Abstract
Advances in donor matching and immunosuppressive therapies have decreased the prevalence of acute rejection of cardiac grafts; however, chronic rejection remains a significant obstacle for long-term allograft survival. While initiating elements of anti-allograft immune responses have been identified, the linkage between these factors and the ultimate development of cardiac fibrosis is not well understood. Tissue fibrosis resembles an exaggerated wound healing response, in which extracellular matrix (ECM) molecules are central. One such ECM molecule is an alternatively spliced isoform of the ubiquitous glycoprotein fibronectin (FN), termed extra domain A-containing cellular fibronectin (EDA cFN). EDA cFN is instrumental in fibrogenesis; thus, we hypothesized that it might also regulate fibrotic remodelling associated with chronic rejection. We compared the development of acute and chronic cardiac allograft rejection in EDA cFN-deficient (EDA−/−) and wild-type (WT) mice. While EDA−/− mice developed acute cardiac rejection in a manner indistinguishable from WT controls, cardiac allografts in EDA−/− mice were protected from fibrosis associated with chronic rejection. Decreased fibrosis was not associated with differences in cardiomyocyte hypertrophy or intra-graft expression of pro-fibrotic mediators. Further, we examined expression of EDA cFN and total FN by whole splenocytes under conditions promoting various T-helper lineages. Conditions supporting regulatory T-cell (Treg) development were characterized by greatest production of total FN and EDA cFN, though EDA cFN to total FN ratios were highest in Th1 cultures. These findings indicate that recipient-derived EDA cFN is dispensable for acute allograft rejection responses but that it promotes the development of fibrosis associated with chronic rejection. Further, conditions favouring the development of regulatory T cells, widely considered graft-protective, may drive production of ECM molecules which enhance deleterious remodelling responses. Thus, EDA cFN may be a therapeutic target for ameliorating fibrosis associated with chronic cardiac allograft rejection.
Keywords: allograft, fibrosis, fibronectin, immune response, transplantation
Introduction
Cardiac transplantation is an important therapeutic option for patients with end-stage cardiac dysfunction due to infiltrative disease, ischaemia, or non-ischaemic cardiomyopathy. While improvements in donor matching and immunosuppressive regimens have greatly decreased the prevalence of acute allograft rejection, they have had much less of an impact on chronic rejection, which remains a leading cause of long-term patient morbidity and mortality. Chronic cardiac allo-graft rejection is characterized by vascular remodelling and occlusion [cardiac allograft vasculopathy (CAV)], myocardial hypertrophy, and excessive extracellular matrix (ECM) deposition leading to interstitial fibrosis, all of which contribute to progressive graft dysfunction [1-3].
Recipient immune responses against donor graft antigens are believed to play a key role in chronic rejection and there is significant potential for interplay between immune cells and ECM molecules [4-6]. The robust mononuclear cell infiltrate associated with unmodified acute rejection response is characterized by a dominant elaboration of Th1 cytokines, especially interferon (IFN)-γ [7,8]. In contrast, chronic transplant rejection is thought to involve a sub-acute inflammatory process that over time gives way to a chronic fibroproliferative response, ultimately resulting in allograft fibrosis and functional insufficiency [9-11]. Fibrotic remodelling in chronic rejection is associated with expression of TGF-β, which in the setting of cardiac transplantation can both dampen inflammation [12,13] and promote ECM production [14,15]. Thus, while TGF-β is important in chronic rejection, its relationship to co-mediators of cardiac fibrosis is not well understood.
Chronic rejection resembles exaggerated wound healing responses, which are by nature driven by ECM molecules. The ubiquitously expressed ECM glycoprotein fibronectin (FN) circulates in soluble form in plasma (plasma FN or pFN), but also constitutes a major fraction of the cell-derived poorly soluble or insoluble FN (cellular FN or cFN) fibrils in fibrotic tissues [16-18]. The distinction between pFN and cFN rests largely on the products of FN pre-mRNA alternative splicing. Specifically, cFN includes one or both of the alternatively spliced domains [termed extra domain A (EDA) and extra domain B (EDB)], whereas pFN lacks both domains (reviewed in ref 19). With respect to tissue fibrosis, EDA-containing cFN (EDA cFN) is better studied, having unique functions in immune signalling and activation, as well as myofibroblast differentiation [16,20,21].
Interestingly, studies have implicated a role for EDA cFN in both acute and chronic rejection of cardiac allografts [22-24]. Early observations associated EDA cFN expression with graft-infiltrating leukocytes, particularly macrophages [22]. More recent studies have demonstrated intra-graft expression of EDA cFN within lesions of CAV and interstitial fibrosis [23,25]. Given these associations between EDA cFN and chronic cardiac allograft rejection, as well as the known role of EDA cFN in promoting fibrosis, we sought to define the role of EDA cFN in chronic cardiac allograft rejection using a vascularized heterotopic mouse model to compare acute and chronic rejection in EDA cFN-deficient (EDA−/−) and wild-type (WT) recipients.
Materials and methods
Mice
Female WT C57BL/6 (H-2b) and BALB/c (H-2d) mice were purchased from Jackson Laboratories (Bar Harbor, ME, USA). All animals were kept under microisolator conditions as reviewed and approved by the University of Michigan’s Committee on the Use and Care of Animals. Female EDA−/− mice on a C57BL/6 background have been described previously [26].
Culture media
Cells were cultured in RPMI 1640 supplemented with 2% FCS, 1 mM sodium pyruvate, 100 U/ml penicillin, 100 μg/ml streptomycin, 1.6 mM L-glutamine, 10 mM HEPES buffer (Invitrogen, Carlsbad, CA, USA), 0.27 mM L-asparagine, 1.4 mM L-arginine HCl, 14 μM folic acid, and 50 μM 2-ME (Sigma-Aldrich, St Louis, MO, USA).
Blood analysis
Peripheral blood was harvested by direct cardiac puncture from 4- to 6-week-old euthanized mice into EDTA-containing tubes. Whole blood was analysed for white cell counts and differentials on a Technicon H1 blood analyzer (Siemens Diagnostics, Deerfield, IL, USA) with veterinary software and mouse parameters.
3H-thymidine proliferation assay
WT and EDA−/− splenocytes (1 × 106 per ml) were harvested and cultured in a 96-well plate for 72 h in the presence or absence of concanavalin A (Con A) (1 μg/ml). Cells were pulsed with 3H-thymidine for the final 18 h of culture. Cultures were then harvested onto fibre mats and radioactive counts per minute (cpm) were assessed using a scintillation counter.
Immunohistochemistry
Immunohistochemistry was performed as previously described [27]. Excised hearts were embedded in paraffin, sectioned, and stained with H&E or trichrome. A Nikon Eclipse 50i microscope was used to visualize and photograph sections.
