Abstract
Aims
To evaluate the killing of Pseudomonas aeruginosa PAO1 on copper cast alloys and the influence of genes on survival on copper containing medium and surfaces.
Methods and Results
Different strains of P. aeruginosa were inoculated on copper containing medium or different copper cast alloys and the survival rate determined. The survival rates were compared to rates on copper-free medium and stainless steel as control. In addition, the effect of temperature on survival was examined.
Conclusions
Copper cast alloys had previously shown to be bactericidal to various bacteria but the mechanism of copper-mediated killing is still not known. In this report we demonstrate that P. aeruginosa PAO1 is rapidly killed on different copper cast alloys and that genes involved in conferring copper resistance in copper-containing medium also influenced survival on copper cast alloys. We also show that the rate of killing is influenced by temperature.
Keywords: Pseudomonas aeruginosa, copper cast alloys, survival assay, copper toxicity
Introduction
The recent renewed interest in the self-sanitizing properties of metallic copper surfaces has resulted in several studies describing the bactericidal effects of copper and copper-cast alloys against a variety of different Gram-positive and Gram-negative bacteria (Noyce et al. 2006, Robine et al. 2002, Wilks et al. 2005, Wilks et al. 2006). Several parameters such as temperature and copper-content of alloys influenced the efficiency of killing of the bacteria tested. So far, however, studies did not address the question how this is accomplished. As a consequence we do not know the underlying mechanism that kills the bacteria, and we are ignorant of potential negative long-term effects of using copper-surface on emerging bacterial resistances.
Previous studies examined the antimicrobial properties of copper cast alloys. Escherichia coli O157 was killed due to exposure to copper cast alloys with a copper content of 85 % to 95 % (Wilks et al. 2005). Additionally the initial number of 107 cells of Listeria monocyogenes exposed to 100% copper cast alloy drops to zero after 60 minutes (Wilks et al 2005).
Copper is required as a cofactor of many enzymes but in excess can have deleterious effects through metal catalyzed protein oxidation and generation of reactive oxygen species. Several mechanisms to handle and regulate intracellular levels of copper have been identified in bacteria (Rensing and Grass, 2003). In addition, sequence comparison revealed an operon that was recently shown to be involved in copper homeostasis and is conserved in P. putida KT2440, Pseudomonas fluorescens PFO-1 and PF-05, Pseudomonas aeruginosa PAO1 and Pseudomonas chlororaphis (Quaranta et al., 2007). The cinRS genes encode a histidine sensor kinase (cinS) and a response regulator (cinR) in the opposite direction of cinAQ (copper induced). Sequence analysis has annotated cinA as encoding for a putative azurin-plastocyanin like protein, and cinQ as encoding for a putative GTP cyclohydrolase/preQ0 reductase (NCBI accession numbers NP_744308 and NP_744309). Recently, we could show that CinQ from Pseudomonas putida is a pyridine nucleotide-dependent nitrile oxidoreductase that catalyzes the conversion of preQ0 to preQ1, in the nucleoside queuosine biosynthetic pathway (Quaranta et al., 2007). CinA is similar to the P. fluorescens DF57 Cot protein, which is involved in copper tolerance. In addition, the P. aeruginosa homolog, cinA (PA2806), is also transcribed in copper exposed cells and its disruption leads to a slight reduction in copper tolerance, such as 12 mm of inhibition radius in disk assay compared to 8 mm of the wild type (Teitzel et al., 2006). P. aeruginosa is of medical importance since it has recently emerged as an opportunistic nosocomial pathogen, which is responsible for a range of mild to severe infections (Livermore 2002.
In this paper, we report the that several genes from P. aeruginosa not only influence survival an copper containing medium but also on copper cast alloys.
Materials and Methods
Bacterial strains and growth conditions
P. aeruginosa PAO1 and its derivative transposon mutants were obtained from the University of Washington Genome Center and cultures prepared in Peptone Yeast liquid medium at 37° C. The strains used in this study were P. aeruginosa PAO1 (wild-type with no gene disruption), P. aeruginosa cinR::ISIacZ/hah (PA2809::ISIacZ/hah PAO1 with gene disruption of cinR::Tetr), P. aeruginosa cinA::ISIacZ::hah (Pa2807::lSpho/hah PAO1 with gene disruption of cinA::Tetr ), and P. aeruginosa cinQ::ISIacZ/hah (PA2806:: lSpho/hah PAO1 with gene disruption of cinQ::Tetr).
Determination of survival rates on copper-containing solid medium
To determine the survival rates of P. aeruginosa PAO1, P. aeruginosa cinR::ISIacZ/hah, P. aeruginosa cinA::ISIacZ::hah, and P. aeruginosa cinQ::ISIacZ/hah on peptone-yeast extract (PY) agar plates containing various copper concentrations, the bacteria were grown to mid log phase at OD600 0.5 in liquid PY medium at 37° C in a shaker at 120 to 150 rpm. Serial dilutions were prepared, and 100μl of each dilution were plated on PY agar containing 0 to 2 mM copper, which was added to the medium from a 1 M CuCl2 stock solution. After spreading the samples over the surface with a sterile glass spreader the plates were incubated at 37° C for 24 hours. Each dilution was performed in triplicate and the mean calculated.
Minimal inhibition concentration (MIC)
To ensure that there is a difference between the four P. aeruginosa strains regarding their sensitivity to copper, agar plates containing different copper concentrations were quartered and in each quarter one strain was streaked out. The different agar plates contained 0, 0.1, 0.5, 1, 1.5, 2, 2.5, 3, 4 and 5 mM copper. The concentrations were reached by adding adequate volumina of 1M CuCl2*2H2O stock solution to the agar while hot.
Preparation of test surfaces
The different copper alloys employed in this study were a gift from the International Copper Association and are listed in Table 1. Prior to use the 1 square inch copper alloy and stainless steel coupons were immersed for 30 seconds in a 3% NaOH solution heated to 70°C, rinsed in DH2O, and followed by a 3– 5 seconds immersion in 10 % H2SO4 and rinsed with DH2O. To prevent re-oxidation of the surfaces the coupons were rinsed in 95 % ethanol, air-dried, and kept in a sterile container until used.
Table 1.
