Abstract
Major histocompatibility complex class I (MHC-I) presented peptide epitopes provide a ‘window’ into the changes occurring in a cell. Conventionally, these peptides are generated by proteolysis of endogenously synthesized proteins in the cytosol, loaded onto MHC-I molecules, and presented on the cell surface for surveillance by CD8+ T cells. MHC-I restricted processing and presentation alerts the immune system to any infectious or tumorigenic processes unfolding intracellularly and provides potential targets for a cytotoxic T cell response. Therefore, therapeutic vaccines based on MHC-I presented peptide epitopes could, theoretically, induce CD8+ T cell responses that have tangible clinical impacts on tumor eradication and patient survival. Three major methods have been used to identify MHC-I restricted epitopes for inclusion in peptide-based vaccines for cancer: genetic, motif prediction and, more recently, immunoproteomic analysis. Although the first two methods are capable of identifying T cell stimulatory epitopes, these have significant disadvantages and may not accurately represent epitopes presented by a tumor cell. In contrast, immunoproteomic methods can overcome these disadvantages and identify naturally processed and presented tumor associated epitopes that induce more clinically relevant tumor specific cytotoxic T cell responses. In this review, we discuss the importance of using the naturally presented MHC-I peptide repertoire in formulating peptide vaccines, the recent application of peptide-based vaccines in a variety of cancers, and highlight the pros and cons of the current state of peptide vaccines.
Keywords: cytotoxic T cells, epitopes, immunoproteomics, mass spectrometry, MHC class I, motif prediction, tumor-associated antigen, vaccine
Introduction
Major histocompatibility complex class I (MHC-I) molecules are present on the surface of all nucleated cells and display a large array of peptide epitopes for surveillance by the CD8+ T cell repertoire. CD8+ T cell responses are essential for the control and clearance of viral infections as well as for the elimination of transformed and tumorigenic cells. CD8+ T cells effectively discriminate between healthy and infected or transformed cells via recognition of peptides associated with MHC-I (pMHC-I) molecules present on the cell surface. These peptides, which range from 8 to 11 amino acids in length, are typically derived from protein antigens in the cytosol that arise from conventional as well as cryptic translational reading frames [Shastri et al. 2002]. Classically, proteins synthesized in the cytosol undergo proteasomal degradation and the resulting peptides are transported into the endoplasmic reticulum (ER) and loaded onto MHC-I molecules [Blum et al. 2013]. Peptide loading results in stabilization of the class I molecules and transit to the cell surface where the complexes can be scanned by circulating CD8+ T cells, a process called ‘immune surveillance’. pMHC-I complexes are constantly shuttled to the cell surface; as such, the peptides bound to MHC-I serve as a readout of cellular events, including viral infection or tumorigenesis. This readout has considerable implications for the design and implementation of effective peptide-based cancer vaccines. In this review, we discuss the importance of using MHC-I presented peptide epitopes as a readout of the internal proteome, or working state, of a cell. We review the recent literature on peptide-based therapeutic vaccines for human cancers highlighting the various delivery methods of these vaccines. Finally, we briefly discuss the pros and cons of pMHC-I based therapeutic vaccines and future directions in this field.
Importance of evaluating MHC-I presented peptide antigens for immunotherapy of cancer
Tumor development and maintenance of malignant phenotypes is driven by a wide range of abnormal cellular events including genetic mutations that result in changes in protein coding sequences, deletions, insertions, and the abnormal expression of critical genes involved in cancer transformation pathways [Hanahan and Weinberg, 2000]. Effective therapeutic cancer vaccines must take advantage of these genetic changes by selecting proteins involved in these cancer pathways in order to induce tumor-specific T cell responses (Figure 1). Identification of new tumor antigens, in general, is limited by certain aspects of the currently available technologies. For example, differential genomic and proteomic approaches identify over- and under-expressed proteins but are unable to identify very low abundant proteins that are often processed and presented by the MHC-I molecules as the true recognition targets for T cells. Indeed, the level of protein expression does not always correlate with MHC processing and presentation in cancer [Shastri et al. 2002]. Therefore, the most appropriate method for identifying truly relevant tumor-associated antigenic peptides is to analyze those actually presented by the MHC-I molecules on tumor cells. Described as ‘nature’s gene chip’ by Shastri and colleagues [Shastri et al. 2002], the peptides displayed by MHC-I molecules represent the ever-changing proteome of the cell, in normal as well as in disease states, that could serve as targets for the CD8+ T cell repertoire. In addition, the MHC-I antigen presentation pathway incorporates cryptic antigenic peptides encoded by alternate reading frames generated by novel translational mechanisms [Starck and Shastri, 2011] and from prespliced mRNAs via a noncanonical translation mechanisms [Apcher et al. 2013], which makes the pMHC-I cellular state specific (i.e. tumor). Therefore, surveying peptides presented by the MHC-I molecules on the cell surface will reveal novel T cell targets for potential immune intervention as tumors have a distinct surface expression of peptides compared with their normal counterparts [Fortier et al. 2008]. Analysis of the peptide repertoire associated with the MHC-I molecules of cancer cells therefore provides a source for new tumor antigens for development of cancer immunotherapy (reviewed by Admon and colleagues [Admon et al. 2003]) and these antigens may serve as targets for the most difficult to treat tumors. Furthermore, the antigens identified by their MHC-I association on tumor cells should be tumor specific. Although normal tissues may express the antigen-coding genes, due to the differences in the regulation of expression and proteasomal processing, normal tissues in general do not present these antigenic epitopes in association with MHC-I molecules [Fortier et al. 2008]. Due to the lack of presentation of the epitopes in the context of MHC molecules in normal cells, the cytotoxic T lymphocytes (CTLs) do not recognize normal tissues, limiting the risk of autoimmunity [Hanada et al. 2004].
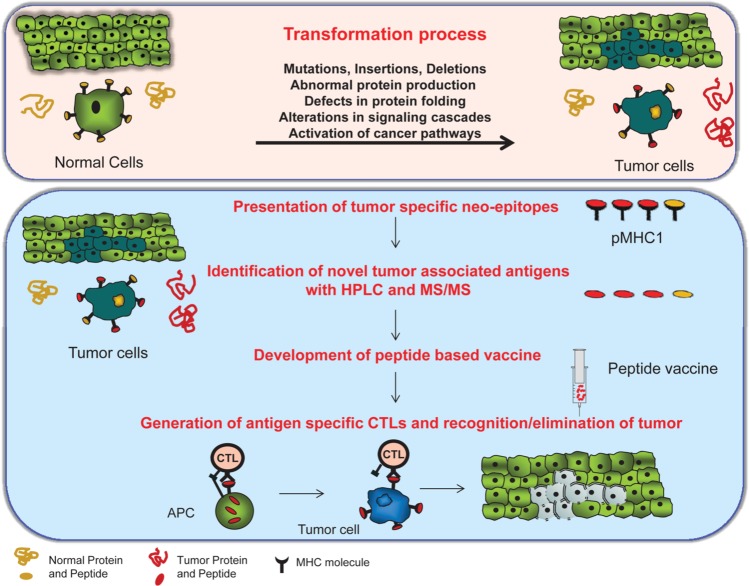
Figure 1.
pMHC-1 (peptides associated with major histocompatibility complex class I) antigens for immunotherapy of cancer. Top: Transformation and subsequent tumorigenesis can be driven by multiple processes, generating abnormal proteins that are available for processing and presentation by the class I machinery. Bottom: The peptide epitopes generated by proteolysis of the abnormal proteins are attractive targets for peptide based vaccines as they represent the epitopes naturally presented by the tumor cell. These ‘neo-epitopes’ are isolated from tumor cells, identified by immunoproteomic methods, validated, and incorporated into a peptide-based vaccine to generate tumor-specific T cell responses.
