Abstract
The incidence of hepatocellular carcinoma is rising due to alcohol drinking, hepatitis C viral infection and metabolic syndrome. Differential expression of CYP2E1 may play a pleiotropic role in the multistep process of liver carcinogenesis. Considerable attention has focused on the antitumor effect of trichostatin A (TSA) as well as CYP2E1 expression-induced apoptosis of cancer cells. However, very few studies have examined the mechanisms by which TSA has an antitumor effect and its association to CYP2E1 expression. The current study examined the action of TSA on CYP2E1 expression and the role of CYP2E1 in inducing apoptosis of HepG2 cells. Our data showed that TSA selectively induced CYP2E1 in four studied human hepatocellular carcinoma (HCC) cell lines (Huh7, PLC/PRF/5, Hep3B and HepG2), but not in normal primary human hepatocytes. TSA-mediated up-regulation of CYP2E1 expression was associated with histone H3 acetylation and the recruitment of HNF-1 and HNF-3β to the CYP2E1 promoter in HepG2 cells. siRNA-mediated knockdown experiments showed that TSA-induced caspase-3 cleavage was decreased due to reduced expression of CYP2E1 in HepG2 cells. Moreover, down-regulation of CYP2E1 was accompanied by decreased production of mitochondrial reactive oxygen species. These results suggest that histone modification is involved in CYP2E1 gene expression and that CYP2E1-dependent mitochondrial oxidative stress plays a role in TSA-induced apoptosis.
Keywords: histone acetylation, CYP2E1, liver cancer, apoptosis, HNF-1, HNF-3β
Introduction
Hepatocellular carcinoma (HCC) is a serious health problem. HCC is the fifth most common cancer worldwide and the third leading cause of cancer mortality.1,2 Twenty years ago, HCC was almost entirely restricted to Asia and sub-Saharan Africa. However, the incidence of HCC is increasing worldwide, with particularly steep increases documented in industrialized countries such as Japan, the USA and Denmark.3–6 A substantial proportion of the increase seen in the USA is due to diabetes, hepatitis C virus and excessive alcohol intake.7,8 Studies conducted in Italy and the USA suggest that alcohol is the most common risk factor contributing to HCC (accounting for 32–45% of HCC cases) as either the primary cause or an important co-factor.9
CYP2E1 plays multiple roles at different stages during alcohol-related hepatocarcinogenesis. In the HCC initiation and promotion stage, the induction of CYP2E1 promoted hepatocarcinogenesis by enhancing the conversion of various pro-carcinogens to carcinogens such as dimethylnitrosamines, aflatoxin B1, vinyl chloride and dimethylhydrazine. 9,10 A recent study showed that the induction of hepatic CYP2E1 by alcohol leads to highly miscoding lipid peroxidation-derived DNA that may play a significant role in hepatocarcinogenesis in patients with alcoholic liver disease (ALD).11 Many studies have showed that CYP2E1 overexpression not only induces reactive oxygen production, toxicity and apoptosis, but also synergizes and increases the susceptibility to different chemicals.12 – 14 However, studies also show the expression of CYP2E1 in human liver tumors was lower than that in non-tumorous tissues.15,16 Therefore, the differential expression of CYP2E1 may play a pleiotropic role in the multistep process of HCC tumorigenesis. The expression of CYP2E1 can be regulated at transcriptional and post-transcriptional levels. Serum shock-induced CYP2E1 expression is related to histone H3 acetylation.17
Epigenetic modifications have emerged as key mechanisms in gene expression regulation. Histone deacetylases (HDACs) play an important role in the epigenetic regulation of gene expression by catalyzing the removal of acetyl groups leading to chromatin condensation and transcriptional repression. However, there is growing evidence that inappropriate gene expression governed by epigenetic changes is also crucial to tumorigenesis and the progression of cancer.18 – 20 The change of acetylated histone was closely related to the expression of specific cancer-related genes in hepatoma cells.21 In addition, HDACs are overexpressed in different types of cancer; therefore, HDACs are becoming promising anticancer targets.22,23 Several classes of histone deacetylase inhibitors (HDACi) have been found to have anticancer activities with remarkable tumor specificity.24 – 26 Trichostatin A (TSA), a natural fungistatic antibiotic, was among the first of a series of hydroxamic acids to be discovered and has become one of the main research tools for probing the function of histone acetylation.27,28 TSA and the development of structurally related HDACi show pronounced anticancer activity at very low concentrations (nanomolar to micromolar range).29,30 Studies have also elucidated TSA’s promising antitumor effects in HCC model systems while not affecting primary hepatocytes; TSA appears only to induce apoptosis in HCC cells, but not in primary hepatocytes.26,31,32
Although studies have showed that apoptosis of cancer cells is caused by TSA and CYP2E1 overexpression, the molecular linkage among TSA, CYP2E1 expression and apoptosis has not been established. We hypothesize that the selective antitumor effect of TSA may be associated with CYP2E1-mediated signaling. The goal of this current study is to examine the effect of TSA on CYP2E1 expression in HCC cells and elucidate the function of CYP2E1 in TSA-induced apoptosis.
Materials and methods
Reagents
All reagents and chemicals used were from Sigma-Aldrich (St Louis, MO) unless noted otherwise. Acetylated histone 3 and 4 antibodies and chromatin immunoprecipitation (ChIP) assay kit were purchased from Upstate Biotechnology (Lake Placid, NY). MitoSOX™ Red mitochondrial superoxide indicator (for live-cell imaging) and Hank’s balanced salt solution (HBSS) with calcium and magnesium were purchased from Molecular Probes, Invitrogen (Carlsbad, CA). VECTASHIELD mounting medium with 4′,6-diamidino-2-phenylindole (DAPI) was purchased from Vector Laboratories (Burlingame, CA). TRIzol reagent and Lipofectamine™ RNAiMAX transfection reagent were purchased from Invitrogen. Polyclonal antibodies to human hepatic nuclear factor 1 (HNF-1), human hepatic nuclear factor 3β (HNF-3β) and goat antirabbit IgG-Texas Red were from Santa Cruz (CA; Louis, MO). Antibody specific for CYP2E1 was purchased from Abcam (Cambridge, MA). Cleaved caspase-3 rabbit mAb was purchased from Cell Signaling (Beverly, MA). TSA was dissolved in dimethyl sulfoxide (DMSO) at 10 mmol/L as the stock solution and stored at −20°C.
Cell cultures and treatment
Human hepatoma cell lines HepG2, Hep3B, Huh-7 and PLC/ PRF/5 were cultured in Dulbecco’s modification of Eagle’s medium (Mediatech). The media were supplemented with 10% charcoal-stripped fetal bovine serum (FBS) (Biomeda) and 1% penicillin/streptomycin (Invitrogen). The cells were cultured at 37°C in 5% CO2 atmosphere with a relative humidity of 95%. The cells were treated with 1 µmol/L of TSA for each studied time point. Primary human hepatocytes were obtained from XenoTech, LLC (Lenexa, KS) as a gift. Primary hepatocytes were cultured in William’s E culture medium supplemented with HEPES buffer (10 mmol/L, pH 7.4), FBS (10%) (Biomeda), l-glutamine (2 mmol/L), 1% penicillin/streptomycin (Invitrogen) and insulin (5 mg/mL).
