Summary
Eukaryotic TIP49a (Pontin) and TIP49b (Reptin) AAA+ ATPases play essential roles in key cellular processes. How their weak ATPase activity contributes to their important functions remains largely unknown and difficult to analyze because of the divergent properties of TIP49a and TIP49b proteins and of their homo- and hetero-oligomeric assemblies. To circumvent these complexities, we have analyzed the single ancient TIP49 ortholog found in the archaeon Methanopyrus kandleri (mkTIP49). All-atom homology modeling and molecular dynamics simulations validated by biochemical assays reveal highly conserved organizational principles and identify key residues for ATP hydrolysis. An unanticipated crosstalk between Walker B and Sensor I motifs impacts the dynamics of water molecules and highlights a critical role of trans-acting aspartates in the lytic water activation step that is essential for the associative mechanism of ATP hydrolysis.
Graphical Abstract
Highlights
-
•
We have studied the single TIP49 ortholog (mkTIP49) from the archaeon M. kandleri
-
•
We propose a model for assembly of the pre-transition state for ATP hydrolysis
-
•
Trans-aspartates downregulate ATP hydrolysis by mkTIP49 hexamers
-
•
Mutational analysis confirms a highly conserved mechanism for lytic water activation
Afanasyeva et al. combine computational and biochemical analysis to reveal conserved organizational principles and residues critical for ATP hydrolysis in the AAA+ ATPase TIP49 ortholog from archaea. They highlight a role of trans-aspartates in the lytic water activation step essential for ATP hydrolysis.
Introduction
Members of the AAA+ (ATPases associated with diverse cellular activities) superfamily facilitate a broad range of cellular processes, but the molecular mechanisms that underpin their activity are in most cases not well characterized. Eukaryotic TIP49a (Pontin, RUVBL1, and Rvb1) and TIP49b (Reptin, RUVBL2, and Rvb2) were initially identified as transcription regulators that interacted with TATA binding protein (TBP), hence the name TBP-interacting proteins (TIPs). Both have subsequently been implicated in several essential pleiotropic functions, including gene expression, ribosomal RNA processing, apoptosis, cell division, and proliferation (recently reviewed in Nano and Houry, 2013 and Rosenbaum et al., 2013). They form oligomers found in multiprotein complexes involved in chromatin remodeling, DNA repair, telomerase, and ribonucleoprotein particle assembly, where they play essential, but yet unknown, role(s) as, most probably, chaperones or adaptor proteins (Bellosta et al., 2005; Etard et al., 2005; Ikura et al., 2000; Jonsson et al., 2001; Machado-Pinilla et al., 2012; McKeegan et al., 2009; Parusel et al., 2006; Shen et al., 2000; Venteicher et al., 2008; Watkins et al., 2004; Weiske and Huber, 2006; Wood et al., 2000). Their ATPase activity is weak but appears to be required for these activities and can be altered according to their homo- or hetero-oligomeric state (Rosenbaum et al., 2013).
TIP49a and TIP49b belong to the classical AAA clade of the AAA+ superfamily (Ammelburg et al., 2006; Erzberger and Berger, 2006; Iyer et al., 2004; Wendler et al., 2012). They contain a conserved core ATPase domain and an additional α-helical, C-terminal domain that often acts as a regulatory moiety for ATP hydrolysis, oligomerization, and co-factor binding. This core includes conserved Walker A and Walker B motifs responsible for coordination of the ATP β- and γ-phosphates and magnesium. TIP49 proteins also contain polar residues in the Sensor I motif that are thought to be involved in the formation of a hydrogen-bonding network required for positioning of the lytic water molecule in the proximity of the γ-phosphate of ATP. The Sensor II motif is an arginine located within the C-terminal α-helical domain and is involved in additional interactions with the ATP γ-phosphate. Another evolutionarily conserved feature is the presence of trans-acting arginines in the vicinity of the catalytic pockets of oligomers. They are often found close to the ATP γ-phosphate bound by the adjacent subunit containing the Walker A and B motifs. Because their configuration resembles that of the nucleotide-binding pocket found in small GTPases, they have been named arginine fingers (R-fingers; recently reviewed in Wendler et al., 2012).
Compared to other members of the AAA+ superfamily, TIP49 proteins have an unusual feature: the presence of an additional 170-amino acid insertion domain that splits the Walker A and Walker B motifs. Structurally organized as an oligonucleotide/oligosaccharide binding fold (OB) and as a partially disordered L2 loop, this insertion domain appears to be involved in nucleic acid binding, allosteric regulation of ATPase activity, and higher-order oligomerization of TIP49 proteins (Gorynia et al., 2011; Lopez-Perrote et al., 2012; Matias et al., 2006; Niewiarowski et al., 2010; Petukhov et al., 2012; Torreira et al., 2008). TIP49 proteins show extensive conformational flexibility; this, together with their weak ATPase activity and strong tendency for higher-order oligomerization, makes them complex objects for mechanistic studies. The pleiotropic effects of TIP49a and TIP49b mutations and of RNA interference experiments in vivo, together with the versatility of the TIP49 proteins in seemingly unrelated biological processes, add multiple layers of complexity for interpretations of their cellular functions that also include their direct implication in carcinogenesis (Kim et al., 2006, 2007; Parusel et al., 2006; Rousseau et al., 2007; Weiske and Huber, 2006).
ATPases—and, more generally, all NTPases—can hydrolyze nucleotide triphosphates according to distinct dissociative or associative pathways (Grigorenko et al., 2011). In the dissociative model, the NTP molecule dissociates into NDP and phosphate before the nucleophile (water) enters the reaction. In the more widely accepted associative mechanism, the water molecule enters the reaction prior to formation of a pentacovalent oxyphosphorane intermediate but can be activated in several different ways: (1) by acceptance of a water proton by polar or negative side chains of the NTPase (Senior et al., 2002), (2) by direct transfer of a proton from the attacking water to the γ-phosphate moiety of NTP as shown for GTPases (Schweins et al., 1994); or (3) according to the “two-water hypothesis,” by another water molecule within the protein active site that relays a water proton to a protein amino-acid side chain or to NTP itself as the general base (Grigorenko et al., 2007, 2008). Regardless of the exact mechanism, these pathways suggest that polar or negatively charged amino acids might be essential for the associative mechanism of ATP hydrolysis and, in the case of eukaryotic TIP49 proteins, remain to be identified.
Archaea are unicellular microorganisms whose machineries for replication, transcription—including the transcription initiation factor TBP (Werner, 2007; Werner and Weinzierl, 2002, 2005)—and translation are more closely related to the eukaryotic information-processing systems than to their bacterial counterparts. Most, but not all, archaeal genomes across the eury- and crenarchaeal phyla harbor one bona-fide homolog of the eukaryotic TIP49a/TIP49b proteins, referred to henceforth as archaeal TIP49. Archaeal TIP49 proteins constitute an attractive model system for mechanistic studies of their biochemical and biophysical properties, which might be extrapolated to their more complex eukaryotic paralogs. We have chosen the single TIP49 protein from the hyperthermophilic methanogen Methanopyrus kandleri (mkTIP49; UniProtKB ID code Q8TZC3_METKA) to further characterize ATP hydrolysis by eukaryotic TIP49 proteins. We reasoned that the study of a single ortholog, free of possible bacterial contamination and that circumvents the complexity inherent to TIP49a and TIP49b hetero-oligomers, could help establish clear principles of catalytic activity in these enigmatic proteins. Our analysis of water dynamics within catalytic pockets of the wild-type and mutant mkTIP49 proteins provides insights into trans-acting water-activating residues, which may serve as proton acceptors in TIP49 oligomers. Our results suggest that the TIP49 AAA+ ATPases evolved a mechanism for ATP hydrolysis that has been conserved from Archaea to eukaryotes.
Results
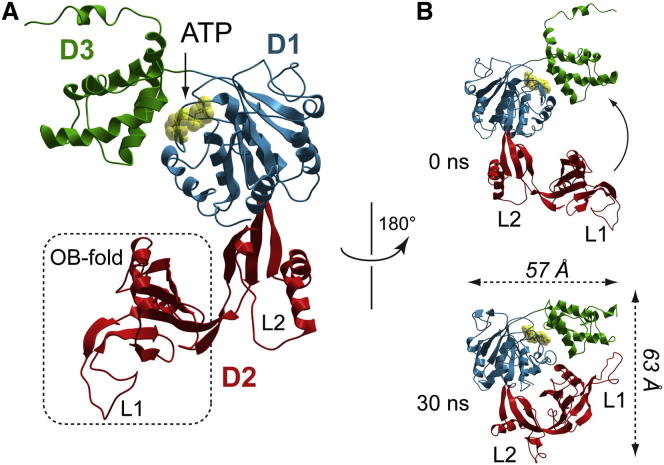
All-atom homology modeling of mkTIP49 relative to human TIP49 proteins is illustrated in Figure 1 and Figure S1 (available online) and was performed as described in Petukhov et al. (2012). Similar to eukaryotic TIP49a and TIP49b proteins, the M. kandleri TIP49 protein (mkTIP49) folds into a tripartite domain structure, whose all-atom homology-based model is illustrated in Figure 1. Apart from short N- and C-terminal tail regions, the AAA+ core D1 (colored in blue) and regulatory D3 (green) domains are well conserved between archaeal and eukaryotic TIP49a and TIP49b paralogs, whereas the D2 insertion domain, which consists of the OB-fold and the L2 loop, forms a predominantly variable region (Figures 1 and S1, colored in red). These analyses show that mkTIP49 shares a highly significant overall similarity with its crystallized eukaryotic TIP49 paralogs that can be used for further comparative studies as investigated below.