In the human immune system, MHC-I molecules are referred to as human leukocyte antigens (HLAs). Within the MHC, located on chromosome six, are three different genetic loci that encode MHC-I molecules; these molecules are referred to as HLA-A, HLA-B, and HLA-C. The genes encoded at each of these loci are extremely polymorphic, and thus, different individuals within the population express different class I MHC molecules on the surface of their cells. In addition, each MHC-I molecule has distinct peptide-binding capabilities determined, in part, by the amino acid composition that makes up the peptide-binding groove. Interestingly, peptides generated by the antigen processing machinery may bind to more than one HLA molecule. This property has allowed the categorization of MHC molecules into HLA supertypes, groups of HLA molecules that present at least one shared epitope. MHC-I associated peptides that have been found to bind to one member of the MHC allele supertype family (Al for example) are thought to be likely to bind to other members of the same supertype family (A32 for example) [Sidney et al. 2008]. As we will explain later, this could have considerable ramifications for peptide-based vaccination strategies.
The large number of pMHC-I complexes expressed at the cell surface combined with multiple pathways to generate epitopes provides a great resource for identifying physiologically and clinically relevant tumor-specific antigens (TSA) or tumor-associated antigens (TAAs). Undoubtedly, an examination of the peptides complexed with MHC-I molecules will reveal novel and highly immunogenic epitopes capable of inducing effective CD8+ T cell responses. However, despite a growing body of literature indicating that CD8+ T cells are naturally activated during an antitumor response [Traversari et al. 1992; Marincola et al. 1996; Nagorsen et al. 2000], these antitumor T cell responses often fail to eradicate tumors, in part due to suppression in the local tumor environment [Woo et al. 2001; Mougiakakos et al. 2010] and/or T cell induced exhaustion from continual antigen stimulation [Wherry, 2011; Baitsch et al. 2012]. Nevertheless, generating tumor-specific T cells capable of inducing tumor regression and/or elimination is a tangible possibility. Stimulating the expansion of new T cells through vaccination and/or reversing the exhaustion phenotypes of CD8+ T cells are both attractive and feasible methods to generate robust antitumor responses [Parmiani et al. 2002; Baitsch et al. 2012; Sliwkowski and Mellman, 2013]. To this end, therapeutic vaccination has the capability to induce tumor-specific T cell responses to a number of TSAs and/or TAAs at once.
Current methods for identifying T cell epitopes for inclusion in peptide cancer vaccines
Currently, one of the major limitations in the development of cancer vaccines is the lack of clearly defined tumor antigens that are capable of being recognized by T cells. The definition of such antigens on tumors could provide the basis for a therapeutic vaccine, or for the stimulation of more effective CTLs for adoptive immunotherapy. One of the first methods used to identify tumor-specific peptides capable of binding to MHC molecules involved transfecting cDNA generated from tumor cells into recipient antigen-presenting cells. In this genetic approach, the proteins expressed from cDNA transfection would be translated and processed into epitopes that could load onto MHC-I molecules. Using this technique, an HLA-A1 restricted epitope from MAGE-1 [Van Der Bruggen et al. 1991], an HLA-A2 restricted epitope from tyrosinase [Brichard et al. 1993], and an HLA-A2 restricted epitope from MART-1 [Kawakami et al. 1994] were identified and capable of inducing robust CD8+ T cell responses in melanoma. This approach was, and to some extent still is, an attractive technique because cDNA can be transfected into cells expressing different MHC molecules allowing for a more broad characterization of tumor-specific peptides. However, genetic approaches to identifying tumor-specific epitopes are not without drawbacks. First, any differences in protein expression between the cDNA transfected cell and the tumor cell from which the cDNA was derived may alter the balance of antigen processing and presentation. This may generate more pMHC-I complexes on the transfected cell than on the natural tumor cell and, potentially, more robust T cell responses. In these cases, the stimulatory impact of these epitopes would be overestimated. Differences in the ability of antigen presenting cells to post-translationally modify proteins may also impact epitope discovery. Skipper and colleagues demonstrated that an epitope generated from tyrosinase is modified, changing an asparagine to aspartic acid which generates a more robust CD8+ T cell response, despite no differences in peptide binding to HLA-A2 [Skipper et al. 1996]. Finally, and perhaps more importantly, cDNA expression in different cell types may not generate physiologically relevant epitopes. It is known that different cell types have different levels of proteolytic activity [Delamarre et al. 2005; Savina et al. 2006]and therefore, epitopes generated in the antigen-presenting cells expressing the cDNA may not be the same as those generated in the tumor cell itself.
A second method to identify potential MHC-I binding peptides from already known tumor antigens (identified by differential genomic and proteomic methods) is motif prediction: using pMHC-I binding algorithms that estimate how well peptides will bind to a specific MHC-I molecule [Schultze and Vonderheide, 2001; Shastri et al. 2002; Admon et al. 2003]. These algorithms are based on patterns obtained from peptides known to bind MHC-I molecules with scores being assessed by evaluating specific anchor residues between the peptide and MHC binding groove. Predictions can be further honed by examining potential proteasomal cleavage events in the parent protein [Nussbaum et al. 2001; Stevanovic, 2002], thus creating an ‘optimal’ epitope. Using an epitope prediction technique, Fisk and colleagues identified 19 peptides within the Her-2 protein sequence that were predicted to bind to HLA-A2 molecules [Fisk et al. 1995]. Interestingly, only one peptide was able to induce tumor-specific CD8+ mediated lysis for all CTL lines tested. Similarly, epitope binding predictions led to the identification of an HLA-A2 restricted epitope from MUC-1 that is presented on a variety of tumors [Brossart et al. 1999] and HLA-A3 restricted epitopes derived from carcinoembryonic antigen (CEA) and Her2/neu [Kawashima et al. 1999]. In fact, the epitopes identified from CEA and Her-2/neu bind to multiple HLA alleles in the A3 superfamily, suggesting that these peptides could overcome some differences in HLA expression in patient-to-patient comparisons [Kawashima et al. 1999]. Since these initial studies, peptide-binding algorithms have been used in an attempt to predict epitopes from virtually all known tumor antigens, including p53 [Papadopoulos et al. 1999], MAGE [Akiyama et al. 2012], HCA587 [Li et al. 2005], TRAG-3 [Zhu et al. 2003], and ALK [Ait-Tahar et al. 2006]. However, like the genetic approach to identifying epitopes, peptide prediction algorithms are not reliable. One major reason is that prediction algorithms do not accurately represent what occurs in an antigen presenting cell. Predicted ‘binders’ to MHC-I may not be generated due to proteolytic events or may not efficiently stimulate CD8+ T cells [Fisk et al. 1995] (also found in the present authors’ unpublished observations). Similarly, those peptides not predicted to bind to a specific HLA molecule with high affinity may in fact induce productive T cell responses. In addition, motif prediction methods may be limited in identifying subdominant epitopes which are likely to escape tolerance mechanism [Thomas et al. 2007]. Comparison of the motif prediction method with direct mass spectrometry analysis of endogenously presented epitopes isolated from virus-infected cells revealed a high number of predicted epitopes were not processed and presented by the infected cells [Zhong et al. 2003]. These findings indicate that the complexity of the motif predicted epitopes combined with CD8+ T cell-based screening of functional epitopes may miss hidden subdominant epitopes.