Differential counts of graft infiltrating cells (GICs)
Portions of at least three transplanted hearts were pooled and minced before digestion with 1 mg/ml collagenase A (Roche Diagnostics, Penzberg, Germany) for 30 min at 37 °C. After tissue debris had settled, suspended GICs were harvested. RBCs were lysed by hypotonic shock and GICs were passed through a 30 μm nylon mesh. Cytocentrifuged GICs were Wright-stained for differential enumeration.
ELISPOT assay
Capture and detection antibody pairs (BD Biosciences, San Jose, CA, USA) were as follows: IFN-γ (R4-6A2, XMG1.2), IL-4 (11B11, BVD6-24G2), and IL-17 (TC11-18H10, TC11-8H4.1). Polyvinylidene difluoride-backed microtitre plates (Millipore, Billerica, MA, USA) were coated with unlabelled mAb and blocked with 1% BSA in PBS. Irradiated (1000 rad) BALB/c splenocytes (4 × 105) and 1 × 106 recipient splenocytes were added to each well for 24 h. After washing, biotinylated detection mAbs were added to the plates. Plates were washed and a 1 : 1000 dilution of anti-biotin alkaline phosphatase conjugate (Vector Laboratories, Burlingame, CA, USA) was added to IFN-γ and IL-17 plates, and a 1 : 2000 dilution of HRP-conjugated streptavidin (Dako, Carpinteria, CA, USA) was added to IL-4 plates. Plates were washed and spots were visualized by addition of NBT (Bio-Rad, Hercules, CA, USA)/3-bromo-4-chloro-indolyl phosphate (Sigma-Aldrich) to IFN-γ and IL-17 plates, or 3-amino-9-ethylcarbazole (Pierce, Rockford, IL, USA) to IL-4 plates. Colour development continued until spots were visible and was stopped by adding H2O. Plates were dried and spots were quantified with an Immunospot Series 1 ELISPOT analyzer (Cellular Technology Ltd, Shaker Heights, OH, USA).
Heterotopic cardiac transplantation
Female C57BL/6 (H-2b) and BALB/c (H-2d) mice were purchased from Jackson Labs. EDA−/− mice were bred and housed at the University of Michigan, and experimental procedures were approved by the Animal Care and Use Committees of our institution. Animals were housed in specific pathogen-free rooms with a 12-h day/night cycle and had ad libitum access to water and chow. Heterotopic cardiac transplantation was performed as previously described [28]. Briefly, the aorta and pulmonary artery of the donor BALB/c heart were anastomosed end-to-side to the recipient’s abdominal aorta and inferior vena cava, respectively. Upon perfusion with the recipient’s blood, the transplanted heart resumes contraction. Graft function was monitored by abdominal palpation.
In vivo mAb therapy
Anti-CD4 (hybridoma GK1.5) was obtained from American Type Culture Collection, (Manassas, VA, USA) and prepared by Bio X Cell (West Lebanon, NH, USA). Allograft recipients were transiently depleted of CD4+ cells by i.p. injection of 1 mg of anti-CD4 mAb on the day prior to, the day of, and 7 days post-transplant [14,15,29].
Morphometric analysis of cardiac graft fibrosis and hypertrophy
Graft fibrosis was quantified by morphometric analysis of Masson’s trichrome-stained sections using iPLab software (Scanalytics Inc, Fairfax, VA, USA). Mean fibrotic area was calculated from 10–12 areas per heart section analysed at 200× magnification [14]. To quantify cardiomyocyte area as a measure of hypertrophy, digital outlines were drawn around at least 80 cardiomyocytes from views of H&E-stained sections at 200× magnification [29]. Areas within outlines were quantified using NIH Image J software (NIH) to measure cardiomyocyte cell size. A minimum of six hearts were analysed per group for both analysis techniques.
Splenocyte Th cultures
WT splenocytes were harvested and cultured at a density of 1 × 106 per ml with 2% anti-CD3 (YCD3-1) supernatant for 72 h. Cultures promoting specific Th lineages were supplemented with the following: Th1: 10 units/ml rIL-2, 10 μg/ml anti-IL-4 (11B11), and 1 ng/ml IL-12; Th2: 10 units/ml rIL-2, 100 ng/ml IL-4, and 10 μg/ml anti-IFN-γ (R46A2); Th17: 5 ng/ml recombinant TGF-β and 25 ng/ml rIL-6; Treg: 10 units/ml rIL-2, 10 ng/ml recombinant TGF-β.
Western blotting
Western blot analysis was performed under reducing conditions from whole-cell lysates prepared in cold RIPA buffer as described previously [27]. Densitometry of visualized bands was performed using Image J software (NIH).
Quantitative real-time PCR
Graft RNA was isolated by homogenizing tissues in TRIzol reagent (Invitrogen) according to the manufacturer’s protocol. Five micrograms of total RNA was reverse-transcribed using Oligo dT, dNTPs, MMLVRT (Invitrogen), and RNAsin (Promega, Madison, WI, USA) in PCR buffer (Roche, Indianapolis, IN, USA). Levels of the various transcripts were determined by quantitative real-time PCR using BioRad SYBR master mix in a Rotor Gene thermocycler. Expression levels were determined relative to GAPDH using the Rotor Gene relative quantitation utility.
RNA from splenocyte cultures was obtained by re-suspending culture pellets in Trizol reagent. Reverse transcription was performed with the Applied Biosystems High Capacity Reverse Transcription Kit and transcript levels were determined using Taq-Man SYBR master mix in an Applied Biosystems 7300 thermocycler (Applied Biosystems, Foster City, CA, USA). Expression levels were determined relative to GAPDH using 2−ΔΔCt normalization. Primers for collagen I, CTGF, IL-6, and IL-17 transcripts were as previously described [14,16]. EDA primers were 5′GATGGTGAAGACGACACTGC and 5′GAATGGCTGTGGACTGGATT.
EDA cFN ELISA
Murine EDA cFN was detected in serum by coating 96-well EIA plates (Costar, Lowell, MA, USA) with 3E2 mouse monoclonal anti-EDA FN antibody (1.4 μg/ml; Sigma) in 0.05 M carbonate–bicarbonate buffer, pH 9.6 (Sigma). Wells were then blocked with 5% BSA in PBS and serum samples were added in triplicate. Serum EDA cFN was detected with H-300 rabbit polyclonal fibronectin antibody (1.4 μg/ml; Santa Cruz Biotechnology) in dilution buffer (PBS, 0.05% Tween 20, 1% BSA) followed by HRP-conjugated goat anti-rabbit IgG secondary antibody (1 : 500; Fisher Scientific) in dilution buffer. Colour development was with Sigma Fast OPD tablets (Sigma) in deionized water. Development was terminated by the addition of an equal volume of 0.5 M sulphuric acid. Plates were read at 490 nm and serum values determined by interpolating against a standard curve. Using purified murine plasma fibronectin, we confirmed the specificity of the ELISA for only EDA cFN (data not shown).
