Abstract
Accumulating evidence supports the role of miR-122 in fatty liver disease. We investigated miR-122 expression in a steatotic hepatocyte model, the effect of miR-122 over-expression and inhibition in the pathogenesis. Human hepatic cell line L02 was induced with oleic acid to establish the steatotic hepatocyte model. Intracellular lipid content was observed with laser scanning confocal microscope (LSCM), and triglyceride content was determined with kits. Total RNA was extracted and reversely transcribed into cDNA. miR-122 expression was measured using qRT-PCR. Subsequently, miR-122 mimic and miR-122 inhibitor were transfected into steatotic hepatocytes to observe their effect on intracellular lipid content. The lipid fluorescence intensity and triglyceride content within the steatotic hepatocytes were significantly higher than those in normal control(860.01±26.52 vs 257.77±29.69 and 3.47±0.12 vs 1.85±0.02 at 24 hours) (p<0.01). miR-122 expression in steatotic hepatocytes was down-regulated compared with that in control (2−ΔCt value: 0.0286±0.0078 vs 0.0075±0.0012) (p<<0.01). After transfection, miR-122 expression (2−ΔCt value) in the miR-122 mimic group increased 2.96-fold compared with that in control, and its lipid fluorescence intensity was significantly lower than that in control(790.92±46.72 vs 1022.16±49.66)(p<0.01). Nevertheless, miR-122 expression decreased 3.45-fold in the miR-122 inhibitor group compared with that in control, and its fluorescence intensity was significantly higher than that in control (1386.49±40.34 vs 1022.16±49.66)(p<<0.01). We concluded that miR-122 was down-regulated in steatotic hepatocytes model. The pathogenesis of hepatocyte steatosis was enhanced by miR-122 mimic and reduced with miR-122 inhibitor.
Keywords: miR-122, Hepatocyte, Steatosis, Fatty liver disease
Introduction
Fatty liver disease (FLD), which includes alcoholic fatty liver disease (ALD) and nonalcoholic fatty liver disease (NAFLD), is one of the most common forms of chronic liver diseases and a cause of elevated serum aminotransferases worldwide. fLD is a clinical syndrome defined by lipid accumulation exceeding 5% of the normal range for liver weight or having more than 5% of liver cells displayed increased adiposity under the microscope. NAFLD refers to a spectrum of histological findings ranging from simple fatty liver (SFL) to non-alcoholic steatohepatitis (NASH) and NASH-related cirrhosis, which can progress to hepatocellular carcinoma (HCC) [1–3]. Due to improved living standards and altered dietary habits, the prevalence of NAFLD has been increasing steadily in recent years in the Asia-Pacific region [4, 5]. Our survey conducted in China revealed a prevalence of 15% in a general population, which was close to the incidence in developed countries (20–30%) [6, 7].
As the pathogenesis of FLD is still unclear, the options for its treatment are limited. MicroRNAs (miRNAs or miRs), which are post-transcriptional regulators, can potentially be used to treat fatty liver. They play important roles in modification of cellular processes e.g. proliferation, differentiation and apoptosis by degrading or inhibiting the translation of target mRNA via RNA interference, so as to alter target protein expression. Disruption of the balance will lead to the development of a wide range of disorders. miRs have been studied more extensively in cancers and immune-mediated diseases than in metabolic disease [8–11]. Recently, some studies have demonstrated that miR-122 is crucial in metabolic-related disorders, but the data concerning miR-122 in NAFLD are not as conclusive [12–14]. In order to explore the roles of miR-122 in the pathogenesis of hepatic steatosis, we examined the expression of miR-122 and the effect of miR-122 over-expression and inhibition on steatotic hepatocytes. .
Material and methods
Establishment of steatotic hepatocyte model
Normal hepatocyte cell line L02(provided by Institute of Cell Biology, Chinese Academy of Sciences, Shanghai, China) [15–18] was cultured in RPMI 1640 media containing 10% fetal bovine serum. As high dose of unsaturated fatty acid including oleic acid might and palmitic acid might cause cell death or decreased cell proliferation [19–21], we tried to screen its optimal concentration of culture. Oleic acid at concentrations of 0, 5µg (17.70µM), 10µg (35.40µM), 20µg (70.81µM) and 40µg (141.62µM) /ml dissolved in dimethyl sulfoxide (DMSO) was supplemented to RPMI 1640 media containing 10% fetal bovine serum in order to induce fat-overloading. The methods were referred to literature [22–25]. After 24, 48, and 72h, the cells were harvested to determine the optimal oleic acid concentration for further steatotic hepatocyte culture. The cell morphology and viability were observed under a microscope (Nicon Co., Japan) with and without trypan blue staining. The cell proliferation was evaluated by MTT assay(Sigma Co. USA). The optimal oleic acid concentration for culture was determined from steatotic hepatocytes survival rate and the least destruction. By comparing the results from different groups, the oleic acid concentration of 20 µg (70.81µM)/ml was established to be suitable for the rest of the study. Intracellular fat content was determined by Nile Red staining and a triglyceride quantification kit (Biovision Co. USA). The methods were referred to literature [16, 17]. The cells were stained with Nile Red and viewed under a laser scanning confocal microscope (LSCM) (Olympus fV-1000, Japan). Three steatotic hepatocytes in each of 10 microscopic fields (10×40) were selected randomly. The fluorescence intensity of intracellular lipid droplets was quantified with the software (Olympus fV-3.0 viewer, Japan). The assay was repeated 5 times to obtain an average value. To quantify triglyceride content, hepatocytes (107) cultured in optimal condition harvested at 24, 48 and 72h were used. The OD570 value was determined after ELISA. Normal hepatocytes were used as controls. The intracellular triglyceride concentration (C) was obtained from the equation: C=TS/SV nmol/µl (TS represents triglyceride content in the standard curve, and SV represents sample volume before dilution).
Total RNA extraction and cDNA synthesis
Total RNA containing miR was extracted using TRIZOL Reagent (Invitrogen Life Technologies, USA). The A260/A280 ratio was determined with a spectrometer. RNA integrity was viewed with formaldehyde degeneration gel electrophoresis. RNA was reverse transcribed into complementary DNA (cDNA) with miRCURYLNA™ Universal cDNA Synthesis Kit(Exiqon, Denmark).