RNA preparation and quantitative realtime PCR
Hepatoma cells were washed with ice-cold polymerase chain reaction PBS and total RNA was isolated using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. Taqman realtime PCR primers and fluorescent probe sequences were designed using Primer Express software (Applied Biosystems). The primer and probe sequences are as follows: CYP2E1, F-5-TCAATCTCT GGACCCCAACTG-3, R-5-GCGCTCTGCACTGTGCTTT-3, 5-6FAM-CCGACTGCCTGCTCGTGGAAATG-BHQ1-30; and β-actin F-5-CCTGGCACCCAGCACAAT-3, R-5-GCCGATC CACACGGAGTACT-30, 5-6FAM-ATCAAGATCATTGCTC CTCCTGAGCGC-BHQ1-3. Probes and primers were synthesized by Sigma-Aldrich and Integrated DNA Technologies. Probes were labeled with the reporter FAM (6-carboxyfluorescein) and quencher BQH1 (black hole quencher 1) at the 5′ and 3′ ends, respectively. Total RNA (1 µg) was reverse-transcribed with random primer and M-MLVRT reverse transcriptase (Invitrogen) for cDNA synthesis. The ABI TaqMan Universal PCR Master Mix (Applied Biosystems, Foster City, CA, USA) was used in a 20 µL realtime PCR reaction. Realtime PCR was conducted using the ABI Prism 7900 Sequence Detection System (Applied Biosystems). Fold inductions were calculated using the ΔΔCt method according to the manufacturer’s instructions.
Chromatin immunoprecipitation assay
ChIP assays were performed using a ChIP assay kit (Upstate Biotechnology) following the manufacturer’s protocol with the following modifications: cells were treated with 1% formaldehyde at 37°C for 15 min, followed by sonication with a Model 500 Sonic Dismembrator (Fisher Scientific). The cross-linked DNA–protein complexes were precipitated with antiacetylated histone H3 (6 µg), antiacetylated histone H4, anti-HNF-1 or anti-HNF-3β. Two regions of the CYP2E1 promoter were analyzed: a proximal region (−290 to −70) and a distal region (−1080 to −836). The following primer pairs were used in the case of histones, HNF-1 and HNF-3β analysis: (−290 to −70 region: sense 5′-ACC CCACGTTCTTAACTAT-3′, antisense 5′-CTTGCTACTCGT CTATCCC-3′; −1080 to −836: sense 5′-CTTCATTTCTCAT CATAT-3′, antisense 5′-GACTAGCTTCTTCTTTCA-3′). PCR products were visualized on 2% agarose gels stained with ethidium bromide and quantified with the Biosystems gel scanner and Quant analysis software.
siRNA transfection
Silencer® select predesigned siRNA for human CYP2E1 gene and scramble siRNA were purchased from Ambion (Austin, TX). HepG2 cells were transfected with siRNA (60 nmol/L per 1 × 105 HepG2 cells per well in a 12-well plate or a chamber slide) using Lipofectamine™ RNAiMAX transfection reagent following the manufacturer’s instruction. Realtime PCR was used to evaluate CYP2E1 knockdown efficiency after cells were incubated in transfection medium for 48 h.
Immunofluorescence microscopy
HepG2 cells were grown in Chamber BD Falcon™ Cultureslides (BD Biosciences, Bedford MA, USA). Following treatment, HepG2 cells were fixed for 15 min at room temperature with freshly prepared 4% paraformaldehyde in PBS. After fixation, cells were rinsed with PBS containing 0.2% Triton X100; cells were then incubated with PBS containing 0.2% Triton X100 and 5% normal goat serum for 30 min at room temperature. Primary antibody specific for acetylated histone H3 (1:200 dilution), acetylated histone H4 (1:100 dilution), CYP2E1 (1:300 dilution), cleaved Caspase 3 (1:200 dilution in PBS containing 0.2% Triton X100 and 1% normal goat serum) was applied to cells overnight at 4°C in a humidified chamber. After washing in PBS containing 0.2% Triton X100 and 1% normal goat serum, cells were incubated with goat antirabbit preadsorbed, Texas Red conjugated secondary antibody (1:400 dilution in PBS containing 0.2% Triton X100 and 1% normal goat serum) overnight at 4°C in a humidified chamber. After washing with PBS, cells were mounted with VECTASHIELD mounting medium with DAPI and analyzed under confocal microscope. Images were then analyzed via Image Pro Plus imaging software (version 5.0, Media Cybernetics, Silver Spring, MD).
Mitochondrial superoxide staining
MitoSOX™ Red mitochondrial superoxide indicator stock solution (5 mmol/L in DMSO, stored at −20°C in dark) was further diluted into 2.5 µM working solution with HBSS right before use. After 48 h of siRNA transfection, cells were incubated with 0.1% DMSO or with TSA (1 µM) in medium for 12 h followed by MitoSOX™ Red mitochondrial superoxide indicator staining according to the manufacturer’s protocol. In brief, medium was replaced with 2.5 µM MitoSOX™ reagent working solution. The cells were incubated for 15 min at 37°C, protected from light, washed gently three times with warm HBSS, and examined under fluorescence microscope in serum-free medium.
Statistical analysis
Data are expressed as mean±SD. Statistical analysis was performed using Student’s t-test or one-way ANOVA. P values of <0.05 were regarded as significant.