Figure 1.
All-Atom Model of the M. kandleri TIP49 Protein
(A) Domain organization of mkTIP49 monomers. The AAA+ D1 core is colored in blue; the D2 insertion domain, composed of the OB-fold (dotted rectangle) and L2, is colored in red; and the D3 regulatory domain is colored in green. ATP (yellow) is located in the catalytic pocket, formed at the interface between D1 and D3. Note the high level of sequence similarity between mkTIP49 and its human paralogs: 45% of identity with hsTIP49a and 42% with hsTIP49b.
(B) Major conformational transitions of mkTIP49 monomers in the course of 30 ns MD simulations are associated with OB-fold flexibility (arrow). Graphics were produced using the ICM-Pro software package. See also Figure S1 for sequence alignments between mkTIP49 and its eukaryotic paralogs TIP49a and TIP49b.
We first analyzed the conformational flexibility of mkTIP49 monomers and compared it with its human TIP49 paralogs by performing MD simulations in a water environment (Petukhov et al., 2012). Because M. kandleri is a hyperthermophilic archaeon, these simulations were performed by ramping up the temperature to 75°C, the optimal reaction temperature for ATP hydrolysis by mkTIP49. This allows the system to equilibrate through thermodynamic barriers, during which the model can progressively proceed through a series of intermediate transitions and rapidly adopt alternative stable conformations. The overall fold of mkTIP49 at 75°C was conserved relative to human TIP49 proteins, together with the high flexibility of the D2 domain. A rapid drift of the OB-fold toward the D3 domain leads to the formation of a stable network of interactions between D2 and D3 (Figure 1B). The resulting dynamic changes in mkTIP49 are very similar to those observed in human TIP49 monomers and are mediated by the formation of Van der Waals and electrostatic contacts between L1 of the OB-fold (residues 144–152) and the hinge region connecting the first and second α helices of the regulatory D3 domain (residue 376). These interactions are stabilized via additional contacts between L1 and the third α helix of D3 (residues 407 and 411). The C-terminal-most part of D3, however, remains unstructured, showing significant flexibility over the 30 ns simulation time (residues 427–455). As previously observed in the likTIP49b mutant, in which an FCR triplet is inserted in the OB-fold (Petukhov et al., 2012; Rottbauer et al., 2002), L2 of mkTIP49 rapidly collapses onto the region that connects the OB-fold to the rest of protein by establishing polar and hydrophobic interactions (residues 253–257 of L2 and hinge residues 126–128 and 237–242). L2 also forms an extensive hydrogen-bond (H-bond) network, resulting in further stabilization of mkTIP49 in a compact configuration (Figure 1B, bottom image). Taken together, our results of homology-based modeling and MD simulations of mkTIP49 show a highly significant overall similarity between the archaeal protein and its crystallized eukaryotic TIP49 paralogs that can be used for further comparative studies.
Structural Organization of the mkTIP49 ATP Binding Pocket in Monomers
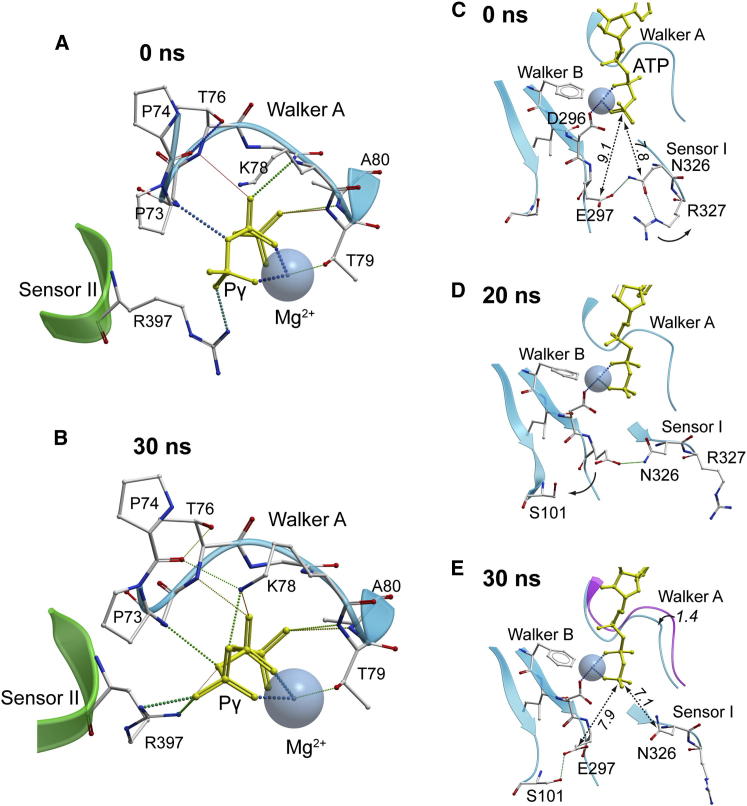
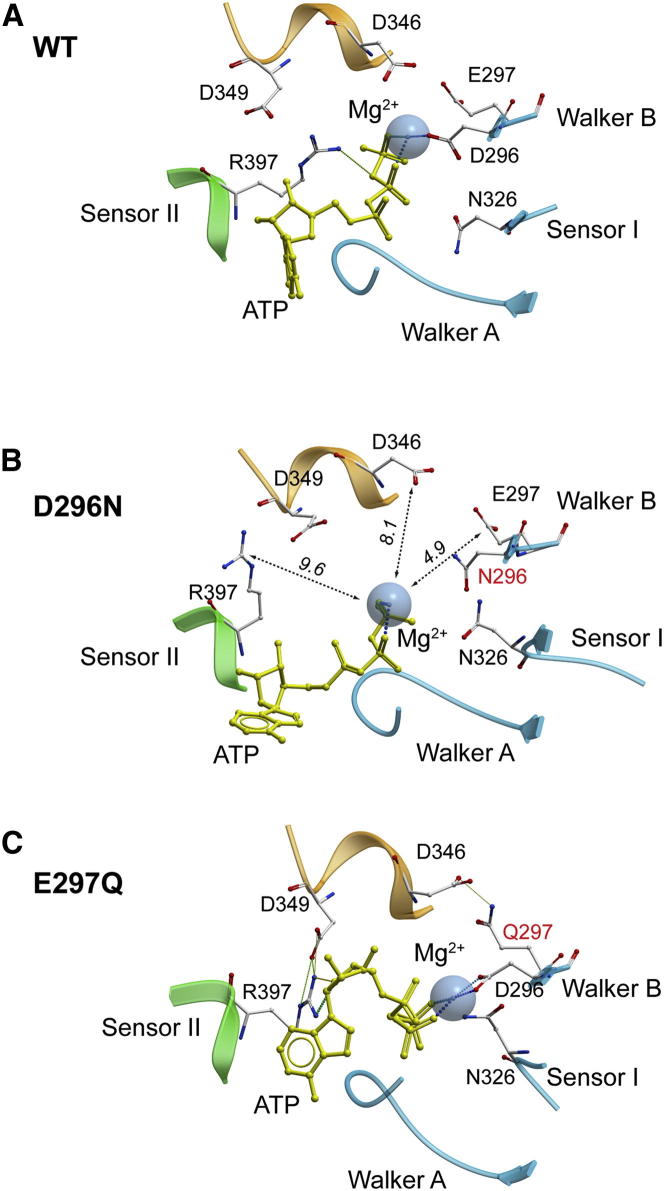
Compared to D2 and D3, the AAA+ core D1 domain is the most stable structural feature of mkTIP49 monomers (average Cα-root-mean-square deviation [rmsd] values for D2, D3, and D1 are 4.1, 4.4, and 3.4 Å, respectively). Analysis of the dynamics of the ATP binding pocket, which is formed at the interface between D1 and D3 (Figure 1A, blue and green colors, respectively), revealed several important organizational features. MkTIP49 contains conserved Walker A (P loop, residues 72-GPPGTGKTA-80) and Walker B (292-VLFIDE-297) motifs as well as other amino acid residues involved in ATP binding and hydrolysis (Figure S1). These include the Sensor I (N326) and II (R397) residues that are hallmarks of AAA+ ATPases (Erzberger and Berger, 2006; Iyer et al., 2004; Wendler et al., 2012). In the regularized mkTIP49 structure modeled with ATP and Mg2+, the phosphate chain of ATP is found within the Walker A motif (Figure 2A). The side chain of Walker A T79 is also found in close proximity to Mg2+. Together with Walker B D296, it coordinates this ion, which is essential for ATP hydrolysis. E297 of Walker B forms an H-bond with N326 (Sensor I) (Figure 2C). Because of this interaction, the distance between the side chain of E297 and the γ-phosphate group of ATP is increased to 9.1 Å. In monomers, this creates a significant impediment for the activation of a water molecule, an essential step in ATP hydrolysis, which, as shown below, can be overcome by oligomerization.