Determination of miR-122 expression
The expression of miR-122 in hepatocytes with and without fat was quantified using a PCR assay kit (Exiqon, Denmark). Briefly, miR-122 expression was measured with a 20 µl final reaction volume, which contained 8µl of cDNA dilution, 10µl of SYBR® Green master mix and 2µl of miR-122 primer or U6 snRNA (as an internal control). Initial denaturation at 95°C for 10 min was followed by 40 cycles of denaturation at 95°C for 10 seconds and then 60°C for 1 min. The products were measured with the PCR software Opticon Monitor 2 (MJ Research Inc. USA). Each sample was tested 3 times. The miR level of each sample was calculated by 2−ΔCt, then 2-ΔΔCt, which represented the fold change between two groups (steatotic hepatocyte/ normal hepatocyte). ΔCt= Ct miR value of target gene–Ct value of internal referee U6RNA, ΔΔCt=ΔCt steatotic hepatocyte−ΔCt normal hepatocyte [26].
Transfections of miR-122 mimic and inhibitor
Transient transfection of the oligonucleotides (provided by Dharmacon Co., USA) into steatotic hepatocytes was conducted 24h after oleic acid induction. In miR-122 mimic group, steatotic hepatocytes were transfected with synthetic pre-miR-122 (miRIDIAN mimic, has-miR-122, UGGAGUGUGACAAUGGUGUUUG); in mimic control group, transfected with pre-miR-122 control (miRIDIAN miR mimic transfection control with Dy547); in inhibitor group, transfected with synthetic anti-miR-122 (miRIDIAN miR-122 Hairpin inhibitor); in inhibitor control group, transfected with anti-miR-122 control (miRIDIAN miR hairpin inhibitor transfection control with Dy547). The sequences of pre-miR-122 control, anti-miR-122 and anti-miR-122 control were all kept confidential by Dharmacon Co. In the blank control group, steatotic hepatocytes were co-incubated with a liposome (Lipofectmine™2000 provided by Invitrogen, Co., USA).
For transfection, miR/lipofectamine compound was prepared. 5µl Lipofectamine-2000 and 245µl opti-MEM culture media were incubated at 25°C for 5 min to produce lipofectamine dilution. To produce miR dilution, 5µl(100pmol)of miRIDIAN miR-122 mimic, miRIDIAN miR-122 hairpin inhibitor, or transfection control were diluted with 245µl opti-MEM culture media respectively. Then, 500µl of lipofectamine dilution and 500µl of each miR dilution were mixed and incubated at 25°C for 20min to generate miR/lipofectamine compound. Before transfection, the hepatocytes were cultured in RPMI 1640 medium containing 10% fetal bovine serum in a CO2 incubator at 37°C overnight. The cells were rinsed twice with phosphate buffered saline (PBS), added into 2ml of opti-MEM culture media, then, mixed with 500µl of miR/lipofectamine compound, incubated in CO2, at 37°C for 6h. Afterwards, RPMI 1640 media containing 10% fetal bovine serum were used to replace the compound. Transfection efficiency was evaluated at 6h after transfection with a fluorescence microscope. miR-122 expression was determined at 72h with qRT-PCR. Intracellular lipid droplets were observed before and 72h after transfection using LSCM. The methods were described in previous sections.
Statistical analysis
