Abstract
We present an Aboriginal Australian genomic sequence obtained from a 100-year-old lock of hair donated by an Aboriginal man from southern Western Australia in the early 20th century. We detect no evidence of European admixture and estimate contamination levels to be below 0.5%. We show that Aboriginal Australians are descendants of an early human dispersal into eastern Asia, possibly 62,000 to 75,000 years ago. This dispersal is separate from the one that gave rise to modern Asians 25,000 to 38,000 years ago. We also find evidence of gene flow between populations of the two dispersal waves prior to the divergence of Native Americans from modern Asian ancestors. Our findings support the hypothesis that present-day Aboriginal Australians descend from the earliest humans to occupy Australia, likely representing one of the oldest continuous populations outside Africa.
The genetic history of Aboriginal Australians is contentious but highly important for understanding the evolution of modern humans. All living non-African populations likely derived from a single dispersal of modern humans out of Africa, followed by subsequent serial founder effects (1, 2). Accordingly, eastern Asia is hypothesized to have been populated by a single early migration wave rather than multiple dispersals (3). In this “single-dispersal model,” Aboriginal Australians are predicted to have diversified from within the Asian cluster [for definitions of human populations and groups, see (4)] (Fig. 1A, top). Recent whole-genome studies reveal a split between Europeans and Asians dating to 17,000 to 43,000 years before the present (B.P.) (5, 6). Because greater Australia (Australia and Melanesia, including New Guinea) has some of the earliest archaeological evidence of anatomically modern humans outside Africa, dating back to ~50,000 years B.P. (7, 8), a divergence of aboriginal Australasians from within the Asian cluster is not compatible with population continuity in Australia. Alternatively, on the basis of archaeological and fossil evidence, it has been proposed that greater Australia was occupied by an early, possibly independent out-of-Africa dispersal, before the population expansion giving rise to the majority of present-day Eurasians (9, 10). According to this “multiple-dispersal model,” the descendants of the earlier migration became assimilated or replaced by the later-dispersing populations, with a few exceptions that include Aboriginal Australians (10, 11) (Fig. 1A, bottom).
Fig 1.
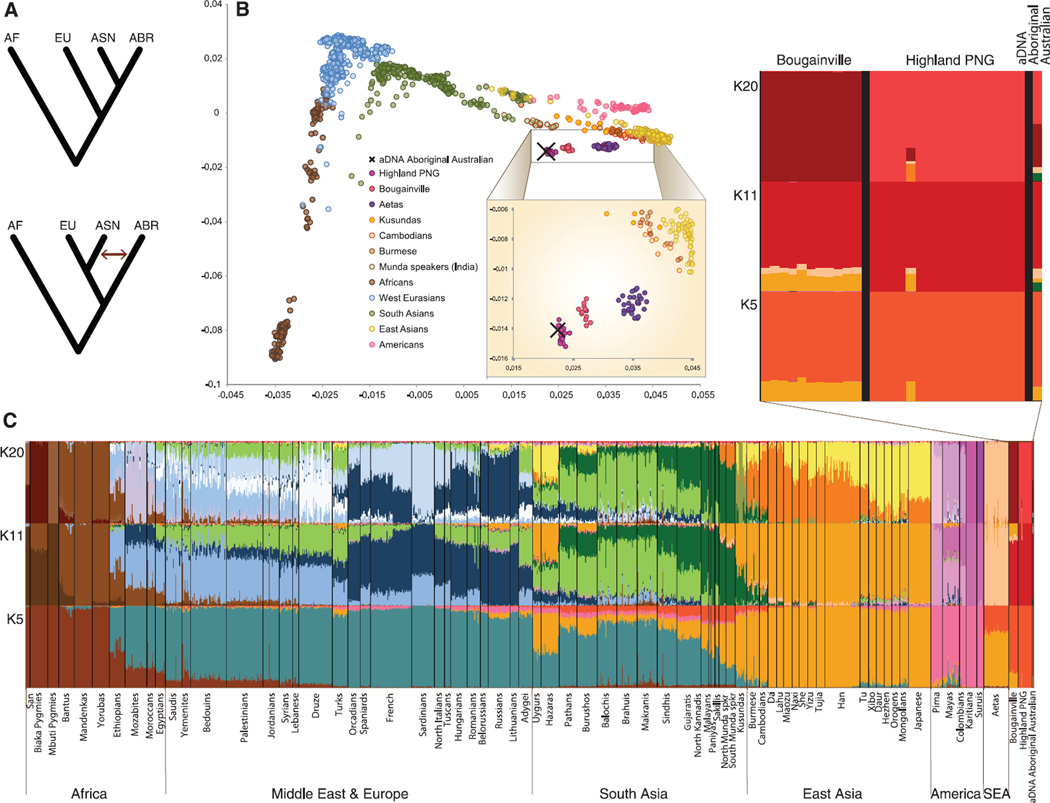
(A) The two models for early dispersal of modern humans into eastern Asia. Top: Single-dispersal model predicting a single early dispersal of modern humans into eastern Asia. Bottom: Multiple-dispersal model predicting separate dispersals into eastern Asia of aboriginal Australasians and the ancestors of most other present-day East Asians. AF, Africans; EU, Europeans; ASN, Asians; ABR, Aboriginal Australians. Arrow symbolizes gene flow. (B) PCA plot (PC1 versus PC2) of the studied populations and the ancient genome of the Aboriginal Australian (marked with a cross). Inset shows the greater Australia populations (4). (C) Ancestry proportions of the studied 1220 individuals from 79 populations and the ancient Aboriginal Australian as revealed by the ADMIXTURE program (28) with K = 5, K = 11, and K = 20. A stacked column of the K proportions represents each individual, with fractions indicated on the y axis [see (4) for the choice of K]. The greater Australia populations are shown in detail at the upper right.