Figure 2.
Snapshots of Dynamic Transitions in the Walker A, Walker B, and Sensor I Motifs in mkTIP49 Monomers Modeled with ATP and Mg2+
(A and B) Initial (A) and 30 ns conformations (B) of the ATP phosphate chain (yellow), of Mg2+ (shown as a Van der Waals sphere), and of the side chains of the Walker A and Sensor II motifs.
(C–E) Dynamics of the glutamate switch in mkTIP49 monomers. Initial (C), 20 ns (D), and 30 ns (E) conformations of Walker B E297 are shown, forming H-bonds (dotted lines) with N326 (Sensor I) and S101, respectively. The initial position of the Walker A motif is shown in purple color in (E). Distances in Å are denoted by italicized numbers.
During MD simulations, we observed a discrete readjustment of the ATP molecule and of the peptidyl backbone and side chains that form the pocket (Figures 2B and 2E). The resulting configuration shows a remarkable coordination of phosphates because of the 1.4 Å shift of the P loop toward ATP and the formation of additional H-bonds between K78 and two available oxygen atoms of the β- and γ-phosphate groups, respectively (Figure 2B). This rearrangement also ruptures the H-bond between N326 (Sensor I) and E297 (Walker B) (Figures 2D and 2E). Consequently, the “glutamate switch” (Zhang and Wigley, 2008) E297 edges closer to the S101, with which it forms an H-bond, and remains 7.9 Å away from the γ-phosphate. The flexible guanidyl group of R397 (Sensor II), protruding from the regulatory D3 domain, makes a stable contact with the γ-phosphate group of ATP (Figure 2B). As a result of these rearrangements, the γ-phosphate oxygens are bound by R397, K78, and Mg2+. Whereas coordination of Mg2+ by D296 is canonical, the behavior of E297 of Walker B is unfavorable for the associative mechanism of ATP hydrolysis, which requires the presence of a negatively charged residue in close proximity of the ATP γ-phosphate (Grigorenko et al., 2007; Senior et al., 2002). This was further investigated in oligomers using in silico and biochemical approaches.
Oligomeric State of mkTIP49 in Solution
The oligomeric state of TIP49 proteins is important for their catalytic activities (Gorynia et al., 2011; Gribun et al., 2008; Lopez-Perrote et al., 2012; Matias et al., 2006; Niewiarowski et al., 2010; Petukhov et al., 2012; Torreira et al., 2008). To study the oligomeric states of mkTIP49, the corresponding gene was amplified from genomic DNA, and recombinant protein was produced in a bacterial expression host (Figure 3). In parallel, a D2-truncated mutant of mkTIP49 (TIPΔD2), lacking amino acid residues 143–290, was included in this study to compare its oligomerization and catalytic properties with analogous deletions of eukaryotic TIP49 paralogs (Gorynia et al., 2011; Gribun et al., 2008; Matias et al., 2006; Niewiarowski et al., 2010; Petukhov et al., 2012).
Figure 3.
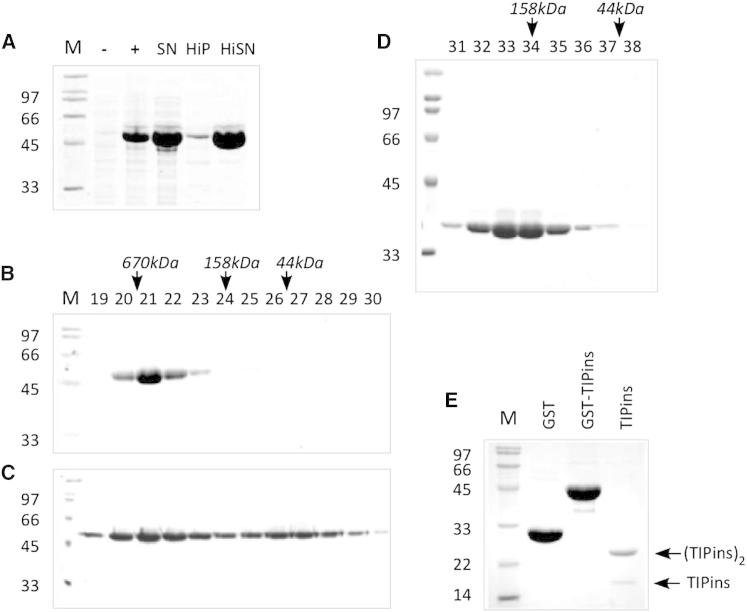
Expression and Purification of Archaeal mkTIP49 Protein
(A) The expression profile and heat stability of mkTIP49 supernatant (SN), heat-inactivated pellet, and supernatant fractions (HiP/HiSN) are shown. ± indicates isopropyl β-D-1-thiogalactopyranoside-induced total cell extracts.
(B and C) The elution profiles of mkTIP49 on a Superdex 200 sizing column in P300 and N1000 buffers, respectively, are shown.
(D) The elution profile of the TIPΔD2 variant on a Superose 6 sizing column is shown. The elution profiles of molecular size standards (670, 158, and 44 kDa) are indicated above the gel in italics (B), (C), and (D).
(E) The purification of the TIP49 D2 insertion domain as a glutathione S-transferase (GST)-fusion protein on glutatione agarose before thrombin cleavage (GST-TIPins) next to purified GST and TIPins domain. SDS-PAGE gels were stained with Coomassie Brilliant blue.
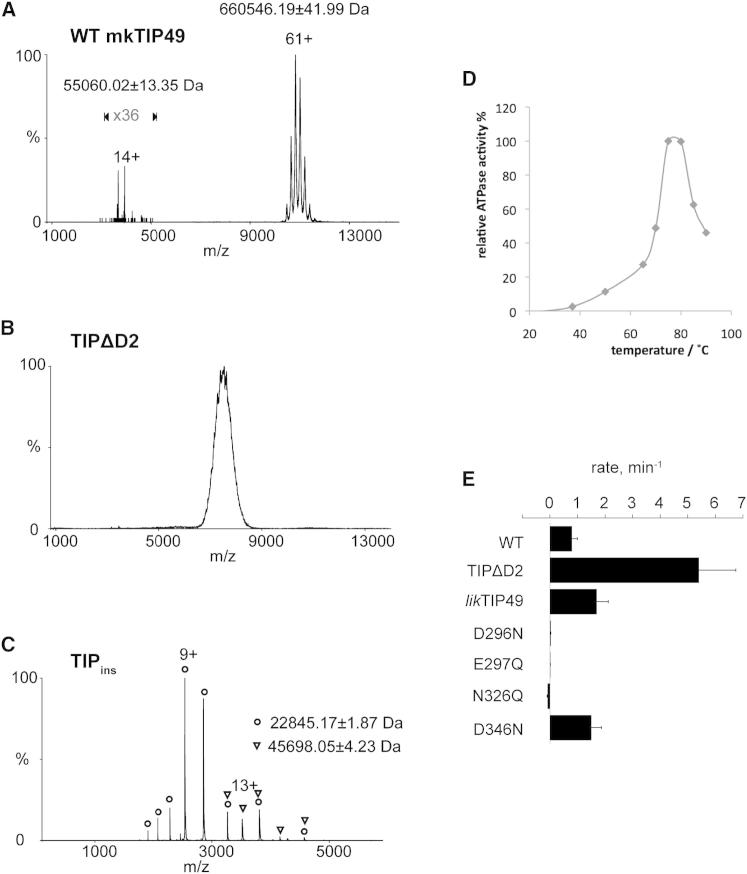
Full-length mkTIP49 and TIPΔD2 proteins were first subjected to nanoelectrospray ionization mass spectrometry, under conditions in which non-covalent interactions are preserved (Figures 4A–4C), and in parallel subjected to gel-filtration analysis (Figures 3B–3D; Figure S2). In agreement with the gel filtration data (Figure S2; 560 kDa), the major and minor species correspond to an mkTIP49 dodecamer (12 × 54,500 Da = 654,000 Da) with 12 bound ADP nucleotides (12 × 501 Da = 6,012 Da) and one mkTIP49 monomer (54,500 Da) bound to one ADP nucleotide (501 Da), respectively, similar to that previously reported for human (hs) hsTIP49a (Matias et al., 2006). We could not obtain a similarly high resolution when we carried out mass spectrometry on the highly purified TIPΔD2 variant: the resulting spectra reproducibly contained one broad signal centered on m/z 7,500 (Figure 4B). We attribute the broadening of the peak to the presence of a heterogeneous population of oligomers. We were able to estimate a mass of ∼340,000 Da for TIPΔD2 oligomers using the relationship between mass and m/z previously determined for a range of proteins and protein complexes (Heck and Van Den Heuvel, 2004; Uetrecht et al., 2008). Such a mass is too small to account for a TIPΔD2 dodecamer (453,600 Da) and too large for a hexamer (226,800 Da). It could correspond to a heterogeneous population with an average composition of nonamers (340,200 Da). Such oligomers can in theory be explained by at least three alternative models: (1) a range of molecular species consisting of one hexameric ring bound to a monomer, a dimer, a trimer, a tetramer, or a pentamer, (2) a population of two stacked “open” rings with an average of 4–5 subunits in each ring, or (3) a mixture of hexamers and dodecamers. The third possibility can be ruled out as we would expect the ion series for dodecamers and hexamers to appear at well-resolved m/z values. This precludes any overlapping of these charge states that might produce a distorted mass measurement corresponding to nonamers (Figure 4).