Results
Differential effects of TSA on CYP2E1 expression in primary hepatocytes and HCC cell lines
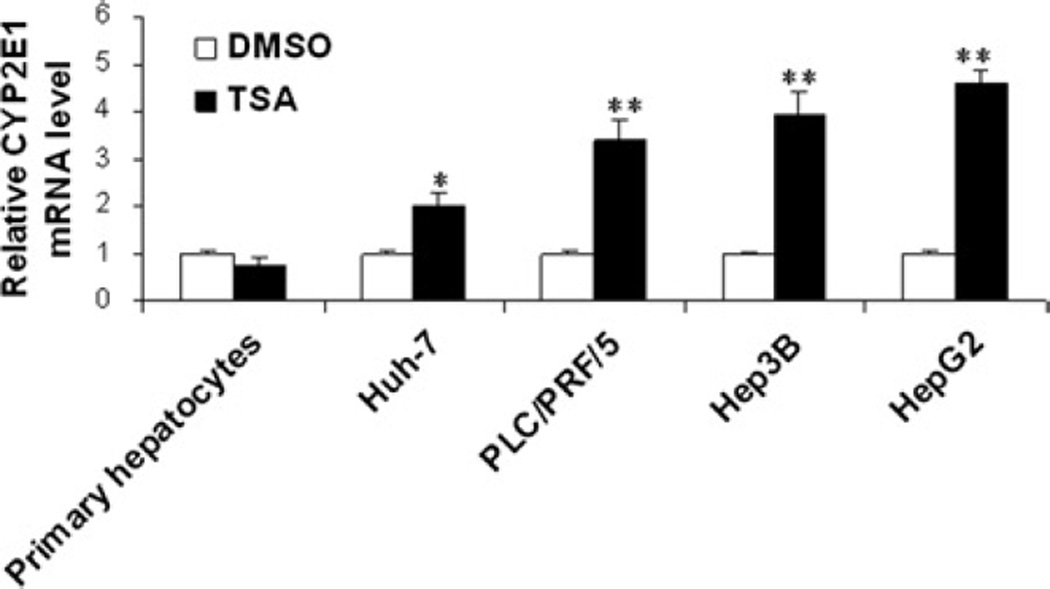
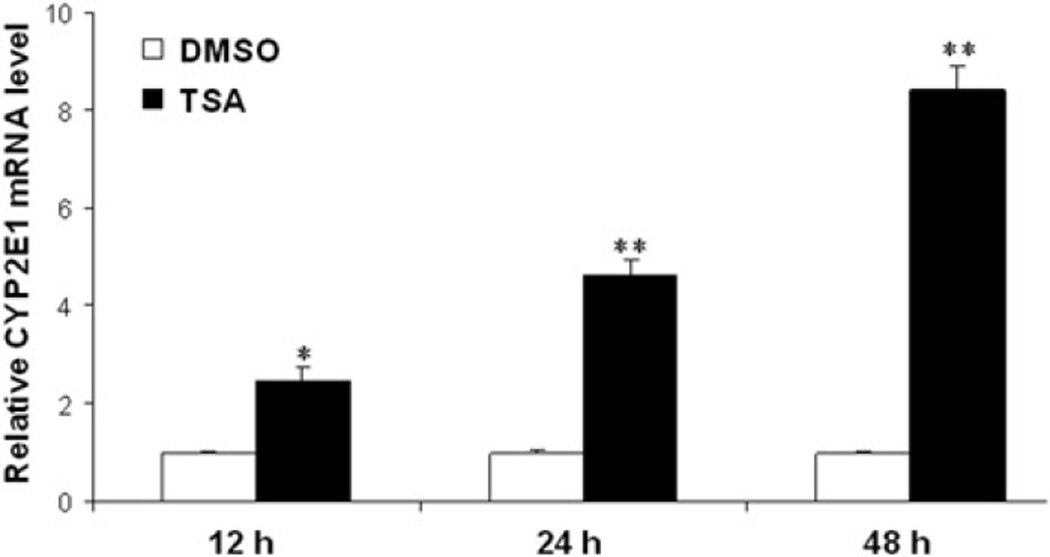
To assess the effect of TSA on CYP2E1 expression in HCC cell lines and primary hepatocytes, CYP2E1 expression was quantitated by realtime PCR. The data demonstrated that TSA induced the mRNA level of CYP2E1 in four studied human HCC cell lines (Huh7, PLC/PRF/5, Hep3B and HepG2), but not in primary hepatocytes (Figure 1). Among the four HCC cell lines, HepG2 cells had the highest induction of CYP2E1 mRNA levels. In addition, the induction of CYP2E1 mRNA in HepG2 cells was time dependent; it occurred within 12 h after TSA treatment (Figure 2).
Figure 1.
TSA induced CYP2E1 mRNA level in HCC cells, but not in primary human hepatocytes. HCC cells and primary human hepatocytes were treated with DMSO or TSA (1 µM) for 24 h. Realtime PCR was used to quantify CYP2E1 mRNA level. Data were expressed as mean ± SD from three independent experiments. *P < 0.05, **P < 0.001 versus DMSO treatment group. TSA, trichostatin A; HCC, hepatocellular carcinoma; DMSO, dimethyl sulfoxide
Figure 2.
The induction of CYP2E1 mRNA levels in HepG2 cells is time dependent. Realtime PCR was used to quantify CYP2E1 mRNA level. Data were expressed as mean ± SD from three independent experiments. *P < 0.05, **P < 0.001 versus DMSO treatment group. DMSO, dimethyl sulfoxide; TSA, trichostatin A
Accumulation of acetylation of histone H3 and H4 in HepG2 cells with TSA
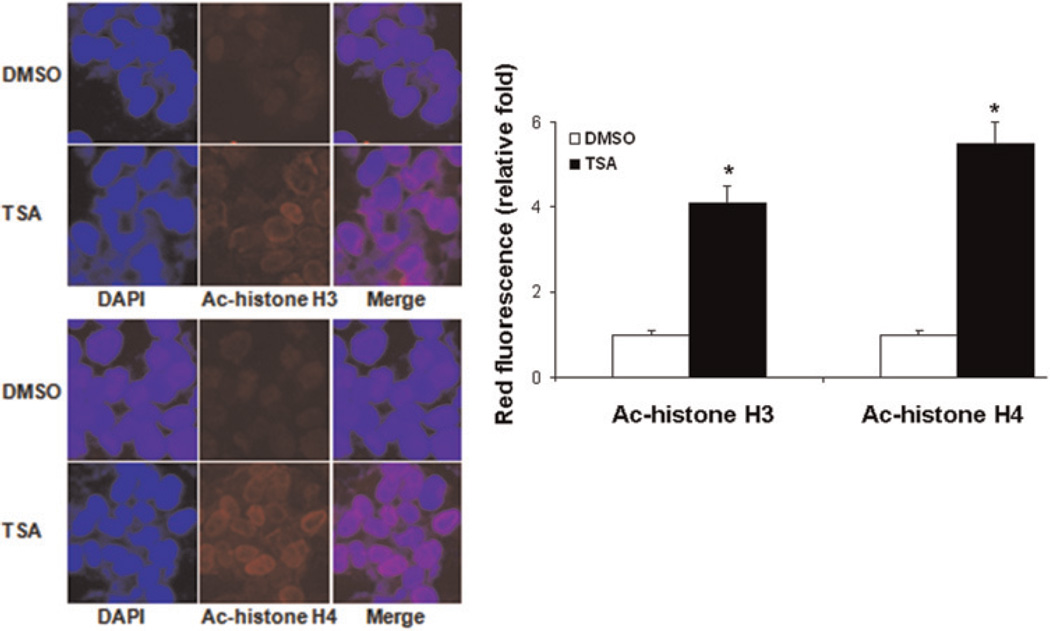
To examine whether TSA affects histone acetylation, TSA-treated HepG2 cells were studied for histone H3 and H4 acetylation by immunocytochemistry using specific antibodies followed by incubation with Texas Red conjugated secondary antibody and imaged by confocal microscopy. An accumulation of acetylated H3 and H4 proteins in HepG2 cells could be detected after TSA treatment for 24 h (Figure 3).
Figure 3.
