris or ModeRNA, also allowed another breakthrough discovery. Warren and colleagues were able to develop a protocol to induce pluripotent stem cells by the repeated transfection of cells with a cocktail of modified mRNAs based on the four canonical stem-cell defining transcription factors (Yamanaka factors) [Takahashi et al. 2007; Takahashi and Yamanaka, 2006]: KLF4, c-MYC, OCT4, and SOX2, as well as a modRNA coding for LIN28 (KMOSL) [Warren et al. 2010]. The latter had previously been shown to facilitate reprogramming [Hanna et al. 2009; Yu et al. 2007]. RNA-induced pluripotent stem cells (RiPSCs) could be generated from multiple independent derivations and all showed robust expression of the pluripotency-associated transcripts OCT4, SOX2, NANOG, and hTERT. Since modRNAs generated iPSCs at very high efficiency, these results could be the starting point for patient-specific therapies. In contrast to these works, it was also shown that five consecutive transfections of in vitro produced, nonmodified mRNA encoding the transcription factors Oct4, Lin28, Sox2, and Nanog into human foreskin fibroblasts resulted in continuous protein expression that led to the formation of induced pluripotent cell colonies that expressed alkaline phosphatase and several embryonic stem cell markers [Yakubov et al. 2010]. A more recent publication even suggested that activation of inflammatory pathways, in particular TLR3 (a sensor of double-stranded RNA), is required for efficient nuclear reprogramming and the induction of pluripotency [Lee et al. 2012]. Hence, nonmodified mRNA might be an alternative to modified mRNA for the induction of RiPSCs.
Therapies based on RiPSCs will most likely require adaptation to diverse cell types [Warren et al. 2010]. The flexibility of the mRNA format and the transient nature of protein expression mediated by mRNA allows one to experimentally activate developmentally important transcriptional programs by expression of relevant transcription factors in a temporally controlled and stage-specific sequence to direct RiPSC cells to diverse fates [Mandal and Rossi, 2013].
The interest in mRNA-based technologies to express proteins in vivo has been further highlighted by a recent call issued by the Defense Advanced Research Projects Agency (USA) searching for technologies able to express antibodies at biologically active levels in vivo that have no risk of genomic integration.
Conclusion
An mRNA-based approach to express biologically active protein levels has principle advantages over DNA-based approaches. Even a minimal risk of genomic integration can be excluded, mRNA has to cross only one cellular membrane to be active in the cytoplasm, there is no need for a promoter and mRNA is also active in nondividing cells. mRNA constructs can be engineered to reduce and even eliminate vector-induced immunogenicity, thus allowing repeated administrations. Impressive preclinical results have been achieved with mRNA-based vaccines in fields as diverse as oncology, infectious diseases, and allergy. The first clinical studies in oncology suggest that it is possible to translate these preclinical results to humans. mRNA-based vaccines can be produced at low costs in a highly flexible and scalable production process and would constitute a revolutionary, disruptive technology in vaccinology. Equally impressive results have been achieved in stem cell research and diseases caused by gene defects. It appears that mRNA technology has now reached a level where protein or gene replacement therapies based on mRNA become conceivable. Hence, mRNA rather than DNA which was favored for decades will be the basis of a new class of drugs based on nucleotides. This might well be the beginning of a revolution in medicine based on mRNA.
Acknowledgments
We thank Ingmar Hoerr, Mariola Fotin-Mleczek, Elmar Maier and Ulrike Gnad-Vogt for their critical review of the manuscript and Philipp Bastian and Birgit Scheel for their kind support.
Footnotes
Funding: This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
Conflict of interest statement: The authors declare no conflicts of interest in preparing this article.
Contributor Information
Karl-Josef Kallen, CureVac GmbH, Paul Ehrlich Str. 15, 72076 Tübingen, Germany.
Andreas Theß, CureVac GmbH, Tübingen, Germany.
References
- Anderson B., Muramatsu H., Nallagatla S., Bevilacqua P., Sansing L., Weissman D., et al. (2010) Incorporation of pseudouridine into mRNA enhances translation by diminishing PKR activation. Nucleic Acids Res 38: 5884–5892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anraku I., Harvey T., Linedale R., Gardner J., Harrich D., Suhrbier A., et al. (2002) Kunjin virus replicon vaccine vectors induce protective CD8+ T-cell immunity. J Virol 76: 3791–3799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoshi T., Koyama S., Kobiyama K., Akira S., Ishii K.J. (2011) Innate and adaptive immune responses to viral infection and vaccination. Curr Opin Virol 1: 226–232 [DOI] [PubMed] [Google Scholar]