Statistical analyses
Statistical analyses were performed using GraphPad Prism 5.01 (GraphPad Software, La Jolla, CA, USA). Differences between groups were evaluated using Student’s t-test. Data were considered significant at p <0.05.
Results
EDA−/− mice display normal anti-graft immune responses
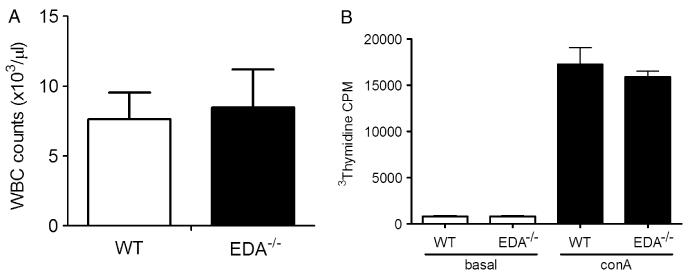
Fibronectin is involved in T-cell development and activation [30,31], and the EDA domain of fibronectin is believed to stimulate TLR4 (either directly or indirectly) to promote the production of inflammatory mediators [21,32]. Thus, we first sought to determine whether EDA−/− mice were immunodeficient. To determine whether EDA−/− mice demonstrated normal white blood cell numbers and differential counts, whole blood was assayed on an automated blood analyser. We found no difference between EDA−/− and WT mice (Figure 1A) in total leukocyte counts, which was consistent with levels reported in normal mice [33]. Differential analysis revealed similar absolute neutrophil, lymphocyte, and mononuclear cell counts (data not shown).
Figure 1.
EDA−/− mice have normal numbers of circulating leukocytes and proliferative capacity. (A) White blood cell counts from whole blood obtained from WT (n = 11) and EDA−/− (n = 12) mice, analysed as described in the Materials and methods section. (B) Proliferative responses of WT and EDA−/− splenocytes cultured in the presence or absence of 1 μg/ml concanavalin A (Con A) for 72 h measured by 3H-thymidine incorporation.
In addition to effects on T-cell development, fibronectin has been reported to promote T-cell proliferation [30]. We therefore examined the proliferative capacity of EDA−/− splenocytes compared with WT splenocytes using 3H-thymidine incorporation following Con A stimulation with unstimulated splenocytes as controls. Scintillation counting revealed no differences between cell types, with both EDA−/− and WT cells demonstrating robust proliferation in response to Con A (Figure 1B). Basal proliferation rates in unstimulated cells were also similar (Figure 1B). Together, these observations suggest that EDA−/− immune cells are competent with regard to homeostatic maintenance of leukocyte populations and proliferative responses.
EDA−/− recipients acutely reject cardiac allografts in a manner indistinguishable from WT mice
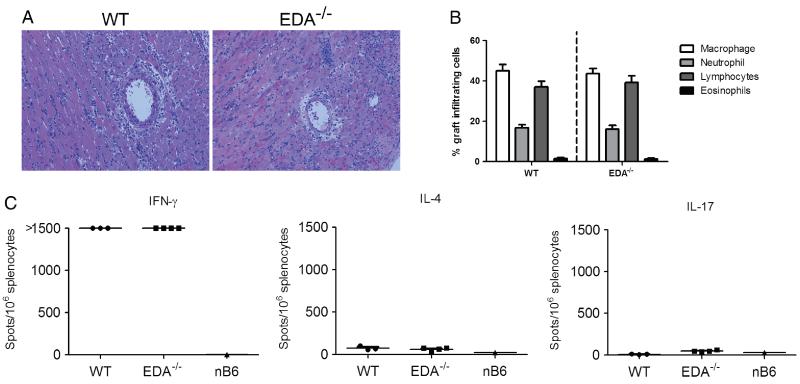
We next assessed the ability of EDA−/− mice to acutely reject cardiac allografts in a similar fashion to WT controls. In the heterotopic cardiac transplant model, C57BL/6 recipients that receive no immunosuppression normally reject BALB/c cardiac grafts within 7–9 days in a manner consistent with dominant Th1 responses and extensive parenchymal cell death [7,8]. EDA cFN up-regulation has been reported in human Th1 cells [34] and has been associated with infiltrating leukocytes in acute rejection [4,22]. Thus, we asked whether EDA−/− mice acutely reject cardiac allografts similarly to WT controls. EDA−/− and WT recipients rejected their grafts within 8 days post-transplant. Histological assessment of the acutely rejected allografts revealed similar degrees of mononuclear infiltration and cellular necrosis between the two genotypes (Figure 2A). Differential counts of GICs revealed no differences between the two genotypes with primarily monocytic and lymphocytic infiltrate (Figure 2B), suggesting equivalent inflammatory cell recruitment.
Figure 2.
EDA−/− recipients acutely reject cardiac allografts. (A) Allograft histology in EDA−/− and WT recipients at 200× (original magnification). H & E stains were performed on sections from allografts that were harvested at the time of rejection. Tissue sections are representative of at least four separate cardiac allografts. (B) Differential cell counts of Wright-stained GICs. (C) Donor reactive IFN-γ, IL-4, and IL-17 production by EDA−/− and WT recipient splenocytes in ELISPOT assays. Grafts and spleens were harvested at the time of rejection. Points represent individual allograft recipients; lines represent mean of group.
Acute allograft rejection in this model is accompanied by a robust Th1 immunological response [7,8]. Thus, we performed ELISPOT analyses to enumerate graft reactive IFN-γ, IL-4, and IL-17 responses in EDA−/− and WT recipients. Comparable, dominant Th1 priming was observed in splenocytes from both groups, with relatively little Th2 or Th17 response (Figure 2C). Collectively, the data suggest that alloimmune responses leading to acute rejection are replete in EDA−/− allograft recipients.