Copper cast alloys and their constituent compositions in percent values.
| Alloy | Cu | Zn | Sn | Ni | Fe | P |
|---|---|---|---|---|---|---|
| Cu 11000 | 99.9 | |||||
| Cu 51000 (Bronze) | 94.8 | 5.0 | 0.2 | |||
| Cu-Ni 70600 | 88.6 | 10.0 | 1.4 | |||
| Cu-Zn 26000 | 70 | 30 | ||||
| Stainless Steel | 0 | 8 | 74 | 18 |
Survival on selected copper cast alloys
To determine the bactericidal effect of copper alloys P. aeruginosa strains were grown in liquid peptone yeast medium (PY). For the experiments 0.1 ml aliquots were taken from overnight cultures and grown in a 37°C shaker at 120 –150 rpm to mid-log phase at OD600 0.5. A 25 μl inoculum of the culture was pipetted on each coupon to be tested and spread with a sterile glass spreader. For experiments testing the influence of copper resistance genes the suspension was not spread but left as a droplet. After defined incubation times at room temperature or 4°C the coupons were transferred into 50 ml centrifuge tubes containing 10 ml sterile phosphate buffered saline (PBS) and ten to twenty sterile 2 mm diameter glass beads. This was mixed thoroughly for thirty seconds to one minute. Then serial dilutions in PBS were made and a 100 μl aliquot of each dilution was plated out on peptone- yeast agar plates and spread with a sterile glass spreader over the surface of the plate. The plates were incubated at 37°C for 24 h. The described experiments were performed in triplicate and the mean calculated.
Experimental Controls
P.aeruginosa PAO1, P. aeruginosa cinR::ISIacZ/hah, P. aeruginosa cinA::ISIacZ::hah, and P. aeruginosa cinQ::ISIacZ/hah were exposed to stainless steel for defined times. Overnight cultures in PY liquid medium were prepared for each strain and a 100 μl aliquot was grown in PY liquid medium to OD600 0.5. A 25 μl aliquot of the culture was placed on each coupon and spread with a sterile glass spreader or left as a droplet. After defined incubation times at room temperature and 4°C the coupons were transferred into 50ml centrifugation tubes which contained 10 ml sterilized PBS and ten to twenty sterilized 2 mm diameter glass beads. After thorough mixing on a vortex mixer serial dilutions were plated out on peptone yeast agar plates and incubated at 37°C for 24 hours.
Results
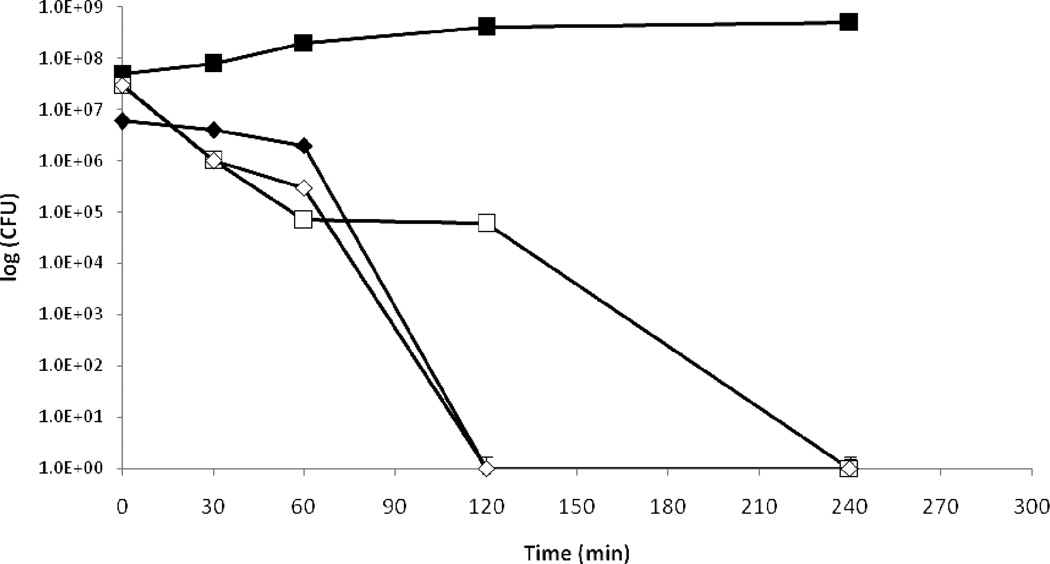
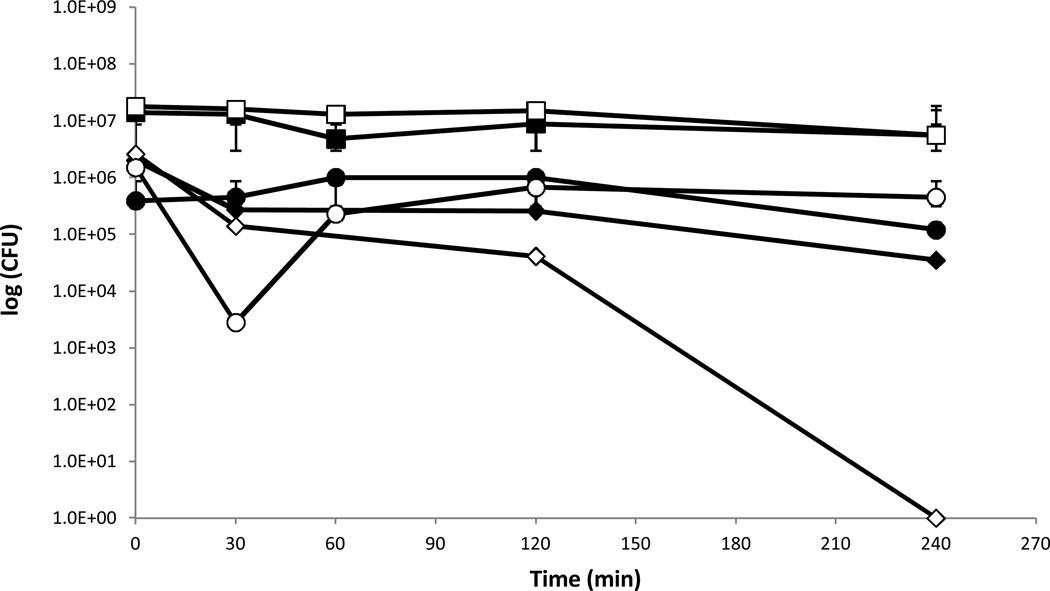
Pseudomonas aeruginosa PAO1 is effectively killed on copper cast alloys. It is evident that when spread on copper cast alloys P.aeruginosa PAO1 cells were killed whereas stainless steel had no bactericidal effect (Figure 1). At time zero 3.5×107 colony-forming-units (CFU) were detected after exposure to stainless steel and after 4 hours 2.6×108 CFU remained. The bactericidal effect of the copper cast alloy was not necessarily linked to the copper content as alloy Cu11000 containing 99.90% copper was slightly less effective than alloy Cu70600 which contains only 88.6% copper in addition to 10% nickel and 1.4% iron. For alloy Cu11000 a drop of 0.44 log units from 4.4×106 CFU per coupon to 1.2×106 CFU per coupon after 60 minutes of exposure could be detected. After 120 minutes no CFU could be detected. For alloy Cu70600 there was a drop of 2.05 log units from 2.5×107 CFU per coupon at time zero to 2.2×106 CFU per coupon after 60 minutes of exposure. A drop to zero countable CFU was reached after 120 minutes. Alloy Cu51000 which contains 94.8% copper was the least effective in reducing P. aeruginosa PAO1 viable counts of the coupons which were tested. A drop from 1.9×107 CFU per coupon at time zero to zero CFU was reached after 240 minutes of incubation at room temperature. After 120 minutes of exposure on alloy Cu51000 3.2×104 CFU remained in contrast to alloys Cu11000 and Cu70600 where a drop to zero was detected.