- Bagarazzi M., Yan J., Morrow M., Shen X., Parker R., Lee J., et al. (2012) Immunotherapy against HPV16/18 generates potent TH1 and cytotoxic cellular immune responses. Sci Transl Med 4: 155ra138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee A. (1980) 5′-terminal cap structure in eucaryotic messenger ribonucleic acids. Microbiol Rev 44: 175–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bargou R., Leo E., Zugmaier G., Klinger M., Goebeler M., Knop S., et al. (2008) Tumor regression in cancer patients by very low doses of a T cell-engaging antibody. Science 321: 974–977 [DOI] [PubMed] [Google Scholar]
- Bauer R., Scheiblhofer S., Kern K., Gruber C., Stepanoska T., Thalhamer T., et al. (2006) Generation of hypoallergenic DNA vaccines by forced ubiquitination: preventive and therapeutic effects in a mouse model of allergy. J Allergy Clin Immunol 118: 269–276 [DOI] [PubMed] [Google Scholar]
- Bedikian A., Richards J., Kharkevitch D., Atkins M., Whitman E., Gonzalez R. (2010) A phase 2 study of high-dose Allovectin-7 in patients with advanced metastatic melanoma. Melanoma Res 20: 218–226 [DOI] [PubMed] [Google Scholar]
- Bendtzen K. (2012) Anti-TNF-alpha biotherapies: perspectives for evidence-based personalized medicine. Immunotherapy 4: 1167–1179 [DOI] [PubMed] [Google Scholar]
- Benteyn D., Van Nuffel A., Wilgenhof S., Corthals J., Heirman C., Neyns B., et al. (2013) Characterization of CD8+ T-cell responses in the peripheral blood and skin injection sites of melanoma patients treated with mRNA electroporated autologous dendritic cells (TriMixDC-MEL). Biomed Res Int 2013: 976383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berglund P., Fleeton M., Smerdou C., Liljeström P. (1999) Immunization with recombinant Semliki Forest virus induces protection against influenza challenge in mice. Vaccine 17: 497–507 [DOI] [PubMed] [Google Scholar]
- Berglund P., Smerdou C., Fleeton M., Tubulekas I., Liljeström P. (1998) Enhancing immune responses using suicidal DNA vaccines. Nat Biotechnol 16: 562–565 [DOI] [PubMed] [Google Scholar]
- Bernstein D., Cardin R., Bravo F., Earwood J., Clark J., Li Y., et al. (2012) Topical SMIP-7.7, a toll-like receptor 7 agonist, protects against genital herpes simplex virus-2 disease in the guinea pig model of genital herpes. Antivir Chem Chemother 12 December (Epub ahead of print). [DOI] [PubMed] [Google Scholar]
- Biankin A., Waddell N., Kassahn K., Gingras M., Muthuswamy L., Johns A., et al. (2012) Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature 491: 399–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boczkowski D., Nair S., Snyder D., Gilboa E. (1996) Dendritic cells pulsed with RNA are potent antigen-presenting cells in vitro and in vivo. J Exp Med 184: 465–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonehill A., Heirman C., Tuyaerts S., Michiels A., Zhang Y., van der Bruggen P., et al. (2003) Efficient presentation of known HLA class II-restricted MAGE-A3 epitopes by dendritic cells electroporated with messenger RNA encoding an invariant chain with genetic exchange of class II-associated invariant chain peptide. Cancer Res 63: 5587–5594 [PubMed] [Google Scholar]
- Bonehill A., Tuyaerts S., Van Nuffel A., Heirman C., Bos T., Fostier K., et al. (2008) Enhancing the T-cell stimulatory capacity of human dendritic cells by co-electroporation with CD40L, CD70 and constitutively active TLR4 encoding mRNA. Mol Ther 16: 1170–1180 [DOI] [PubMed] [Google Scholar]
- Breckpot K., Dullaers M., Bonehill A., van Meirvenne S., Heirman C., de Greef C., et al. (2003) Lentivirally transduced dendritic cells as a tool for cancer immunotherapy. J Gene Med 5: 654–667 [DOI] [PubMed] [Google Scholar]
- Breckpot K., Heirman C., Neyns B., Thielemans K. (2004) Exploiting dendritic cells for cancer immunotherapy: genetic modification of dendritic cells. J Gene Med 6: 1175–1188 [DOI] [PubMed] [Google Scholar]
- Castle J., Kreiter S., Diekmann J., Lower M., van de Roemer N., de Graaf J., et al. (2012) Exploiting the mutanome for tumor vaccination. Cancer Res 72: 1081–1091 [DOI] [PubMed] [Google Scholar]
- Christensen C. (1997) The innovator’s dilemma: when new technologies cause great firms to fail (management of innovation and change). Watertown, MA: Harvard Business Review Press [Google Scholar]
- Conry R., LoBuglio A., Wright M., Sumerel L., Pike M., Johanning F., et al. (1995) Characterization of a messenger RNA polynucleotide vaccine vector. Cancer Res 55: 1397–1400 [PubMed] [Google Scholar]
- Davis B., Chang G., Cropp B., Roehrig J., Martin D., Mitchell C., et al. (2001) West Nile virus recombinant DNA vaccine protects mouse and horse from virus challenge and expresses in vitro a noninfectious recombinant antigen that can be used in enzyme-linked immunosorbent assays. J Virol 75: 4040–4047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day K., Sweeny L., Kulbersh B., Zinn K., Rosenthal E. (2013) Preclinical comparison of near-infrared-labeled cetuximab and panitumumab for optical imaging of head and neck squamous cell carcinoma. Mol Imaging Biol 29 May (Epub ahead of print). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desmet C., Ishii K. (2012) Nucleic acid sensing at the interface between innate and adaptive immunity in vaccination. Nat Rev Immunol 12: 479–491 [DOI] [PubMed] [Google Scholar]