EDA deficiency prevents fibrosis associated with chronic rejection
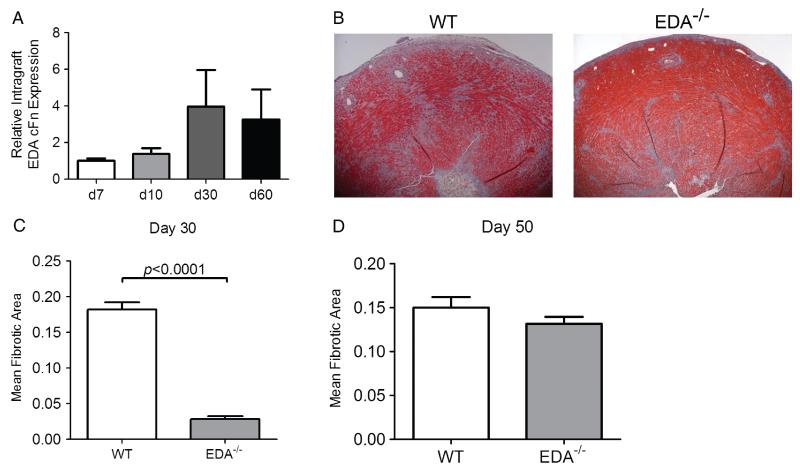
Although acute rejection responses were fully apparent in EDA−/− mice, we considered that allograft fibrosis associated with chronic rejection might be compromised in these animals given the described role for EDA cFN in other models of fibrosis [16,35-37]. To better ascertain the contribution of EDA cFN in cardiac allograft fibrosis, we compared cardiac allografts in EDA−/− and WT recipients in an established model of chronic rejection. In this model, allograft recipients are transiently depleted of CD4+ cells and chronic rejection ensues as CD4+ cells begin to repopulate the periphery [14,15,29,38,39]. We first examined the kinetics of intra-graft EDA cFN expression in this model, which suggested an increase in intra-graft EDA cFN transcript levels at day 30 (Figure 3A). Previous investigations by our group revealed that day 30 post-transplant is a critical time point in this model, with peak levels of interstitial fibrosis and pathological cardiac hypertrophy [14,29]. Thus, increased intra-graft EDA cFN expression coincides with pathological manifestations of chronic rejection.
Figure 3.
EDA cFN promotes fibrotic remodelling associated with chronic cardiac allograft rejection. (A) Kinetic time course of intra-graft EDA cFN transcript levels in WT recipients transiently depleted of CD4+ cells. Bars represent mean and SEM of three individual grafts. (B) Representative Masson’s trichrome-stained sections of cardiac allografts harvested on day 30 post-transplant. (C) Morphometric quantification of allograft fibrosis on day 30 post-transplant in WT and EDA−/− graft recipients. Bars illustrate mean and SEM of a minimum of ten frames of view from at least eight independent transplants. (D) Morphometric quantification of allograft fibrosis on day 50 post-transplant in WT and EDA−/− graft recipients. Bars illustrate mean and SEM of a minimum of ten frames of view from at least six independent hearts.
Masson’s trichrome staining revealed marked differences in cardiac allograft fibrosis between genotypes at day 30 post-transplant, with EDA−/− recipients demonstrating significantly less fibrotic tissue deposition than WT controls (Figure 3B). Quantitative scoring of these sections using morphometric analysis of graft fibrosis revealed a striking and significant decrease in allograft fibrosis in EDA−/− recipients compared with WT recipients (Figure 3C). Because WT BALB/c cardiac allografts used in these experiments can produce EDA cFN, we considered that graft origin EDA cFN might eventually compensate for the lack in EDA−/− recipients. To this end, we followed some grafts to day 50 post-transplant and although statistically significant differences were seen between EDA−/− and WT counterparts, the degree of fibrosis was much more similar (Figure 3D). This indicates that recipient-derived EDA cFN promotes fibrosis associated with chronic rejection, but is ultimately not required for the evolution of graft fibrosis, possibly because graft-derived EDA cFN can compensate for the absence of recipient EDA cFN.
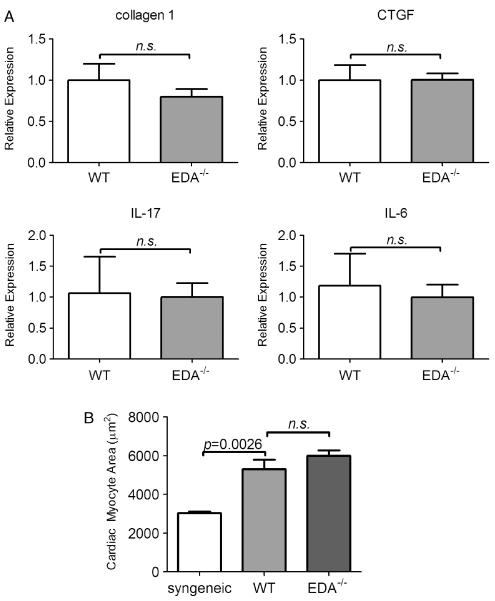
To better understand how cardiac allografts in EDA−/− recipients might be protected from fibrosis, we assessed levels of factors that promote fibrosis in this model [15,29,40]. Intra-graft transcripts of connective tissue growth factor (CTGF), IL-6, and IL-17 were not different between EDA−/− and WT recipients (Figure 4A). This may suggest that EDA cFN promotes cardiac allograft fibrosis downstream or independently of these cytokines.
Figure 4.
Intra-graft transcripts of fibrotic markers and cardiomyocyte hypertrophy are unchanged in EDA−/− recipients. (A) Intra-graft expression of CTGF, collagen 1, IL-17, and IL-6 transcripts. Bars represent mean and SEM of a minimum of six individual cardiac grafts. (B) Morphometric quantitation of cardiomyocyte area from H & E stains of day 30 post-transplant cardiac allografts from WT or EDA−/− recipients. Bars represent mean and SEM of area measurements taken from ≥80 cardiac myocytes per heart from a minimum of six hearts per group.
EDA cFN is unnecessary for cardiomyocyte hypertrophy
Chronic rejection of cardiac grafts is concomitant with the development of pathological hypertrophy in both this model [29] and human cardiac grafts [41]. Since EDA cFN deficiency protected grafts from fibrotic remodelling, we sought to determine its effects on cardiomyocyte hypertrophy. Morphometric determination of the cardiac myocyte area demonstrated significant hypertrophy in allografts from both EDA−/− and WT recipients compared with syngeneic transplanted hearts (Figure 4B). As EDA−/− recipients display equivalent mean cardiomyocyte areas, these data indicate that recipient EDA cFN is dispensable for hypertrophic remodelling associated with chronic rejection of cardiac grafts.