Accumulation of acetylated histone H3 and H4 in HepG2 cells after TSA treatment. HepG2 cells were treated with TSA (1 µM) for 24 h and immunostained with antiacetylated histone H3 (a) and H4 (b) antibodies and imaged by confocal microscopy as described in the Materials and methods. TSA increased acetylated histone H3 and H4 in the nuclei. Nuclei are shown counterstained with DAPI. Representative images of three independent experiments are shown. *P < 0.01 versus DMSO treatment group. TSA, trichostatin A; DAPI, 4′,6-diamidino-2-phenylindole; DMSO, dimethyl sulfoxide (A color version of this figure is available in the online journal)
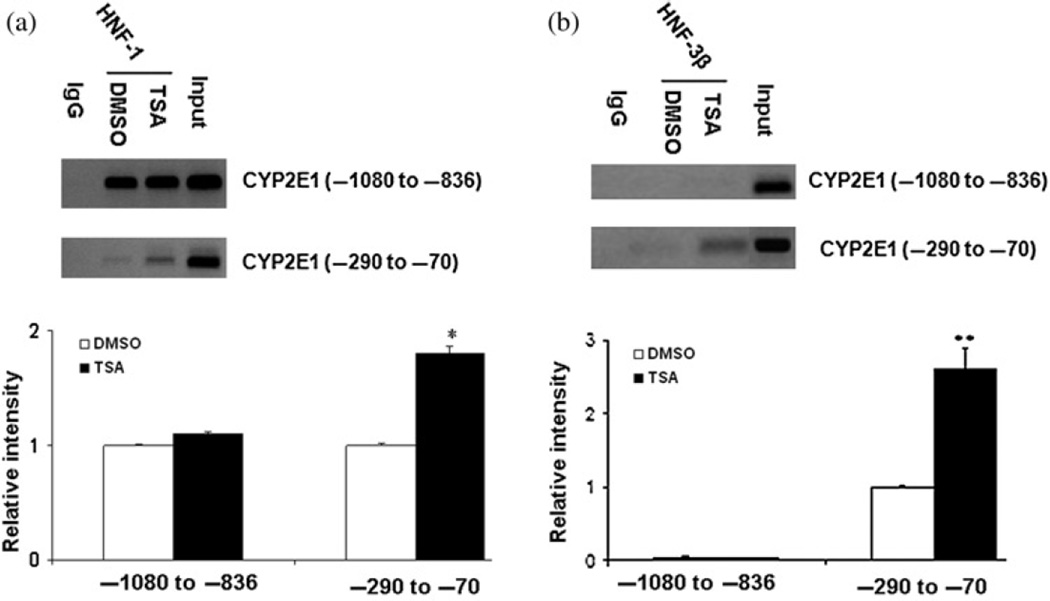
TSA increases the recruitment of acetylated histone H3 and transcription factors on the proximal CYP2E1 promoter
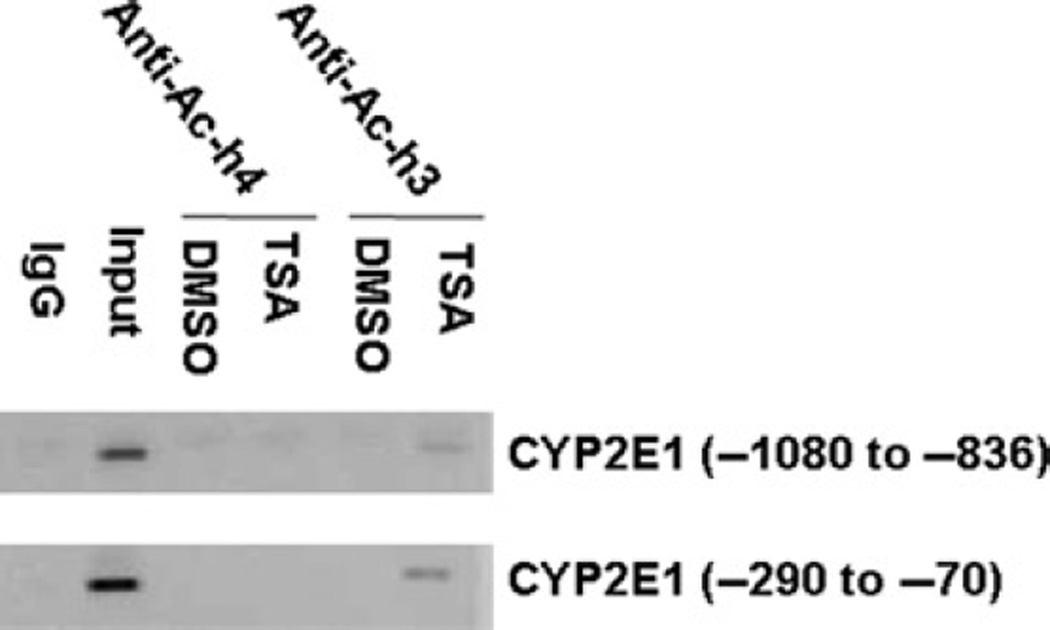
To study the molecular mechanisms whereby TSA mediates the induction of CYP2E1 expression, we performed ChIP assays to study the factors recruited in vivo to the promoter as well as chromatin modifications. Using antibodies against acetylated histone H3 and H4, the immunoprecipitated genomic DNA was amplified by PCR using primers spanning positions −1080 to −836 and −290 to −70, which contain HNF-1 and HNF-3β and predicted transcriptional binding sites. TSA treatment increased the levels of acetylated histone H3, but not H4, recruited to these two regions (Figure 4). We next sought to determine whether histone modification correlated with the recruitment of transcription factors to each specified region. Antibodies specific for HNF-1 and HNF-3β were used. The results showed that TSA treatment enhanced the recruitment of HNF-1 and HNF-3β to the promoter (−290 to −70) region of the CYP2E1 gene (Figure 5). HNF-1, but not HNF-3β, was constitutively bound to the −1080 to −836 region of the CYP2E1 gene, and TSA treatment did not further increase the binding (Figure 5). These findings suggest that TSA-mediated up-regulation of CYP2E1 expression is associated with histone H3 acetylation and the recruitment of HNF-1 and HNF-3β to the −290 to −70 region within the promoter.
Figure 4.
Induction of CYP2E1 by TSA is associated with the recruitment of acetylated H3 to the CYP2E1 promoter. HepG2 cells were treated with TSA (1 µM) for 24 h followed by chromatin immunoprecipitation (ChIP) assay as described in the Materials and methods. CYP2E1 promoter fragments were immunoprecipitated with antiacetylated H3 or antiacetylated H4 antibody and amplified by PCR. Results are representative of three independent experiments. TSA, trichostatin A; DMSO, dimethyl sulfoxide
Figure 5.