The data were analyzed with the SPSS 17.0 for Windows statistical package (Chicago, IL, USA). Continuous data with normal distribution were expressed as mean ± standard deviation and examined using the Student’s t-test. Continuous data with skewed distribution were examined using rank sum test. Categorical variables were expressed as a percentage and examined using the Chi-square and fisher’s exact tests. Statistical significance was set at p < 0.05 (two-tailed).
Result
Establishment of a steatotic hepatocyte model
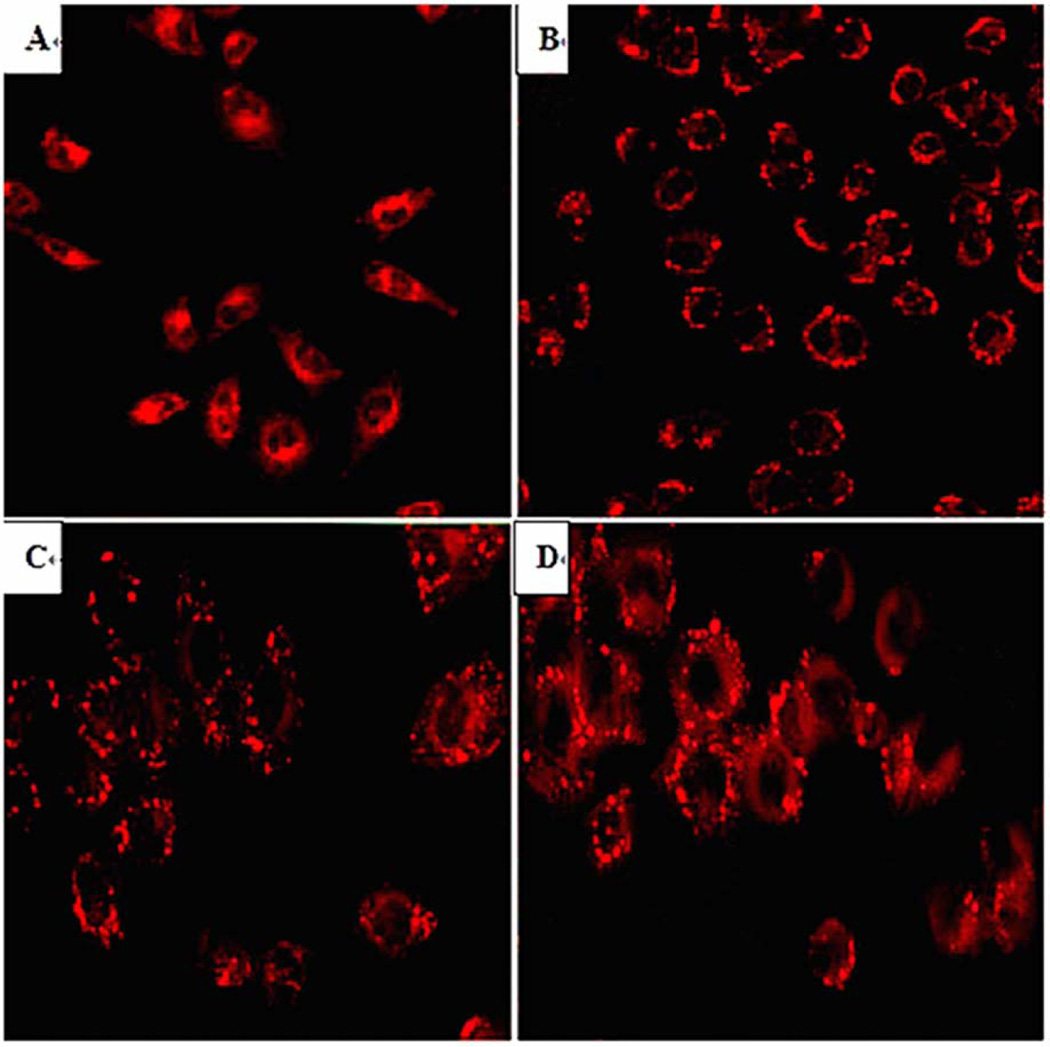
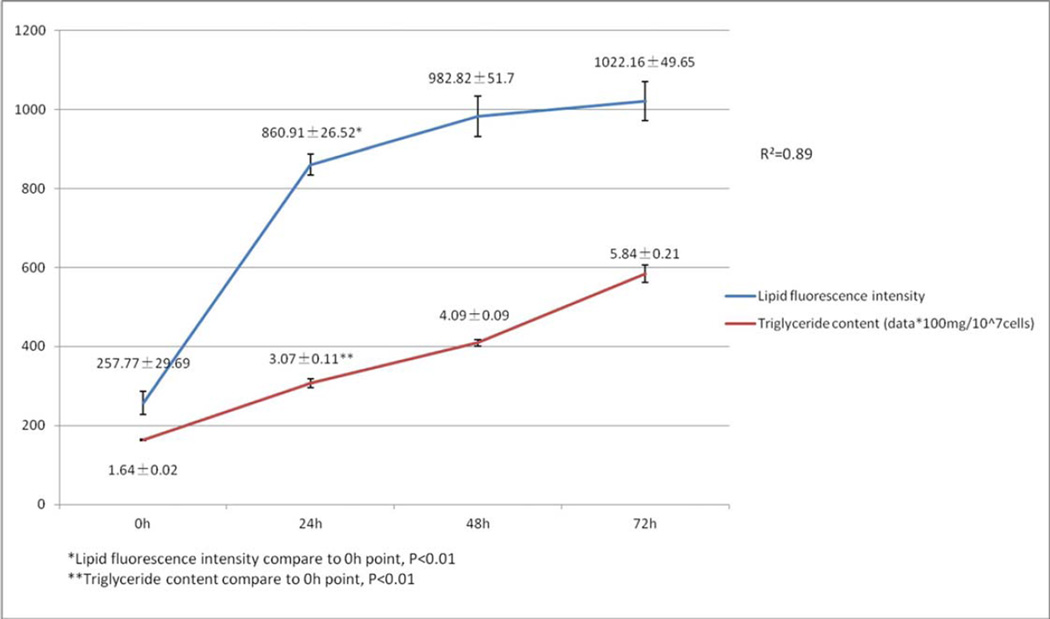
Under light microscope, the hepatocyte cells (L02) had a spindle-shaped with clear cytoplasm. The optional time for cell passage was on day 3 after oleic acid treatment when the cells have become round with blurred margins. More than 95% of cells survived after the treatments. The OD values derived from MTT assay, which reflected cell proliferation, in different oleic acid concentration groups were shown in Table 1. The optional culture condition for this steatotic hepatocyte model was treating with the highest oleic acid concentration since these hepatocytes had the highest cell survival rate and showed proliferation. Oleic acid treatment did not change cell proliferation with the exception of when 40µg (141.62µM)/ml was used. Thus, 20µg (70.81µM)/ml oleic acid was used for subsequent experiments. Before and after oleic acid induction, hepatocyte lipid droplets stained by Nile Red fluorescence were clearly seen under light microscope and LSCM (Fig 1). Both the intracellular mean lipid fluorescence intensity tested by LSCM and the intracellular triglyceride content measured by triglyceride kit increased gradually with prolonged treatment (Fig 2). Because data from the fluorescence intensity correlated well with the triglyceride content (R2=0.89, p<0.01) (Fig 2), fluorescence intensity test alone was used for the rest of the studies. This method was reported in literature [20–22].
Table 1.
Steatotic hepatocyte proliferation in culture media with different oleic acid concentrations
| 24h |
48h |
72h |
||||
|---|---|---|---|---|---|---|
| χ̄±s * | P | χ̄±s * | P | χ̄±s * | P | |
| 0µg/ml (0µM) | 0.528±0.029 | 0.660±0.020 | 0.925±0.022 | |||
| 5µg/ml (17.70µM) | 0.542±0.016 | 0.229 | 0.657±0.017 | 0.704 | 0.922±0.017 | 0.755 |
| 10µg/ml (35.40µM) | 0.546±0.049 | 0.352 | 0.669±0.025 | 0.437 | 0.923±0.017 | 0.776 |
| 20µg/ml (70.81µM) | 0.544±0.036 | 0.343 | 0.653±0.022 | 0.497 | 0.925±0.008 | 0.946 |
| 40µg/ml (141.62µM) | 0.232±0.025 | <0.01 | 0.261±0.022 | <0.01 | 0.305±0.049 | <0.01 |
expressed as OD values
Fig 1.
Hepatocytes stained with Nile Red under LSCM (10×40) at different time points after oleic acid induction
A Normal hepatocytes (0h)
B Steatotic hepatocytes (24h)
C Steatotic hepatocytes (48h)
D Steatotic hepatocytes (72h)
Fig 2.
Intracellular lipid fluorescence intensity and triglyceride content in steatotic hepatocyte model after oleic acid induction
Expression of miR-122 in steatotic hepatocytes
The miR-122 expression in steatotic hepatocytes expressed with 2−ΔCt(0.0075±0.0012) tested by qRT-PCR was significantly decreased than that (0.0286±0.0078) in control of normal hepatocytes (p<0.01).