Figure 4.
mkTIP49 Oligomeric State and ATP Turnover
(A–C) Mass spectrometry analysis of wild-type mkTIP49, of TIPΔD2, and of the isolated D2 domain (TIPins, residues 124–290) is shown in (A), (B), and (C), respectively.
(D) The optimal temperature range for ATP hydrolysis by the wild-type mkTIP49 protein.
(E) Comparison of ATP turnover between the wild-type, TIPΔD2, likTIP49, and catalytic-site mutant proteins. The ATP turnover of mkTIP was determined at temperatures of 37°C, 50°C, 65°C, 70°C, 75°C, 80°C, 85°C, and 90°C as described in the Supplemental Experimental Procedures. Measurements were repeated 2–4 times, and the plotted values show a representative experiment within the full temperature range. See also Figure S1 for the location of mutations within the amino-acid sequence of mkTIP49.
To conclude these comparisons, we also analyzed the isolated mkTIP49 D2 domain (TIPins, residues 124–290; Figure 4C), which yielded two ion series. The major ion series centered on m/z 2,539 had a measured mass of 22,845 ± 2 Da, whereas the minor ion series centered on m/z 3,516 had a measured mass of 45,698 ± 4 Da. These masses correspond to monomers and dimers (monomeric TIPins Mw = 23,018 Da), respectively. The presence of stable dimers confirms that SDS-resistant dimers of the mkTIP49 D2 domain can form (Figure 3). In summary, this analysis demonstrates that (1) archaeal mkTIP49, like eukaryotic TIP49a/TIP49b protein assemblies, is able to form dodecamers (i.e., double hexamers) and (2) that the formation of stable dodecamers, compromised in the TIPΔD2 deletion mutant, is likely to be dependent on the D2 domain. Both full-length mkTIP49 and TIPΔD2 contain a tightly bound ADP cofactor scavenged from the bacterial expression host. We conclude from these observations that the archaeal mkTIP49 protein behaves like its eukaryotic paralogs in terms of oligomerization and ADP trapping within the ATP binding and hydrolysis pocket.
Do Previously Identified TIP49 Mutations Affect ATP Hydrolysis by mkTIP49?
Mutations that affect in vivo functions of eukaryotic TIP49 proteins have been previously described: the liebeskummer (lik) insertion within the OB-fold of zebrafish TIP49b and an active-site mutation within the Walker B motif (Gorynia et al., 2011; Grigoletto et al., 2013; Jonsson et al., 2001; Rottbauer et al., 2002). We asked whether the corresponding mutations would similarly affect the ATPase activity of mkTIP49. To address this question, we measured the intrinsic ATPase activity of mkTIP49 and mutants thereof. We first tested a broad range of temperatures from 25°C to 90°C and found an optimal reaction temperature of 75°C–80°C, congruent with the temperatures that are conducive to M. kandleri growth (Figure 4D). The observed rate of ATP hydrolysis was similar to that of human TIP49a/TIP49b heterohexameric assemblies (Gribun et al., 2008; Niewiarowski et al., 2010; Puri et al., 2007). As for eukaryotic TIP49 proteins, the deletion of D2 had a dramatic stimulating effect on the catalytic activity of mkTIP49 (Figure 4E), recalling the autoinhibitory effect of D2 on ATP hydrolysis observed in eukaryotic TIP49 proteins (Gorynia et al., 2011; Matias et al., 2006). A lik-like insertion of an FCR triplet in the OB-fold of mkTIP49 (see Figure S1) resulted in a modest, but reproducible, stimulation of ATP hydrolysis and is also in line with the properties of likTIP49b and with the results of our structural analysis (Petukhov et al., 2012; see also Rottbauer et al., 2002). Again, as judged by the ATPase activity of mkTIP49, the protein behaves like its eukaryotic paralogs in terms of allosteric regulation of the ATP hydrolysis by the D2 (OB-fold) insertion domain. Taken together, these results again support the conclusion that the archaeal mkTIP49 protein follows the same principles of domain flexibility as its human paralogs, TIP49a and TIP49b (Petukhov et al., 2012).
Effect of Walker B Mutations on ATP Hydrolysis
To rule out that the measured ATP hydrolysis was due to contamination of recombinant mkTIP49 protein with copurifying heat-stable ATPase or phosphatase activities from Escherichia coli, we mutated the Walker B motif to eliminate the negative charge of residues D296 and E297 that are essential for Mg2+ and water coordination in RuvB (D296N and E297Q, respectively) (Mezard et al., 1997). These mutants had only a background level of activity (Figure 4E), whereas their expression level, solubility, and heat stability were not affected. These Walker B mkTIP49 variants serve as key negative controls for our interpretations of ATP hydrolysis data. For this reason, their effects on the configuration of the mkTIP49 catalytic pocket were analyzed and compared to wild-type protein in the MD simulations presented below.
ATP Binding Pocket Dynamics within mkTIP49 Hexamers
We built atomic models of wild-type, D296N, and E297Q mkTIP49 hexamers bound to ATP and Mg2+ and analyzed their conformational flexibility by performing additional MD simulations (Figure 5). Consistent with results shown above, the most pronounced domain movements within mkTIP49 hexamers were associated with the C-terminal-most part of D3 and the D2 insertion domain. Maximal drifts from initial positions as estimated from Cα-rmsd values were 2.9 Å for D3, 5.2 Å for the OB-fold and 2.1 Å for D1, the most stable domain. Multiple D2/D2 interactions between adjacent protomers were detected in mkTIP49 hexamers, recalling the interprotomer D2-mediated interactions described for the TIP49 hetero- and likTIP49b homohexamers (Petukhov et al., 2012). These observations are consistent with the possible implication of D2 in high-order oligomerization of the mkTIP49 protein and further validate our atomic models of the mkTIP49 protein.
Figure 5.
Dilatation of Catalytic Pockets in the Walker B Mutants in mkTIP49 Hexamers
The 30 ns conformation of the wild-type protein (A) is aligned with that of the D296N (B) and E297Q (C) mutant proteins. The GDLLDR motif from the adjacent protomer is colored in orange. See also Figure S4 for details of D296, E297, D346, and R397 dynamics. Note that during MD simulations the phosphate chain remains the most stably bound part of ATP within the catalytic pockets of mkTIP49. The nucleoside moiety of ATP, however, can also undergo significant thermal fluctuations (compare A, B, and C). This could be attributed to the smaller number of H-bonds that are formed with the nucleoside part of ATP relative to the phosphate chain, which is remarkably well coordinated by Walker A, Sensor II, and Mg2+ (see also Figure 2).
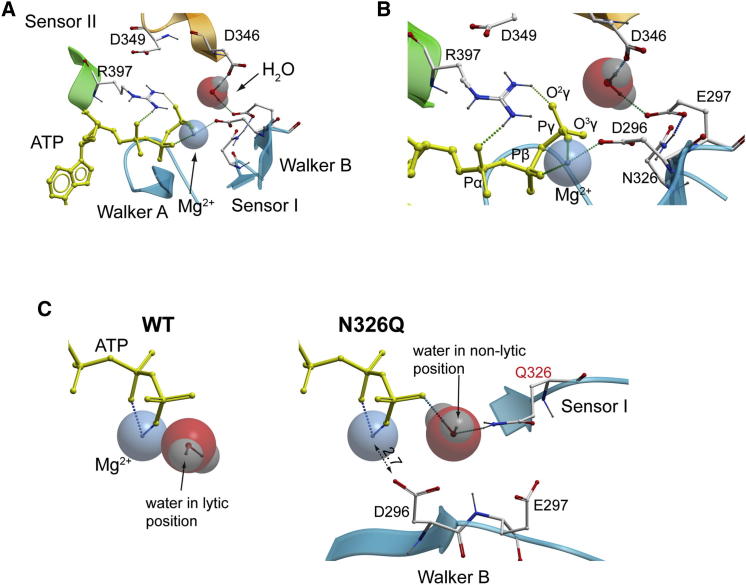
In TIP49 oligomers, a short conserved α-helical GDLLDR motif (residues 345–350 in mkTIP49, α-helical residues are underlined; see also Figures S1 and S3) is involved in the formation of TIP49 homo- and heterohexamers (Gorynia et al., 2011; Matias et al., 2006; Petukhov et al., 2012). Provided by an adjacent protomer, this motif contributes D346 and D349 to the catalytic pocket of mkTIP49 (Figure 5A, colored in orange). The side chain of D346 is located in the vicinity of the ATP γ-phosphate group, closer than the Walker B E297 residue as estimated by MD simulations (Cγ346-Pγ of 5.9 ± 0.48 Å and Cδ297-Pγ of 6.92 ± 0.54 Å, respectively; see Figure S3). As in mkTIP49 monomers, the side chain of E297 adopts a variety of conformations in hexamers. Accordingly, it can reach N326 (Sensor I) or adjacent R327 residues and be trapped away from the γ-phosphate via H-bonding with these residues. In contrast, D346 moves toward ATP due to the presence of positively charged R397 (Sensor II). These dynamics of the catalytic pocket within mkTIP49 hexamers suggest that D346, together with cis-E297, may serve as a trans-acting residue that can accept a proton from a water molecule, thus initiating a nucleophilic attack of the covalent bond formed between the β- and γ-phosphates of ATP.
