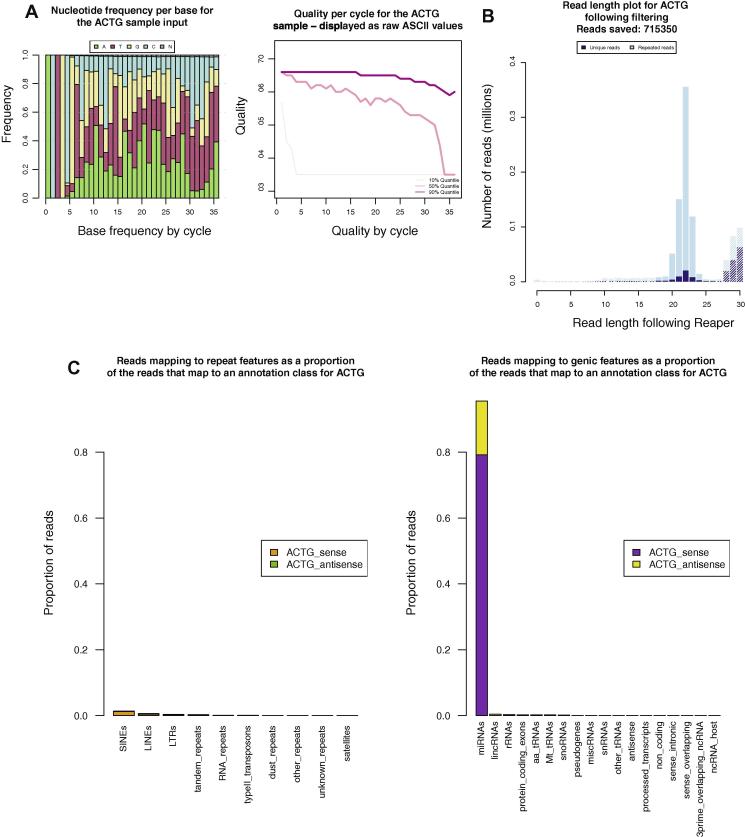
Fig. 3.
Analysis of a barcoded small RNA cloning experiment. Examples of the plots produced by the SequenceImp pipeline to summarise the analysis of small RNA. (A) Two plots taken from the reaper stage of the SequenceImp pipeline. These plots describe the ACTG-barcoded sample before Reaper trims and cleans the reads. (B) The length of reads for the ACTG barcoded sample, at the filter stage. Trimming in the reaper step defines a clear 20–23 nt peak in this sample. At the filter step reads can be selected for downstream analysis based on length. Solid bars correspond to those reads passed to the later stages of the pipeline. Hashed bars represent reads removed, falling outside the maximum and minimum length criteria. (C) Reads passed from the filter step which map to Ensembl annotation at the align stage of the pipeline are separated into individual annotation classes.