The relationship between miR-122 and fat content in hepatocytes
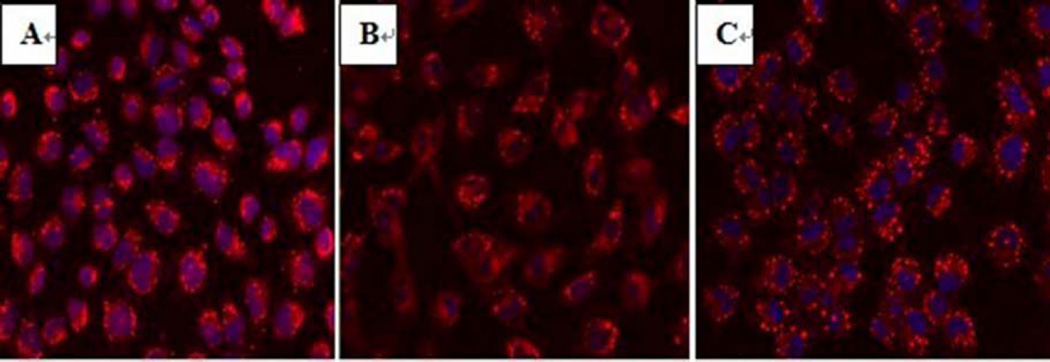
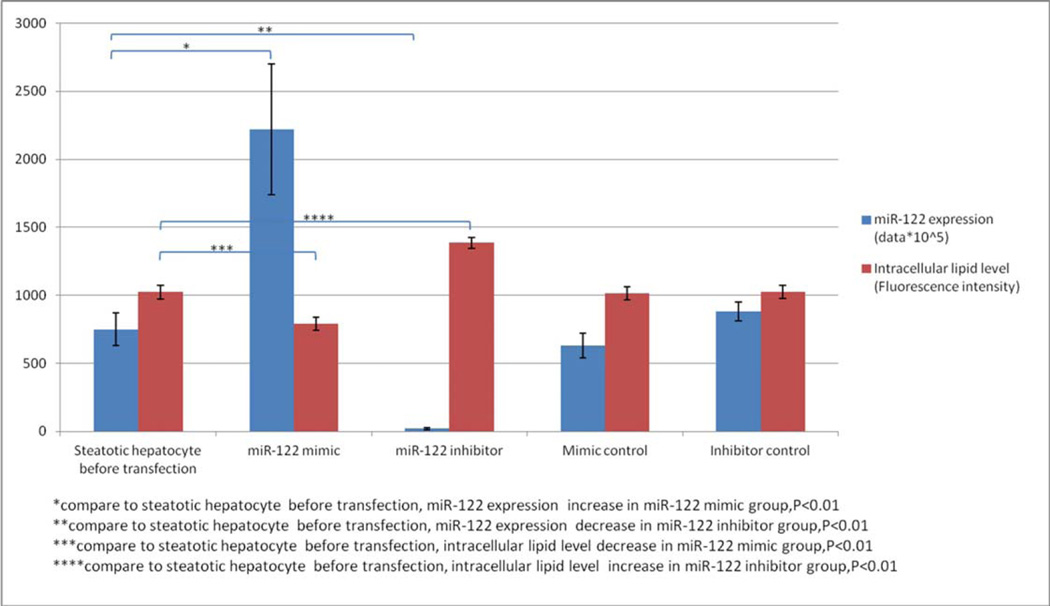
miR-122 level decreased significantly (0.0286±0.0078) after 3 day oleic acid treatment. The miR-122 mimic (pre-miR-122), mimic control (pre-miR-122 control), miR-122 inhibitor (anti-miR-122) and inhibitor control (anti-miR-122 control) were transfected into steatotic hepatocytes respectively. High transfection rate (>70%) determined by oligonucleotides (marked by Dy-547) confirmed successful transfections. Intracellular lipid fluorescence intensity of the steatotic hepatocytes was observed by LSCM (Fig 3). After transfection with miR-122 mimic, intracellular lipid fluorescence intensity decreased significantly (Fig 3B) compared with that before transfection (Fig 3A). In contrast, the fluorescence intensity was increased significantly after transfection with miR-122 inhibitor (Fig 3C). The miR-122 expression (2−ΔCt value) had a 2.96-fold increase in the miR-122 mimic group (0.0222±0.0048) compared with that (0.0075±0.0012) in the steatotic hepatocytes before transfection (control) (p<0.01). Its intracellular lipid fluorescence intensity (790.92±46.72)was significantly lower than that (1022.16±49.66) in control (p<0.01). The miR-122 expression (2−ΔCt value) decreased 3.75-fold in the miR-122 inhibitor group (0.0002±0.0001) compared with that (0.0075±0.0012) in control (p<0.01), and its intracellular lipid droplet fluorescence intensity(1386.49±403.44) was significantly higher than that(1022.16±49.66)in control (p<0.01). The miR-122 expression and fluorescence intensity in miR-122 mimic control and inhibitor control groups did not differ from those in control(p>0.05). The miR-122 expression and intracellular lipid levels before and after transfection with miR-122 mimic, miR-122 inhibitor and their controls were shown in Table 2 and Figure 4.
Fig 3.
Hepatocytes stained with Nile Red under LSCM (10×40) before and after transfection
A Steatotic hepatocytes before transfection (control)
B Steatotic hepatocytes transfected with miR-122 mimic
C Steatotic hepatocytes transfected with miR-122 inhibitor
Table 2.
miR-122 expression and intracellular lipid levels after transfection with miR-122 mimic, miR-122 inhibitor and their controls
| miR-122 expression | Intracellular lipid level (Fluorescence intensity) |
P | |
|---|---|---|---|
| miR-122 mimic | 0.0222±0.0048 | 790.92±46.72 | <0.01 |
| miR-122 inhibitor | 0.0002±0.0001 | 1386.49±40.34 | <0.01 |
| Mimic control | 0.0063±0.0009 | 1015.65±47.34 | >0.05 |
| Inhibitor control | 0.0088±0.0007 | 1025.12±47.52 | >0.05 |
| Steatotic hepatocyte before transfection | 0.0075±0.0012 | 1022.16±49.65 | - |
Fig 4.
miR-122 expression and intracellular lipid levels after transfection with miR-122 mimic, miR-122 inhibitor and their controls
Discussion
During the past decade, the role of epigenetic mechanisms in the pathogenesis of diseases has been increasingly recognized. Epigenetic modification, mainly including miRs, DNA methylation and histone modification, refers to phenotypic changes caused by the mechanisms unrelated to changes in the underlying DNA sequence. Among epigenetic modifications, miRs are studied most extensively in liver diseases. miRs are a class of endogenously expressed small regulatory noncoding RNAs regulating mRNA degradation or translation inhibition by binding with imperfect complementarity in their 3' untranslated region (UTR), subsequently altering protein expression of target genes [27–29].