In the last decade, direct identification of MHC-I presented epitopes from tumors or infected cells has emerged as an alternate to the motif prediction method, a process termed immunoproteomics [Purcell and Gorman, 2004]. The analytical challenge lies in the discrimination between the tumor-related peptides among a majority of nondisease-related peptides that are presented on the cell surface [De Jong, 1998]. This could be overcome by cancer specific database search of the identified peptides to select those that are derived from tumorigenesis pathway involved proteins [Hanahan and Weinberg, 2000]. Immunoproteomic analysis is generally based on the isolation of the MHC-peptide complexes from tumor cells and elution of the bound peptides from the MHC molecules followed by offline high-performance liquid chromatography (HPLC) fractionation [Rotzschke et al. 1990; Falk et al. 1991] and online HPLC fractionation combined with mass spectrometry [Hunt et al. 1992; Di Marzo Veronese et al. 1996; Van Els et al. 2000; Berzofsky et al. 2001; Hickman et al. 2003; Lemmel et al. 2004]. The peptides are then validated by both in vitro and in vivo assays. Elution of peptides from both mouse and human MHC-I molecules identified MHC-I restricted epitopes from tumor (i.e. P815 and JY cells) [Falk et al. 1991] and influenza-infected cells [Rotzschke et al. 1990] and nine HLA-A2 restricted epitopes from the human B-cell lymphoblastoid line C1R.A21 [Hunt et al. 1992]. Since these pioneering studies, our group [Shetty et al. 2011, 2012; Testa et al. 2012a, 2012b] and others [Skipper et al. 1999; Hogan et al. 2003, 2004; Zarling et al. 2006; Hawkins et al. 2008; Feyerabend et al. 2009; Haen and Rammensee, 2013] have applied this technique to identify naturally processed epitopes from various tumor or infected cells capable of inducing CD8+ T cell responses. This immunoproteomic approach to epitope identification has significant advantages. First and foremost, naturally processed epitopes present on the surface of tumor cells represent the most clinically relevant targets for vaccination or immunotherapy design. Differences in protein expression levels and antigen processing are minimized greatly in comparison with other identification techniques. Second, the same tumor cell sample can be used to identify epitopes that will bind to multiple MHC-I alleles, either via superfamily mapping or the use of allele specific antibodies during the discovery process. Importantly however, after identification of the epitope, validation must ensure that the epitopes are not present on normal tissues either by similar immunoproteomic analysis or cellular assays demonstrating no CD8+ T cell reactivity to normal cells.
Peptide-based vaccines in the clinical setting
Peptide-based vaccines have enjoyed minimal success thus far in the clinical setting. In this section, we will review recent developments in peptide-based cancer vaccines for a select number of malignancies focusing on peptide composition and the antitumor immune response generated. To date, most of the peptide-based vaccines tested in the late stage clinical studies include peptides identified by motif prediction methodology with fewer exceptions mainly in melanoma and renal carcinoma.
Melanoma
The vast majority of research into peptide-based therapeutic vaccines has centered on melanoma, as there are many well described MHC-I restricted epitopes available for testing. An epitope derived from the MAGE-1 protein was the first to be tested in a peptide-based clinical trial. Although epitope specific CD8+ T cells could be generated and expanded in vitro post-vaccination, no clinical responses in patients were observed [Hu et al. 1996]. Despite these results, this study was important as it reinforced the idea that CD8+ T cells could be induced to generate an antitumor response. More recent studies of peptide-based (most of them identified by immunoproteomic methods) vaccines have utilized a multi-epitope approach in order to induce a broader range of T cell specificities and potentially overcome the problem of antigen loss variants that arise during cancer progression [Admon et al. 2003; Slingluff, 2011]. In a randomized phase II clinical trial of patients with stage IIB to IV melanoma, Slingluff and colleagues compared the effectiveness of a 12 versus a 4 MHC-I peptide-based vaccine [Slingluff et al. 2007]. Vaccines contained tetanus helper peptide, granulocyte-macrophage colony-stimulating factor (GM-CSF), and Montanide ISV-51 and were given intradermal (i.d.) and subcutaneous (s.c.) CD8+ T cell responses induced after vaccination with the 12 peptide vaccine were more broad and robust as characterized by CD8+ interferon (IFN)-γ secretion, however no clinical efficacy was observed in either vaccine [Slingluff et al. 2007]. Importantly however, the data demonstrated that multiple peptides could be injected safely and at the same site with no effect on competition for class I binding. In contrast, clinical efficacy was observed in a trial of a three peptide vaccine given s.q. with Montanide ISV-51 and containing GM-CSF, IFNα2b, or both [Kirkwood et al. 2009]. Data from 115 patients with stage IV melanoma were analyzed and demonstrated that functional responses to the peptides (as judged by IFN-γ secretion) were correlated with a roughly 8-month increase in overall survival with two complete remission and six partial remission cases, with no differences observed between any of the cytokine groups [Kirkwood et al. 2009]. The inclusion of cytokines in vaccines needs to be explored further in order to enhance the antitumor effectiveness of the CD8+ T cells. Indeed much of the data to date indicates that certain cytokines, at least at the doses used currently, are not effective at enhancing antitumor responses and in fact decrease CD4+ and CD8+ T cell responses [Slingluff et al. 2009] and may induce the accumulation of regulatory T cells (TREGs) [Block et al. 2011].
Because CD4+ T cell responses can potentiate CD8+ T cell responses, multiepitope vaccines may also need to include CD4+ activating peptides. Slingluff and colleagues monitored CD4+ and CD8+ T cell responses in 175 patients with stage IV melanoma after administration of a 12 peptide vaccine alone, with tetanus peptide, with 6 confirmed melanoma helper epitopes, or a vaccine of the 6 melanoma helper epitopes alone [Slingluff et al. 2013]. Vaccines were administered i.d. and s.q. emulsified in Montanide ISV-51. Although including tetanus helper peptide in vaccines enhanced CD8+ T cell responses, it did not have any impact on overall survival. In direct contrast, including melanoma-specific helper peptides did not enhance CD8+ T cell responses but was associated with increase in survival [Slingluff et al. 2013]. This data suggest that absolute numbers of CD8+ T cells might not be the most appropriate way of assessing vaccine-induced responses and that there exists an optimal ratio between the CD8+ and CD4+ compartment for effective antitumor responses. Nevertheless, continuous exploration of vaccine strategies to incorporate class II epitopes is of high priority.
Dendritic cells (DCs) are considered one of the most important cells in initiating an immune response and as such have received much attention in designing peptide-based vaccines for cancers. Lesterhuis and colleagues evaluated the ability of a peptide pulsed DC vaccine to induce clinical responses in metastatic melanoma patients [Lesterhuis et al. 2011]. DCs were generated from peripheral blood mononuclear cells (PBMCs) and pulsed with tyrosinase and wildtype gp100 peptides or modified versions with higher binding affinity to HLA-A2 and injected intravenously (i.v.) and i.d. into patients. Although clinical responses were limited, 2 out of 27 patients had responses lasting at least 8 months [Lesterhuis et al. 2011]. Oshita and colleagues evaluated DC induced clinical responses in a phase II trial of metastatic melanoma patients. Melanoma-specific HLA-A2 and HLA-A24 peptides were loaded onto DCs generated from patient blood and administered subcutaneously (s.c.) over a period of 5 months [Oshita et al. 2012]. A total of 18 (out of 24; 75%) patients mounted specific CD8+ T cell responses as assessed by IFN-γ ELISpot and the majority of these patients had TH1 type cytokine skewing. Despite most patients progressing clinically, six patients experienced stable disease and one patient experienced a partial response.
Colon cancer
In contrast to melanoma vaccines, peptide vaccines for colorectal cancer have typically relied on a single peptide injected with adjuvant, usually Montanide ISA-51. In 2004, an HLA-A24 restricted CD8+ T cell epitope from the survivin protein, called survivin-2B80-88, was injected s.q. into patients with colon cancer [Tsuruma et al. 2004]. No adjuvant appeared to be used so it is not surprising that no clinical response were observed except for a minor increase in survivin tetramer positive CD8+ T cells in a handful of patients. Building off of this study, the group then combined survivin peptide with Montanide ISA-51 with or without IFN-α in patients with unresectable colon cancer [Kameshima et al. 2011]. Of the five patients that received only peptide and Montanide, one had stable disease. In contrast, four out of the eight patients receiving peptide and Montanide with IFN-α had stable disease that was accompanied by decreased levels of the colon cancer tumor marker CEA [Kameshima et al. 2011]. Other peptide-based vaccines have been tested clinically but these do not induce CD8+ T cell responses. Notably, vaccination of patients with an extended p53 peptide induced sustained CD4+ T cell responses [Speetjens et al. 2009] that were enhanced (i.e. higher levels of IFN-γ) when administered with IFN-α [Zeestraten et al. 2013].