EDA cFN is produced by splenocytes under conditions promoting specific Th lineages
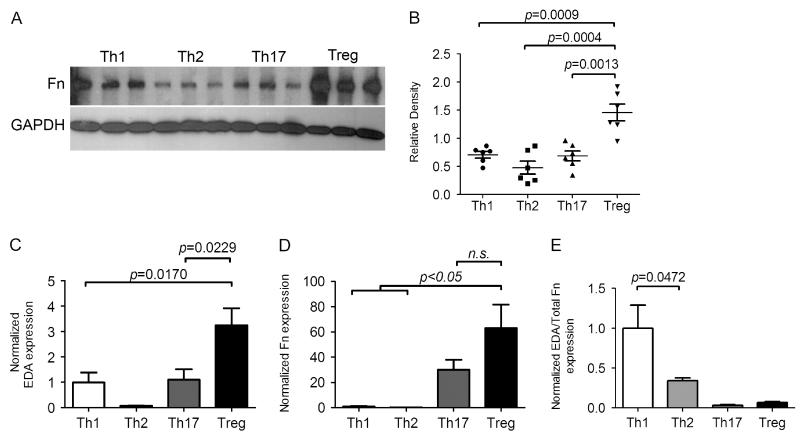
Recipient-derived GICs are a likely source of fibrosis-promoting EDA cFN in chronic rejection of cardiac allografts. Furthermore, as EDA−/− recipients acutely reject WT cardiac allografts in a fashion similar to WT recipients, differences in alloimmune responses are unlikely to account for attenuated allograft fibrosis seen in EDA−/− recipients. Chronic rejection in CD4+ cell depleted recipients is associated with diminutive but detectable levels of graft reactive Th1 and Th2 priming [42]. Furthermore, EDA cFN has been described as a Th1-specific molecule in human patients [34]. Hence, we considered that specific Th1 cells may preferentially produce EDA cFN and total FN. WT splenocytes were isolated and cultured in media supplemented to promote Th1, Th2, Th17, and Treg phenotypes. Examination of cellular transcripts revealed similar amounts of EDA cFN production by both Th1 and Th17 cells, with significantly greater EDA cFN expression levels in Treg cultures (Figure 5A). When analysis of cellular transcripts was extended to total FN, significantly increased levels of total FN were observed in Th17 and Treg cultures, with minimal FN transcripts expressed by Th1 and Th2 cultures (Figure 5B). Previous findings have suggested that the ratio of EDA containing FN to total FN production impacts the effects of FN on cellular responses [26]. Therefore, we determined the EDA to total FN transcript ratio (Figure 5C). Interestingly, Treg cultures had the highest transcript levels of EDA and total FN, although the EDA/total FN ratio was greatest in Th1 culture cells. Thus, while Treg makes more FN than other Th lineages, Th1 cells appear to transcribe EDA in a more concentrated fashion.
Figure 5.
Characterization of splenocyte production of fibronectin under conditions promoting specific Th lineages. (A) Detection of total fibronectin content in cell lysates from splenocytes cultured under conditions promoting Th1, Th2, Th17, or Treg conditions. Bands are representative of three independent cultures; GAPDH is shown as a loading control. Results shown are representative of six independent cultures for each condition. For a detailed description of culture conditions see the Materials and methods section. (B) Quantification of total fibronectin protein expression as relative band density of bands represented by panel A. (C–E) Real-time PCR quantitation of EDA cFN (C), total Fn (D), and EDA cFN/total Fn ratio (E), from splenocytes cultured under conditions promoting Th1, Th2, Th17, or Treg differentiation. Bars represent mean and SEM of six independent cultures.
We extended our analysis of FN production by Th-differentiated splenocyte cultures by measuring cellular protein levels with western blotting. Consistent with RT-PCR data (Figure 5B), Treg cultures produced significantly higher levels of total FN than Th1 and Th2 cultures (Figures 5D and 5E).
Increased levels of EDA cFN in serum of allograft recipients chronically rejecting their grafts
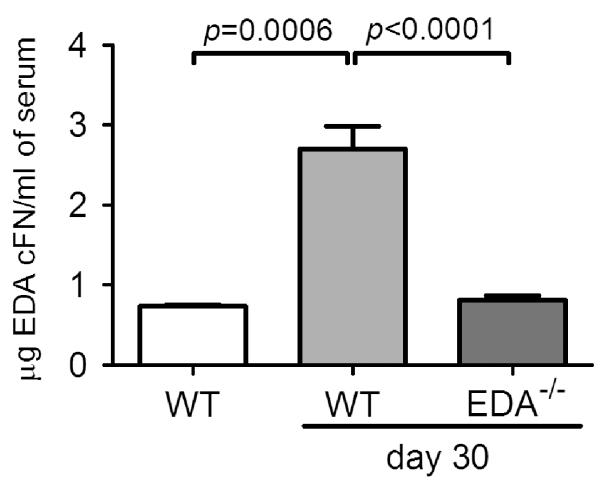
Given that a percentage of tissue fibronectin is derived from the circulation [43], EDA cFN in cardiac allografts may derive from this source as well as graft-infiltrating cells. We therefore assessed levels of EDA cFN in the serum of anti-CD4-treated WT and EDA−/− cardiac allograft recipients 30 days post-transplant. WT recipients exhibited significantly higher amounts of EDA cFN in serum compared with non-transplanted WT controls (Figure 6). In contrast, EDA−/− recipients of WT cardiac grafts exhibited similar levels of EDA cFN in the serum to non-transplanted WT mice (Figure 6). These findings suggest that circulating EDA cFN (perhaps derived from the engrafted vasculature) may be another important source of EDA cFN in cardiac allografts.
Figure 6.
Serum EDA cFN levels in recipients chronically rejecting cardiac allografts. Serum was isolated from WT or EDA−/− CD4+ cell-depleted allograft recipients 30 days following transplantation and compared with naive, non-transplanted WT controls. Bars represent mean and SEM of a minimum of six independent allograft recipients or four non-transplanted controls.
Discussion
In this article, we highlight an important role for recipient-derived EDA cFN in driving cardiac allograft fibrosis in the setting of chronic rejection. Recent work has described EDA cFN expression in areas of chronic allograft vasculopathy and interstitial fibrosis in cardiac transplants [23,25], and has suggested that EDA cFN may be deposited by graft-infiltrating leukocytes [22]. We now extend these observations by demonstrating that recipient-derived EDA cFN, rather than graft-derived EDA cFN, plays an active part in promoting the fibrotic response, as EDA-competent allografts in EDA−/− recipients exhibited significant delay in the onset of fibrosis. Moreover, our data suggest that Th1 cells and Treg may be the important cellular sources of EDA cFN and total FN in this and other systems.
Fibrotic remodelling is a hallmark of chronic rejection pathology which is believed to be driven by immune responses to donor alloantigens. However, the interrelationship between anti-graft immune responses and graft fibrosis is not known. Many have postulated that these processes are linked by immune cell-derived cytokines, namely TGF-β. However, TGF-β is known to exert multiple graft-protective effects, making it a tenuous therapeutic target. Furthermore, our previous work in models of chronic cardiac allograft rejection has revealed that TGF-β alone is insufficient to drive allograft fibrosis and requires other co-mediators and downstream factors for its pro-fibrotic effects [14,40].