Since the first discovery in 1993, many miRs in a variety of organisms have been determined. In 2011, more than 1420 miRs have been identified in humans and miR-122 expression has been detected in 18 vertebrates so far (miRBase v17). (http://www.mirbase.org/) [9]. miR-122 was first identified in 2002. By using cloning approaches in various mouse tissues, the study uncovered a number of novel miRs including miR-122, which was highly enriched in the liver but absent from other tissues [30]. The precise molecular function of miR-122 in humans is largely unknown. It is one of many tissue-specific miRs important for establishing patterns of gene expression that may be responsible for maintaining the differentiated state of a tissue [30–32]. Accumulating evidence supports the effects of miR-122 in lipid and cholesterol metabolism, and adipocyte differentiation [33]. In humans, miR-122 is expressed in the developing liver and at high levels in the adult liver, where it makes up 70% of all miRs. RNase protection analysis indicated that miR-122 was present at approximately 66,000 copies per cell in adult liver [34]. In NASH patients, miR-122 was significantly under-expressed (63%) compared to normal controls [34, 35]. In the first clinical study regarding miR-122 with NASH, the miR profiles of 15 patients with biopsy proven NASH and 15 normal controls were investigated. Out of a total of 474 tested miRs, 46 were differentially expressed in NASH with 23 being down-regulated (in particular, miR-122), and 23 being up-regulated (in particular, miR-34a and miR-146b). These differentially expressed miRs were further validated by quantitative real-time PCR [35]. Serum levels of miR-122 were significantly elevated in NAFLD patients than in controls, and positively correlated with disease severity from simple steatosis to steatohepatitis. miR-122 was also associated with liver enzyme levels, fibrosis stages, and inflammation activities [36]. Inhibition studies have further our understanding of the critical role played by miR-122 in lipid synthesis modulation and NAFLD. In normal mice, miR-122 inhibition with antisense oligonucleotide (ASO) resulted in reduced plasma cholesterol level, increased hepatic fatty-acid oxidation, and decreased rates of hepatic fatty-acid and cholesterol synthesis. Activation of the central metabolic sensor AMPK was also increased due to the inhibition of miR-122. In a diet-induced obesity mouse model, miR-122 inhibition with ASO caused decreased plasma cholesterol levels, significant improvement of liver steatosis and reduced expression of several lipogenic genes [37]. All these findings strongly suggested the significance of miR-122 in the regulation of lipid metabolism and the contribution to NAFLD development. However, the phenotypes of miR-122 in hepatic and non-hepatic tissues might not always be concordant. In liver-specific knockouts (LKO) and germline knockouts (KO) mice, miR-122 inhibition was shown to reduce 30% serum total cholesterol via down-regulation of genes involved in cholesterol biosynthesis such as the rate-limiting enzyme 3-hydroxy-3-methylglutaryl coenzyme A (HMGCoA) reductase [37], but the livers developed progressive steatohepatitis [38, 39]. In animal study, genes involved in lipid synthesis in the liver e.g. Agpat1, Mogat1, Agpat3, Agpat9, Ppap2a, Ppap2c and Cidec were found to be the direct targets of miR-122 [40]. miR-122 was shown to link to the output system of the circadian clock by regulating circadianly expressed genes [41].
The roles of miR-122 in other liver diseases have also been documented. miR-122 is required for hepatitis C virus (HCV) replication in cultured human hepatic cell line Huh7 [42, 43]. In patients with chronic hepatitis C (CHC), serum levels of miR-122 were correlated with liver enzyme levels, fibrosis stage and inflammatory activity. miR-122 enhanced the replication of HCV and influenced the efficiency of interferon therapy [36]. miR-122 levels were frequently reduced in hepatocellular carcinoma (HCC) compared with those in normal liver [44] and were correlated with poor prognosis [45]. Over-expression of miR-122 reduced tumorigenic properties of HCC cell lines [46].
Besides miR-122, other miRs have also been demonstrated to be involved in NAFLD development. miR-34a and miR-146b were shown to be significantly over-expressed (99% and 80%, respectively) in human NASH [35]. The expression of miR-335 in the liver was up-regulated in mice. The increased miR-335 expression was associated with increased body, liver and white adipose tissue weight, as well as elevated hepatic triglyceride and cholesterol levels. furthermore, hepatic miR-335 level was closely correlated with the expression of adipocyte differentiation markers, i.e. PPAR-α and fAS in adipocyte [47]. The presence of miR-181d significantly decreased lipid droplets in the liver (60%), and subsequently reduced cellular triglyceride and cholesterol [48]. miR-10b regulated steatosis level through PPAR-α pathway in a steatotic hepatocyte (L02 cell line) model. Post-transcriptional regulation of PPAR-α by miR-10b was maintained by a single binding site [49]. In a smaller sample clinical study (n=12/group), many miRs (-132, -150, -433, -28, -511, -517a, -671) were found to be differentially expressed in patients with NAFLD [50]. Other miRs involved in NAFLD included miR-16, -29c, -33 (up-regulated) and miR-99b, -150 (down-regulated) [14]. So far, there is insufficient evidence to support the relation of these miRs with NAFLD.
To our knowledge, this is the first attempt to observe the effect miR-122 transfection on steatotic hepatocytes in vitro. We find that the miR-122 expression increased, but lipid content decreased significantly after miR-122 mimic transfection. On the contrary, the miR-122 expression decreased, but the lipid content increased significantly after miR-122 inhibitor transfection. These findings were consistent with the data observed in animal studies with ASO inhibition, and demonstrated the importance of miR-122 in the pathogenesis of fatty liver disease. miR-122 may be an attractive target for NAFLD diagnosis and treatment.
Footnotes
This article has been accepted for publication and undergone full peer review but has not been through the copyediting, typesetting, pagination and proofreading process, which may lead to differences between this version and the Version of Record.