Figure 1.
a. Survival of P. aeruginosa PAO1 on different copper cast alloys. P.aeruginosa was grown to OD600 0.5. A 25 μl aliquot of the culture was spread with a sterile glass spreader on stainless steel (■), C11000 (♦), C51000 (□), C70600 (◊) for defined incubation times at room temperature (21°C) the copper coupons were transferred into 50 ml centrifugation tubes containing 10 ml sterile PBS and 10 to 20 glass beads. This was mixed thoroughly on a vortex mixer for about 30 s to 1 min. Serial dilutions were prepared and plated out on Peptone Yeast agar plates. The plates were incubated, lid down, for 24h at 37°C. This experiment was done in triplicate and the mean and the standard deviation were calculated.
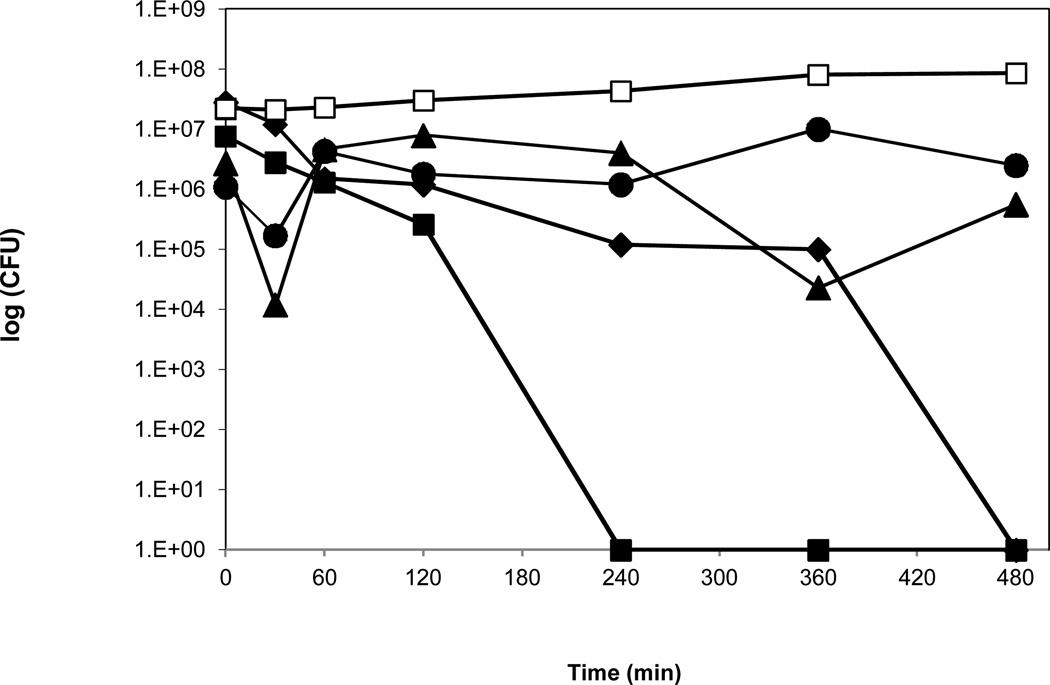
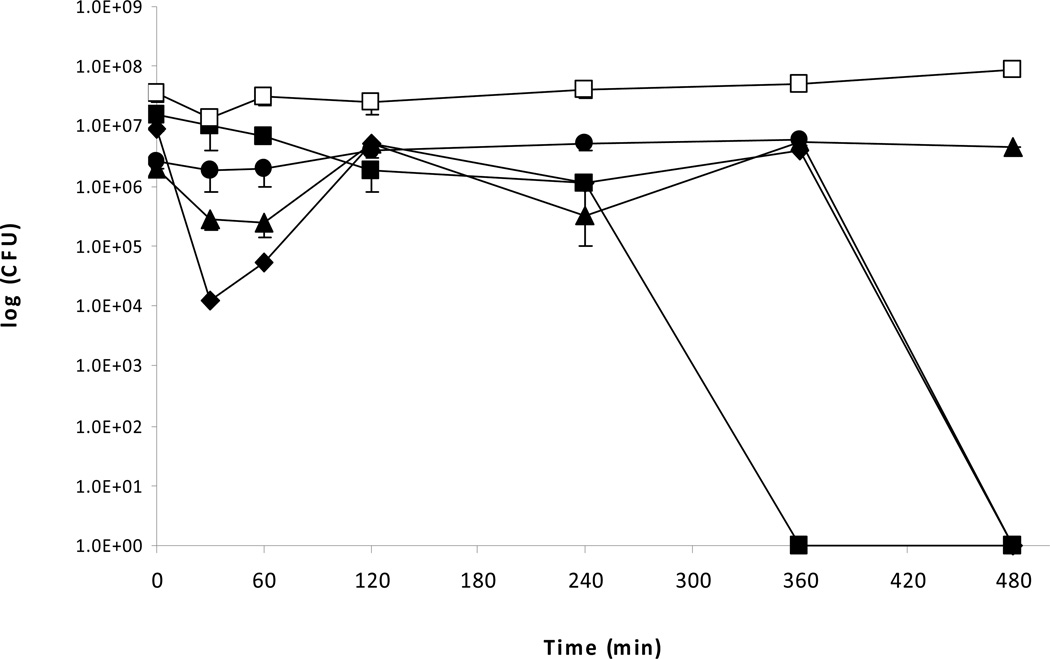
b. Survival of P. aeruginosa PAO1 on copper cast alloys with droplet inoculation. From overnight cultures P. aeruginosa PAO1 was grown to OD600 0.5. A 25 μl aliquot of the culture was placed as a droplet on stainless steel (□), Cu 11000 (■), Cu 51000 (▲), Cu 70600 (♦), and Cu 26000 (●) for defined incubation times at room temperature (21°C). Serial dilutions were prepared in PBS, plated on PY agar, and incubated at 37°C for 24 hours.