A potential downstream mediator of TGF-β effects could be EDA cFN, which is induced by TGF-β in certain cell types, particularly fibroblasts [44]. We observed no significant differences in intra-graft expression of several pro-fibrotic mediators in EDA−/− recipients. This indicates that EDA cFN promotes fibrosis downstream or outside of these fibrotic mediators. One possibility is that EDA cFN promotes fibrosis by preventing matrix degradation rather than promoting matrix synthesis, as the balance of these pathways determines fibrotic tissue deposition [11,45].
Our current findings reveal that recipient-derived EDA cFN drives cardiac allograft fibrosis. We considered that recipient-derived EDA cFN could enter the graft through multiple routes, including GICs. This mechanism is supported by multiple immunohistochemical studies that delineate a clear association of EDA cFN expression in association with GICs [23,24]. As the majority of GICs are recipient leukocytes, EDA cFN produced by recipient immune cells appears to be an important immunological mediator of fibrosis in chronic cardiac allograft rejection. Our investigations into EDA cFN production by immune cells revealed that both Th1 and Treg cells can produce significant amounts of both EDA and total FN. The production of matrix molecules by immune cells of these lineages could provide a link between alloimmune responses and fibrotic remodelling. However, further investigation is necessary to understand if and how Th1 and Treg cells, generally considered of opposing functional lineages, promote fibrosis.
It is conceivable that cellular damage by alloreactive Th1 cells is accompanied by deposition of EDA cFN and provides an initial fibrotic stimulus. In contrast, Treg could simultaneously dampen immune responses against cardiac grafts and produce FN that contributes to fibrotic responses. One explanation for the production of matrix molecules by Treg could be the immune cytokine TGF-β, well known as a fibrotic mediator. However, it should also be noted that differences between Th17 and Treg cultures (which are both supplemented with TGF-β) may highlight the context-dependent signalling of TGF-β. Furthermore, Th1 differentiation cultures, which were not supplemented with TGF-β, exhibited the most concentrated production of EDA cFN, as evidenced by the EDA/total FN ratio. This suggests that regulation of EDA cFN production by immune cells may also occur independently of TGF-β.
We observed increased levels of EDA cFN in the serum of WT recipients who were chronically rejecting their grafts. Hepatocytes secrete significant amounts of fibronectin into the peripheral circulation, though plasma fibronectin does not typically contain EDA cFN [46]. Indeed, hepatocytes have been shown to selectively retain and degrade EDA cFN when it is produced, rather than secreting it into the circulation [43]. Alternatively, significant evidence suggests that EDA cFN may be produced by vascular endothelium in response to inflammation or injury [47-49]. Hence, the injury resulting from the anastomosis of graft aorta and pulmonary artery to recipient aorta and inferior vena cava in the heterotopic allograft model may be a source of the circulating EDA cFN that we observed. Interestingly, however, it is more likely that the recipient vessels (rather than the graft vessels) are the source since EDA−/− recipients did not display an increase in serum EDA cFN. Regardless, although the cellular origin of EDA cFN in the circulation is unclear, it is likely that it plays a substantial role in the development of cardiac allograft fibrosis.
Our prior work has illustrated the obligate role for EDA cFN in normal skin wound healing [26] and lung fibrosis following injury [16]. Although the reasons for this have not been fully elucidated, it appears that EDA cFN is necessary for in vivo fibroblast-to-myofibroblast differentiation; EDA−/− mice developed fewer α-SMA-expressing myofibroblasts in injured areas compared with WT animals [16,26]. Moreover, in vitro studies in both rodent and human cells show that in the absence of EDA cFN, fibroblasts are unable to transdifferentiate into myofibroblasts, even in the presence of the pro-fibrotic cytokine TGF-β [16,20]. Myofibroblast transdifferentiation is a complex process requiring both soluble factors (such as TGF-β) and integrin signals via focal adhesion kinase [50]. Indeed, our recent work demonstrated that α4β7 integrins, previously thought to be restricted to leukocytes, are expressed on the fibroblast cell surface and play an important role in transducing EDA cFN signals, via MAP and ERK1/2 kinases, to effect myofibroblast transdifferentiation [51].
Together, our data describe a function for EDA cFN as a downstream factor mediating fibrotic remodelling responses in cardiac allografts. While current immunosuppressive therapeutics obviate acute rejection and prolong organ survival, they are generally ineffective at preventing chronic graft rejection. This highlights the need for mechanistic evaluation to identify other targets for potential therapeutic intervention. In addition to its potential contributions to the development of graft fibrosis, increased EDA cFN in the serum of graft recipients suggests that EDA cFN could be explored as a non-invasive biomarker for the development of chronic rejection in cardiac allografts. Furthermore, EDA cFN and elements mediating EDA cFN signal transduction may be therapeutic targets for limiting fibrosis in cardiac, and potentially other solid organ, allografts.
Acknowledgment
This work was funded by grants from the National Institutes of Health (R01 HL085083 to ESW; R01 HL070613 and R56 AI061469 to DKB).
Footnotes
No conflicts of interest were declared.
Author contribution statement
AB conceptualized the project, performed experiments, analysed data, and wrote the manuscript. SCW performed splenocyte cultures and ELISPOT experiments. GL performed all cardiac transplants. AMC and AAD performed EDA cFN ELISAs. ESW intellectually guided experiments, provided reagents, analysed data, and wrote/edited the manuscript. DKB intellectually guided experiments, provided reagents, and edited the manuscript.