DC-based vaccines for colon cancer have also been tested in the clinical setting. In a phase I/II clinical trial, Kavanagh and colleagues evaluated the ability of matured DCs to activate CD8+ T cells in colon cancer patients [Kavanagh et al. 2007]. DCs were pulsed with peptides derived from CEA, Her2-neu, MAGE-2, and MAGE-3 and injected over a period of 3 weeks. Only 3 out of 21 patients made specific CD8+ T cell responses that were directed at a single CEA epitope, though expansion of other peptide specific T cells was observed after in vitro T cell stimulation [Kavanagh et al. 2007]. Despite the ability to induce T cell responses, no significant clinical benefits were observed. Lesterhuis and colleagues also evaluated DCs as a vaccine candidate comparing peptide pulsing with mRNA electroporation [Lesterhuis et al. 2010]. DCs were pulsed with the CEA peptide CAP-1 or electroporated with CEA mRNA and delivered i.d. and i.v. a total of three times. A total of 8 out of 11 patients receiving peptide pulsed DCs mounted a CD8+ T cell response detectable by tetramer staining compared with 2 out of 5 patients in the electroporated group [Lesterhuis et al. 2010]. This latter study reinforces the need to identify naturally processed epitopes presented on tumor cells as it is not clear that the electroporated cells generated the CAP-1 epitope efficiently.
Breast cancer
Tsuruma and colleagues tested a survivin peptide vaccine with or without Montanide ISA-51 in a phase I trial of patients with breast cancer [Tsuruma et al. 2008]. As in previous studies, no clinical responses were observed, but the four patients receiving the peptide with Montanide vaccine had more survivin tetramer positive CD8+ T cells with one patient making a specific, IFN-γ functional response [Tsuruma et al. 2008]. A more common target of breast cancer peptide vaccines is the Her2-neu antigen. Two recent phase I or phase II clinical trials evaluated immune responses after vaccination of the E75 or GP2 peptide vaccine in HLA-A2 expressing patients with disease-free breast cancer. Together, the studies indicated that both the E75 and GP2 epitopes were immunogenic, induced epitope specific CD8+ T cells [Carmichael et al. 2010; Mittendorf et al. 2012] and, in a subset of patients, potentially prolong disease-free survival states [Mittendorf et al. 2012]. Multi-epitope breast cancer vaccines have also been tested in clinical trials. A mixture of 12 HLA-A2 restricted epitopes identified by the immunoproteomic method in ovarian cancers [Ramakrishna et al. 2003] was combined with Montanide ISA-51 and GM-CSF and delivered s.q. and i.d. into patients with resected breast cancer [Morse et al. 2011]. Patients that received a high-dose vaccine made broader CD8+ T cell responses than patients that received a low-dose vaccine (as assessed by IFN-γ secretion in an ELISpot assay; >9 responses in high dose, 0–4 response in low dose) suggesting that a multi-epitope vaccine can induce specific T cell responses, but that the effectiveness of these may depend on dose of peptide given.
Generating Her2-neu specific T cell responses in breast cancer is also possible via DC-based vaccines. Patients with confirmed ductal carcinoma in situ (DCIS) were injected with DCs pulsed with a group of Her-2/neu peptides (six MHC class II peptides and two MHC class I restricted peptides) [Sharma et al. 2012]. A total of 85% of patients enrolled had detectable CD4+ and CD8+ T cell responses to the vaccine, and it seems likely that these responses led to a decrease in Her2-neu expression in these patients [Sharma et al. 2012] although a decrease in antigen expression is not necessarily indicative of complete elimination of the cancer. In a second study of patients with DCIS, DCs were pulsed with a mixture of class I and II binding peptides, matured in vitro with IFN-γ and lipopolysaccharide (LPS), and injected into the patient [Koski et al. 2012]. This immunization strategy resulted in functional (IFNγ secreting) CD8+ T cells in 11/13 patients expressing the HLA-A2 allele and functional CD4+ T cell responses in 22/25 patients enrolled in the study.
Renal cancer
Renal cell carcinoma (RCC) is one of the most common types of cancers that occur in the adult population with metastatic RCC having a 5-year survival rate of less than 10% [Schrader et al. 2006]. Vascular endothelial growth factor receptor 1 (VEGFR1) plays a key role in the progression of RCC and therefore peptides derived from this protein could serve as an attractive target for T cell based therapies. To this end, Yoshimura and colleagues investigated the effectiveness of a two-peptide VEGFR1 vaccine (one HLA-A2 and one HLA-A24 restricted peptide) delivered s.q. in Montanide ISA-51 [Yoshimura et al. 2013]. A total of 15 out of 18 patients had specific CD8+ T cell responses, complete with IFN-γ secretion. Clinically, two patients had a partial response and nine patients had stable disease for at least 5 months [Yoshimura et al. 2013]. Using the immunoproteomic approach, Walter and colleagues identified nine HLA-A2 restricted epitopes from RCC patient samples [Walter et al. 2012]. These epitopes were incorporated into a vaccine called IMA901 that was synthesized and injected i.d. along with GM-CSF into patients with RCC. CD8+ T cell responses to multiple antigens were associated with control of the disease. Further, inclusion of cyclophosphamide 3 days before IMA901 injection prolonged survival and reduced the number of regulatory T cells [Walter et al. 2012]. This latter point is critical: because TREGs are well represented in the tumor microenvironment, peptide-based vaccines may need a TREG depleting step prior to injection or other modulation of the anti-inflammatory environment by concomitant cytokine treatment. However, not all cytokines are ideal in this application. In trials of DC-based vaccines combined with interleukin (IL)-2 administration, TREGs were induced to significantly higher levels than before treatment, albeit transiently [Lemoine et al. 2009; Berntsen et al. 2010].
Other malignancies
Peptide-based vaccines have also been evaluated in many other clinical settings. In a phase 1 clinical trial, 15 HLA-A2+ patients with stage III–IV non-small cell lung cancer were vaccinated with a peptide vaccine derived from indoleamine 2,3 dioxygenase (IDO) [Zeeberg Iversen et al. 2013]. A total of 6 out of 15 of the patients had stable disease and overall survival was increased ~18 months compared with HLA-A2-negative patients who were unvaccinated. Sawada and colleagues demonstrated that vaccination of patients with hepatocellular carcinoma using a peptide derived from glypican-3 resulted in CD8+ T cell expansion with an improvement in overall survival in patients with robust GPC3 responses [Sawada et al. 2012]. In phase I clinical studies, a multi-epitope-based vaccine demonstrated CD8+ T cell responses and delay in progression of disease in ovarian and breast [Morse et al. 2011] and prostate cancer [Berinstein et al. 2012]. Finally, a multi-epitope vaccination approach was used in a phase I trial of patients with biliary tract cancer and resulted in a detectable clinical response in six of the nine patients [Aruga et al. 2013].
Advantages and disadvantages of peptide vaccines: where do we go from here?