References
- 1.Argo CK, Caldwell SH. Epidemiology and natural history of non-alcoholic steatohepatitis. Clinics in liver disease. 2009;13(4):511–531. doi: 10.1016/j.cld.2009.07.005. [PMID: 19818302 DOI: 10.1016/j.cld.2009.07.005] [DOI] [PubMed] [Google Scholar]
- 2.Bellentani S, Scaglioni F, Marino M, Bedogni G. Epidemiology of non-alcoholic fatty liver disease. Dig Dis. 2010;28(1):155–161. doi: 10.1159/000282080. [PMID: 20460905 DOI: 10.1159/000282080] [DOI] [PubMed] [Google Scholar]
- 3.Vernon G, Baranova A, Younossi ZM. Systematic review: the epidemiology and natural history of non-alcoholic fatty liver disease and non-alcoholic steatohepatitis in adults. Aliment Pharmacol Ther. 2011;34(3):274–285. doi: 10.1111/j.1365-2036.2011.04724.x. [PMID: 21623852 DOI: 10.1111/j.1365-2036.2011.04724.x] [DOI] [PubMed] [Google Scholar]
- 4.Fan JG, Zhu J, Li XJ, Chen L, Lu YS, Li L, Dai F, Li F, Chen SY. Fatty liver and the metabolic syndrome among Shanghai adults. Journal of gastroenterology and hepatology. 2005;20(12):1825–1832. doi: 10.1111/j.1440-1746.2005.04058.x. [PMID: 16336439 DOI: 10.1111/j.1440-1746.2005.04058.x] [DOI] [PubMed] [Google Scholar]
- 5.Amarapurkar DN, Hashimoto E, Lesmana LA, Sollano JD, Chen PJ, Goh KL. How common is non-alcoholic fatty liver disease in the Asia-Pacific region and are there local differences? Journal of gastroenterology and hepatology. 2007;22(6):788–793. doi: 10.1111/j.1440-1746.2007.05042.x. [PMID: 17565631 DOI: 10.1111/j.1440-1746.2007.05042.x] [DOI] [PubMed] [Google Scholar]
- 6.Zhou YJ, Li YY, Nie YQ, Huang CM, Cao CY. Natural course of nonalcoholic fatty liver disease in southern China: a prospective cohort study. Journal of digestive diseases. 2012;13(3):153–160. doi: 10.1111/j.1751-2980.2011.00571.x. [PMID: 22356310 DOI: 10.1111/j.1751-2980.2011.00571.x] [DOI] [PubMed] [Google Scholar]
- 7.Zhou YJ, Li YY, Nie YQ, Ma JX, Lu LG, Shi SL, Chen MH, Hu PJ. Prevalence of fatty liver disease and its risk factors in the population of South China. World journal of gastroenterology : WJG. 2007;13(47):6419–6424. doi: 10.3748/wjg.v13.i47.6419. [PMID: 18081233] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tili E, Michaille JJ, Croce CM. MicroRNAs play a central role in molecular dysfunctions linking inflammation with cancer. Immunological reviews. 2013;253(1):167–184. doi: 10.1111/imr.12050. [PMID: 23550646 DOI: 10.1111/imr.12050] [DOI] [PubMed] [Google Scholar]
- 9.Tomankova T, Petrek M, Gallo J, Kriegova E. MicroRNAs: emerging regulators of immune-mediated diseases. Scandinavian journal of immunology. 2011 doi: 10.1111/j.1365-3083.2011.02650.x. [PMID: 21988491 DOI: 10.1111/j.1365-3083.2011.02650.x] [DOI] [PubMed] [Google Scholar]
- 10.Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic acids research. 2011;39(Database issue):D152–D157. doi: 10.1093/nar/gkq1027. [PMID: 21037258 PMCID: 3013655 DOI: 10.1093/nar/gkq1027] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feinberg AP. Phenotypic plasticity and the epigenetics of human disease. Nature. 2007;447(7143):433–440. doi: 10.1038/nature05919. [PMID: 17522677 DOI: 10.1038/nature05919] [DOI] [PubMed] [Google Scholar]
- 12.Fernandez-Hernando C, Suarez Y, Rayner KJ, Moore KJ. MicroRNAs in lipid metabolism. Current opinion in lipidology. 2011;22(2):86–92. doi: 10.1097/MOL.0b013e3283428d9d. [PMID: 21178770 PMCID: 3096067 DOI: 10.1097/MOL.0b013e3283428d9d] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheung O, Sanyal AJ. Role of microRNAs in non-alcoholic steatohepatitis. Current pharmaceutical design. 2010;16(17):1952–1957. doi: 10.2174/138161210791208866. [PMID: 20370674] [DOI] [PubMed] [Google Scholar]
- 14.Ceccarelli S, Panera N, Gnani D, Nobili V. Dual Role of MicroRNAs in NAFLD. International journal of molecular sciences. 2013;14(4):8437–8455. doi: 10.3390/ijms14048437. [PMID: 23594995 PMCID: 3645753 DOI: 10.3390/ijms14048437] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang X, Wang D, Jing P, Wu Y, Xia Y, Chen M, Hong L. A novel Golgi retention signal RPWS for tumor suppressor UBIAD1. PloS one. 2013;8(8):e72015. doi: 10.1371/journal.pone.0072015. [PMID: 23977195 PMCID: 3747158 DOI: 10.1371/journal.pone.0072015] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang XH, Tian Y, Guo ZJ, Fan ZP, Qiu de K, Zeng MD. Cholesterol metabolism and expression of its relevant genes in cultured steatotic hepatocytes. Journal of digestive diseases. 2009;10(4):310–314. doi: 10.1111/j.1751-2980.2009.00401.x. [PMID: 19906111 DOI: 10.1111/j.1751-2980.2009.00401.x] [DOI] [PubMed] [Google Scholar]
- 17.Yang LH, Chen DF. [Effects of TNF alpha on the expression of SCAP and triglyceride contents in cultured steatotic hepatocytes] Zhonghua gan zang bing za zhi = Zhonghua ganzangbing zazhi = Chinese journal of hepatology. 2007;15(10):767–770. [PMID: 17963605] [PubMed] [Google Scholar]
- 18.Chen H, Luo Z, Sun W, Zhang C, Sun H, Zhao N, Ding J, Wu M, Li Z, Wang H. Low glucose promotes CD133mAb-elicited cell death via inhibition of autophagy in hepatocarcinoma cells. Cancer letters. 2013;336(1):204–212. doi: 10.1016/j.canlet.2013.04.031. [PMID: 23652197 DOI: 10.1016/j.canlet.2013.04.031] [DOI] [PubMed] [Google Scholar]