- Diamond M., Kinder M., Matsushita H., Mashayekhi M., Dunn G., Archambault J., et al. (2011) Type I interferon is selectively required by dendritic cells for immune rejection of tumors. J Exp Med 208: 1989–2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diebold S., Schulz O., Alexopoulou L., Leitner W., Flavell R., Reis e Sousa C. (2009) Role of TLR3 in the immunogenicity of replicon plasmid-based vaccines. Gene Ther 16: 359–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diken M., Kreiter S., Selmi A., Britten C., Huber C., Tureci O., et al. (2011) Selective uptake of naked vaccine RNA by dendritic cells is driven by macropinocytosis and abrogated upon DC maturation. Gene Ther 18: 702–708 [DOI] [PubMed] [Google Scholar]
- Diken M., Kreiter S., Selmi A., Tureci O., Sahin U. (2013) Antitumor vaccination with synthetic mRNA: strategies for in vitro and in vivo preclinical studies. Methods Mol Biol 969: 235–246 [DOI] [PubMed] [Google Scholar]
- Dubensky T., Jr, Driver D., Polo J., Belli B., Latham E., Ibanez C., et al. (1996) Sindbis virus DNA-based expression vectors: utility for in vitro and in vivo gene transfer. J Virol 70: 508–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn G., Old L., Schreiber R. (2004) The three Es of cancer immunoediting. Annu Rev Immunol 22: 329–360 [DOI] [PubMed] [Google Scholar]
- Ferraro B., Talbott K., Balakrishnan A., Cisper N., Morrow M., Hutnick N., et al. (2013) Inducing humoral and cellular responses to multiple sporozoite and liver stage malaria antigens using pDNA. Infect Immun 81: 3709–3720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnegan G. (2012) Europe risks ‘vaccine fatigue’. Vaccines today 8 October [Google Scholar]
- Fleeton M., Chen M., Berglund P., Rhodes G., Parker S., Murphy M., et al. (2001) Self-replicative RNA vaccines elicit protection against influenza A virus, respiratory syncytial virus, and a tickborne encephalitis virus. J Infect Dis 183: 1395–1398 [DOI] [PubMed] [Google Scholar]
- Fleeton M., Liljeström P., Sheahan B., Atkins G. (2000) Recombinant Semliki Forest virus particles expressing louping ill virus antigens induce a better protective response than plasmid-based DNA vaccines or an inactivated whole particle vaccine. J Gen Virol 81: 749–758 [DOI] [PubMed] [Google Scholar]
- Fotin-Mleczek M., Duchardt K., Lorenz C., Pfeiffer R., Ojkic-Zrna S., Probst J., et al. (2011) Messenger RNA-based vaccines with dual activity induce balanced TLR-7 dependent adaptive immune responses and provide antitumor activity.J Immunother 34: 1–15 [DOI] [PubMed] [Google Scholar]
- Fotin-Mleczek M., Zanzinger K., Heidenreich R., Lorenz C., Thess A., Duchardt K., et al. (2012) Highly potent mRNA based cancer vaccines represent an attractive platform for combination therapies supporting an improved therapeutic effect. J Gene Med 14: 428–439 [DOI] [PubMed] [Google Scholar]
- Garver K., LaPatra S., Kurath G. (2005) Efficacy of an infectious hematopoietic necrosis (IHN) virus DNA vaccine in Chinook Oncorhynchus tshawytscha and sockeye O. nerka salmon. Dis Aquat Organ 64: 13–22 [DOI] [PubMed] [Google Scholar]
- Geall A., Mandl C., Ulmer J. (2013) RNA: The new revolution in nucleic acid vaccines. Semin Immunol 25: 152–159 [DOI] [PubMed] [Google Scholar]
- Geall A., Verma A., Otten G., Shaw C., Hekele A., Banerjee K., et al. (2012) Nonviral delivery of self-amplifying RNA vaccines. Proc Natl Acad Sci U S A 109: 14604–14609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilboa E. (2007) DC-based cancer vaccines. J Clin Invest 117: 1195–1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilboa E. (2012) mRNA leapfrogs DNA to show promise for therapeutic gene transfer. Mol Ther 20: 694–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilboa E., Vieweg J. (2004) Cancer immunotherapy with mRNA-transfected dendritic cells. Immunol Rev 199: 251–263 [DOI] [PubMed] [Google Scholar]
- Gould H., Sutton B. (2008) IgE in allergy and asthma today. Nat Rev Immunol 8: 205–217 [DOI] [PubMed] [Google Scholar]
- Grosenbaugh D., Leard A., Bergman P., Klein M., Meleo K., Susaneck S., et al. (2011) Safety and efficacy of a xenogeneic DNA vaccine encoding for human tyrosinase as adjunctive treatment for oral malignant melanoma in dogs following surgical excision of the primary tumor. Am J Vet Res 72: 1631–1638 [DOI] [PubMed] [Google Scholar]
- Gulley J., Drake C. (2011) Immunotherapy for prostate cancer: recent advances, lessons learned, and areas for further research. Clin Cancer Res 17: 3884–3891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S., Asoyan A., Rabenstein H., Nakano N., Obst R. (2010) Role of antigen persistence and dose for CD4+ T-cell exhaustion and recovery. Proc Natl Acad Sci U S A 107: 20453–20458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna J., Saha K., Pando B., van Zon J., Lengner C., Creyghton M., et al. (2009) Direct cell reprogramming is a stochastic process amenable to acceleration. Nature 462: 595–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hariharan M., Driver D., Townsend K., Brumm D., Polo J., Belli B., et al. (1998) DNA immunization against herpes simplex virus: enhanced efficacy using a Sindbis virus-based vector. J Virol 72: 950–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann E., Graefe H., Hopert A., Pries R., Rothenfusser S., Poeck H., et al. (2006) Analysis of plasmacytoid and myeloid dendritic cells in nasal epithelium. Clin Vaccine Immunol 13: 1278–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann E., Wollenberg B., Rothenfusser S., Wagner M., Wellisch D., Mack B., et al. (2003) Identification and functional analysis of tumor-infiltrating plasmacytoid dendritic cells in head and neck cancer. Cancer Res 63: 6478–6487 [PubMed] [Google Scholar]