MD analysis of D296N hexamers showed significant dilatation of the catalytic pockets, occurring during the equilibration step that precedes MD simulations (Figures 5B and S3). Compared to wild-type hexamers, the distance between the mutated N296 residue and Mg2+ increases from 3.16 ± 0.09 Å to 5.02 ± 0.36 Å (mean ± SD), respectively, leading to complete loss of Mg2+ coordination by the mutated Walker B motif (Figures 5 and S3). Consequently, the side chain of R397 (Sensor II) moves away from the γ-phosphate toward D349, which protrudes from the adjacent protomer (compare Figures 5A and 5B), whereas R397 remains trapped by two H-bonds formed with D349 (Figure 5B). As a result of this rearrangement, the distance between the guanidyl group of R397 and the γ-phosphorus atom of ATP in the D296N mutant increases more than twice relative to the wild-type protein: 9.38 ± 0.74 Å versus 4.30 ± 0.20 Å (mean ± SD), respectively. The putative water-activating trans-D346 residue also follows a similar pattern of behavior and drifts away from the γ-phosphate (8.7 ± 0.71 Å in the mutant versus 5.9 ± 0.48 Å (mean ± SD) in the wild-type protein; see Figures 5B and S3). Analysis of E297Q hexamers also revealed a distinct disorganization of the mkTIP49 catalytic pocket (Figure 5C). This mutation resulted in a shift of negatively charged D346 and D349 residues away from the phosphate chain of ATP and in the formation of an H-bond with the mutated residue 297. Remarkably, the guanidyl group of the Sensor II R397 can be found in close contact with the sugar moiety instead of the phosphate chain, where it forms H-bonds with the O4’ oxygen of the ribose. This interaction can be further stabilized by the trans-D349 residue (Figures 5C and S3). Taken together, these results demonstrate, at the atomic level, the drastic consequences that may arise from simple charge eliminations within the Walker B motif on the overall configuration of the catalytic pocket of mkTIP49 and highlight the importance of key residues that can activate water molecules.
Dynamics of Lytic Water Molecules inside the ATP Binding Pockets of TIP49 Hexamers
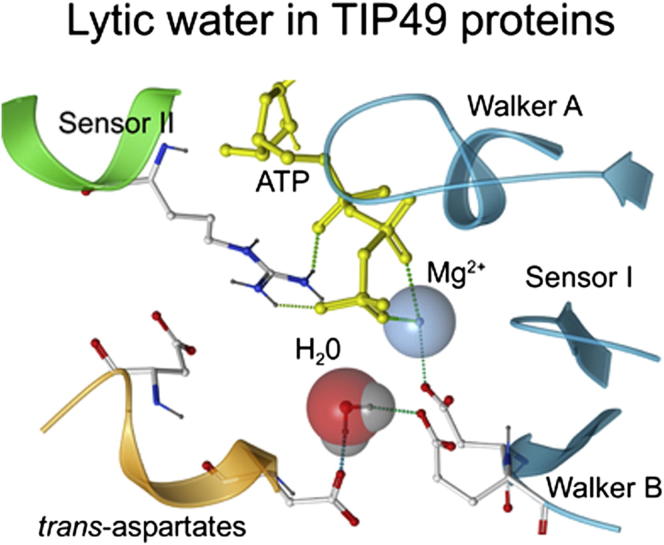
Can cis-E297 or trans-D346 act as water-activating residues for ATP hydrolysis by mkTIP49? We answered this question by first determining the minimal volume that can accommodate a lytic water molecule in the immediate vicinity of the ATP γ-phosphate. We then correlated the dynamics of water molecules in this volume with H-bond formation with E297 and D346. Over 30 ns simulation times, the cumulative probability of the minimal volume occupancy by water molecules located within 2.5 Å of the γ-phosphorus atom was found to be similar in the wild-type mkTIP49 protein and in the D296N variant (0.22 versus 0.19, respectively). This indicates that the γ-phosphate of ATP may be similarly accessible to water in both proteins. However, the fraction of water molecules that form appropriate H-bonds (see the Supplemental Experimental Procedures) with E297 or D346 decreased approximately seven times over the simulation period, dropping from 0.105 in the wild-type protein to 0.014 in the mutant, which is consistent with the dilatation of the mutant catalytic pockets (Figure 5B). From a total of 165 appropriate H-bonds formed with water molecules in the wild-type catalytic pockets, 97 were formed with E297, 67 with D346, and 1 with D349, suggesting that both cis-acting E297 and trans-acting D346 can function as proton-accepting residues during ATP hydrolysis by mkTIP49 hexamers (Figures 6A and 6B). Both of these interactions can orient the water molecule to the appropriate position, in which the oxygen atom of water points to the γ-phosphorus atom of ATP and to the last γ-phosphate oxygen that does not form H-bonds with the protein (O3γ in Figure 6B). Accordingly, this model illustrates the pre-transition state assembly of all components required for ATP hydrolysis to occur in TIP49 hexamers.
Figure 6.
Lytic Water Coordination in the Catalytic Pockets of mkTIP49 Hexamers
(A and B) An overall view of the pocket (A) and the zoom on the γ-phosphate region (B) are shown with ATP (yellow), Mg2+, and a water molecule in the attacking position.
(C) Lytic water in the attacking position in the wild-type mkTIP49 protein (left) and in the N326Q variant (right). The N- to Q substitution in the Sensor I motif of mkTIP49 may result in reorientation of water molecules from lytic to non-lytic positions by the mutated Q326 residue and interfere with coordination of Mg2+ by D296 of Walker B.
ATP Hydrolysis by TIP49 Catalytic Site Mutants
We tested this model by introducing point mutations in the catalytic pockets of mkTIP49 and studied these mutants in biochemical assays (Figure 4E). To investigate the influence of the Sensor I motif on water dynamics and ATP hydrolysis by mkTIP49, we replaced N326 by glutamine (N326Q). This substitution brings the polar amino group of the mutated 326 residue closer to ATP compared to the wild-type protein and may lead to the formation of a donor H-bond with the water oxygen (Figure 6C). Mechanistically, this interaction is the opposite of the formation of an acceptor H-bond by water’s hydrogen with negatively charged residues, which is required for water activation: it reorients the water molecule into a non-lytic position and may also interfere with Mg2+ coordination by D296 of Walker B (Figure 6C). As predicted from such a rearrangement, the ATPase activity of the purified N326Q mkTIP49 variant was undetectable compared to the wild-type protein (Figure 4E). Indeed, as judged by the reproducible negative rate values obtained in independent experiments, this mutant might actually protect ATP from spontaneous hydrolysis measured in parallel at 75°C, in agreement with results of our MD and water dynamics simulations within the catalytic pocket of mkTIP49.
To investigate the effects of mutations within the GDLLDR motif, we eliminated the negative charge of D346 in mkTIP49 by replacing it with asparagine. We then purified the corresponding D346N mutant and measured its ATPase activity. We observed a moderate ∼2-fold increase in ATP hydrolysis for the D346N variant relative to wild-type proteins (Figure 4E). Because we did not detect any significant defects in the oligomerization properties of these mutants, such an increased level of ATP turnover can be explained by reconfiguration of remaining intact proton-accepting residues in the mutant TIP49 catalytic pockets. The charge elimination due to the D- to N346 substitution results in a decrease of local negative electrostatic potential in TIP49 oligomers. This, in turn, can decrease the activation barrier of the proton transfer from the lytic water molecule to the next available negatively charged proton-accepting residue, such as the Walker B E297 residue in mkTIP49, thus increasing the rate of ATP hydrolysis. Such possible rearrangements of lytic water molecules in the vicinity of the ATP γ-phosphates in this mutant may be similar to the positioning of lytic water in RuvB hexamers modeled with ATP and Mg2+ (see Figure 7).
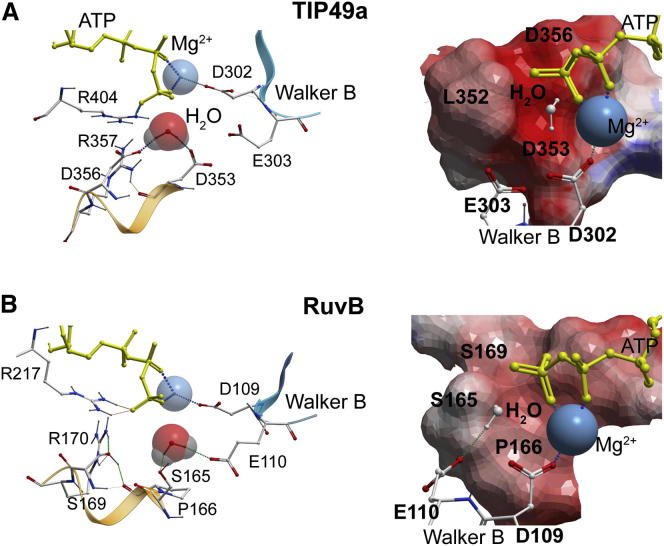
Figure 7.