The rate of survival of P. aeruginosa PAO1 on copper cast alloys was prolonged when the 25 μl inoculum was placed as a small droplet on the copper alloy and stainless steel surfaces. For alloy Cu11000 a drop to zero countable CFU was reached after 240 minutes, however, for alloy Cu70600 it took 480 minutes to achieve no detectable CFU. On alloy Cu51000 CFU counts dropped from 2.7×106 at time zero to 5.6×105 at 480 minutes, and on alloy Cu26000 with 1.1×106 CFU at time zero 2.5×106 CFU remained after 480 minutes exposure time. No zero countable CFU was achieved over the 8 hour time period. CFU count on stainless steel using droplets was compatible with the results obtained by spreading the inoculum over the copper coupons: the initial CFU was 2.8×107 and after 480 minutes 8.6×107.
Survival of different strains of P. aeruginosa PAO1 on copper-containing medium
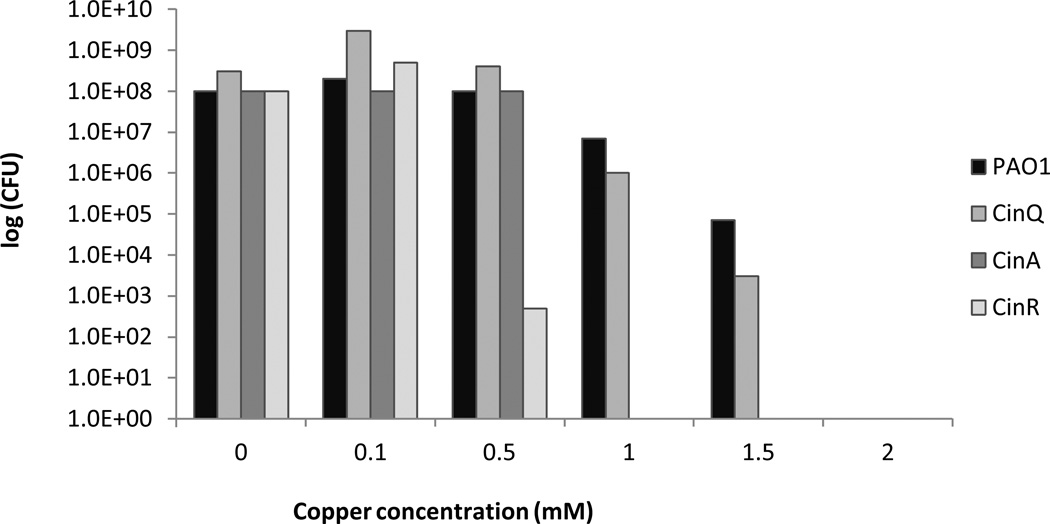
A number of genes were shown to be involved in copper resistance in previous studies (Quaranta et al., 2007; Teitzel et al., 2006). In this study the four strains P.aeruginosa PAO1 (wildtype), P. aeruginosa cinR::ISIacZ/hah, P. aeruginosa cinA::ISIacZ/hah and P. aeruginosa cinQ::ISIacZ/hah were more thoroughly characterized concerning their role in the survival of P.aeruginosa PAO1 on copper containing medium by using a survival assay. Serial dilutions of the different strains of P. aeruginosa PAO1 in Peptone Yeast medium were plated out on agar plates containing different copper concentrations (Figure 2). P. aeruginosa PAO1 (wildtype) was observed to be the most copper resistant strain. About 6.4×104 colony forming units (CFU) could be detected at a concentration of 1.5 mM copper. At no copper addition 1.22×108 CFU were counted. The number of countable cells dropped to zero at 2 mM copper. P. aeruginosa cinQ::ISIacZ/hah is similar to P. aeruginosa PAO1 regarding its resistance to copper. Here about 2×103 cells of an initial number of 2×108 CFU could be observed at a concentration of 1.5 mM copper. The drop to zero countable bacteria was reached at a concentration of 2 mM CuCl2. P.aeruginosa PAO1 was therefore slightly more resistant than P. aeruginosa cinQ::ISIacZ/hah (Figure 2). However, P. aeruginosa cinA::ISIacZ/hah and P. aeruginosa cinR::ISIacZ/hah were far more sensitive to copper. There were no countable CFU detected for P. aeruginosa cinR::ISIacZ/hah at a 1 mM copper concentration and at 0.5 mM there already was a drop of about 5.5 log units from about 1.6×108 initial CFU to 4.8×102 CFU. For P. aeruginosa cinA::ISIacZ/hah as well, zero viable CFU could be detected at a CuCl2 concentration of 1.0 mM. However, in contrast to P. aeruginosa cinR::ISIacZ/hah, the number of cells stayed constant at about 1×108 CFU at concentrations from 0 mM copper to 0.5 mM CuCl2 before it dropped to zero at a concentration of 1 mM copper. Therefore, these experiments could clearly show that the genes cinA, cinR and cinQ influence the ability of Pseudomonas aeruginosa PAO1 to resist the bactericidal effect of copper in solid medium.
Figure 2.