- 19.Cui W, Chen SL, Hu KQ. Quantification and mechanisms of oleic acid-induced steatosis in HepG2 cells. American journal of translational research. 2010;2(1):95–104. [PMID: 20182586 PMCID: 2826826] [PMC free article] [PubMed] [Google Scholar]
- 20.Gomez-Lechon MJ, Donato MT, Martinez-Romero A, Jimenez N, Castell JV, O'Connor JE. A human hepatocellular in vitro model to investigate steatosis. Chemico-biological interactions. 2007;165(2):106–116. doi: 10.1016/j.cbi.2006.11.004. [PMID: 17188672 DOI: 10.1016/j.cbi.2006.11.004] [DOI] [PubMed] [Google Scholar]
- 21.Wu HR, Chen SH, Lu Y, Li YM. [Comparison of two nonalcoholic hepatocellular steatosis models] Zhonghua gan zang bing za zhi = Zhonghua ganzangbing zazhi = Chinese journal of hepatology. 2010;18(4):297–299. doi: 10.3760/cma.j.issn.1007-3418.2010.04.015. [PMID: 20460052 DOI: 10.3760/cma.j.issn.1007-3418.2010.04.015] [DOI] [PubMed] [Google Scholar]
- 22.Malhi H, Bronk SF, Werneburg NW, Gores GJ. Free fatty acids induce JNK-dependent hepatocyte lipoapoptosis. The Journal of biological chemistry. 2006;281(17):12093–12101. doi: 10.1074/jbc.M510660200. [PMID: 16505490 DOI: 10.1074/jbc.M510660200] [DOI] [PubMed] [Google Scholar]
- 23.De Gottardi A, Vinciguerra M, Sgroi A, Moukil M, Ravier-Dall'Antonia F, Pazienza V, Pugnale P, Foti M, Hadengue A. Microarray analyses and molecular profiling of steatosis induction in immortalized human hepatocytes. Laboratory investigation; a journal of technical methods and pathology. 2007;87(8):792–806. doi: 10.1038/labinvest.3700590. [PMID: 17558421 DOI: 10.1038/labinvest.3700590] [DOI] [PubMed] [Google Scholar]
- 24.Malhi H, Barreyro FJ, Isomoto H, Bronk SF, Gores GJ. Free fatty acids sensitise hepatocytes to TRAIL mediated cytotoxicity. Gut. 2007;56(8):1124–1131. doi: 10.1136/gut.2006.118059. [PMID: 17470478 PMCID: 1955518 DOI: 10.1136/gut.2006.118059] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao JJ, Zhao LF, Yang H, Zhang L. [Effects of PPAR-alpha activation on oleic acid-induced steatosis and expression of heme oxygenase-1 in HepG2 cells] Zhonghua gan zang bing za zhi = Zhonghua ganzangbing zazhi = Chinese journal of hepatology. 2013;21(3):218–221. [PMID: 23967745] [PubMed] [Google Scholar]
- 26.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nature protocols. 2008;3(6):1101–1108. doi: 10.1038/nprot.2008.73. [PMID: 18546601] [DOI] [PubMed] [Google Scholar]
- 27.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [PMID: 14744438] [DOI] [PubMed] [Google Scholar]
- 28.Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annual review of biochemistry. 2010;79:351–379. doi: 10.1146/annurev-biochem-060308-103103. [PMID: 20533884 DOI: 10.1146/annurev-biochem-060308-103103] [DOI] [PubMed] [Google Scholar]
- 29.Ambros V. The functions of animal microRNAs. Nature. 2004;431(7006):350–355. doi: 10.1038/nature02871. [PMID: 15372042 DOI: 10.1038/nature02871] [DOI] [PubMed] [Google Scholar]
- 30.Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Current biology : CB. 2002;12(9):735–739. doi: 10.1016/s0960-9822(02)00809-6. [PMID: 12007417] [DOI] [PubMed] [Google Scholar]
- 31.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433(7027):769–773. doi: 10.1038/nature03315. [PMID: 15685193 DOI: 10.1038/nature03315] [DOI] [PubMed] [Google Scholar]
- 32.Jopling C. Liver-specific microRNA-122: Biogenesis and function. RNA biology. 2012;9(2):137–142. doi: 10.4161/rna.18827. [PMID: 22258222 PMCID: 3346312 DOI: 10.4161/rna.18827] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. Silencing of microRNAs in vivo with 'antagomirs'. Nature. 2005;438(7068):685–689. doi: 10.1038/nature04303. [PMID: 16258535 DOI: 10.1038/nature04303] [DOI] [PubMed] [Google Scholar]
- 34.Chang J, Nicolas E, Marks D, Sander C, Lerro A, Buendia MA, Xu C, Mason WS, Moloshok T, Bort R, Zaret KS, Taylor JM. miR-122, a mammalian liver-specific microRNA, is processed from hcr mRNA and may downregulate the high affinity cationic amino acid transporter CAT-1. RNA biology. 2004;1(2):106–113. doi: 10.4161/rna.1.2.1066. [PMID: 17179747] [DOI] [PubMed] [Google Scholar]
- 35.Cheung O, Puri P, Eicken C, Contos MJ, Mirshahi F, Maher JW, Kellum JM, Min H, Luketic VA, Sanyal AJ. Nonalcoholic steatohepatitis is associated with altered hepatic MicroRNA expression. Hepatology. 2008;48(6):1810–1820. doi: 10.1002/hep.22569. [PMID: 19030170 PMCID: 2717729 DOI: 10.1002/hep.22569] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cermelli S, Ruggieri A, Marrero JA, Ioannou GN, Beretta L. Circulating microRNAs in patients with chronic hepatitis C and non-alcoholic fatty liver disease. PloS one. 2011;6(8):e23937. doi: 10.1371/journal.pone.0023937. [PMID: 21886843 PMCID: 3160337 DOI: 10.1371/journal.pone.0023937] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Esau C, Davis S, Murray SF, Yu XX, Pandey SK, Pear M, Watts L, Booten SL, Graham M, McKay R, Subramaniam A, Propp S, Lollo BA, Freier S, Bennett CF, Bhanot S, Monia BP. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell metabolism. 2006;3(2):87–98. doi: 10.1016/j.cmet.2006.01.005. [PMID: 16459310 DOI: 10.1016/j.cmet.2006.01.005] [DOI] [PubMed] [Google Scholar]