Comparison of Lytic Water Coordination in Wild-Type hsTIP49a and tmRuvB
(A) hsTIP49a (PDB ID code 2C9O).
(B) tmRuvB (PDB ID code 1IN4).
TIP49 and RuvB ATPases
TIP49 proteins are often called RuvB-like on the basis of sequence similarities (Ammelburg et al., 2006; Erzberger and Berger, 2006; Iyer et al., 2004; Wendler et al., 2012). Does such a comparison hold in terms of lytic water activation? Aside from the presence of D2 insertion domains in TIP49 proteins, the ATP binding and hydrolysis pockets are composite and similarly organized in both TIP49 and Thermotoga maritima (tm) RuvB hexamers (Figures 7 and S4) (Gorynia et al., 2011; Matias et al., 2006; Petukhov et al., 2012; Putnam et al., 2001). Dense packing of the catalytic pockets in both hsTIP49a and tmRuvB hexamers, together with the necessity for magnesium, leads to only one possible position for the γ-phosphate group of ATP. In this conformation, two γ-phosphate oxygens form H-bonds with the Walker A (K64 and K76 in hsTIP49a and tmRuvB, respectively) and Sensor II residues (R404 and R217 in hsTIP49a and tmRuvB, respectively). In the regularized structures modeled with ATP, Mg2+, and a lytic water molecule, aspartate residues from Walker B (D302 in hsTIP49a and D109 in tmRuvB) coordinate Mg2+, whereas the glutamate residues of Walker B (E303 in hsTIP49a and E110 in tmRuvB) show different behaviors. In tmRuvB hexamers, E110 is located within 3 Å of the lytic water oxygen, sufficiently close to form the H-bond required for direct proton transfer from water to the side-chain carboxyl oxygen atom of E110. In contrast, E303 of hsTIP49a is found in an unfavorable configuration for the formation of the required H-bond, because of its possible trapping by the side chains of N332 (Sensor I) or S99 (S101 in mkTIP49; Figure 2). These observations suggest that, compared to RuvB, E303 of Walker B (E301 in hsTIP49b) may play a lesser role in the mechanisms that activate water within TIP49 hexamers.
Discussion
In this study, we have extended the mechanistic analysis of ATP hydrolysis by AAA+ TIP49 ATPases by addressing the dynamics of lytic water molecules that are a critical but little understood step in ATP hydrolysis. To do so, we have characterized their archaeal mkTIP49 ortholog, which is largely free of the ambiguities associated with eukaryotic TIP49 proteins and their oligomeric assemblies. Our results demonstrate that mkTIP49 behaves as its crystallized eukaryotic TIP49 paralogs in terms of oligomerization and regulation of ATP hydrolysis, validating the use of molecular modeling and MD simulations for the comparative structural analysis of TIP49 proteins and biochemical verification thereof.
Within the composite catalytic pockets of both archaeal and human TIP49 hexamers, we show that a complex H-bond network connects the Walker B and Sensor I motifs and contributes to Mg2+ and lytic water coordination. Our focus on lytic water dynamics provides insights into the associative mechanisms of ATP hydrolysis by TIP49 proteins, which have remained somewhat intractable. We propose that trans-acting aspartate D346 in mkTIP49 and the corresponding residues in hsTIP49 proteins (Figure S1), protruding from the next adjacent protomer, participate in water coordination and also serve as cryptic proton-acceptors that downregulate ATP hydrolysis by wild-type TIP49 hexamers. These conclusions are fully supported by mutational analysis of purified archaeal TIP49 proteins. Taken together, our data highlight a distinctive organization of TIP49 catalytic pockets within oligomeric assemblies, helping explain a variety of previous biochemical results obtained in different experimental systems (see Rosenbaum et al., 2013 and the references therein).
As suggested in the pioneering study of the hsTIP49a hexameric X-ray structure, cooperativity between TIP49 monomers plays an important role in ATP hydrolysis (Matias et al., 2006). However, the complex mechanisms that lead to this cooperativity have remained elusive. For example, the structurally conserved short GDLLDR helix of mkTIP49 corresponds to the α4 SPLRSR helix of tmRuvB (the α-helical part is underlined; see Figure S4). In both proteins, this helix is involved in oligomerization and contributes to the catalytic pocket in hexamers. The flanking arginine (R350 in mkTIP49 and R170 in tmRuvB) can be aligned as a trans-acting R-finger by sequence and structural similarity with other members of the AAA+ superfamily (see Figure S4 for the alignment with RuvB) (Ammelburg et al., 2006; Erzberger and Berger, 2006; Iyer et al., 2004; Wendler et al., 2012). However, in the TIP49a, TIP49b, and TIP49a/b hexameric crystal structures (Gorynia et al., 2011; Matias et al., 2006; Petukhov et al., 2012), as well as in the course of MD simulations of TIP49a/b homo- and heterohexamers (Petukhov et al., 2012) and of the archaeal mkTIP49 protein (this study), this arginine remains rotated away from the catalytic pocket because of H-bond formation with the backbone carbonyl group of D353 (hsTIP49a). Similarly, R170 of tmRuvB can form an H-bond with adjacent S169 (Figure 6), which results in the same effect as in TIP49 hexamers: its guanidyl group is kept 8.0 Å away (8.8 Å in TIP49a) from the γ-phosphate of ATP. These observations suggest that trans-arginines are not involved in direct transactions with ATP γ-phosphates in TIP49 and tmRuvB hexamers.
The xDLLDR helical motif (90% consensus for TIP49a and TIP49b) contributes two negatively charged residues, D353 and D356 (hsTIP49a), to the catalytic pockets of hsTIP49 hexamers. We believe that this structural feature may be important for downregulation of ATP hydrolysis in TIP49 hexamers. Indeed, the presence of two additional aspartates generates a strong local negative electrostatic potential in the vicinity of the ATP γ-phosphate group. This may not only impact water dynamics in this region but also impact the height of the activation barrier of proton transfer from lytic water to an acceptor. In the D346N mkTIP49 variant, however, the lytic water molecules can be redirected to the next available glutamate residue of Walker B and then coordinated in a RuvB-like manner (Figure 7). In agreement with our conclusions from in silico water dynamics experiments and biochemical data, the increased rate of ATP turnover by the D346N mkTIP49 variant is similar to that of RuvB, as observed in the absence of stimuli by RuvA and Holliday junctions (Hishida et al., 2004; Mezard et al., 1997).
We note the resemblance between the xDLLDR motif found in TIP49 proteins and the recently described trans-acting “R-hand” (162-RADLLDR-168) motif found in the E. coli PspF AAA+ ATPase (Joly et al., 2012; Rappas et al., 2005; Wendler et al., 2012). Despite the identical DLLDR sequence shared by TIP49 and PspF proteins, the D164 mutation (D346 in mkTIP49) drastically reduces the rate of ATP hydrolysis by PspF, but not by mkTIP49. This discrepancy with the gain-of-function phenotype we report here can be explained by a mechanism implicating trans-acting aspartates and lytic water positioning. According to the PspF X-ray structures (Protein Data Bank [PDB] ID codes 2C96 and 2C9C), the second trans-aspartate of the R-hand, D167 (D349 in mkTIP49), as well as Walker B D107 and E108 residues, are found in unfavorable positions with respect to the γ-phosphate of ATP to serve as water activators. The only remaining trans-acting D164 residue would then be the best candidate as a proton-acceptor, explaining the loss-of-function phenotypes of the D164A/N/Q variants of PspF. In contrast, the drastic decrease in ATP hydrolysis by the Sensor I N64Q PspF variant is similar to the effect of the N326Q mutation in mkTIP49, suggesting the existence of shared mechanisms that underlie the regulation of ATP hydrolysis by the Sensor I motif in TIP49 and PspF proteins (Joly et al., 2012).
Size-exclusion chromatography and nanoelectrospray ionisation mass spectrometry of archaeal mkTIP49 demonstrate that it is predominantly in a dodecameric form, reminiscent of the assemblies formed by its purified eukaryotic TIP49a/TIP49b counterparts (Gorynia et al., 2011; Lopez-Perrote et al., 2012; Niewiarowski et al., 2010; Puri et al., 2007; Torreira et al., 2008). The principles of conformational flexibility and the allosteric link between the D2 insertion and AAA+ core ATPase domains are very similar between the archaeal and human TIP49 proteins. Similarities also extend to the configuration of catalytic pockets, where possible mechanisms of ATP/Mg2+ coordination and activation of attacking water molecules appear to be conserved, further supporting the hypothesis that TIP49 AAA+ ATPases evolved a mechanism for ATP hydrolysis that has been conserved from Archaea to eukaryotes. This would suggest that an ancestral form of contemporary TIP49 underwent gene duplication in eukaryotes after the split of the archaeal and eukaryotic lineages, which in all likelihood expanded its functional repertoire.