- Hekele A., Bertholet S., Archer J., Gibson D., Palladino G., Brito L., et al. (2013) Rapidly produced SAMH vaccine against H7N9 influenza is immunogenic in mice. Emerging Microbes and Infections 2: e52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrickson S., Mempel T., Mazo I., Liu B., Artyomov M., Zheng H., et al. (2008) T cell sensing of antigen dose governs interactive behavior with dendritic cells and sets a threshold for T cell activation. Nat Immunol 9: 282–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoerr I., Obst R., Rammensee H., Jung G. (2000) In vivo application of RNA leads to induction of specific cytotoxic T lymphocytes and antibodies. Eur J Immunol 30: 1–7 [DOI] [PubMed] [Google Scholar]
- Holtkamp S., Kreiter S., Selmi A., Simon P., Koslowski M., Huber C., et al. (2006) Modification of antigen-encoding RNA increases stability, translational efficacy, and T-cell stimulatory capacity of dendritic cells. Blood 108: 4009–4017 [DOI] [PubMed] [Google Scholar]
- Iavarone C., Ramsauer K., Kubarenko A., Debasitis J., Leykin I., Weber A., et al. (2011) A point mutation in the amino terminus of TLR7 abolishes signaling without affecting ligand binding. J Immunol 186: 4213–4222 [DOI] [PubMed] [Google Scholar]
- Johanning F., Conry R., LoBuglio A., Wright M., Sumerel L., Pike M., et al. (1995) A Sindbis virus mRNA polynucleotide vector achieves prolonged and high level heterologous gene expression in vivo. Nucleic Acids Res 23: 1495–1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson D., Ljungberg K., Kakoulidou M., Liljeström P. (2012) Intradermal electroporation of naked replicon RNA elicits strong immune responses. PLoS One 7: e29732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julkunen I., Sareneva T., Pirhonen J., Ronni T., Melen K., Matikainen S. (2001) Molecular pathogenesis of influenza A virus infection and virus-induced regulation of cytokine gene expression. Cytokine Growth Factor Rev 12: 171–180 [DOI] [PubMed] [Google Scholar]
- Kallen K., Heidenreich R., Schnee M., Petsch B., Schlake T., Thess A., et al. (2013) A novel, disruptive vaccination technology: self-adjuvanted RNActive® vaccines. Hum Vaccin Immunother 9: 2263–2276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kariko K., Buckstein M., Ni H., Weissman D. (2005) Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity 23: 165–175 [DOI] [PubMed] [Google Scholar]
- Kariko K., Muramatsu H., Keller J., Weissman D. (2012) Increased erythropoiesis in mice injected with submicrogram quantities of pseudouridine-containing mRNA encoding erythropoietin. Mol Ther 20: 948–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kariko K., Muramatsu H., Welsh F., Ludwig J., Kato H., Akira S., et al. (2008) Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Mol Ther 16: 1833–1840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kariko K., Weissman D. (2007) Naturally occurring nucleoside modifications suppress the immunostimulatory activity of RNA: implication for therapeutic RNA development. Curr Opin Drug Discov Devel 10: 523–532 [PubMed] [Google Scholar]
- Ketterer T., von der Mülbe F., Reidel L., Mutzke T. (2008) Method for purifying RNA on a preparative scale by means of HPLC. Patent WO2008077592. [Google Scholar]
- Kharfan-Dabaja M., Boeckh M., Wilck M., Langston A., Chu A., Wloch M., et al. (2012) A novel therapeutic cytomegalovirus DNA vaccine in allogeneic haemopoietic stem-cell transplantation: a randomised, double-blind, placebo-controlled, phase 2 trial. Lancet Infect Dis 12: 290–299 [DOI] [PubMed] [Google Scholar]
- Killela P., Reitman Z., Jiao Y., Bettegowda C., Agrawal N., Diaz L., Jr, et al. (2013) TERT promoter mutations occur frequently in gliomas and a subset of tumors derived from cells with low rates of self-renewal. Proc Natl Acad Sci U S A 110: 6021–6026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirman J., Turon T., Su H., Li A., Kraus C., Polo J., et al. (2003) Enhanced immunogenicity to Mycobacterium tuberculosis by vaccination with an alphavirus plasmid replicon expressing antigen 85A. Infect Immun 71: 575–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kormann M., Hasenpusch G., Aneja M., Nica G., Flemmer A., Herber-Jonat S., et al. (2011) Expression of therapeutic proteins after delivery of chemically modified mRNA in mice. Nat Biotechnol 29: 154–157 [DOI] [PubMed] [Google Scholar]
- Koski G., Kariko K., Xu S., Weissman D., Cohen P., Czerniecki B. (2004) Cutting edge: innate immune system discriminates between RNA containing bacterial versus eukaryotic structural features that prime for high-level IL-12 secretion by dendritic cells. J Immunol 172: 3989–3993 [DOI] [PubMed] [Google Scholar]