- 38.Tsai WC, Hsu SD, Hsu CS, Lai TC, Chen SJ, Shen R, Huang Y, Chen HC, Lee CH, Tsai TF, Hsu MT, Wu JC, Huang HD, Shiao MS, Hsiao M, Tsou AP. MicroRNA-122 plays a critical role in liver homeostasis and hepatocarcinogenesis. The Journal of clinical investigation. 2012;122(8):2884–2897. doi: 10.1172/JCI63455. [PMID: 22820290 PMCID: 3408747 DOI: 10.1172/JCI63455] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hsu SH, Wang B, Kota J, Yu J, Costinean S, Kutay H, Yu L, Bai S, La Perle K, Chivukula RR, Mao H, Wei M, Clark KR, Mendell JR, Caligiuri MA, Jacob ST, Mendell JT, Ghoshal K. Essential metabolic, anti-inflammatory, and anti-tumorigenic functions of miR-122 in liver. The Journal of clinical investigation. 2012;122(8):2871–2883. doi: 10.1172/JCI63539. [PMID: 22820288 PMCID: 3408748 DOI: 10.1172/JCI63539] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim YJ, Cho SY, Yun CH, Moon YS, Lee TR, Kim SH. Transcriptional activation of Cidec by PPARgamma2 in adipocyte. Biochemical and biophysical research communications. 2008;377(1):297–302. doi: 10.1016/j.bbrc.2008.09.129. [PMID: 18845124 DOI: 10.1016/j.bbrc.2008.09.129] [DOI] [PubMed] [Google Scholar]
- 41.Gatfield D, Le Martelot G, Vejnar CE, Gerlach D, Schaad O, Fleury-Olela F, Ruskeepaa AL, Oresic M, Esau CC, Zdobnov EM, Schibler U. Integration of microRNA miR-122 in hepatic circadian gene expression. Genes & development. 2009;23(11):1313–1326. doi: 10.1101/gad.1781009. [PMID: 19487572 PMCID: 2701584 DOI: 10.1101/gad.1781009] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jopling CL, Schutz S, Sarnow P. Position-dependent function for a tandem microRNA miR-122-binding site located in the hepatitis C virus RNA genome. Cell host & microbe. 2008;4(1):77–85. doi: 10.1016/j.chom.2008.05.013. [PMID: 18621012 PMCID: 3519368 DOI: 10.1016/j.chom.2008.05.013] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309(5740):1577–1581. doi: 10.1126/science.1113329. [PMID: 16141076 DOI: 10.1126/science.1113329] [DOI] [PubMed] [Google Scholar]
- 44.Kutay H, Bai S, Datta J, Motiwala T, Pogribny I, Frankel W, Jacob ST, Ghoshal K. Downregulation of miR-122 in the rodent and human hepatocellular carcinomas. Journal of cellular biochemistry. 2006;99(3):671–678. doi: 10.1002/jcb.20982. [PMID: 16924677 PMCID: 3033198 DOI: 10.1002/jcb.20982] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Coulouarn C, Factor VM, Andersen JB, Durkin ME, Thorgeirsson SS. Loss of miR-122 expression in liver cancer correlates with suppression of the hepatic phenotype and gain of metastatic properties. Oncogene. 2009;28(40):3526–3536. doi: 10.1038/onc.2009.211. [PMID: 19617899 PMCID: 3492882 DOI: 10.1038/onc.2009.211] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bai S, Nasser MW, Wang B, Hsu SH, Datta J, Kutay H, Yadav A, Nuovo G, Kumar P, Ghoshal K. MicroRNA-122 inhibits tumorigenic properties of hepatocellular carcinoma cells and sensitizes these cells to sorafenib. The Journal of biological chemistry. 2009;284(46):32015–32027. doi: 10.1074/jbc.M109.016774. [PMID: 19726678 PMCID: 2797273 DOI: 10.1074/jbc.M109.016774] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nakanishi N, Nakagawa Y, Tokushige N, Aoki N, Matsuzaka T, Ishii K, Yahagi N, Kobayashi K, Yatoh S, Takahashi A, Suzuki H, Urayama O, Yamada N, Shimano H. The up-regulation of microRNA-335 is associated with lipid metabolism in liver and white adipose tissue of genetically obese mice. Biochemical and biophysical research communications. 2009;385(4):492–496. doi: 10.1016/j.bbrc.2009.05.058. [PMID: 19460359 DOI: 10.1016/j.bbrc.2009.05.058] [DOI] [PubMed] [Google Scholar]
- 48.Whittaker R, Loy PA, Sisman E, Suyama E, Aza-Blanc P, Ingermanson RS, Price JH, McDonough PM. Identification of MicroRNAs that control lipid droplet formation and growth in hepatocytes via high-content screening. Journal of biomolecular screening. 2010;15(7):798–805. doi: 10.1177/1087057110374991. [PMID: 20639500 DOI: 10.1177/1087057110374991] [DOI] [PubMed] [Google Scholar]
- 49.Zheng L, Lv GC, Sheng J, Yang YD. Effect of miRNA-10b in regulating cellular steatosis level by targeting PPAR-alpha expression, a novel mechanism for the pathogenesis of NAFLD. Journal of gastroenterology and hepatology. 2010;25(1):156–163. doi: 10.1111/j.1440-1746.2009.05949.x. [PMID: 19780876 DOI: 10.1111/j.1440-1746.2009.05949.x] [DOI] [PubMed] [Google Scholar]
- 50.Estep M, Armistead D, Hossain N, Elarainy H, Goodman Z, Baranova A, Chandhoke V, Younossi ZM. Differential expression of miRNAs in the visceral adipose tissue of patients with non-alcoholic fatty liver disease. Aliment Pharmacol Ther. 2010;32(3):487–497. doi: 10.1111/j.1365-2036.2010.04366.x. [PMID: 20497147 DOI: 10.1111/j.1365-2036.2010.04366.x] [DOI] [PubMed] [Google Scholar]