The precise biological function of TIP49 in the archaea remains obscure. MkTIP49 neither forms stable complexes with mkTBP nor with the cognate TATA-TBP or with TATA-TBP-TFB transcription initiation complexes (F.W., unpublished data), which does not support a role in the regulation of transcription similar to its eukaryotic homologs. However, the archaeal TIP49 gene is induced under heat-shock conditions; the protein is stable at elevated temperatures (Iqbal and Qureshi, 2010; Kanai et al., 2010); and it has been found to associate with ribosomes in a mass spectrometry approach (Marquez et al., 2011). These observations are congruent with a role of TIP49 as a protein chaperone and/or ATP-dependent remodeling factor of higher-order complexes. Future studies will delineate the function of this “ancestral” singular version of TIP49 in extant members of the archaeal domain.
Experimental Procedures
Experimental procedures are described in detail in the Supplemental Experimental Procedures. Molecular modeling was performed as described in Petukhov et al. (2012). Molecular dynamic simulations of mkTIP49 were performed using the GROMACS (v. 4.5.5) software package on the multiprocessor clusters of the St. Petersburg State Polytechnical University and of the “Kurchatov Institute” National Research Centre (Moscow). Cloning, expression, and purification of untagged recombinant mkTIP49 protein from Methanopyrus kandleri genomic DNA were performed using standard procedures. Single mutations were generated in the mkTIP49 expression plasmid (pET151D-TOPO-mkTIP49) by site-directed mutagenesis (QuikChange II Site-Directed Mutagenesis Kit). Size-exclusion chromatography of the various mkTIP49 proteins was carried out on Superdex 200 and Superose 6 columns (GE Healthcare). Nanoelectrospray ionization mass spectrometry was performed on an LCT Premier XE mass spectrometer (Waters) modified for high-mass operation. ATP hydrolysis was measured by phosphate released in a malachite green-based chromogenic assay (PiColorLock ALS, Innova Biosciences).
Acknowledgments
We thank the St. Petersburg State Polytechnical University and the “Kurchatov Institute” National Research Centre (Moscow) for allocation of computer time on multiprocessor clusters. M.G. thanks Martine Defais for stimulating discussions. G.P. is a recipient of the Scholarship of the President of the Russian Federation for PhD Students (no. 5651.2013.4). A.R.M. is funded by the Research Councils UK Academic Fellowship. This work was supported by the Federal Program of the Russian Ministry of Education and Science for invited foreign scientists (no. 11.519.11.2002 to M.G.), intramural LBME and CNRS funding (to M.G. and E.K.), and Wellcome Trust (079351/Z/06/Z) and BBSRC (BB/E008232/1) grants (to F.W.).
Footnotes
This is an open access article under the CC BY license (http://creativecommons.org/licenses/by/3.0/).
Contributor Information
Emmanuel Käs, Email: kas@ibcg.biotoul.fr.
Mikhail Grigoriev, Email: grigor@biotoul.fr.
Supplemental Information
References
- Ammelburg M., Frickey T., Lupas A.N. Classification of AAA+ proteins. J. Struct. Biol. 2006;156:2–11. doi: 10.1016/j.jsb.2006.05.002. [DOI] [PubMed] [Google Scholar]
- Bellosta P., Hulf T., Balla Diop S., Usseglio F., Pradel J., Aragnol D., Gallant P. Myc interacts genetically with Tip48/Reptin and Tip49/Pontin to control growth and proliferation during Drosophila development. Proc. Natl. Acad. Sci. USA. 2005;102:11799–11804. doi: 10.1073/pnas.0408945102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erzberger J.P., Berger J.M. Evolutionary relationships and structural mechanisms of AAA+ proteins. Annu. Rev. Biophys. Biomol. Struct. 2006;35:93–114. doi: 10.1146/annurev.biophys.35.040405.101933. [DOI] [PubMed] [Google Scholar]
- Etard C., Gradl D., Kunz M., Eilers M., Wedlich D. Pontin and Reptin regulate cell proliferation in early Xenopus embryos in collaboration with c-Myc and Miz-1. Mech. Dev. 2005;122:545–556. doi: 10.1016/j.mod.2004.11.010. [DOI] [PubMed] [Google Scholar]
- Gorynia S., Bandeiras T.M., Pinho F.G., McVey C.E., Vonrhein C., Round A., Svergun D.I., Donner P., Matias P.M., Carrondo M.A. Structural and functional insights into a dodecameric molecular machine - the RuvBL1/RuvBL2 complex. J. Struct. Biol. 2011;176:279–291. doi: 10.1016/j.jsb.2011.09.001. [DOI] [PubMed] [Google Scholar]
- Gribun A., Cheung K.L., Huen J., Ortega J., Houry W.A. Yeast Rvb1 and Rvb2 are ATP-dependent DNA helicases that form a heterohexameric complex. J. Mol. Biol. 2008;376:1320–1333. doi: 10.1016/j.jmb.2007.12.049. [DOI] [PubMed] [Google Scholar]
- Grigoletto A., Neaud V., Allain-Courtois N., Lestienne P., Rosenbaum J. The ATPase activity of reptin is required for its effects on tumor cell growth and viability in hepatocellular carcinoma. Mol. Cancer Res. 2013;11:133–139. doi: 10.1158/1541-7786.MCR-12-0455. [DOI] [PubMed] [Google Scholar]
- Grigorenko B.L., Kaliman I.A., Nemukhin A.V. Minimum energy reaction profiles for ATP hydrolysis in myosin. J. Mol. Graph. Model. 2011;31:1–4. doi: 10.1016/j.jmgm.2011.07.005. [DOI] [PubMed] [Google Scholar]
- Grigorenko B.L., Rogov A.V., Topol I.A., Burt S.K., Martinez H.M., Nemukhin A.V. Mechanism of the myosin catalyzed hydrolysis of ATP as rationalized by molecular modeling. Proc. Natl. Acad. Sci. USA. 2007;104:7057–7061. doi: 10.1073/pnas.0701727104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigorenko B.L., Shadrina M.S., Topol I.A., Collins J.R., Nemukhin A.V. Mechanism of the chemical step for the guanosine triphosphate (GTP) hydrolysis catalyzed by elongation factor Tu. Biochim. Biophys. Acta. 2008;1784:1908–1917. doi: 10.1016/j.bbapap.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heck A.J., Van Den Heuvel R.H. Investigation of intact protein complexes by mass spectrometry. Mass Spectrom. Rev. 2004;23:368–389. doi: 10.1002/mas.10081. [DOI] [PubMed] [Google Scholar]
- Hishida T., Han Y.W., Fujimoto S., Iwasaki H., Shinagawa H. Direct evidence that a conserved arginine in RuvB AAA+ ATPase acts as an allosteric effector for the ATPase activity of the adjacent subunit in a hexamer. Proc. Natl. Acad. Sci. USA. 2004;101:9573–9577. doi: 10.1073/pnas.0403584101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikura T., Ogryzko V.V., Grigoriev M., Groisman R., Wang J., Horikoshi M., Scully R., Qin J., Nakatani Y. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell. 2000;102:463–473. doi: 10.1016/s0092-8674(00)00051-9. [DOI] [PubMed] [Google Scholar]
- Iqbal J., Qureshi S.A. Selective depletion of Sulfolobus solfataricus transcription factor E under heat shock conditions. J. Bacteriol. 2010;192:2887–2891. doi: 10.1128/JB.01534-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer L.M., Leipe D.D., Koonin E.V., Aravind L. Evolutionary history and higher order classification of AAA+ ATPases. J. Struct. Biol. 2004;146:11–31. doi: 10.1016/j.jsb.2003.10.010. [DOI] [PubMed] [Google Scholar]
- Joly N., Zhang N., Buck M. ATPase site architecture is required for self-assembly and remodeling activity of a hexameric AAA+ transcriptional activator. Mol. Cell. 2012;47:484–490. doi: 10.1016/j.molcel.2012.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson Z.O., Dhar S.K., Narlikar G.J., Auty R., Wagle N., Pellman D., Pratt R.E., Kingston R., Dutta A. Rvb1p and Rvb2p are essential components of a chromatin remodeling complex that regulates transcription of over 5% of yeast genes. J. Biol. Chem. 2001;276:16279–16288. doi: 10.1074/jbc.M011523200. [DOI] [PubMed] [Google Scholar]
- Kanai T., Takedomi S., Fujiwara S., Atomi H., Imanaka T. Identification of the Phr-dependent heat shock regulon in the hyperthermophilic archaeon, Thermococcus kodakaraensis. J. Biochem. 2010;147:361–370. doi: 10.1093/jb/mvp177. [DOI] [PubMed] [Google Scholar]
- Kim J.H., Choi H.J., Kim B., Kim M.H., Lee J.M., Kim I.S., Lee M.H., Choi S.J., Kim K.I., Kim S.I. Roles of sumoylation of a reptin chromatin-remodelling complex in cancer metastasis. Nat. Cell Biol. 2006;8:631–639. doi: 10.1038/ncb1415. [DOI] [PubMed] [Google Scholar]