- Kreiter S., Castle J., Tureci O., Sahin U. (2012) Targeting the tumor mutanome for personalized vaccination therapy. Oncoimmunology 1: 768–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreiter S., Diken M., Selmi A., Tureci O., Sahin U. (2011) Tumor vaccination using messenger RNA: prospects of a future therapy. Curr Opin Immunol 23: 399–406 [DOI] [PubMed] [Google Scholar]
- Kreiter S., Selmi A., Diken M., Koslowski M., Britten C., Huber C., et al. (2010) Intranodal vaccination with naked antigen-encoding RNA elicits potent prophylactic and therapeutic antitumoral immunity. Cancer Res 70: 9031–9040 [DOI] [PubMed] [Google Scholar]
- Kreiter S., Selmi A., Diken M., Sebastian M., Osterloh P., Schild H., et al. (2008) Increased antigen presentation efficiency by coupling antigens to MHC class I trafficking signals. J Immunol 180: 309–318 [DOI] [PubMed] [Google Scholar]
- Kübler H., Maurer T., Stenzl A., Feyerabend S., Steiner U., Schostak M., et al. (2011) Final analysis of a phase I/IIa study with CV9103, an intradermally administered prostate cancer immunotherapy based on self-adjuvanted mRNA. J Clin Oncol 29(Suppl.): abstract 4535. [Google Scholar]
- Kuhn A., Diken M., Kreiter S., Selmi A., Kowalska J., Jemielity J., et al. (2010) Phosphorothioate cap analogs increase stability and translational efficiency of RNA vaccines in immature dendritic cells and induce superior immune responses in vivo. Gene Ther 17: 961–971 [DOI] [PubMed] [Google Scholar]
- Kurath G., Garver K., Corbeil S., Elliott D., Anderson E., LaPatra S. (2006) Protective immunity and lack of histopathological damage two years after DNA vaccination against infectious hematopoietic necrosis virus in trout. Vaccine 24: 345–354 [DOI] [PubMed] [Google Scholar]
- Lee J., Sayed N., Hunter A., Au K., Wong W., Mocarski E., et al. (2012) Activation of innate immunity is required for efficient nuclear reprogramming. Cell 151: 547–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitner W., Hwang L., deVeer M., Zhou A., Silverman R., Williams B., et al. (2003) Alphavirus-based DNA vaccine breaks immunological tolerance by activating innate antiviral pathways. Nat Med 9: 33–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitner W., Ying H., Driver D., Dubensky T., Restifo N. (2000) Enhancement of tumor-specific immune response with plasmid DNA replicon vectors. Cancer Res 60: 51–55 [PMC free article] [PubMed] [Google Scholar]
- Lindsay C., Roxburgh P., Graham J. (2013) How do we optimally use cetuximab in first-line treatment for metastatic colorectal cancer? Future Oncol 9: 825–829 [DOI] [PubMed] [Google Scholar]
- Ljung R. (2013) Hemophilia and prophylaxis. Pediatr Blood Cancer 60(Suppl. 1): S23–S26 [DOI] [PubMed] [Google Scholar]
- Lorenz C., Fotin-Mleczek M., Roth G., Becker C., Dam T., Verdurmen W., et al. (2011) Protein expression from exogenous mRNA: uptake by receptor-mediated endocytosis and trafficking via the lysosomal pathway. RNA Biol 8: 627–636 [DOI] [PubMed] [Google Scholar]
- Madan R., Bilusic M., Heery C., Schlom J., Gulley J. (2012) Clinical evaluation of TRICOM vector therapeutic cancer vaccines. Semin Oncol 39: 296–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal P., Rossi D. (2013) Reprogramming human fibroblasts to pluripotency using modified mRNA. Nat Protoc 8: 568–582 [DOI] [PubMed] [Google Scholar]
- Marichal T., Ohata K., Bedoret D., Mesnil C., Sabatel C., Kobiyama K., et al. (2011) DNA released from dying host cells mediates aluminum adjuvant activity. Nat Med 17: 996–1002 [DOI] [PubMed] [Google Scholar]
- Martinon F., Krishnan S., Lenzen G., Magne R., Gomard E., Guillet J., et al. (1993) Induction of virus-specific cytotoxic T lymphocytes in vivo by liposome-entrapped mRNA. Eur J Immunol 23: 1719–1722 [DOI] [PubMed] [Google Scholar]
- Mbow M., De Gregorio E., Valiante N., Rappuoli R. (2010) New adjuvants for human vaccines. Curr Opin Immunol 22: 411–416 [DOI] [PubMed] [Google Scholar]
- Moots R., Naisbett-Groet B. (2012) The efficacy of biologic agents in patients with rheumatoid arthritis and an inadequate response to tumour necrosis factor inhibitors: a systematic review. Rheumatology (Oxford) 51: 2252–2261 [DOI] [PubMed] [Google Scholar]
- Morse M., Hobeika A., Osada T., Berglund P., Hubby B., Negri S., et al. (2010) An alphavirus vector overcomes the presence of neutralizing antibodies and elevated numbers of Tregs to induce immune responses in humans with advanced cancer. J Clin Invest 120: 3234–3241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naik S., Metcalf D., van Nieuwenhuijze A., Wicks I., Wu L., O’Keeffe M., et al. (2006) Intrasplenic steady-state dendritic cell precursors that are distinct from monocytes. Nat Immunol 7: 663–671 [DOI] [PubMed] [Google Scholar]
- Neyns B., Wilgenhof S., Van Nuffel A., Benteyn D., Corthals J., Heirman C, et al. (2012) Phase IB study on combined intradermal (ID) and intravenous (IV) administration of autologous mRNA electroporated dendritic cells (DC) as a single-agent cellular immunotherapy or combined with ipilimumab. J Clin Oncol 30(Suppl.): abstract 2507. [Google Scholar]
