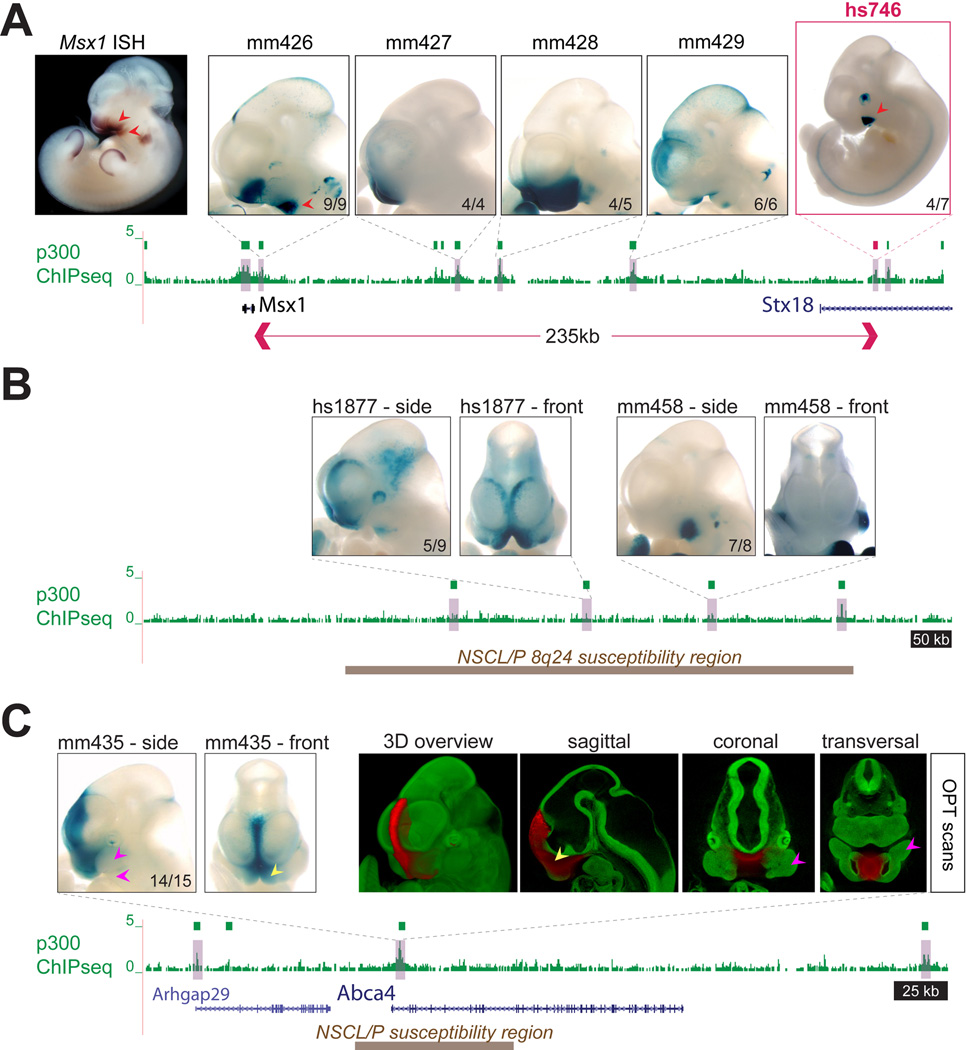
Fig. 4. Regulatory landscapes of craniofacial loci.
(A) Craniofacial enhancers near Msx1, a major craniofacial gene, were identified by p300 ChIP-seq (green boxes). This included the re-identification of a region proximal to Msx1 with previously described enhancer activity (mm426, (50)), as well as four additional, more distal enhancers with complementary activity patterns. For each enhancer, only one representative embryo is shown, numbers indicate reproducibility. Red arrows indicate selected correlations between Msx1 RNA expression (ISH) and individual enhancers (see main text). Red box indicates enhancer hs746 which was further studied by knockout analysis. Msx1 ISH: Embrys database (http://embrys.jp) (51). (B) Identification of craniofacial enhancers in the cleft- and morphology-associated gene desert at human chromosome 8q24 (orthologous mouse region shown, (35)). Brown box indicates the region corresponding to a 640kb human region associated with orofacial clefts (non-syndromic cleft lip with or without cleft palate, NSCL/P) and devoid of protein-coding genes. Two of four candidate enhancers within the region drove craniofacial expression. For each enhancer, lateral and frontal views of one representative embryo are shown. (C) Identification of a craniofacial midline enhancer at the cleft-associated susceptibility interval at the ABCA4 locus (36). The enhancer is highly active in the nasal prominences (yellow arrows), but not the maxillary or mandible (pink arrows).