Overall, the data discussed above indicate that peptide vaccines are capable of inducing robust CD8+ T cell responses that, in some cases, provide clinical benefit to patients. Peptide based vaccines have significant advantages as a cancer immunotherapy option. First, these vaccines are flexible in their design and can accommodate many peptide epitopes in a single dose. This allows for multiple MHC-I epitopes to be included to initiate a T cell response. This is an important feature because not all individuals share the same MHC alleles; peptides that bind to single alleles (i.e. HLA-A2 or HLA-A24) and peptides that bind to multiple alleles (i.e. HLA-A2 and HLA-A24) can be included in the same formulation. Thus, a vaccine derived from naturally processed peptides can be given to individuals with a wide diversity in their MHC alleles and still be effective. Second, a multi-epitope vaccine may protect against tumor resistance due to antigen downregulation by inducing a more broad, oligoclonal response. Although multiple epitopes from a single antigen have been identified and might overcome HLA restriction (i.e. MAGE-n [Zhang et al. 2010], survivin [Tsuruma et al. 2008; Shen et al. 2013], and CEA [Nukaya et al. 1999; Keogh et al. 2001]), it is important that the epitopes included in such a vaccine be derived from different parent proteins. This not only will increase the clonality of the T cell response but also prevent tumor cells from downregulating a single protein and escaping the T cell response induced by the vaccine. Finally, peptide-based vaccines can also incorporate MHC class II restricted epitopes to activate CD4+ T cells and/or B cell epitopes to activate T helper and antibody-mediated responses. Together, a complete adaptive immune response could prove to be a more effective and robust way by which to eliminate tumors. While a protein-based vaccine might be attractive for similar reasons, antigen processing can be markedly different from cell to cell. Downregulation of proteasomal subunits, including the IFN-γ inducible immunoproteasome, occurs in numerous cancers, such as B cell lymphoma and breast cancer [Seliger et al. 2000]. This downregulation alters the cleavage specificities of the tumor proteasome; therefore, epitopes generated in antigen presenting cells that process the protein vaccine via a ‘normal’ proteasome may not accurately reflect epitopes generated by the class I machinery of tumor cells thereby limiting the effectiveness of the CD8+ T cell response. Despite these advantages, peptide-based vaccine strategies are not without their downfalls. First and foremost, in order for the vaccine to be effective the tumors must be expressing the antigens included in the vaccine formulation. Ideally, the tumors should be presenting the epitopes included in the vaccine, which is a major reason for using an immunoproteomic approach for the discovery and selection of antigens in vaccine development. Second, peptide-based vaccination has been shown to induce the accumulation of immunosuppressive regulatory T cells [Lemoine et al. 2009; Berntsen et al. 2010; Block et al. 2011] which would limit vaccine utility in vivo. Finally, in some instances peptide vaccines may not be enough to eradicate tumors from patients, depending on staging of the disease. Importantly, potential solutions exist to prevent or mitigate each of these limitations.
In addition to identifying novel peptides, there are several avenues of research needed to improve the effectiveness of peptide vaccines. First, it is possible that improvements in adjuvant technology will enhance the T cell responses generated during vaccination. One active area of research in this regard is including TLR agonists in vaccine formulations, as these have been shown to heighten protective immune responses [Mahla et al. 2013]. Second, inclusion of cytokines in the vaccine formulation to enhance the immune responses may also improve vaccine effectiveness. As described above, cytokines included in some formulations induced the formation of TREGs [Lemoine et al. 2009; Berntsen et al. 2010; Block et al. 2011]. It will be critical to understand the appropriate cytokines or adjuvants in the form of antigen delivery (i.e. viral or bacterial vector or biodegradable nanoparticle based) to include that will enhance responses without inducing an immunosuppressive environment. Along these lines, and perhaps most critical to inducing effective response after vaccination, is determining how to limit the formation of TREGS either by including a cytokine or adjuvant in the vaccine or via pretreatment with certain drugs as demonstrated by Walter and colleagues [Walter et al. 2012].
Peptide-based vaccines, despite their limited effectiveness to date, have shown promise and progress in the clinic. Identifying novel and perhaps more immunogenic peptides through an immunoproteomics approach combined with a better understanding of adjuvant and cytokine therapy should result in more clinically effective vaccine regimens.
Acknowledgments
The authors wish to thank Mr Aykan Karabudak for his critical editing of the article.
Footnotes
Funding: The work was supported by the Immunotope corporate funding source.
Conflict of interest statement: None of the authors have relevant financial interests related to this manuscript.
Contributor Information
Joseph D. Comber, Immunotope, Inc., Pennsylvania Biotechnology Center, USA
Ramila Philip, Immunotope, Inc., Pennsylvania Biotechnology Center, 3805 Old Easton Road, Doylestown, PA 18902, USA.
References
- Admon A., Barnea E., Ziv T. (2003) Tumor antigens and proteomics from the point of view of the major histocompatibility complex peptides. Mol Cell Proteomics 2: 388–398 [DOI] [PubMed] [Google Scholar]
- Ait-Tahar K., Cerundolo V., Banham A., Hatton C., Blanchard T., Kusec R., et al. (2006) B and CTL responses to the Alk protein in patients with Alk-positive ALCL. Int J Cancer 118: 688–695 [DOI] [PubMed] [Google Scholar]
- Akiyama Y., Komiyama M., Nakamura Y., Iizuka A., Oshita C., Kume A., et al. (2012) Identification of novel MAGE-A6- and MAGE-A12-derived HLA-A24-restricted cytotoxic T lymphocyte epitopes using an in silico peptide-docking assay. Cancer Immunol Immunother 61: 2311–2319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apcher S., Millot G., Daskalogianni C., Scherl A., Manoury B., Fahraeus R. (2013) Translation of pre-spliced RNAS in the nuclear compartment generates peptides for the MHC class I pathway. Proc Natl Acad Sci U S A 110: 17951–17956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aruga A., Takeshita N., Kotera Y., Okuyama R., Matsushita N., Ohta T., et al. (2013) Long-term vaccination with multiple peptides derived from cancer-testis antigens can maintain a specific T cell response and achieve disease stability in advanced biliary tract cancer. Clin Cancer Res 19: 2224–2231 [DOI] [PubMed] [Google Scholar]
- Baitsch L., Fuertes-Marraco S., Legat A., Meyer C., Speiser D. (2012) The three main stumbling blocks for anticancer T cells. Trends Immunol 33: 364–372 [DOI] [PubMed] [Google Scholar]
- Berinstein N., Karkada M., Morse M., Nemunaitis J., Chatta G., Kaufman H., et al. (2012) First-in-man application of a novel therapeutic cancer vaccine formulation with the capacity to induce multi-functional T cell responses in ovarian, breast and prostate cancer patients. J Transl Med 10: 156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berntsen A., Brimnes M., Thor Straten P., Svane I. (2010) Increase of circulating CD4+CD25HIGHFOXP3+ regulatory T cells in patients with metastatic renal cell carcinoma during treatment with dendritic cell vaccination and low-dose interleukin-2. J Immunother 33: 425–434 [DOI] [PubMed] [Google Scholar]