Survival of P. aeruginosa PAO1 and its mutant strains P. aeruginosa cinR::ISIacZ/hah, P. aeruginosa cinA::ISIacZ::hah, and P. aeruginosa cinQ::ISIacZ/hah on solid media containing various copper concentrations. Cells from overnight cultures were grown to OD600 0.5, serial dilutions in PY medium prepared, plated on PY agar plates containing different copper concentrations, and incubated at 37°C for 24 hours.
The minimal inhibitory concentrations of copper required to completely inhibit the growth of P. aeruginosa PAO1 and its mutant strains were determined by preparing streak plates of peptone yeast agar containing various copper concentrations.(Table 2). For the wildtype P. aeruginosa PAO1 and P. aeruginosa cinQ::ISIacZ/hah a minimal inhibition concentration of 2 mM copper was determined. In contrast, a minimal inhibition concentration of 0.5 mM was determined for P. aeruginosa cinR::ISIacZ/hah and for P. aeruginosa cinA::ISIacZ/hah it was 1 mM copper with an already significant decrease of growth at 0.5 mM. The experiment was performed in duplicate and CinA only grew on one of the two plates with a decreased colony size. The results of this experiment corroborate the previous results looking at survival rates. The minimal inhibitory concentration of copper was different for the strains tested. P.aeruginosa PAO1 and CinQ were both inhibited by a copper concentration of 2 mM. Interestingly, we could detect a small but significant difference between the wildtype and the strain disrupted in cinQ in the survival assay but not when looking at the MIC. Therefore, we could clearly establish that the queuosine reductase CinQ is involved in handling excess copper, however, the exact mechanism is still not known (Quaranta et al., 2007). An insertion in the gene encoding the response regulator CinR was most sensitive for copper, and an insertion in cinA made cells significantly less resistant to copper than the wildtype P. aeruginosa PAO1 and CinQ but was not as sensitive as CinR (Figure 2, Table 2). In general the results of the gene disruption on copper sensitivity were similar to those reported by Teitzel et al (2006).
Table 2.
MIC of different strains of P. aeruginosa on copper-containing solid medium
| Cu-conc in mM | PAO 1 | CinR | CinQ | CinA |
|---|---|---|---|---|
| 0 | ++ | ++ | ++ | ++ |
| 0.1 | ++ | ++ | ++ | ++ |
| 0.5 | ++ | − | ++ | +− |
| 1 | ++ | − | ++ | − |
| 1.5 | ++ | − | ++ | − |
| 2 | − | − | − | − |
| 2.5 | − | − | − | − |
| 3 | − | − | − | − |
| 4 | − | − | − | − |
| 5 | − | − | − | − |
P. aeruginosa strains PAO1, CinR, CinQ and CinA were streaked out on plates containing different copper concentrations. The plates were incubated for 24 h at 37°C after which growth was monitored (− no colonies; +− small colonies; ++ normal colonies).