TSA enhanced the recruitment of HNF-1 and HNF-3β to the −290 to −70 region of the CYP2E1 gene. HepG2 cells were treated by TSA (1 µM) for 24 h. ChIP assay was performed using anti-HNF-1 (a) and anti-HNF-3β (b) antibody as described in the Materials and methods. Results were representative of three independent experiments. *P < 0.05 versus DMSO treatment group. TSA, trichostatin A; DMSO, dimethyl sulfoxide
TSA-induced CYP2E1 is involved in reactive oxygen species generation and apoptosis in HepG2 cells
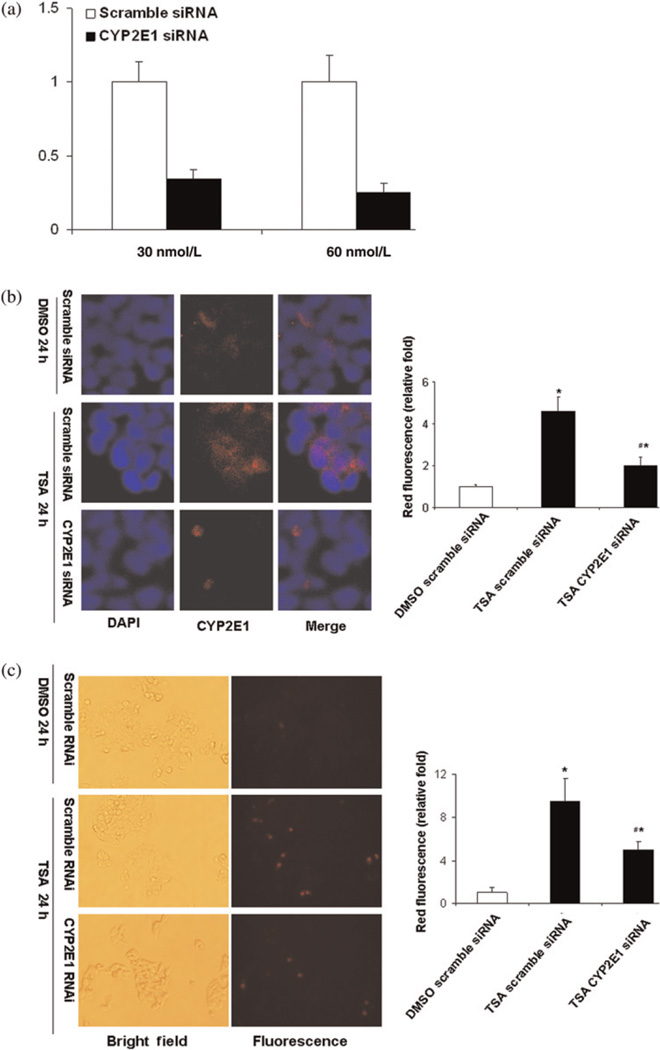
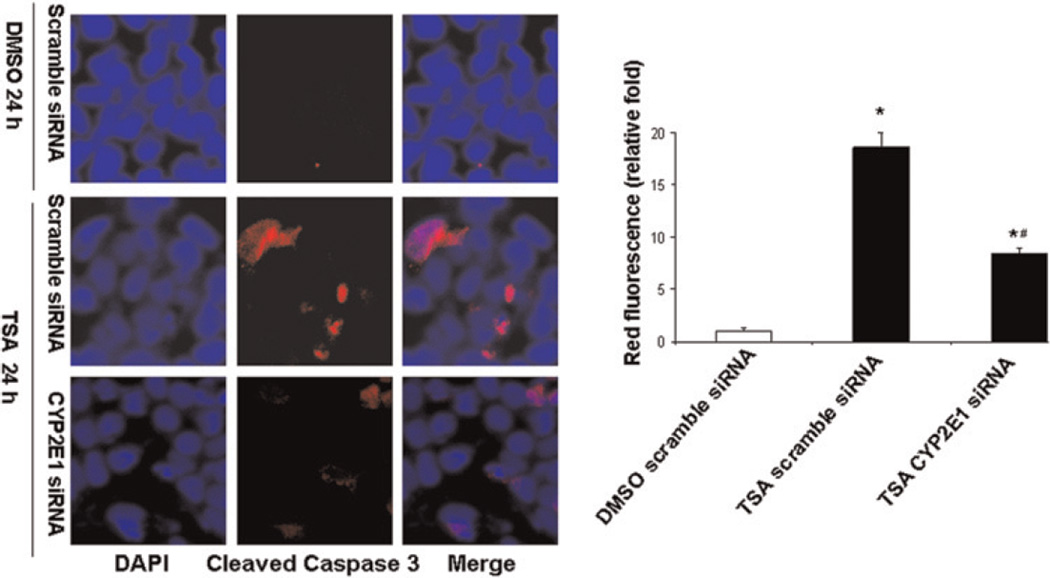
In order to determine the role of TSA-induced CYP2E1 in mediating cell death, siRNA-mediated knockdown was performed. HepG2 cells were transfected with predesigned siRNA for human CYP2E1 or scramble siRNA. The CYP2E1 siRNA transfection caused >70% reduction of CYP2E1 mRNA level (Figure 6a) and blocked TSA-induced CYP2E1 protein expression in HepG2 cells (Figure 6b). In addition, down-regulation of CYP2E1 was accompanied by decreased production of mitochondrial reactive oxygen species (ROS) (Figure 6c). Moreover, TSA-induced caspase-3 cleavage was also reduced due to reduced expression of CYP2E1 in HepG2 cells (Figure 7).
Figure 6.
CYP2E1 siRNA blocked TSA-induced CYP2E1 expression and mitochondria ROS generation induced by TSA in HepG2 cells. (a) HepG2 cells were transfected with either scramble siRNA or CYP2E1 siRNA at concentrations of 30 and 60 nM for 48 h as described in the Materials and methods. Endogenous CYP2E1 mRNA level was quantified by realtime PCR and expressed as relative fold compared to cells transfected with scramble siRNA transfection. (b) HepG2 cells were transfected with either scramble siRNA or CYP2E1 siRNA (60 nm) for 48 h and then cells were treated with TSA (1 µM) for 24 h followed by immunofluorescence staining for CYP2E1 and nuclear counterstaining. Representative results are from at least two independent experiments. (c) CYP2E1 was knocked down by siRNA in HepG2 cells and then cells were treated with TSA (1 µM) for 12 h. ROS was detected by MitoSOX™ Red mitochondrial superoxide indicator for live-cell imaging staining. MitoSOX™ Red reagent is highly selective for detection of mitochondrial superoxide in live cells. Once the compound is oxidized by superoxide in the mitochondria, red fluorescence is visible under fluorescence. The positive cells were shown in red with MitoSOX™ Red mitochondrial superoxide indicator staining. Four random fields of more than 200 cells each were counted under a fluorescence microscope. The results were generated from three independent experiments. *P < 0.01 versus DMSO treatment group, #P < 0.05 versus TSA (scramble siRNA). TSA, trichostatin A; ROS, reactive oxygen species; DMSO, dimethyl sulfoxide (A color version of this figure is available in the online journal)
Figure 7.
TSA-induced cleaved caspase-3 is mediated through CYP2E1 in HepG2 cells. CYP2E1 expression was knocked down by siRNA in HepG2 cells and then cells were treated with TSA (1 µM) for 24 h followed by immunofluorescence staining with a specific antibody against cleaved caspase-3 and nuclear counterstaining with DAPI and viewed by confocal microscopy. Representative images of three independent experiments are shown. *P < 0.01 versus DMSO treatment group, #P < 0.05 versus TSA (scramble siRNA). TSA, trichostatin A; DAPI, 4′,6-diamidino-2-phenylindole; DMSO, dimethyl sulfoxide (A color version of this figure is available in the online journal)
Discussion
CYP2E1 has important functions in the metabolism of a wide variety of endogenous and exogenous compounds and is relevant to chemical toxicity and carcinogenesis in liver.33,34 Clinical studies have showed that decreased expression of CYP2E1 was associated with poorly differentiated HCC and poor prognosis in HCC patients. These findings suggested that high expression of CYP2E1 might be cytotoxic to HCC cells and inhibit their rapid proliferation. 35,36 TSA also induced apoptosis of HCC cells and the expression of many genes was altered after exposure to TSA in HCC cell lines.37 Therefore, it is of interest to study the potential impact of TSA on CYP2E1 gene expression and its function in apoptosis. The current study provides novel information, which demonstrates chromatin modification and the recruitment of transcription factors lead to enhanced expression of CYP2E1 gene as well as ROS generation, which ultimately cause HepG2 cell death.