- Berzofsky J., Ahlers J., Belyakov I. (2001) Strategies for designing and optimizing new generation vaccines. Nat Rev Immunol 1: 209–219 [DOI] [PubMed] [Google Scholar]
- Block M., Suman V., Nevala W., Kottschade L., Creagan E., Kaur J., et al. (2011) Pilot study of granulocyte-macrophage colony-stimulating factor and interleukin-2 as immune adjuvants for a melanoma peptide vaccine. Melanoma Res 21: 438–445 [DOI] [PubMed] [Google Scholar]
- Blum J., Wearsch P., Cresswell P. (2013) Pathways of antigen processing. Annu Rev Immunol 31: 443–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brichard V., Van Pel A., Wolfel T., Wolfel C., De Plaen E., Lethe B., et al. (1993) The tyrosinase gene codes for an antigen recognized by autologous cytolytic T lymphocytes on HLA-A2 melanomas. J Exp Med 178: 489–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brossart P., Heinrich K., Stuhler G., Behnke L., Reichardt V., Stevanovic S., et al. (1999) Identification of HLA-A2-restricted T cell epitopes derived from the MUC1 tumor antigen for broadly applicable vaccine therapies. Blood 93: 4309–4317 [PubMed] [Google Scholar]
- Carmichael M., Benavides L., Holmes J., Gates J., Mittendorf E., Ponniah S., et al. (2010) Results of the first phase 1 clinical trial of the Her-2/neu peptide (GP2) vaccine in disease-free breast cancer patients: United States Military Cancer Institute Clinical Trials Group Study I-04. Cancer 116: 292–301 [DOI] [PubMed] [Google Scholar]
- De Jong A. (1998) Contribution of mass spectrometry to contemporary immunology. Mass Spectrom Rev 17: 311–335 [DOI] [PubMed] [Google Scholar]
- Delamarre L., Pack M., Chang H., Mellman I., Trombetta E. (2005) Differential lysosomal proteolysis in antigen-presenting cells determines antigen fate. Science 307: 1630–1634 [DOI] [PubMed] [Google Scholar]
- Di Marzo Veronese F., Arnott D., Barnaba V., Loftus D., Sakaguchi K., Thompson C., et al. (1996) Autoreactive cytotoxic T lymphocytes in human immunodeficiency virus type 1-infected subjects. J Exp Med 183: 2509–2516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk K., Rotzschke O., Stevanovic S., Jung G., Rammensee H. (1991) Allele-specific motifs revealed by sequencing of self-peptides eluted from MHC molecules. Nature 351: 290–296 [DOI] [PubMed] [Google Scholar]
- Feyerabend S., Stevanovic S., Gouttefangeas C., Wernet D., Hennenlotter J., Bedke J., et al. (2009) Novel multi-peptide vaccination in HLA-A2+ hormone sensitive patients with biochemical relapse of prostate cancer. Prostate 69: 917–927 [DOI] [PubMed] [Google Scholar]
- Fisk B., Blevins T., Wharton J., Ioannides C. (1995) Identification of an immunodominant peptide of Her-2/neu protooncogene recognized by ovarian tumor-specific cytotoxic T lymphocyte lines. J Exp Med 181: 2109–2117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortier M., Caron E., Hardy M., Voisin G., Lemieux S., Perreault C., et al. (2008) The MHC class I peptide repertoire is molded by the transcriptome. J Exp Med 205: 595–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haen S., Rammensee H. (2013) The repertoire of human tumor-associated epitopes - identification and selection of antigens and their application in clinical trials. Curr Opin Immunol 25: 277–283 [DOI] [PubMed] [Google Scholar]
- Hanada K., Yewdell J., Yang J. (2004) Immune recognition of a human renal cancer antigen through post-translational protein splicing. Nature 427: 252–256 [DOI] [PubMed] [Google Scholar]
- Hanahan D., Weinberg R. (2000) The hallmarks of cancer. Cell 100: 57–70 [DOI] [PubMed] [Google Scholar]
- Hawkins O., Vangundy R., Eckerd A., Bardet W., Buchli R., Weidanz J., et al. (2008) Identification of breast cancer peptide epitopes presented by HLA-A*0201. J Proteome Res 7: 1445–1457 [DOI] [PubMed] [Google Scholar]
- Hickman H., Luis A., Bardet W., Buchli R., Battson C., Shearer M., et al. (2003) Cutting edge: class I presentation of host peptides following HIV infection. J Immunol 171: 22–26 [DOI] [PubMed] [Google Scholar]
- Hogan K., Coppola M., Gatlin C., Thompson L., Shabanowitz J., Hunt D., et al. (2003) Identification of a shared epitope recognized by melanoma-specific, HLA-A3-restricted cytotoxic T lymphocytes. Immunol Lett 90: 131–135 [DOI] [PubMed] [Google Scholar]
- Hogan K., Coppola M., Gatlin C., Thompson L., Shabanowitz J., Hunt D., et al. (2004) Identification of novel and widely expressed cancer/testis gene isoforms that elicit spontaneous cytotoxic T-lymphocyte reactivity to melanoma. Cancer Res 64: 1157–1163 [DOI] [PubMed] [Google Scholar]
- Hu X., Chakraborty N., Sporn J., Kurtzman S., Ergin M., Mukherji B. (1996) Enhancement of cytolytic T lymphocyte precursor frequency in melanoma patients following immunization with the MAGE-1 peptide loaded antigen presenting cell-based vaccine. Cancer Res 56: 2479–2483 [PubMed] [Google Scholar]
- Hunt D., Henderson R., Shabanowitz J., Sakaguchi K., Michel H., Sevilir N., et al. (1992) Characterization of peptides bound to the class I MHC molecule HLA-A2.1 by mass spectrometry. Science 255: 1261–1263 [DOI] [PubMed] [Google Scholar]
- Kameshima H., Tsuruma T., Torigoe T., Takahashi A., Hirohashi Y., Tamura Y., et al. (2011) Immunogenic enhancement and clinical effect by type-I interferon of anti-apoptotic protein, survivin-derived peptide vaccine, in advanced colorectal cancer patients. Cancer Sci 102: 1181–1187 [DOI] [PubMed] [Google Scholar]
- Kavanagh B., Ko A., Venook A., Margolin K., Zeh H., Lotze M., et al. (2007) Vaccination of metastatic colorectal cancer patients with matured dendritic cells loaded with multiple major histocompatibility complex class I peptides. J Immunother 30: 762–772 [DOI] [PubMed] [Google Scholar]
- Kawakami Y., Eliyahu S., Sakaguchi K., Robbins P., Rivoltini L., Yannelli J., et al. (1994) Identification of the immunodominant peptides of the MART-1 human melanoma antigen recognized by the majority of HLA-A2-restricted tumor infiltrating lymphocytes. J Exp Med 180: 347–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashima I., Tsai V., Southwood S., Takesako K., Sette A., Celis E. (1999) Identification of HLA-A3-restricted cytotoxic T lymphocyte epitopes from carcinoembryonic antigen and Her-2/neu by primary in vitro immunization with peptide-pulsed dendritic cells. Cancer Res 59: 431–435 [PubMed] [Google Scholar]
- Keogh E., Fikes J., Southwood S., Celis E., Chesnut R., Sette A. (2001) Identification of new epitopes from four different tumor-associated antigens: recognition of naturally processed epitopes correlates with HLA-A*0201-binding affinity. J Immunol 167: 787–796 [DOI] [PubMed] [Google Scholar]
- Kirkwood J., Lee S., Moschos S., Albertini M., Michalak J., Sander C., et al. (2009) Immunogenicity and antitumor effects of vaccination with peptide vaccine+/-granulocyte-monocyte colony-stimulating factor and/or IFN-Alpha2b in advanced metastatic melanoma: Eastern Cooperative Oncology Group Phase II Trial E1696. Clin Cancer Res 15: 1443–1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koski G., Koldovsky U., Xu S., Mick R., Sharma A., Fitzpatrick E., et al. (2012) A novel dendritic cell-based immunization approach for the induction of durable Th1-polarized anti-Her-2/neu responses in women with early breast cancer. J Immunother 35: 54–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmel C., Weik S., Eberle U., Dengjel J., Kratt T., Becker H., et al. (2004) Differential quantitative analysis of MHC ligands by mass spectrometry using stable isotope labeling. Nat Biotechnol 22: 450–454 [DOI] [PubMed] [Google Scholar]