Temperature has an impact on the efficiency of copper cast alloys
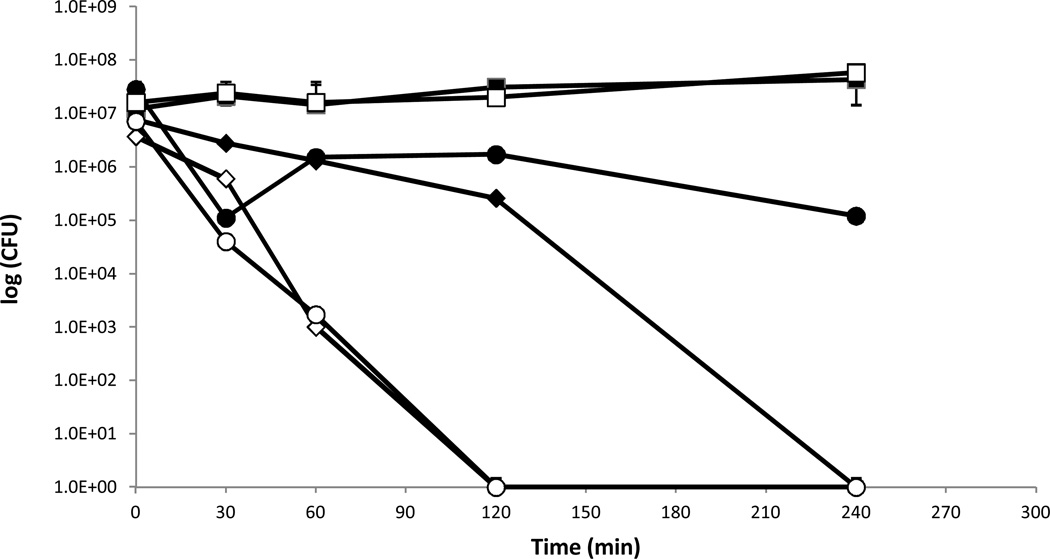
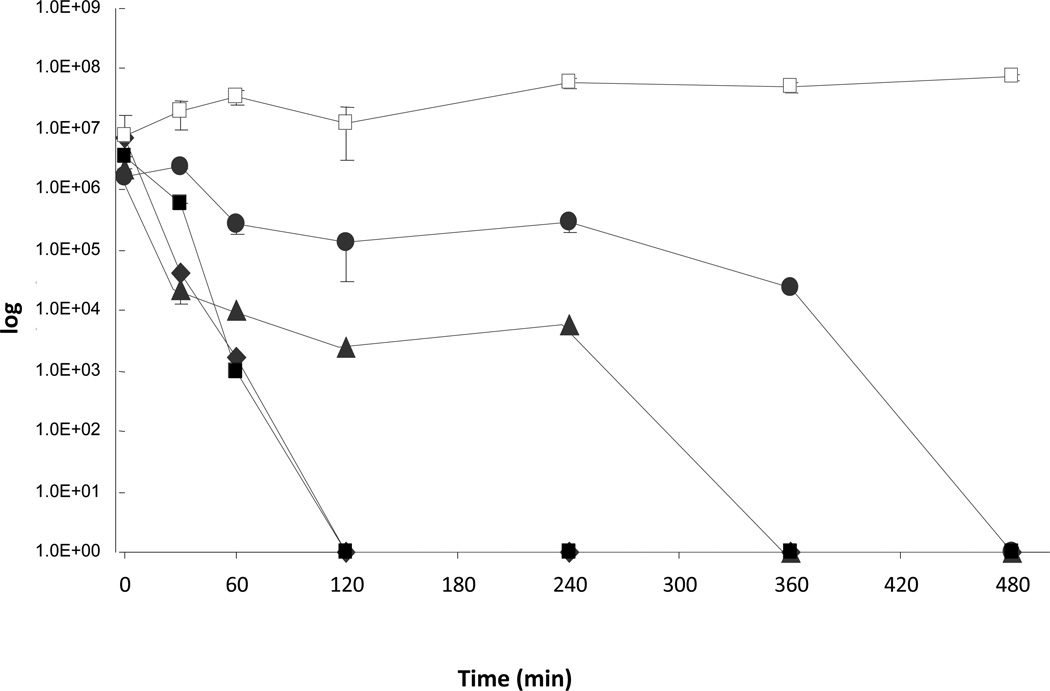
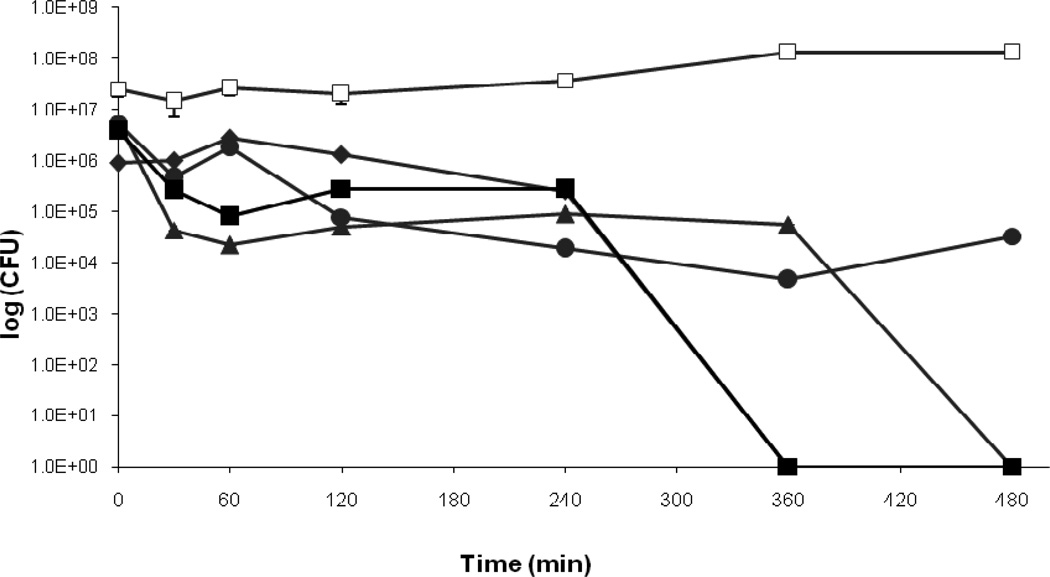
Previous studies showed that temperature affects the bactericidal properties of copper cast alloys (Wilks et al., 2005; 2006; Noyce et al., 2006). Therefore, the influence of temperature on the survival of P. aeruginosa PAO1 and P. aeruginosa cinR::ISIacZ/hah when placed as droplets on copper cast alloys was tested. At room temperature (21°C) CFU counts for P. aeruginosa PAO1 on alloy Cu11000 dropped from 7.7×106 to zero countable CFU after 240 minutes, while CFU on alloy Cu70600 dropped from 2.8×107 to 1.2×105, and CFU on stainless steel was 1.2×107 at time zero and 4.3×107 after 240 minutes (Figure 3a). For P. aeruginosa cinR::ISIacZ/hah CFU on alloy Cu11000 dropped from 3.7×106 to zero countable CFU after 120 minutes, on alloy 70600 from 7.1×107 to zero countable CFU after 120 minutes, and on stainless steel the initial CFU were 1.6×107 and 5.8×107 after 240 minutes (Figure 3b). At 4°C CFU count for P. aeruginosa PAO1 on alloy Cu11000 dropped from 2×106 to 3.5×104 after 240 minutes, on alloy Cu70600 from 3.9×105 to 1.2×105, and on stainless steel from 1.4×107 to 5.6×106 (Figure 3b). At 4°C survival on alloy Cu11000 was prolonged for P. aeruginosa cinR::ISIacZ/hah with CFU dropping from 2.6×106 to zero after 240 minutes, on Cu70600 from 1.5×106 to 4.5×105, and on stainless steel from 1.8×107 to 5.6×106 (Figure 3b). It could be convincingly demonstrated that the bactericidal effect of copper cast alloys could also be seen at incubation at 4°C. However, cells clearly survived longer on copper cast alloys at 4°C than at RT at 21°C.
Figure 3.
a. Survival of P. aeruginosa PAO1 and P. aeruginosa cinR::ISIacZ/hah at room temperature on copper cast alloys. Cells from overnight cultures were grown to OD600 0.5. A 25μl aliquot of the culture was placed as a droplet on each copper cast alloy and incubated at room temperature (21°C) for defined times. P. aeruginosa PAO1 Cu11000 (♦), P. aeruginosa PAO1 Cu70600 (●), and P. aeruginosa PAO1 stainless steel (■). P. aeruginosa cinR::ISIacZ/hah Cu11000 (◊), P. aeruginosa cinR::ISIacZ/hah Cu70600 (○), and P. aeruginosa cinR::ISIacZ/hah stainless steel (□).
b. Survival of P. aeruginosa PAO1 and P. aeruginosa cinR::ISIacZ/hah at 4°C on copper cast alloys. Cells from overnight cultures were grown to OD600 0.5. A 25μl aliquot of the culture was placed as a droplet on each copper cast alloy and incubated at 4°C for defined times. P. aeruginosa PAO1 Cu11000 (♦), P. aeruginosa PAO1 Cu70600 (●), and P. aeruginosa PAO1 stainless steel (■). P. aeruginosa cinR::ISIacZ/hah Cu11000 (◊), P. aeruginosa cinR::ISIacZ/hah Cu70600 (○), and P. aeruginosa cinR::ISIacZ/hah stainless steel (□).