- Kim J.H., Lee J.M., Nam H.J., Choi H.J., Yang J.W., Lee J.S., Kim M.H., Kim S.I., Chung C.H., Kim K.I. SUMOylation of pontin chromatin-remodeling complex reveals a signal integration code in prostate cancer cells. Proc. Natl. Acad. Sci. USA. 2007;104:20793–20798. doi: 10.1073/pnas.0710343105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Perrote A., Munoz-Hernandez H., Gil D., Llorca O. Conformational transitions regulate the exposure of a DNA-binding domain in the RuvBL1-RuvBL2 complex. Nucleic Acids Res. 2012;40:11086–11099. doi: 10.1093/nar/gks871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado-Pinilla R., Liger D., Leulliot N., Meier U.T. Mechanism of the AAA+ ATPases pontin and reptin in the biogenesis of H/ACA RNPs. RNA. 2012;18:1833–1845. doi: 10.1261/rna.034942.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquez V., Frohlich T., Armache J.P., Sohmen D., Donhofer A., Mikolajka A., Berninghausen O., Thomm M., Beckmann R., Arnold G.J. Proteomic characterization of archaeal ribosomes reveals the presence of novel archaeal-specific ribosomal proteins. J. Mol. Biol. 2011;405:1215–1232. doi: 10.1016/j.jmb.2010.11.055. [DOI] [PubMed] [Google Scholar]
- Matias P.M., Gorynia S., Donner P., Carrondo M.A. Crystal structure of the human AAA+ protein RuvBL1. J. Biol. Chem. 2006;281:38918–38929. doi: 10.1074/jbc.M605625200. [DOI] [PubMed] [Google Scholar]
- McKeegan K.S., Debieux C.M., Watkins N.J. Evidence that the AAA+ proteins TIP48 and TIP49 bridge interactions between 15.5K and the related NOP56 and NOP58 proteins during box C/D snoRNP biogenesis. Mol. Cell. Biol. 2009;29:4971–4981. doi: 10.1128/MCB.00752-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mezard C., Davies A.A., Stasiak A., West S.C. Biochemical properties of RuvBD113N: a mutation in helicase motif II of the RuvB hexamer affects DNA binding and ATPase activities. J. Mol. Biol. 1997;271:704–717. doi: 10.1006/jmbi.1997.1225. [DOI] [PubMed] [Google Scholar]
- Nano N., Houry W.A. Chaperone-like activity of the AAA+ proteins Rvb1 and Rvb2 in the assembly of various complexes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2013;368:20110399. doi: 10.1098/rstb.2011.0399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niewiarowski A., Bradley A.S., Gor J., McKay A.R., Perkins S.J., Tsaneva I.R. Oligomeric assembly and interactions within the human RuvB-like RuvBL1 and RuvBL2 complexes. Biochem. J. 2010;429:113–125. doi: 10.1042/BJ20100489. [DOI] [PubMed] [Google Scholar]
- Parusel C.T., Kritikou E.A., Hengartner M.O., Krek W., Gotta M. URI-1 is required for DNA stability in C. elegans. Development. 2006;133:621–629. doi: 10.1242/dev.02235. [DOI] [PubMed] [Google Scholar]
- Petukhov M., Dagkessamanskaja A., Bommer M., Barrett T., Tsaneva I., Yakimov A., Queval R., Shvetsov A., Khodorkovskiy M., Käs E. Large-scale conformational flexibility determines the properties of AAA+ TIP49 ATPases. Structure. 2012;20:1321–1331. doi: 10.1016/j.str.2012.05.012. [DOI] [PubMed] [Google Scholar]
- Puri T., Wendler P., Sigala B., Saibil H., Tsaneva I.R. Dodecameric structure and ATPase activity of the human TIP48/TIP49 complex. J. Mol. Biol. 2007;366:179–192. doi: 10.1016/j.jmb.2006.11.030. [DOI] [PubMed] [Google Scholar]
- Putnam C.D., Clancy S.B., Tsuruta H., Gonzalez S., Wetmur J.G., Tainer J.A. Structure and mechanism of the RuvB Holliday junction branch migration motor. J. Mol. Biol. 2001;311:297–310. doi: 10.1006/jmbi.2001.4852. [DOI] [PubMed] [Google Scholar]
- Rappas M., Schumacher J., Beuron F., Niwa H., Bordes P., Wigneshweraraj S., Keetch C.A., Robinson C.V., Buck M., Zhang X. Structural insights into the activity of enhancer-binding proteins. Science. 2005;307:1972–1975. doi: 10.1126/science.1105932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum J., Baek S.H., Dutta A., Houry W.A., Huber O., Hupp T.R., Matias P.M. The Emergence of the Conserved AAA+ ATPases Pontin and Reptin on the Signaling Landscape. Sci. Signal. 2013;6:mr1. doi: 10.1126/scisignal.2003906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottbauer W., Saurin A.J., Lickert H., Shen X., Burns C.G., Wo Z.G., Kemler R., Kingston R., Wu C., Fishman M. Reptin and pontin antagonistically regulate heart growth in zebrafish embryos. Cell. 2002;111:661–672. doi: 10.1016/s0092-8674(02)01112-1. [DOI] [PubMed] [Google Scholar]
- Rousseau B., Menard L., Haurie V., Taras D., Blanc J.F., Moreau-Gaudry F., Metzler P., Hugues M., Boyault S., Lemiere S. Overexpression and role of the ATPase and putative DNA helicase RuvB-like 2 in human hepatocellular carcinoma. Hepatology. 2007;46:1108–1118. doi: 10.1002/hep.21770. [DOI] [PubMed] [Google Scholar]
- Schweins T., Langen R., Warshel A. Why have mutagenesis studies not located the general base in ras p21. Nat. Struct. Biol. 1994;1:476–484. doi: 10.1038/nsb0794-476. [DOI] [PubMed] [Google Scholar]
- Senior A.E., Nadanaciva S., Weber J. The molecular mechanism of ATP synthesis by F1F0-ATP synthase. Biochim. Biophys. Acta. 2002;1553:188–211. doi: 10.1016/s0005-2728(02)00185-8. [DOI] [PubMed] [Google Scholar]
- Shen X., Mizuguchi G., Hamiche A., Wu C. A chromatin remodelling complex involved in transcription and DNA processing. Nature. 2000;406:541–544. doi: 10.1038/35020123. [DOI] [PubMed] [Google Scholar]
- Torreira E., Jha S., Lopez-Blanco J.R., Arias-Palomo E., Chacon P., Canas C., Ayora S., Dutta A., Llorca O. Architecture of the pontin/reptin complex, essential in the assembly of several macromolecular complexes. Structure. 2008;16:1511–1520. doi: 10.1016/j.str.2008.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetrecht C., Versluis C., Watts N.R., Roos W.H., Wuite G.J., Wingfield P.T., Steven A.C., Heck A.J. High-resolution mass spectrometry of viral assemblies: molecular composition and stability of dimorphic hepatitis B virus capsids. Proc. Natl. Acad. Sci. USA. 2008;105:9216–9220. doi: 10.1073/pnas.0800406105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venteicher A.S., Meng Z., Mason P.J., Veenstra T.D., Artandi S.E. Identification of ATPases pontin and reptin as telomerase components essential for holoenzyme assembly. Cell. 2008;132:945–957. doi: 10.1016/j.cell.2008.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins N.J., Lemm I., Ingelfinger D., Schneider C., Hossbach M., Urlaub H., Luhrmann R. Assembly and maturation of the U3 snoRNP in the nucleoplasm in a large dynamic multiprotein complex. Mol. Cell. 2004;16:789–798. doi: 10.1016/j.molcel.2004.11.012. [DOI] [PubMed] [Google Scholar]
- Weiske J., Huber O. The histidine triad protein Hint1 triggers apoptosis independent of its enzymatic activity. J. Biol. Chem. 2006;281:27356–27366. doi: 10.1074/jbc.M513452200. [DOI] [PubMed] [Google Scholar]
- Wendler P., Ciniawsky S., Kock M., Kube S. Structure and function of the AAA+ nucleotide binding pocket. Biochim. Biophys. Acta. 2012;1823:2–14. doi: 10.1016/j.bbamcr.2011.06.014. [DOI] [PubMed] [Google Scholar]
- Werner F. Structure and function of archaeal RNA polymerases. Mol. Microbiol. 2007;65:1395–1404. doi: 10.1111/j.1365-2958.2007.05876.x. [DOI] [PubMed] [Google Scholar]
- Werner F., Weinzierl R.O. A recombinant RNA polymerase II-like enzyme capable of promoter-specific transcription. Mol. Cell. 2002;10:635–646. doi: 10.1016/s1097-2765(02)00629-9. [DOI] [PubMed] [Google Scholar]
- Werner F., Weinzierl R.O. Direct modulation of RNA polymerase core functions by basal transcription factors. Mol. Cell. Biol. 2005;25:8344–8355. doi: 10.1128/MCB.25.18.8344-8355.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood M.A., McMahon S.B., Cole M.D. An ATPase/helicase complex is an essential cofactor for oncogenic transformation by c-Myc. Mol. Cell. 2000;5:321–330. doi: 10.1016/s1097-2765(00)80427-x. [DOI] [PubMed] [Google Scholar]
- Zhang X., Wigley D.B. The ‘glutamate switch’ provides a link between ATPase activity and ligand binding in AAA+ proteins. Nat. Struct. Mol. Biol. 2008;15:1223–1227. doi: 10.1038/nsmb.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.