- O’Neill D., Adams S., Bhardwaj N. (2004) Manipulating dendritic cell biology for the active immunotherapy of cancer. Blood 104: 2235–2246 [DOI] [PubMed] [Google Scholar]
- Okada T., Miller M., Parker I., Krummel M., Neighbors M., Hartley S., et al. (2005) Antigen-engaged B cells undergo chemotaxis toward the T zone and form motile conjugates with helper T cells. PLoS Biol 3: e150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panjarian S., Iacob R., Chen S., Engen J., Smithgall T. (2013) Structure and dynamic regulation of Abl kinases. J Biol Chem 288: 5443–5450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascolo S. (2004) Messenger RNA-based vaccines. Expert Opin Biol Ther 4: 1285–1294 [DOI] [PubMed] [Google Scholar]
- Pascolo S. (2006) Vaccination with messenger RNA. Methods Mol Med 127: 23–40 [DOI] [PubMed] [Google Scholar]
- Petsch B., Schnee M., Vogel A., Lange E., Hoffmann B., Voss D., et al. (2012) Protective efficacy of in vitro synthesized, specific mRNA vaccines against influenza A virus infection. Nat Biotechnol 30: 1210–1216 [DOI] [PubMed] [Google Scholar]
- Pichlmair A., Schulz O., Tan C., Naslund T., Liljeström P., Weber F., et al. (2006) RIG-I-mediated antiviral responses to single-stranded RNA bearing 5’-phosphates. Science 314: 997–1001 [DOI] [PubMed] [Google Scholar]
- Pollard C., Rejman J., De Haes W., Verrier B., Van Gulck E., Naessens T., et al. (2013) Type I IFN counteracts the induction of antigen-specific immune responses by lipid-based delivery of mRNA vaccines. Mol Ther 21: 251–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintas-Cardama A., Jabbour E. (2013) Considerations for early switch to nilotinib or dasatinib in patients with chronic myeloid leukemia with inadequate response to first-line imatinib. Leuk Res 37: 487–495 [DOI] [PubMed] [Google Scholar]
- Rehwinkel J., Tan C., Goubau D., Schulz O., Pichlmair A., Bier K., et al. (2010) RIG-I detects viral genomic RNA during negative-strand RNA virus infection. Cell 140: 397–408 [DOI] [PubMed] [Google Scholar]
- Rittig S., Haentschel M., Weimer K., Heine A., Muller M., Brugger W., et al. (2011) Intradermal vaccinations with RNA coding for TAA generate CD8+ and CD4+ immune responses and induce clinical benefit in vaccinated patients. Mol Ther 19: 990–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robak T. (2012) Rituximab for chronic lymphocytic leukemia. Expert Opin Biol Ther 12: 503–515 [DOI] [PubMed] [Google Scholar]
- Roesler E., Weiss R., Weinberger E., Fruehwirth A., Stoecklinger A., Mostbock S., et al. (2009) Immunize and disappear-safety-optimized mRNA vaccination with a panel of 29 allergens. J Allergy Clin Immunol 124: 1070–1077 e1071-1011. [DOI] [PubMed] [Google Scholar]
- Russo C., Cornella-Taracido I., Galli-Stampino L., Jain R., Harrington E., Isome Y., et al. (2011) Small molecule toll-like receptor 7 agonists localize to the MHC class II loading compartment of human plasmacytoid dendritic cells. Blood 117: 5683–5691 [DOI] [PubMed] [Google Scholar]
- Sardesai N., Weiner D. (2011) Electroporation delivery of DNA vaccines: prospects for success. Curr Opin Immunol 23: 421–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schadendorf D., Ugurel S., Schuler-Thurner B., Nestle F., Enk A., Brocker E., et al. (2006) Dacarbazine (DTIC) versus vaccination with autologous peptide-pulsed dendritic cells (DC) in first-line treatment of patients with metastatic melanoma: a randomized phase III trial of the DC study group of the DeCOG. Ann Oncol 17: 563–570 [DOI] [PubMed] [Google Scholar]
- Scheel B., Teufel R., Probst J., Carralot J., Geginat J., Radsak M., et al. (2005) Toll-like receptor-dependent activation of several human blood cell types by protamine-condensed mRNA. Eur J Immunol 35: 1557–1566 [DOI] [PubMed] [Google Scholar]
- Scheiblhofer S., Gabler M., Leitner W., Bauer R., Zoegg T., Ferreira F., et al. (2006a) Inhibition of type I allergic responses with nanogram doses of replicon-based DNA vaccines. Allergy 61: 828–835 [DOI] [PubMed] [Google Scholar]
- Scheiblhofer S., Thalhamer J., Weiss R. (2013) Laser microporation of the skin: prospects for painless application of protective and therapeutic vaccines. Expert Opin Drug Deliv 10: 761–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheiblhofer S., Weiss R., Gabler M., Leitner W., Thalhamer J. (2006b) Replicase-based DNA vaccines for allergy treatment. Methods Mol Med 127: 221–235 [DOI] [PubMed] [Google Scholar]
- Schlake T., Thess A., Fotin-Mleczek M., Kallen K. (2012) Developing mRNA-vaccine technologies. RNA Biol 9: 1319–1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz O., Diebold S., Chen M., Naslund T., Nolte M., Alexopoulou L., et al. (2005) Toll-like receptor 3 promotes cross-priming to virus-infected cells. Nature 433: 887–892 [DOI] [PubMed] [Google Scholar]
- Sebastian M., von Boehmer L., Zippelius A., Mayer F., Reck M., Atanackovic, et al. (2011) Messenger RNA vaccination in NSCLC: findings from a phase I/IIa clinical trial. J Clin Oncol 29(Suppl.): abstract 2584. [Google Scholar]