Genes involved in copper resistance influence survival on copper surfaces
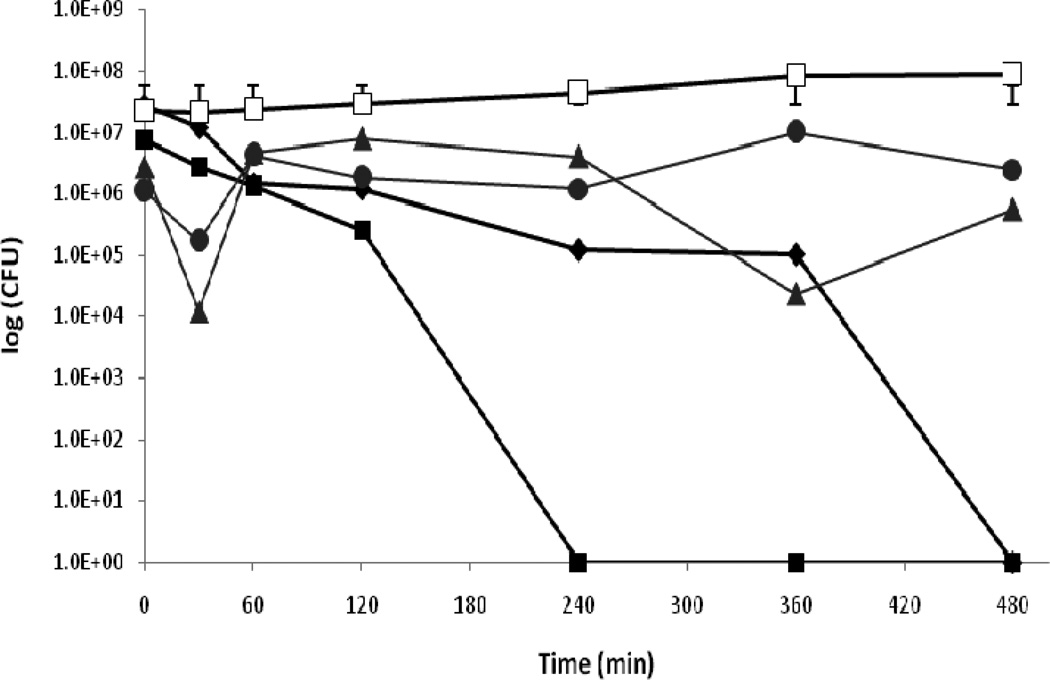
In this study it could be convincingly shown that genes involved in copper resistance influence the survival of P. aeruginosa PAO1 and its three mutants on copper containing solid medium. It was then examined if the same effect is detectable when the bacteria are exposed to copper cast alloys, since copper cast alloys could be an alternative to combat the problem of the proliferation of antibiotic resistant bacteria. The previously characterized strains P. aeruginosa cinR::ISIacZ/hah, P. aeruginosa cinA::ISIacZ/hah and P. aeruginosa cinQ::ISIacZ/hah were placed on copper cast alloys to determine the influence of genes involved in copper resistance on survival on copper surfaces. The results demonstrated that genes involved in copper resistance also influence the survival on copper alloys. Colony forming units of P. aeruginosa cinR::ISIacZ/hah on Cu11000 and Cu70600 were no longer detectable after 120 minutes, on Cu51000 a drop to zero count CFU was observed at 360 minutes, and on Cu26000 after 480 minutes. However, no decrease in CFU was detected on stainless steel after 480 minutes (Figure 4). Moreover, on alloys Cu11000 and Cu70600 CFU of P. aeruginosa cinA::ISIacZ/hah were no longer detectable after 360 minutes, on alloy 51000 CFU count dropped to zero after 480 minutes, and on alloy 26000 CFU count dropped from 5.3×106 to 3.3×104 after 480 minutes (Figure 5). For P. aeruginosa cinQ::ISIacZ/hah on alloy Cu11000 no CFU were detected after 360 minutes incubation time, on alloy 70600 and 26000 a drop to zero count CFU was reached after 480 min, while on alloy Cu51000 no zero count CFU was attained after 480 minutes. In all assays tested it could be demonstrated that P. aeruginosa PAO1 and P. aeruginosa cinQ::ISIacZ/hah cells were the most copper resistant strain and that the mutant P. aeruginosa cinR::ISIacZ/hah has the lowest ability to survive on copper containing medium and on copper cast alloys.
Figure 4.
a. Survival of P. aeruginosa PAO1 on copper cast alloys. Cells from overnight cultures were grown to OD600 0.5. A 25μl aliquot of the culture was placed as a droplet on each copper cast alloy and incubated at room temperature (21°C) for defined times. Cu11000 (■), Cu51000 (▲), Cu70600 (♦), Cu26000 (●), and stainless steel (□).
b. Survival of P. aeruginosa cinR::ISIacZ/hah on copper cast alloys. Cells from overnight cultures were grown to OD600 0.5. A 25μl aliquot of the culture was placed as a droplet on each copper cast alloy and incubated at room temperature (21°C) for defined times. Cu11000 (■), Cu51000 (▲), Cu70600 (♦), Cu26000 (●), and stainless steel (□).
c. Survival of P. aeruginosa cinA::ISIacZ::hah on copper cast alloys. Cells from overnight cultures were grown to OD600 0.5. A 25μl aliquot of the culture was placed as a droplet on each copper cast alloy and incubated at room temperature (21°C) for defined times. Cu11000 (■), Cu51000 (▲), Cu70600 (♦), Cu26000 (●), and stainless steel (□).
d. Survival of P. aeruginosa cinQ::ISIacZ/hah on copper cast alloys. Cells from overnight cultures were grown to OD600 0.5. A 25μl aliquot of the culture was placed as a droplet on each copper cast alloy and incubated at room temperature (21°C) for defined times. Cu11000 (■), Cu51000 (▲), Cu70600 (♦), Cu26000 (●), and stainless steel (□).