- Lemoine F., Cherai M., Giverne C., Dimitri D., Rosenzwajg M., Trebeden-Negre H., et al. (2009) Massive expansion of regulatory T cells following interleukin 2 treatment during a phase I–II dendritic cell-based immunotherapy of metastatic renal cancer. Int J Oncol 35: 569–581 [DOI] [PubMed] [Google Scholar]
- Lesterhuis W., De Vries I., Schreibelt G., Schuurhuis D., Aarntzen E., De Boer A., et al. (2010) Immunogenicity of dendritic cells pulsed with CEA peptide or transfected with CEA MRNA for vaccination of colorectal cancer patients. Anticancer Res 30: 5091–5097 [PubMed] [Google Scholar]
- Lesterhuis W., Schreibelt G., Scharenborg N., Brouwer H., Gerritsen M., Croockewit S., et al. (2011) Wild-type and modified Gp100 peptide-pulsed dendritic cell vaccination of advanced melanoma patients can lead to long-term clinical responses independent of the peptide used. Cancer Immunol Immunother 60: 249–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Wang Y., Chen J., Wu H., Chen W. (2005) Identification of a new HLA-A*0201-restricted CD8+ T cell epitope from hepatocellular carcinoma-associated antigen HCA587. Clin Exp Immunol 140: 310–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahla R., Reddy M., Prasad D., Kumar H. (2013) Sweeten PAMPs: role of sugar complexed PAMPs in innate immunity and vaccine biology. Front Immunol 4: 248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marincola F., Rivoltini L., Salgaller M., Player M., Rosenberg S. (1996) Differential anti-MART-1/MELANA CTL activity in peripheral blood of HLA-A2 melanoma patients in comparison to healthy donors: evidence of in vivo priming by tumor cells. J Immunother Emphasis Tumor Immunol 19: 266–277 [DOI] [PubMed] [Google Scholar]
- Mittendorf E., Clifton G., Holmes J., Clive K., Patil R., Benavides L., et al. (2012) Clinical trial results of the Her-2/neu (E75) vaccine to prevent breast cancer recurrence in high-risk patients: from US Military Cancer Institute Clinical Trials Group Study I-01 and I-02. Cancer 118: 2594–2602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse M., Secord A., Blackwell K., Hobeika A., Sinnathamby G., Osada T., et al. (2011) MHC class I-presented tumor antigens identified in ovarian cancer by immunoproteomic analysis are targets for T cell responses against breast and ovarian cancer. Clin Cancer Res 17: 3408–3419 [DOI] [PubMed] [Google Scholar]
- Mougiakakos D., Choudhury A., Lladser A., Kiessling R., Johansson C. (2010) Regulatory T cells in cancer. Adv Cancer Res 107: 57–117 [DOI] [PubMed] [Google Scholar]
- Nagorsen D., Keilholz U., Rivoltini L., Schmittel A., Letsch A., Asemissen A., et al. (2000) Natural T cell response against MHC class I epitopes of epithelial cell adhesion molecule, Her-2/neu, and carcinoembryonic antigen in patients with colorectal cancer. Cancer Res 60: 4850–4854 [PubMed] [Google Scholar]
- Nukaya I., Yasumoto M., Iwasaki T., Ideno M., Sette A., Celis E., et al. (1999) Identification of HLA-A24 epitope peptides of carcinoembryonic antigen which induce tumor-reactive cytotoxic T lymphocyte. Int J Cancer 80: 92–97 [DOI] [PubMed] [Google Scholar]
- Nussbaum A., Kuttler C., Hadeler K., Rammensee H., Schild H. (2001) PAPROC: a prediction algorithm for proteasomal cleavages available on the WWW. Immunogenetics 53: 87–94 [DOI] [PubMed] [Google Scholar]
- Oshita C., Takikawa M., Kume A., Miyata H., Ashizawa T., Iizuka A., et al. (2012) Dendritic cell-based vaccination in metastatic melanoma patients: phase II clinical trial. Oncol Rep 28: 1131–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadopoulos K., Hesdorffer C., Suciu-Foca N., Hibshoosh H., Harris P. (1999) Wild-type P53 epitope naturally processed and presented by an HLA-B haplotype on human breast carcinoma cells. Clin Cancer Res 5: 2089–2093 [PubMed] [Google Scholar]
- Parmiani G., Sensi M., Castelli C., Rivoltini L., Anichini A. (2002) T cell response to unique and shared antigens and vaccination of cancer patients. Cancer Immun 2: 6. [PubMed] [Google Scholar]
- Purcell A., Gorman J. (2004) Immunoproteomics: mass spectrometry-based methods to study the targets of the immune response. Mol Cell Proteomics 3: 193–208 [DOI] [PubMed] [Google Scholar]
- Ramakrishna V., Ross M., Petersson M., Gatlin C., Lyons C., Miller C., et al. (2003) Naturally occurring peptides associated with HLA-A2 in ovarian cancer cell lines identified by mass spectrometry are targets of HLA-A2-restricted cytotoxic T cells. Int Immunol 15: 751–763 [DOI] [PubMed] [Google Scholar]
- Rotzschke O., Falk K., Deres K., Schild H., Norda M., Metzger J., et al. (1990) Isolation and analysis of naturally processed viral peptides as recognized by cytotoxic T cells. Nature 348: 252–254 [DOI] [PubMed] [Google Scholar]
- Savina A., Jancic C., Hugues S., Guermonprez P., Vargas P., Moura I., et al. (2006) NOX2 controls phagosomal Ph to regulate antigen processing during crosspresentation by dendritic cells. Cell 126: 205–218 [DOI] [PubMed] [Google Scholar]
- Sawada Y., Yoshikawa T., Nobuoka D., Shirakawa H., Kuronuma T., Motomura Y., et al. (2012) Phase I trial of a glypican-3-derived peptide vaccine for advanced hepatocellular carcinoma: immunologic evidence and potential for improving overall survival. Clin Cancer Res 18: 3686–3696 [DOI] [PubMed] [Google Scholar]
- Schrader A., Varga Z., Hegele A., Pfoertner S., Olbert P., Hofmann R. (2006) Second-line strategies for metastatic renal cell carcinoma: classics and novel approaches. J Cancer Res Clin Oncol 132: 137–149 [DOI] [PubMed] [Google Scholar]
- Schultze J., Vonderheide R. (2001) From cancer genomics to cancer immunotherapy: toward second-generation tumor antigens. Trends Immunol 22: 516–523 [DOI] [PubMed] [Google Scholar]
- Seliger B., Maeurer M., Ferrone S. (2000) Antigen-processing machinery breakdown and tumor growth. Immunol Today 21: 455–464 [DOI] [PubMed] [Google Scholar]
- Sharma A., Koldovsky U., Xu S., Mick R., Roses R., Fitzpatrick E., et al. (2012) Her-2 pulsed dendritic cell vaccine can eliminate Her-2 expression and impact ductal carcinoma in situ. Cancer 118: 4354–4362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shastri N., Schwab S., Serwold T. (2002) Producing nature’s gene-chips: the generation of peptides for display by MHC class I molecules. Annu Rev Immunol 20: 463–493 [DOI] [PubMed] [Google Scholar]
- Shen H., Shao H., Chen X., Wu F., Wang H., Huang Z., et al. (2013) Identification of a novel HLA-A2-restricted mutated survivin epitope and induction of specific anti-HCC CTLs that could effectively cross-recognize wild-type survivin antigen. Cancer Immunol Immunother 62: 393–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shetty V., Nickens Z., Testa J., Hafner J., Sinnathamby G., Philip R. (2012) Quantitative immunoproteomics analysis reveals novel MHC class I presented peptides in cisplatin-resistant ovarian cancer cells. J Proteomics 75: 3270–3290 [DOI] [PubMed] [Google Scholar]
- Shetty V., Sinnathamby G., Nickens Z., Shah P., Hafner J., Mariello L., et al. (2011) MHC class I-presented lung cancer-associated tumor antigens identified by immunoproteomics analysis are targets for cancer-specific T cell response. J Proteomics 74: 728–743 [DOI] [PubMed] [Google Scholar]