- Sebastian M., von Boehmer L., Zippelius A., Mayer F., Reck M., Atanackovic D, et al. (2012) Messenger RNA vaccination and B-cell responses in NSCLC patients. J Clin Oncol 30(Suppl.): abstract 2573. [Google Scholar]
- Sellam J., Rouanet S., Hendel-Chavez H., Miceli-Richard C., Combe B., Sibilia J., et al. (2013) CCL19, a B-cell chemokine, is related to the decrease of blood memory B-cells and predicts the clinical response to rituximab in rheumatoid arthritis. Arthritis Rheum 65: 2253–2261 [DOI] [PubMed] [Google Scholar]
- Shin H., Wherry E. (2007) CD8 T cell dysfunction during chronic viral infection. Curr Opin Immunol 19: 408–415 [DOI] [PubMed] [Google Scholar]
- Slovin S., Kehoe M., Durso R., Fernandez C., Olson W., Gao J., et al. (2013) A phase I dose escalation trial of vaccine replicon particles (VRP) expressing prostate-specific membrane antigen (PSMA) in subjects with prostate cancer. Vaccine 31: 943–949 [DOI] [PubMed] [Google Scholar]
- Specenier P., Vermorken J. (2013) Cetuximab: its unique place in head and neck cancer treatment. Biologics 7: 77–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spranger S., Frankenberger B., Schendel D. (2012) NOD/scid IL-2Rg(null) mice: a preclinical model system to evaluate human dendritic cell-based vaccine strategies in vivo. J Transl Med 10: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spranger S., Javorovic M., Burdek M., Wilde S., Mosetter B., Tippmer S., et al. (2010) Generation of Th1-polarizing dendritic cells using the TLR7/8 agonist CL075. J Immunol 185: 738–747 [DOI] [PubMed] [Google Scholar]
- Sullenger B., Gilboa E. (2002) Emerging clinical applications of RNA. Nature 418: 252–258 [DOI] [PubMed] [Google Scholar]
- Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., et al. (2007) Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131: 861–872 [DOI] [PubMed] [Google Scholar]
- Takahashi K., Yamanaka S. (2006) Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126: 663–676 [DOI] [PubMed] [Google Scholar]
- Takatsu K., Nakajima H. (2008) IL-5 and eosinophilia. Curr Opin Immunol 20: 288–294 [DOI] [PubMed] [Google Scholar]
- Tang D., DeVit M., Johnston S. (1992) Genetic immunization is a simple method for eliciting an immune response. Nature 356: 152–154 [DOI] [PubMed] [Google Scholar]
- Thalhamer T., Dobias H., Stepanoska T., Proll M., Stutz H., Dissertori O., et al. (2010) Designing hypoallergenic derivatives for allergy treatment by means of in silico mutation and screening. J Allergy Clin Immunol 125: 926–934 e910. [DOI] [PubMed] [Google Scholar]
- Ulmer J., Donnelly J., Parker S., Rhodes G., Felgner P., Dwarki V., et al. (1993) Heterologous protection against influenza by injection of DNA encoding a viral protein. Science 259: 1745–1749 [DOI] [PubMed] [Google Scholar]
- Ulmer J., Mason P., Geall A., Mandl C. (2012) RNA-based vaccines. Vaccine 30: 4414–4418 [DOI] [PubMed] [Google Scholar]
- Vaine M., Wang S., Crooks E., Jiang P., Montefiori D., Binley J., et al. (2008) Improved induction of antibodies against key neutralizing epitopes by human immunodeficiency virus type 1 gp120 DNA prime-protein boost vaccination compared to gp120 protein-only vaccination. J Virol 82: 7369–7378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Bosch G., Van Gulck E., Ponsaerts P., Nijs G., Lenjou M., Apers L., et al. (2006) Simultaneous activation of viral antigen-specific memory CD4+ and CD8+ T-cells using mRNA-electroporated CD40-activated autologous B-cells. J Immunother 29: 512–523 [DOI] [PubMed] [Google Scholar]
- Van Gulck E., Vlieghe E., Vekemans M., Van Tendeloo V., Van De Velde A., Smits E., et al. (2012) mRNA-based dendritic cell vaccination induces potent antiviral T-cell responses in HIV-1-infected patients. AIDS 26: F1–F12 [DOI] [PubMed] [Google Scholar]
- Vanham G., Van Gulck E. (2012) Can immunotherapy be useful as a ’functional cure‘ for infection with human immunodeficiency virus-1? Retrovirology 9: 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Lint S., Goyvaerts C., Maenhout S., Goethals L., Disy A., Benteyn D., et al. (2012) Preclinical evaluation of TriMix and antigen mRNA-based antitumor therapy. Cancer Res 72: 1661–1671 [DOI] [PubMed] [Google Scholar]
- Van Lint S., Heirman C., Thielemans K., Breckpot K. (2013) mRNA: From a chemical blueprint for protein production to an off-the-shelf therapeutic. Hum Vaccin Immunother 9: 265–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Nuffel A., Benteyn D., Wilgenhof S., Corthals J., Heirman C., Neyns B., et al. (2012a) Intravenous and intradermal TriMix-dendritic cell therapy results in a broad T-cell response and durable tumor response in a chemorefractory stage IV-M1c melanoma patient. Cancer Immunol Immunother 61: 1033–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Nuffel A., Benteyn D., Wilgenhof S., Pierret L., Corthals J., Heirman C., et al. (2012b) Dendritic cells loaded with mRNA encoding full-length tumor antigens prime CD4+ and CD8+ T cells in melanoma patients. Mol Ther 20: 1063–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Nuffel A., Wilgenhof S., Thielemans K., Bonehill A. (2012c) Overcoming HLA restriction in clinical trials: Immune monitoring of mRNA-loaded DC therapy. Oncoimmunology 1: 