Discussion
The use of copper surfaces as a bactericidal agent has shown significant promise (Faundez et al., 2004; Noyce et al., 2006). However, one concern often voiced is the potential of development of resistance as has happened with antibiotics (Summers, 2002; Walsh, 2006). It was therefore an important aim of this study to determine the influence, if any, of genes involved in copper resistance on survival on copper alloy surfaces. This study could clearly demonstrate that genes involved in copper resistance influence the survival of P. aeruginosa on both medium containing copper and on copper cast alloys. However, this effect is not as clear-cut as appears at first glance and could best be observed if the culture was not left to dry or spread thinly on copper alloys. The best results were obtained when the 25 μl inoculum was left as a droplet on the metallic copper surfaces, thus maintaining a relatively small, consistent contact area and a uniform cell suspension. Under these conditions killing is probably due to slow accumulation of copper ions in the droplet due to corrosion. The rate of copper in solubilisation is clearly influenced by the metabolic activity since killing is much more rapid at higher than at lower temperatures as shown with the prolonged survival of P.aeruginosa PAO1 and P. aeruginosa cinR::ISIacZ/hah at 4° C on alloys Cu11000 and Cu70600. P. aeruginosa cinR::ISIacZ/hah, however, upheld its greater sensitivity to copper at lower temperature.
When grown on copper containing solid medium mutant strains of P.aeruginosa PAO1 demonstrated different levels of sensitivity to copper. The most sensitive strain P. aeruginosa cinR::ISIacZ/hah was also killed most rapidly on copper cast alloys. Here the alloys with higher copper concentrations were most effective in killing P. aeruginosa cinR::ISIacZ/hah, which suggests that copper ions in the droplet may accumulate faster and in greater concentrations than with exposure to alloys containing 70 % copper or less. The mutant strain P. aeruginosa cinA::ISIacZ/hah had an overall greater sensitivity to copper when grown on copper containing solid medium and on copper cast alloys. The decrease in CFU count over time on all alloys may also result from increasing levels of copper ions in the droplet, but the mutant is able to maintain copper homeostasis better than P. aeruginosa cinR::ISIacZ/hah. However, P. aeruginosa cinQ::ISIacZ/hah did not demonstrate any increase in sensitivity to copper on copper containing solid medium when compared to the wild type P.aeruginosa PAO1. In addition, the mutation in this strain did not significantly influence the survival rates on copper cast alloys under the conditions tested.
The findings of this study illustrate the contributions of individual genetic components of the cin operon for the survival of P. aeruginosa on metallic copper surfaces. These genes are known to be involved in copper homeostasis in Pseudomonas species, and their effect could be demonstrated under the environmental conditions in the experiments described. While the exact pathways leading to copper resistance in P. aeruginosa remain unknown, the genes influencing its survival on copper cast alloys are part of the resistance mechanisms to copper. The results obtained provide important information for the utilization of copper cast alloys as bactericidal surfaces in the healthcare industry.
Significance and Impact of the Study.
In order to more widely use copper surfaces as bactericidal agents in various settings it is important to understand how genes influence survival on these surfaces. Here we show that genes shown to be involved in copper resistance in P. aeruginosa PAO1 can have an impact on the length of survival time on copper cast alloys under certain conditions. This is an important first step for evaluation of future use of copper surfaces as bactericidal agents.
Acknowledgement
Research in the lab of C.R. was supported by NIH grant GM079192 and a grant from the International Copper Association (ICA). Transposon mutants were a gift from the University of Washington Genome Center and the different copper cast alloys from ICA.
References
- Faundez G, Trancoso M, Navarete P, Figueroa G. Antimicrobial activity of copper surfaces against suspensions of Salmonella enterica and Camylobacter jejuni. BMC Microbiol. 2004;4:19. doi: 10.1186/1471-2180-4-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livermore DM. Multiple mechanisms of antimicrobial resistance in Pseudomonas aeruginosa: our worst nightmare? Clin. Infect. Dis. 2002;34:634–640. doi: 10.1086/338782. [DOI] [PubMed] [Google Scholar]
- Noyce JO, Michels H, Keevil CW. Use of copper cast alloys to control Escherichia coli O157 cross-contamination during food processing. Appl. Environ. Microbiol. 2006;72:4239–4244. doi: 10.1128/AEM.02532-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noyce JO, Michels H, Keevil CW. Potential use of copper surfaces to reduce survival of epidemic meticillin-resistant Staphylococcus aureus in the healthcare environment. J. Hosp. Inf. 2006;63:289–297. doi: 10.1016/j.jhin.2005.12.008. [DOI] [PubMed] [Google Scholar]
- Quaranta D, McCarty R, Bandarian V, Rensing C. The copper-inducible cin operon encodes an unusual methionine-rich azurin and a queuosine reductase in Pseudomonas putida KT 2440. J. Bacteriol. 2007;189:5361–5371. doi: 10.1128/JB.00377-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rensing C, Grass G. Escherichia coli mechanisms of copper homeostasis in a changing environment. FEMS Microbiol. Rev. 2003;27:197–213. doi: 10.1016/S0168-6445(03)00049-4. [DOI] [PubMed] [Google Scholar]
- Robine E, Boulange-Petermann L, Derangere D. Assessing bactericidal properties of materials: the case of metallic surfaces in contact with air. J. Microbiol Methods. 2002;49:225–234. doi: 10.1016/s0167-7012(01)00371-2. [DOI] [PubMed] [Google Scholar]
- Summers AO. Generally overlooked fundamentals of bacterial genetics and ecology. Clin. Infect. Dis. 2002;3:S85–S92. doi: 10.1086/340245. [DOI] [PubMed] [Google Scholar]
- Teitzel GM, Geddie A, De Long SK, Kirisits MJ, Whiteley M, Parsek MR. Survival and Growth in the presence of elevated copper: transcriptional profiling of copper stressed Pseudomonas aeruginosa. J. Bacteriol. 2006;188:7242–7256. doi: 10.1128/JB.00837-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh TR. Combinatorial genetic evolution of multiresistance. Curr. Opin. Microbiol. 2006;9:476–482. doi: 10.1016/j.mib.2006.08.009. [DOI] [PubMed] [Google Scholar]
- Wilks SA, Michels H, Keevil CW. The survival of Escherichia coli O157 on a range of metal surfaces. Int. J. Food Microbiol. 2005;105:445–454. doi: 10.1016/j.ijfoodmicro.2005.04.021. [DOI] [PubMed] [Google Scholar]
- Wilks SA, Michels HT, Keevil CW. Survival of Listeria monocytogenes Scott A on metal surfaces: implications for cross-contamination. Int. J. Food Microbiol. 2006;111:93–98. doi: 10.1016/j.ijfoodmicro.2006.04.037. [DOI] [PubMed] [Google Scholar]