The selective antitumor activity of HDACi is due to the alteration of gene expression by accumulating acetylated histones and reordering of transcription of target genes and components of the transcriptional machinery.38 – 40 CYP2E1 can be regulated at different levels via various mechanisms. For example, many investigators have reported that post-transcriptional regulation plays a major role in ethanol-mediated CYP2E1 expression.34,41 Some studies showed that CYP2E1 transcription is influenced by a variety of compounds such as IL-6, T3 and insulin, but the molecular mechanism is still poorly understood.42 – 44 The present study showed that TSA could increase CYP2E1 transcription by increased enrichment of acetylated histone H3, but not H4. Histone H3 acetylation usually occurs in the proximal promoter; in contrast to H3, acetylated histone H4 is found to occur in distal regions.45 The main CYP2E1 transcription activity is dependent on the proximal promoter. Deletion analysis showed that the proximal promoter of CYP2E1 was responsible for 90% of the in vitro transcription activity of rat adult liver extracts.46 Human CYP2E1 transcription activity can be affected by the polymorphism at nucleotide −1019. This polymorphism affects its ability to bind HNF-1 and changes its transcriptional regulation.47,48 Our data suggest that the binding of HNF-1 to the −1080 to −836 region maintains the basal transcription activity of CYP2E1, but the inducibility of CYP2E1 by TSA is mainly due to additional recruitment of HNF-1 and HNF-3β to the −290 to −70 region. Our data are consistent with the literature indicating that HNF-1 plays a major role in controlling the liver-specific expression of CYP2E1 gene in vitro and in vitro.46,49 Taken together, TSA treatment results in HNF-1 and HNF-3β recruitment to the CYP2E1 promoter (−290 to −70) by acetylated histone H3. Thus, HNF-1 and HNF-3β played an important role in TSA-induced CYP2E1 expression in HepG2 cells.
TSA not only induced CYP2E1 mRNA, but also CYP2E1 protein levels in HepG2cells. The catalytic cycle of CYP2E1 plays an important role in ROS generation, in which hydrogen peroxide and superoxide can be produced from the ferrous-dioxygen complex or released from the peroxy ferric intermediate (peroxy P450).50 CYP2E1-generated ROS could increase lipid peroxidation and mitochondrial membrane permeability, cause the release of pro-apoptotic factors and activate caspase 3 to induce apoptosis.51 Our data showed that the TSA-induced CYP2E1 expression was associated with the induction cleaved caspase 3 in HepG2 cells. Furthermore, knockdown CYP2E1 expression not only inhibited ROS generation but also the induction of cleaved caspase 3. Activation of the caspase cascade plays an important role in TSA- and suberoylanilide hydroxamic acid-induced apoptosis in different cancer cell lines.52,53 CYP2E1 overexpression is involved in the production of lipid peroxidation or ROS and activation of caspase 1, 2 and 3, followed by mitochondrial injury and apoptosis.54–56 CYP2E1 is mainly located in the membrane of the endoplasmic reticulum (ER) and mitochondrion; the damage to mitochondrion produced by mitochondrial CYP2E1 might be an early event in cell injury and mitochondrial CYP2E1 might play an important role in biochemical and toxicological effects just as CYP2E1 does in the ER.50,57–59 Taken together, our data established the linkage between TSA-induced CYP2E1 expression and ROS generation as well as program cell death of HepG2 cells.
In summary, TSA induces apoptosis through a mechanism that involves increased CYP2E1 gene transcription due to acetylation of histone H3 at the proximal promoter and recruitment of HNF-1 and HNF-3β to the same region. CYP2E1-dependent mitochondrial oxidative stress plays a role in TSA-induced apoptosis of HepG2 cells. Given the importance of CYP2E1 in HCC, induction of CYP2E1 by TSA might provide a basis for a therapeutic approach in patients with HCC.
ACKNOWLEDGMENTS
We thank Drs Don Warn and Eric Grimes from the Confocal Core at the University of Kansas Medical Center for technical assistance with the confocal microscope. We thank Barbara Brede for proofreading this manuscript. This work was supported by NIH grants CA 53596 and P20RR021940 Molecular Biology Core.
REFERENCES
- 1.Llovet JM, Burroughs A, Bruix J. Hepatocellular carcinoma. Lancet. 2003;362:1907–1917. doi: 10.1016/S0140-6736(03)14964-1. [DOI] [PubMed] [Google Scholar]
- 2.El-Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557–2576. doi: 10.1053/j.gastro.2007.04.061. [DOI] [PubMed] [Google Scholar]
- 3.El-Serag HB. Hepatocellular carcinoma: recent trends in the United States. Gastroenterology. 2004;127:S27–S34. doi: 10.1053/j.gastro.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 4.Llovet JM. Updated treatment approach to hepatocellular carcinoma. J Gastroenterol. 2005;40:225–235. doi: 10.1007/s00535-005-1566-3. [DOI] [PubMed] [Google Scholar]
- 5.Varela M, Bruix J. Hepatocellular carcinoma in the United States. Lessons from a population-based study in Medicare recipients. J Hepatol. 2006;44:8–10. doi: 10.1016/j.jhep.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 6.Jepsen P, Vilstrup H, Tarone RE, Friis S, Sorensen HT. Incidence rates of hepatocellular carcinoma in the U.S. and Denmark: recent trends. Int J Cancer. 2007;121:1624–1626. doi: 10.1002/ijc.22860. [DOI] [PubMed] [Google Scholar]
- 7.Yuan JM, Govindarajan S, Arakawa K, Yu MC. Synergism of alcohol, diabetes, and viral hepatitis on the risk of hepatocellular carcinoma in blacks and whites in the U.S. Cancer. 2004;101:1009–1017. doi: 10.1002/cncr.20427. [DOI] [PubMed] [Google Scholar]
- 8.Davila JA, Morgan RO, Shaib Y, McGlynn KA, El-Serag HB. Hepatitis C infection and the increasing incidence of hepatocellular carcinoma: a population-based study. Gastroenterology. 2004;127:1372–1380. doi: 10.1053/j.gastro.2004.07.020. [DOI] [PubMed] [Google Scholar]
- 9.Morgan TR, Mandayam S, Jamal MM. Alcohol and hepatocellular carcinoma. Gastroenterology. 2004;127:S87–S96. doi: 10.1053/j.gastro.2004.09.020. [DOI] [PubMed] [Google Scholar]