References
- 1.Hornick P, Rose M. Chronic rejection in the heart. Methods Mol Biol. 2006;333:131–144. doi: 10.1385/1-59745-049-9:131. [DOI] [PubMed] [Google Scholar]
- 2.Mehra MR. Contemporary concepts in prevention and treatment of cardiac allograft vasculopathy. Am J Transplant. 2006;6:1248–1256. doi: 10.1111/j.1600-6143.2006.01314.x. [DOI] [PubMed] [Google Scholar]
- 3.Torre-Amione G. Cardiac allograft hypertrophy: a new target for therapy, a surrogate marker for survival? Am J Transplant. 2009;9:7–8. doi: 10.1111/j.1600-6143.2008.02503.x. [DOI] [PubMed] [Google Scholar]
- 4.Coito AJ, Onodera K, Kato H, et al. Fibronectin–mononuclear cell interactions regulate type 1 helper T cell cytokine network in tolerant transplant recipients. Am J Pathol. 2000;157:1207–1218. doi: 10.1016/S0002-9440(10)64636-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Adair-Kirk TL, Senior RM. Fragments of extracellular matrix as mediators of inflammation. Int J Biochem Cell Biol. 2008;40:1101–1110. doi: 10.1016/j.biocel.2007.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Korpos E, Wu C, Song J, et al. Role of the extracellular matrix in lymphocyte migration. Cell Tissue Res. 2010;339:47–57. doi: 10.1007/s00441-009-0853-3. [DOI] [PubMed] [Google Scholar]
- 7.Chan SY, DeBruyne LA, Goodman RE, et al. In vivo depletion of CD8+ T cells results in Th2 cytokine production and alternate mechanisms of allograft rejection. Transplantation. 1995;59:1155–1161. [PubMed] [Google Scholar]
- 8.Piccotti JR, Chan SY, Goodman RE, et al. IL-12 antagonism induces T helper 2 responses, yet exacerbates cardiac allograft rejection. Evidence against a dominant protective role for T helper 2 cytokines in alloimmunity. J Immunol. 1996;157:1951–1957. [PubMed] [Google Scholar]
- 9.Libby P, Pober JS. Chronic rejection. Immunity. 2001;14:387–397. doi: 10.1016/s1074-7613(01)00119-4. [DOI] [PubMed] [Google Scholar]
- 10.Boehler A, Kesten S, Weder W, et al. Bronchiolitis obliterans after lung transplantation: a review. Chest. 1998;114:1411–1426. doi: 10.1378/chest.114.5.1411. [DOI] [PubMed] [Google Scholar]
- 11.Mannon RB. Therapeutic targets in the treatment of allograft fibrosis. Am J Transplant. 2006;6:867–875. doi: 10.1111/j.1600-6143.2006.01261.x. [DOI] [PubMed] [Google Scholar]
- 12.Csencsits K, Wood SC, Lu G, et al. Transforming growth factor-beta1 gene transfer is associated with the development of regulatory cells. Am J Transplant. 2005;5:2378–2384. doi: 10.1111/j.1600-6143.2005.01042.x. [DOI] [PubMed] [Google Scholar]
- 13.Chan SY, Goodman RE, Szmuszkovicz JR, et al. DNA-liposome versus adenoviral mediated gene transfer of transforming growth factor beta1 in vascularized cardiac allografts: differential sensitivity of CD4+ and CD8+ T cells to transforming growth factor beta1. Transplantation. 2000;70:1292–1301. doi: 10.1097/00007890-200011150-00006. [DOI] [PubMed] [Google Scholar]
- 14.Booth AJ, Csencsits-Smith K, Wood SC, et al. Connective tissue growth factor promotes fibrosis downstream of TGFbeta and IL-6 in chronic cardiac allograft rejection. Am J Transplant. 2010;10:220–230. doi: 10.1111/j.1600-6143.2009.02826.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Csencsits K, Wood SC, Lu G, et al. Transforming growth factor beta-induced connective tissue growth factor and chronic allograft rejection. Am J Transplant. 2006;6:959–966. doi: 10.1111/j.1600-6143.2006.01292.x. [DOI] [PubMed] [Google Scholar]
- 16.Muro AF, Moretti FA, Moore BB, et al. An essential role for fibronectin extra type III domain A in pulmonary fibrosis. Am J Respir Crit Care Med. 2008;177:638–645. doi: 10.1164/rccm.200708-1291OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Broekelmann TJ, Limper AH, Colby TV, et al. Transforming growth factor beta 1 is present at sites of extracellular matrix gene expression in human pulmonary fibrosis. Proc Natl Acad Sci U S A. 1991;88:6642–6646. doi: 10.1073/pnas.88.15.6642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuhn C, 3rd, Boldt J, King TE, Jr, et al. An immunohistochemical study of architectural remodeling and connective tissue synthesis in pulmonary fibrosis. Am Rev Respir Dis. 1989;140:1693–1703. doi: 10.1164/ajrccm/140.6.1693. [DOI] [PubMed] [Google Scholar]
- 19.White ES, Baralle FE, Muro AF. New insights into form and function of fibronectin splice variants. J Pathol. 2008;216:1–14. doi: 10.1002/path.2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Serini G, Bochaton-Piallat ML, Ropraz P, et al. The fibronectin domain ED-A is crucial for myofibroblastic phenotype induction by transforming growth factor-beta1. J Cell Biol. 1998;142:873–881. doi: 10.1083/jcb.142.3.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Okamura Y, Watari M, Jerud ES, et al. The extra domain A of fibronectin activates Toll-like receptor 4. J Biol Chem. 2001;276:10229–10233. doi: 10.1074/jbc.M100099200. [DOI] [PubMed] [Google Scholar]
- 22.Coito AJ, Brown LF, Peters JH, et al. Expression of fibronectin splicing variants in organ transplantation: a differential pattern between rat cardiac allografts and isografts. Am J Pathol. 1997;150:1757–1772. [PMC free article] [PubMed] [Google Scholar]
- 23.Franz M, Berndt A, Grun K, et al. Expression of extra domain A containing fibronectin in chronic cardiac allograft rejection. J Heart Lung Transplant. 2011;30:86–94. doi: 10.1016/j.healun.2010.08.015. [DOI] [PubMed] [Google Scholar]
- 24.Franz M, Brehm BR, Richter P, et al. Changes in extra cellular matrix remodelling and re-expression of fibronectin and tenascin-C splicing variants in human myocardial tissue of the right atrial auricle: implications for a targeted therapy of cardiovascular diseases using human SIP format antibodies. J Mol Histol. 2010;41:39–50. doi: 10.1007/s10735-010-9260-z. [DOI] [PubMed] [Google Scholar]
- 25.Franz M, Grun K, Richter P, et al. Extra cellular matrix remodelling after heterotopic rat heart transplantation: gene expression profiling and involvement of ED-A+ fibronectin, alpha-smooth muscle actin and B+ tenascin-C in chronic cardiac allograft rejection. Histochem Cell Biol. 2010;134:503–517. doi: 10.1007/s00418-010-0750-6. [DOI] [PubMed] [Google Scholar]