- Sidney J., Peters B., Frahm N., Brander C., Sette A. (2008) HLA class I supertypes: a revised and updated classification. BMC Immunol 9: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skipper J., Gulden P., Hendrickson R., Harthun N., Caldwell J., Shabanowitz J., et al. (1999) Mass-spectrometric evaluation of HLA-A*0201-associated peptides identifies dominant naturally processed forms of CTL epitopes from MART-1 and Gp100. Int J Cancer 82: 669–677 [DOI] [PubMed] [Google Scholar]
- Skipper J., Hendrickson R., Gulden P., Brichard V., Van Pel A., Chen Y., et al. (1996) An HLA-A2-restricted tyrosinase antigen on melanoma cells results from posttranslational modification and suggests a novel pathway for processing of membrane proteins. J Exp Med 183: 527–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slingluff C., Jr (2011) The present and future of peptide vaccines for cancer: single or multiple, long or short, alone or in combination? Cancer J 17: 343–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slingluff C., Jr, Lee S., Zhao F., Chianese-Bullock K., Olson W., Butterfield L., et al. (2013) A randomized phase II trial of multiepitope vaccination with melanoma peptides for cytotoxic T cells and helper T cells for patients with metastatic melanoma (E1602). Clin Cancer Res 19: 4228–4238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slingluff C., Jr, Petroni G., Chianese-Bullock K., Smolkin M., Hibbitts S., Murphy C., et al. (2007) Immunologic and clinical outcomes of a randomized phase II trial of two multipeptide vaccines for melanoma in the adjuvant setting. Clin Cancer Res 13: 6386–6395 [DOI] [PubMed] [Google Scholar]
- Slingluff C., Jr, Petroni G., Olson W., Smolkin M., Ross M., Haas N., et al. (2009) Effect of granulocyte/macrophage colony-stimulating factor on circulating CD8+ and CD4+ T cell responses to a multipeptide melanoma vaccine: outcome of a multicenter randomized trial. Clin Cancer Res 15: 7036–7044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sliwkowski M., Mellman I. (2013) Antibody therapeutics in cancer. Science 341: 1192–1198 [DOI] [PubMed] [Google Scholar]
- Speetjens F., Kuppen P., Welters M., Essahsah F., Voet Van Den Brink A., Lantrua M., et al. (2009) Induction of P53-specific immunity by a P53 synthetic long peptide vaccine in patients treated for metastatic colorectal cancer. Clin Cancer Res 15: 1086–1095 [DOI] [PubMed] [Google Scholar]
- Starck S., Shastri N. (2011) Non-conventional sources of peptides presented by MHC class I. Cell Mol Life Sci 68: 1471–1479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevanovic S. (2002) Identification of tumour-associated T cell epitopes for vaccine development. Nat Rev Cancer 2: 514–520 [DOI] [PubMed] [Google Scholar]
- Testa J., Shetty V., Hafner J., Nickens Z., Kamal S., Sinnathamby G., et al. (2012a) MHC class I-presented T cell epitopes identified by immunoproteomics analysis are targets for a cross reactive influenza-specific T cell response. PLoS One 7: e48484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Testa J., Shetty V., Sinnathamby G., Nickens Z., Hafner J., Kamal S., et al. (2012b) Conserved MHC class I-presented dengue virus epitopes identified by immunoproteomics analysis are targets for cross-serotype reactive T cell response. J Infect Dis 205: 647–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas P., Brown S., Keating R., Yue W., Morris M., So J., et al. (2007) Hidden epitopes emerge in secondary influenza virus-specific CD8+ T cell responses. J Immunol 178: 3091–3098 [DOI] [PubMed] [Google Scholar]
- Traversari C., Van Der Bruggen P., Luescher I., Lurquin C., Chomez P., Van Pel A., et al. (1992) A nonapeptide encoded by human gene MAGE-1 is recognized on HLA-A1 by cytolytic T lymphocytes directed against tumor antigen MZ2-E. J Exp Med 176: 1453–1457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuruma T., Hata F., Torigoe T., Furuhata T., Idenoue S., Kurotaki T., et al. (2004) Phase I clinical study of anti-apoptosis protein, survivin-derived peptide vaccine therapy for patients with advanced or recurrent colorectal cancer. J Transl Med 2: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuruma T., Iwayama Y., Ohmura T., Katsuramaki T., Hata F., Furuhata T., et al. (2008) Clinical and immunological evaluation of anti-apoptosis protein, survivin-derived peptide vaccine in phase I clinical study for patients with advanced or recurrent breast cancer. J Transl Med 6: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Bruggen P., Traversari C., Chomez P., Lurquin C., De Plaen E., Van Den Eynde B., et al. (1991) A gene encoding an antigen recognized by cytolytic T lymphocytes on a human melanoma. Science 254: 1643–1647 [DOI] [PubMed] [Google Scholar]
- Van Els C., Herberts C., Van Der Heeft E., Poelen M., Van Gaans-Van Den Brink J., Van Der Kooi A., et al. (2000) A single naturally processed measles virus peptide fully dominates the HLA-A*0201-associated peptide display and is mutated at its anchor position in persistent viral strains. Eur J Immunol 30: 1172–1181 [DOI] [PubMed] [Google Scholar]
- Walter S., Weinschenk T., Stenzl A., Zdrojowy R., Pluzanska A., Szczylik C., et al. (2012) Multipeptide immune response to cancer vaccine IMA901 after single-dose cyclophosphamide associates with longer patient survival. Nat Med 18: 1254–1261 [DOI] [PubMed] [Google Scholar]
- Wherry E. (2011) T cell exhaustion. Nat Immunol 12: 492–499 [DOI] [PubMed] [Google Scholar]
- Woo E., Chu C., Goletz T., Schlienger K., Yeh H., Coukos G., et al. (2001) Regulatory CD4(+)CD25(+) T cells in tumors from patients with early-stage non-small cell lung cancer and late-stage ovarian cancer. Cancer Res 61: 4766–4772 [PubMed] [Google Scholar]
- Yoshimura K., Minami T., Nozawa M., Uemura H. (2013) Phase I clinical trial of human vascular endothelial growth factor receptor 1 peptide vaccines for patients with metastatic renal cell carcinoma. Br J Cancer 108: 1260–1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarling A., Polefrone J., Evans A., Mikesh L., Shabanowitz J., Lewis S., et al. (2006) Identification of class I MHC-associated phosphopeptides as targets for cancer immunotherapy. Proc Natl Acad Sci U S A 103: 14889–14894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeeberg Iversen T., Engell-Noerregaard L., Ellebaek E., Andersen R., Kiaer Larsen S., Bjoern J., et al. (2013) Long-lasting disease stabilization in the absence of toxicity in metastatic lung cancer patients vaccinated with an epitope derived from indoleamine 2,3 dioxygenase. Clin Cancer Res: [DOI] [PubMed] [Google Scholar]
- Zeestraten E., Speetjens F., Welters M., Saadatmand S., Stynenbosch L., Jongen R., et al. (2013) Addition of interferon-alpha to the P53-SLP(R) vaccine results in increased production of interferon-gamma in vaccinated colorectal cancer patients: a phase I/II clinical trial. Int J Cancer 132: 1581–1591 [DOI] [PubMed] [Google Scholar]
- Zhang X., Huang Y., Li Z., Lin H., Sui Y. (2010) Prediction and analysis of HLA-A2/A24-restricted cytotoxic T-lymphocyte epitopes of the tumor antigen MAGE-N using the artificial neural networks method on NETCTL1.2 Server. Oncol Lett 1: 1097–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong W., Reche P., Lai C., Reinhold B., Reinherz E. (2003) Genome-wide characterization of a viral cytotoxic T lymphocyte epitope repertoire. J Biol Chem 278: 45135–45144 [DOI] [PubMed] [Google Scholar]
- Zhu B., Chen Z., Cheng X., Lin Z., Guo J., Jia Z., et al. (2003) Identification of HLA-A*0201-restricted cytotoxic T lymphocyte epitope from TRAG-3 antigen. Clin Cancer Res 9: 1850–1857 [PubMed] [Google Scholar]