- 10.Stickel F, Schuppan D, Hahn EG, Seitz HK. Cocarcinogenic effects of alcohol in hepatocarcinogenesis. Gut. 2002;51:132–139. doi: 10.1136/gut.51.1.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Y, Millonig G, Nair J, Patsenker E, Stickel F, Mueller S, Bartsch H, Seitz HK. Ethanol-induced cytochrome P4502E1 causes carcinogenic etheno-DNA lesions in alcoholic liver disease. Hepatology. 2009;50:453–461. doi: 10.1002/hep.22978. [DOI] [PubMed] [Google Scholar]
- 12.Schattenberg JM, Wang Y, Rigoli RM, Koop DR, Czaja MJ. CYP2E1 overexpression alters hepatocyte death from menadione and fatty acids by activation of ERK1/2 signaling. Hepatology. 2004;39:444–455. doi: 10.1002/hep.20067. [DOI] [PubMed] [Google Scholar]
- 13.Wang X, Lu Y, Cederbaum AI. Induction of cytochrome P450 2E1 increases hepatotoxicity caused by Fas agonistic Jo2 antibody in mice. Hepatology. 2005;42:400–410. doi: 10.1002/hep.20792. [DOI] [PubMed] [Google Scholar]
- 14.Hodges NJ, Green RM, Chipman JK, Graham M. Induction of DNA strand breaks and oxidative stress in HeLa cells by ethanol is dependent on CYP2E1 expression. Mutagenesis. 2007;22:189–194. doi: 10.1093/mutage/gem001. [DOI] [PubMed] [Google Scholar]
- 15.Hirose Y, Naito Z, Kato S, Onda M, Sugisaki Y. Immunohistochemical study of CYP2E1 in hepatocellular carcinoma carcinogenesis: examination with newly prepared anti-human CYP2E1 antibody. J Nippon Med Sci. 2002;69:243–251. doi: 10.1272/jnms.69.243. [DOI] [PubMed] [Google Scholar]
- 16.Kinoshita M, Miyata M. Underexpression of mRNA in human hepatocellular carcinoma focusing on eight loci. Hepatology. 2002;36:433–438. doi: 10.1053/jhep.2002.34851. [DOI] [PubMed] [Google Scholar]
- 17.Matsunaga N, Ikeda M, Takiguchi T, Koyanagi S, Ohdo S. The molecular mechanism regulating 24-hour rhythm of CYP2E1 expression in the mouse liver. Hepatology. 2008;48:240–251. doi: 10.1002/hep.22304. [DOI] [PubMed] [Google Scholar]
- 18.Lund AH, van Lohuizen M. Epigenetics and cancer. Genes Dev. 2004;18:2315–2335. doi: 10.1101/gad.1232504. [DOI] [PubMed] [Google Scholar]
- 19.Minucci S, Pelicci PG. Histone deacetylase inhibitors and the promise of epigenetic (and more) treatments for cancer. Nat Rev Cancer. 2006;6:38–51. doi: 10.1038/nrc1779. [DOI] [PubMed] [Google Scholar]
- 20.Esteller M. The necessity of a human epigenome project. Carcinogenesis. 2006;27:1121–1125. doi: 10.1093/carcin/bgl033. [DOI] [PubMed] [Google Scholar]
- 21.Chiba T, Yokosuka O, Fukai K, Kojima H, Tada M, Arai M, Imazeki F, Saisho H. Cell growth inhibition and gene expression induced by the histone deacetylase inhibitor, trichostatin A, on human hepatoma cells. Oncology. 2004;66:481–491. doi: 10.1159/000079503. [DOI] [PubMed] [Google Scholar]
- 22.Conley BA, Wright JJ, Kummar S. Targeting epigenetic abnormalities with histone deacetylase inhibitors. Cancer. 2006;107:832–840. doi: 10.1002/cncr.22064. [DOI] [PubMed] [Google Scholar]
- 23.Carew JS, Giles FJ, Nawrocki ST. Histone deacetylase inhibitors: mechanisms of cell death and promise in combination cancer therapy. Cancer Lett. 2008;269:7–17. doi: 10.1016/j.canlet.2008.03.037. [DOI] [PubMed] [Google Scholar]
- 24.Vanhaecke T, Papeleu P, Elaut G, Rogiers V. Trichostatin A-like hydroxamate histone deacetylase inhibitors as therapeutic agents: toxicological point of view. Curr Med Chem. 2004;11:1629–1643. doi: 10.2174/0929867043365099. [DOI] [PubMed] [Google Scholar]
- 25.Dashwood RH, Myzak MC, Ho E. Dietary HDAC inhibitors: time to rethink weak ligands in cancer chemoprevention? Carcinogenesis. 2006;27:344–349. doi: 10.1093/carcin/bgi253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Venturelli S, Armeanu S, Pathil A, Hsieh CJ, Weiss TS, Vonthein R, Wehrmann M, Gregor M, Lauer UM, Bitzer M. Epigenetic combination therapy as a tumor-selective treatment approach for hepatocellular carcinoma. Cancer. 2007;109:2132–2141. doi: 10.1002/cncr.22652. [DOI] [PubMed] [Google Scholar]
- 27.Yoshida M, Kijima M, Akita M, Beppu T. Potent and specific inhibition of mammalian histone deacetylase both in vivo and in vitro by trichostatin A. J Biol Chem. 1990;265:17174–17179. [PubMed] [Google Scholar]
- 28.Monneret C. Histone deacetylase inhibitors. Eur J Med Chem. 2005;40:1–13. doi: 10.1016/j.ejmech.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 29.Finnin MS, Donigian JR, Cohen A, Richon VM, Rifkind RA, Marks PA, Breslow R, Pavletich NP. Structures of a histone deacetylase homologue bound to the TSA and SAHA inhibitors. Nature. 1999;401:188–193. doi: 10.1038/43710. [DOI] [PubMed] [Google Scholar]
- 30.Bouchain G, Delorme D. Novel hydroxamate and anilide derivatives as potent histone deacetylase inhibitors: synthesis and antiproliferative evaluation. Curr Med Chem. 2003;10:2359–2372. doi: 10.2174/0929867033456585. [DOI] [PubMed] [Google Scholar]
- 31.Papeleu P, Loyer P, Vanhaecke T, Elaut G, Geerts A, Guguen-Guillouzo C, Rogiers V. Trichostatin A induces differential cell cycle arrests but does not induce apoptosis in primary cultures of mitogen-stimulated rat hepatocytes. J Hepatol. 2003;39:374–382. doi: 10.1016/s0168-8278(03)00288-5. [DOI] [PubMed] [Google Scholar]
- 32.Armeanu S, Pathil A, Venturelli S, Mascagni P, Weiss TS, Gottlicher M, Gregor M, Lauer UM, Bitzer M. Apoptosis on hepatoma cells but not on primary hepatocytes by histone deacetylase inhibitors valproate and ITF2357. J Hepatol. 2005;42:210–217. doi: 10.1016/j.jhep.2004.10.020. [DOI] [PubMed] [Google Scholar]
- 33.Caro AA, Cederbaum AI. Oxidative stress, toxicology, and pharmacology of CYP2E1. Annu Rev Pharmacol Toxicol. 2004;44:27–42. doi: 10.1146/annurev.pharmtox.44.101802.121704. [DOI] [PubMed] [Google Scholar]
- 34.Gonzalez FJ. The 2006 Bernard B. Brodie Award Lecture. Cyp2e1. Drug Metab Dispos. 2007;35:1–8. doi: 10.1124/dmd.106.012492. [DOI] [PubMed] [Google Scholar]
- 35.Ng IO, Lai EC, Ng MM, Fan ST. Tumor encapsulation in hepatocellular carcinoma. A pathologic study of 189 cases. Cancer. 1992;70:45–49. doi: 10.1002/1097-0142(19920701)70:1<45::aid-cncr2820700108>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 36.Ho JC, Cheung ST, Leung KL, Ng IO, Fan ST. Decreased expression of cytochrome P450 2E1 is associated with poor prognosis of hepatocellular carcinoma. Int J Cancer. 2004;111:494–500. doi: 10.1002/ijc.20282. [DOI] [PubMed] [Google Scholar]