1392–1394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein B., Papadopoulos N., Velculescu V., Zhou S., Diaz L., Jr, Kinzler K. (2013) Cancer genome landscapes. Science 339: 1546–1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallner M., Hauser M., Himly M., Zaborsky N., Mutschlechner S., Harrer A., et al. (2011) Reshaping the Bet v 1 fold modulates T(H) polarization. J Allergy Clin Immunol 127: 1571–1578 e1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh K., Teijaro J., Zuniga E., Welch M., Fremgen D., Blackburn S., et al. (2012) Toll-like receptor 7 is required for effective adaptive immune responses that prevent persistent virus infection. Cell Host Microbe 11: 643–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter S., Weinschenk T., Stenzl A., Zdrojowy R., Pluzanska A., Szczylik C., et al. (2012) Multipeptide immune response to cancer vaccine IMA901 after single-dose cyclophosphamide associates with longer patient survival. Nat Med 18: 1254–1261 [DOI] [PubMed] [Google Scholar]
- Wang S., Kishko M., Wan S., Wang Y., Brewster F., Gray G., et al. (2012) Pilot study on the immunogenicity of paired Env immunogens from mother-to-child transmitted HIV-1 isolates. Hum Vaccin Immunother 8: 1638–1647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren L., Manos P., Ahfeldt T., Loh Y., Li H., Lau F., et al. (2010) Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell 7: 618–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weide B., Carralot J., Reese A., Scheel B., Eigentler T., Hoerr I., et al. (2008) Results of the first phase I/II clinical vaccination trial with direct injection of mRNA. J Immunother 31: 180–188 [DOI] [PubMed] [Google Scholar]
- Weide B., Pascolo S., Scheel B., Derhovanessian E., Pflugfelder A., Eigentler T., et al. (2009) Direct injection of protamine-protected mRNA: results of a phase 1/2 vaccination trial in metastatic melanoma patients. J Immunother 32: 498–507 [DOI] [PubMed] [Google Scholar]
- Weiss R., Scheiblhofer S., Roesler E., Weinberger E., Thalhamer J. (2012) mRNA vaccination as a safe approach for specific protection from type I allergy. Expert Rev Vaccines 11: 55–67 [DOI] [PubMed] [Google Scholar]
- Weiss R., Scheiblhofer S., Thalhamer J. (2006) DNA vaccines for allergy treatment. Methods Mol Med 127: 253–267 [DOI] [PubMed] [Google Scholar]
- Wherry E., Blattman J., Murali-Krishna K., van der Most R., Ahmed R. (2003) Viral persistence alters CD8 T-cell immunodominance and tissue distribution and results in distinct stages of functional impairment. J Virol 77: 4911–4927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickens M. (1990) How the messenger got its tail: addition of poly(A) in the nucleus. Trends Biochem Sci 15: 277–281 [DOI] [PubMed] [Google Scholar]
- Wilgenhof S., Pierret L., Corthals J., Van Nuffel A., Heirman C., Roelandt T., et al. (2011a) Restoration of tumor equilibrium after immunotherapy for advanced melanoma: three illustrative cases. Melanoma Res 21: 152–159 [DOI] [PubMed] [Google Scholar]
- Wilgenhof S., Van Nuffel A., Corthals J., Heirman C., Tuyaerts S., Benteyn D., et al. (2011b) Therapeutic vaccination with an autologous mRNA electroporated dendritic cell vaccine in patients with advanced melanoma. J Immunother 34: 448–456 [DOI] [PubMed] [Google Scholar]
- Wilgenhof S., Van Nuffel A., Benteyn D., Corthals J., Aerts C., Heirman C., et al. (2013) Phase IB study on intravenous synthetic mRNA electroporated dendritic cell immunotherapy in pretreated advanced melanoma patients. Ann Oncol 24: 2686–2693 [DOI] [PubMed] [Google Scholar]
- Wolff J., Malone R., Williams P., Chong W., Acsadi G., Jani A., et al. (1990) Direct gene transfer into mouse muscle in vivo. Science 247: 1465–1468 [DOI] [PubMed] [Google Scholar]
- Wollenberg A., Wagner M., Gunther S., Towarowski A., Tuma E., Moderer M., et al. (2002) Plasmacytoid dendritic cells: a new cutaneous dendritic cell subset with distinct role in inflammatory skin diseases. J Invest Dermatol 119: 1096–1102 [DOI] [PubMed] [Google Scholar]
- Xiong C., Levis R., Shen P., Schlesinger S., Rice C., Huang H. (1989) Sindbis virus: an efficient, broad host range vector for gene expression in animal cells. Science 243: 1188–1191 [DOI] [PubMed] [Google Scholar]
- Yakubov E., Rechavi G., Rozenblatt S., Givol D. (2010) Reprogramming of human fibroblasts to pluripotent stem cells using mRNA of four transcription factors. Biochem Biophys Res Commun 394: 189–193 [DOI] [PubMed] [Google Scholar]
- Yu J., Vodyanik M., Smuga-Otto K., Antosiewicz-Bourget J., Frane J., Tian S., et al. (2007) Induced pluripotent stem cell lines derived from human somatic cells. Science 318: 1917–1920 [DOI] [PubMed] [Google Scholar]
- Zangi L., Lui K., von Gise A., Ma Q., Ebina W., Ptaszek L., et al. (2013) Modified mRNA directs the fate of heart progenitor cells and induces vascular regeneration after myocardial infarction. Nat Biotechnol 8 September (Epub ahead of print). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Berglund P., Rhodes G., Parker S., Jondal M., Liljeström P. (1994) Self-replicating Semliki Forest virus RNA as recombinant vaccine. Vaccine 12: 1510–1514 [DOI] [PubMed] [Google Scholar]