- 26.Muro AF, Chauhan AK, Gajovic S, et al. Regulated splicing of the fibronectin EDA exon is essential for proper skin wound healing and normal lifespan. J Cell Biol. 2003;162:149–160. doi: 10.1083/jcb.200212079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.White ES, Atrasz RG, Hu B, et al. Negative regulation of myofibroblast differentiation by PTEN (Phosphatase and Tensin Homolog Deleted on chromosome 10) Am J Respir Crit Care Med. 2006;173:112–121. doi: 10.1164/rccm.200507-1058OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Corry RJ, Winn HJ, Russell PS. Primarily vascularized allografts of hearts in mice. The role of H-2D, H-2K, and non-H-2 antigens in rejection. Transplantation. 1973;16:343–350. doi: 10.1097/00007890-197310000-00010. [DOI] [PubMed] [Google Scholar]
- 29.Diaz JA, Booth AJ, Lu G, et al. Critical role for IL-6 in hypertrophy and fibrosis in chronic cardiac allograft rejection. Am J Transplant. 2009;9:1773–1783. doi: 10.1111/j.1600-6143.2009.02706.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Davis LS, Oppenheimer-Marks N, Bednarczyk JL, et al. Fibronectin promotes proliferation of naive and memory T cells by signaling through both the VLA-4 and VLA-5 integrin molecules. J Immunol. 1990;145:785–793. [PubMed] [Google Scholar]
- 31.Utsumi K, Sawada M, Narumiya S, et al. Adhesion of immature thymocytes to thymic stromal cells through fibronectin molecules and its significance for the induction of thymocyte differentiation. Proc Natl Acad Sci U S A. 1991;88:5685–5689. doi: 10.1073/pnas.88.13.5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saito S, Yamaji N, Yasunaga K, et al. The fibronectin extra domain A activates matrix metalloproteinase gene expression by an interleukin-1-dependent mechanism. J Biol Chem. 1999;274:30756–30763. doi: 10.1074/jbc.274.43.30756. [DOI] [PubMed] [Google Scholar]
- 33.Nemzek JA, Bolgos GL, Williams BA, et al. Differences in normal values for murine white blood cell counts and other hematological parameters based on sampling site. Inflamm Res. 2001;50:523–527. doi: 10.1007/PL00000229. [DOI] [PubMed] [Google Scholar]
- 34.Sandig H, McDonald J, Gilmour J, et al. Fibronectin is a TH1-specific molecule in human subjects. J Allergy Clin Immunol. 2009;124:528–535. doi: 10.1016/j.jaci.2009.04.036. [DOI] [PubMed] [Google Scholar]
- 35.Arslan F, Smeets MB, Riem Vis PW, et al. Lack of fibronectin-EDA promotes survival and prevents adverse remodeling and heart function deterioration after myocardial infarction. Circ Res. 2011;108:582–592. doi: 10.1161/CIRCRESAHA.110.224428. [DOI] [PubMed] [Google Scholar]
- 36.Hernnas J, Nettelbladt O, Bjermer L, et al. Alveolar accumulation of fibronectin and hyaluronan precedes bleomycin-induced pulmonary fibrosis in the rat. Eur Respir J. 1992;5:404–410. [PubMed] [Google Scholar]
- 37.Jarnagin WR, Rockey DC, Koteliansky VE, et al. Expression of variant fibronectins in wound healing: cellular source and biological activity of the EIIIA segment in rat hepatic fibrogenesis. J Cell Biol. 1994;127:2037–2048. doi: 10.1083/jcb.127.6.2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bishop DK, Li W, Chan SY, et al. Helper T lymphocyte unresponsiveness to cardiac allografts following transient depletion of CD4-positive cells. Implications for cellular and humoral responses. Transplantation. 1994;58:576–584. doi: 10.1097/00007890-199409150-00009. [DOI] [PubMed] [Google Scholar]
- 39.Piccotti JR, Li K, Chan SY, et al. Cytokine regulation of chronic cardiac allograft rejection: evidence against a role for Th1 in the disease process. Transplantation. 1999;67:1548–1555. doi: 10.1097/00007890-199906270-00008. [DOI] [PubMed] [Google Scholar]
- 40.Booth AJ, Bishop DK. TGF-beta, IL-6, IL-17 and CTGF direct multiple pathologies of chronic cardiac allograft rejection. Immunotherapy. 2010;2:511–520. doi: 10.2217/imt.10.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Raichlin E, Villarraga HR, Chandrasekaran K, et al. Cardiac allograft remodeling after heart transplantation is associated with increased graft vasculopathy and mortality. Am J Transplant. 2009;9:132–139. doi: 10.1111/j.1600-6143.2008.02474.x. [DOI] [PubMed] [Google Scholar]
- 42.Wood SC, Lu G, Burrell BE, et al. Transplant acceptance following anti-CD4 versus anti-CD40L therapy: evidence for differential maintenance of graft-reactive T cells. Am J Transplant. 2008;8:2037–2048. doi: 10.1111/j.1600-6143.2008.02372.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moretti FA, Chauhan AK, Iaconcig A, et al. A major fraction of fibronectin present in the extracellular matrix of tissues is plasma-derived. J Biol Chem. 2007;282:28057–28062. doi: 10.1074/jbc.M611315200. [DOI] [PubMed] [Google Scholar]
- 44.Borsi L, Castellani P, Risso AM, et al. Transforming growth factor-beta regulates the splicing pattern of fibronectin messenger RNA precursor. FEBS Lett. 1990;261:175–178. doi: 10.1016/0014-5793(90)80664-5. [DOI] [PubMed] [Google Scholar]
- 45.Thickett DR, Poole AR, Millar AB. The balance between collagen synthesis and degradation in diffuse lung disease. Sarcoidosis Vasc Diffuse Lung Dis. 2001;18:27–33. [PubMed] [Google Scholar]
- 46.Kornblihtt AR, Umezawa K, Vibe-Pedersen K, et al. Primary structure of human fibronectin: differential splicing may generate at least 10 polypeptides from a single gene. EMBO J. 1985;4:1755–1759. doi: 10.1002/j.1460-2075.1985.tb03847.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Peters JH, Sporn LA, Ginsberg MH, et al. Human endothelial cells synthesize, process, and secrete fibronectin molecules bearing an alternatively spliced type III homology (ED1) Blood. 1990;75:1801–1808. [PubMed] [Google Scholar]
- 48.Dubin D, Peters JH, Brown LF, et al. Balloon catheterization induced arterial expression of embryonic fibronectins. Arterioscler Thromb Vasc Biol. 1995;15:1958–1967. doi: 10.1161/01.atv.15.11.1958. [DOI] [PubMed] [Google Scholar]
- 49.Peters JH, Ginsberg MH, Bohl BP, et al. Intravascular release of intact cellular fibronectin during oxidant-induced injury of the in vitro perfused rabbit lung. J Clin Invest. 1986;78:1596–1603. doi: 10.1172/JCI112752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thannickal VJ, Lee DY, White ES, et al. Myofibroblast differentiation by transforming growth factor-beta1 is dependent on cell adhesion and integrin signaling via focal adhesion kinase. J Biol Chem. 2003;278:12384–12389. doi: 10.1074/jbc.M208544200. [DOI] [PubMed] [Google Scholar]
- 51.Kohan M, Muro AF, White ES, et al. EDA-containing cellular fibronectin induces fibroblast differentiation through binding to alpha4beta7 integrin receptor and MAPK/Erk 1/2-dependent signaling. FASEB J. 2010;24:4503–4512. doi: 10.1096/fj.10-154435. [DOI] [PMC free article] [PubMed] [Google Scholar]