- 37.Chiba T, Yokosuka O, Arai M, Tada M, Fukai K, Imazeki F, Kato M, Seki N, Saisho H. Identification of genes up-regulated by histone deacetylase inhibition with cDNA microarray and exploration of epigenetic alterations on hepatoma cells. J Hepatol. 2004;41:436–445. doi: 10.1016/j.jhep.2004.05.018. [DOI] [PubMed] [Google Scholar]
- 38.Dokmanovic M, Marks PA. Prospects: histone deacetylase inhibitors. J Cell Biochem. 2005;96:293–304. doi: 10.1002/jcb.20532. [DOI] [PubMed] [Google Scholar]
- 39.Pan L, Lu J, Wang X, Han L, Zhang Y, Han S, Huang B. Histone deacetylase inhibitor trichostatin a potentiates doxorubicin-induced apoptosis by up-regulating PTEN expression. Cancer. 2007;109:1676–1688. doi: 10.1002/cncr.22585. [DOI] [PubMed] [Google Scholar]
- 40.Smith CL. A shifting paradigm: histone deacetylases and transcriptional activation. Bioessays. 2008;30:15–24. doi: 10.1002/bies.20687. [DOI] [PubMed] [Google Scholar]
- 41.Gonzalez FJ, Ueno T, Umeno M, Song BJ, Veech RL, Gelboin HV. Microsomal ethanol oxidizing system: transcriptional and posttranscriptional regulation of cytochrome P450, CYP2E1. Alcohol Alcohol Suppl. 1991;1:97–101. [PubMed] [Google Scholar]
- 42.Johansson I, Lindros KO, Eriksson H, Ingelman-Sundberg M. Transcriptional control of CYP2E1 in the perivenous liver region and during starvation. Biochem Biophys Res Commun. 1990;173:331–338. doi: 10.1016/s0006-291x(05)81061-7. [DOI] [PubMed] [Google Scholar]
- 43.Lagadic-Gossmann D, Lerche C, Rissel M, Joannard F, Galisteo M, Guillouzo A, Corcos L. The induction of the human hepatic CYP2E1 gene by interleukin 4 is transcriptional and regulated by protein kinase C. Cell Biol Toxicol. 2000;16:221–233. doi: 10.1023/a:1007625925095. [DOI] [PubMed] [Google Scholar]
- 44.Woodcroft KJ, Hafner MS, Novak RF. Insulin signaling in the transcriptional and posttranscriptional regulation of CYP2E1 expression. Hepatology. 2002;35:263–273. doi: 10.1053/jhep.2002.30691. [DOI] [PubMed] [Google Scholar]
- 45.Rada-Iglesias A, Enroth S, Ameur A, Koch CM, Clelland GK, Respuela-Alonso P, Wilcox S, Dovey OM, Ellis PD, Langford CF, Dunham I, Komorowski J, Wadelius C. Butyrate mediates decrease of histone acetylation centered on transcription start sites and down-regulation of associated genes. Genome Res. 2007;17:708–719. doi: 10.1101/gr.5540007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ueno T, Gonzalez FJ. Transcriptional control of the rat hepatic CYP2E1 gene. Mol Cell Biol. 1990;10:4495–4505. doi: 10.1128/mcb.10.9.4495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hayashi S, Watanabe J, Kawajiri K. Genetic polymorphisms in the 5′-flanking region change transcriptional regulation of the human cytochrome P450IIE1 gene. J Biochem. 1991;110:559–565. doi: 10.1093/oxfordjournals.jbchem.a123619. [DOI] [PubMed] [Google Scholar]
- 48.Watanabe J, Hayashi S, Kawajiri K. Different regulation and expression of the human CYP2E1 gene due to the RsaI polymorphism in the 5′-flanking region. J Biochem. 1994;116:321–326. doi: 10.1093/oxfordjournals.jbchem.a124526. [DOI] [PubMed] [Google Scholar]
- 49.Liu SY, Gonzalez FJ. Role of the liver-enriched transcription factor HNF-1 alpha in expression of the CYP2E1 gene. DNA Cell Biol. 1995;14:285–293. doi: 10.1089/dna.1995.14.285. [DOI] [PubMed] [Google Scholar]
- 50.Lu Y, Cederbaum AI. CYP2E1 and oxidative liver injury by alcohol. Free Radic Biol Med. 2008;44:723–738. doi: 10.1016/j.freeradbiomed.2007.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee HC, Wei YH. Oxidative stress, mitochondrial DNA mutation, and apoptosis in aging. Exp Biol Med (Maywood) 2007;232:592–606. [PubMed] [Google Scholar]
- 52.Gillenwater AM, Zhong M, Lotan R. Histone deacetylase inhibitor suberoylanilide hydroxamic acid induces apoptosis through both mitochondrial and Fas (Cd95) signaling in head and neck squamous carcinoma cells. Mol Cancer Ther. 2007;6:2967–2975. doi: 10.1158/1535-7163.MCT-04-0344. [DOI] [PubMed] [Google Scholar]
- 53.Taghiyev AF, Guseva NV, Sturm MT, Rokhlin OW, Cohen MB. Trichostatin A (TSA) sensitizes the human prostatic cancer cell line DU145 to death receptor ligands treatment. Cancer Biol Ther. 2005;4:382–390. doi: 10.4161/cbt.4.4.1615. [DOI] [PubMed] [Google Scholar]
- 54.Sakurai K, Cederbaum AI. Oxidative stress and cytotoxicity induced by ferric-nitrilotriacetate in HepG2 cells that express cytochrome P450 2E1. Mol Pharmacol. 1998;54:1024–1035. doi: 10.1124/mol.54.6.1024. [DOI] [PubMed] [Google Scholar]
- 55.Wu D, Cederbaum AI. Cyclosporine A protects against arachidonic acid toxicity in rat hepatocytes: role of CYP2E1 and mitochondria. Hepatology. 2002;35:1420–1430. doi: 10.1053/jhep.2002.33639. [DOI] [PubMed] [Google Scholar]
- 56.Lee M, Bae MA. Docosahexaenoic acid induces apoptosis in CYP2E1-containing HepG2 cells by activating the c-Jun N-terminal protein kinase related mitochondrial damage. J Nutr Biochem. 2007;18:348–354. doi: 10.1016/j.jnutbio.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 57.Neve EP, Ingelman-Sundberg M. A soluble NH(2)-terminally truncated catalytically active form of rat cytochrome P450 2E1 targeted to liver mitochondria(1) FEBS Lett. 1999;460:309–304. doi: 10.1016/s0014-5793(99)01361-7. [DOI] [PubMed] [Google Scholar]
- 58.Robin MA, Anandatheerthavarada HK, Fang JK, Cudic M, Otvos L, Avadhani NG. Mitochondrial targeted cytochrome P450 2E1 (P450 MT5) contains an intact N terminus and requires mitochondrial specific electron transfer proteins for activity. J Biol Chem. 2001;276:24680–24689. doi: 10.1074/jbc.M100363200. [DOI] [PubMed] [Google Scholar]
- 59.Raza H, Prabu SK, Robin MA, Avadhani NG. Elevated mitochondrial cytochrome P450 2E1 and glutathione S-transferase A4-4 in streptozotocin-induced diabetic rats: tissue-specific variations and roles in oxidative stress. Diabetes. 2004;53:185–194. doi: 10.2337/diabetes.53.1.185. [DOI] [PubMed] [Google Scholar]