We sequenced the genome of an Aboriginal Australian male from the early 20th century to overcome problems of recent European admixture and contamination (4).We used 0.6 g of hair for DNA extraction (4, 12). Despite its relatively young age, the genomic sequence showed a high degree of fragmentation, with an average length of 69 base pairs. The genome was sequenced to an overall depth of 6.4×; the ~ 60% of the genomic regions covered was sequenced to an average depth of 11× (4) [theoretical maximum is ~85% (12)]. Cytosine-to-thymine misincorporation levels typical of ancient DNA (13) were low (maximum3% of all cytosines) and were restricted to a 5-nucleotide region at each read terminus. For this reason, read termini were trimmed to improve single-nucleotide polymorphism (SNP) call quality (4).
The genome was mapped and genotyped, identifying 2,782,401 SNPs, of which 449,115 were considered high-confidence, with a false-positive rate of <2.4%, and were used in further analyses (4). Of these, 28,395 (6.3%) have not been previously reported (4). Despite extensive handling of the hair by people of European ancestry, contamination levels based on the level of X-chromosome heterozygosity were estimated to be less than 0.5% (4). These findings are in agreement with studies showing that ancient human hair can be decontaminated by pretreatment (12, 14). Furthermore, no evidence of recent European admixture or contamination could be detected at the genotype level (4).
The Australian individual’s mitochondrial genome (mtDNA) was sequenced to an average depth of 338×. It belongs to a new subclade of haplogroup O (hg O) that we term hg O1a (4). Haplogroup O is one of the four major lineage groups specific to Australia and has been reported from various parts of the Northern Territory (15 to 16%) (15–17). From high-confidence Y-chromosome SNPs, we assigned his Y chromosome to the K-M526* macro-haplogroup (4). Although the O and P branches of haplogroup K-M526 account for the majority of East and West Eurasian Y chromosomes, the unresolved K-M526* lineages are more common (>5%) only among contemporary populations of Australasia (15, 18). Both uniparental markers fall within the known pattern found among contemporary Aboriginal Australians (15), providing further evidence that the genomic sequence obtained is not contaminated.
We compared our high-confidence SNPs with Illumina SNP chip data from 1220 individuals belonging to 79 populations (4). Among these are individuals from the Kusunda and Aeta, two populations of hunter-gatherers from Nepal and the Philippines, respectively. Both groups have been hypothesized to be possible relict populations from the proposed early wave of dispersal across eastern Asia (19, 20).
Principal components analysis (PCA) results illustrated genetic differentiation among Africans, Asians, and populations of greater Australia. The Australian genome clusters together with Highland Papua New Guinea (PNG) samples and is thus positioned roughly between South and East Asians. Apart from the neighboring Bougainville Papuans, the closest populations to the Aboriginal Australian are the Munda speakers of India and the Aeta from the Philippines (Fig. 1B). This pattern is confirmed from542 individuals from43 Asian and greater Australia populations (4) and by including an additional 25 populations from India (21) that all fall on the Eurasian axis, including those of the Great Andamanese and Onge from the Andaman Islands (21). The PCA and admixture results (Fig. 1C) further confirm the lack of European contamination or recent admixture in the genome sequence.
We used the D test (22, 23) on the SNP chip data and genomes to look for shared ancestry between Aboriginal Australians and other groups (4). We found significantly larger proportions of shared derived alleles between the Aboriginal Australian and Asians (Cambodian, Japanese, Han, and Dai) than between the Aboriginal Australian and Europeans (French) (Table 1, rows 1 to 4). We also found a significantly larger proportion of shared derived alleles between the French and the Asians than between the French and the Aboriginal Australian (Table 1, rows 5 to 8). These findings do not allow us to discriminate between the two models of origin, but they do rule out simple models of complete isolation of populations since divergence. Our data do not provide consistent evidence of gene flow between populations of greater Australia (Aboriginal Australian/PNG Highlands) and Asian ancestors after the latter split from Native Americans under various models (4) (there may still be some gene flow between Bougainville and some Asian ancestors after that time; Table 1). This suggests that before European contact occurred, Aboriginal Australian and PNG Highlands ancestors had been genetically isolated from other populations (except possibly each other) since at least 15,000 to 30,000 years B.P. (24).
Table 1.
Results of D test
| Ingroup 1 | Ingroup 2 | Outgroup | Difference* | Total† | D‡ | SD§ | Z‖ | |
|---|---|---|---|---|---|---|---|---|
| 1 | French | Cambodian | Australian | 461 | 8,035 | 0.06 | 0.013 | 4.5 |
| 2 | French | Japanese | Australian | 463 | 8,107 | 0.06 | 0.013 | 4.5 |
| 3 | French | Han | Australian | 674 | 7,908 | 0.09 | 0.012 | 7.0 |
| 4 | French | Dai | Australian | 636 | 8,214 | 0.08 | 0.013 | 6.0 |
| 5 | Australian | Cambodian | French | 435 | 8,009 | 0.05 | 0.013 | 4.3 |
| 6 | Australian | Japanese | French | 357 | 7,991 | 0.04 | 0.012 | 3.6 |
| 7 | Australian | Han | French | 487 | 7,713 | 0.06 | 0.012 | 5.1 |
| 8 | Australian | Dai | French | 343 | 7,919 | 0.04 | 0.012 | 3.5 |
| 9 | Surui | Cambodian | Australian | −4 | 7,644 | 0.00 | 0.012 | 0.0 |
| 10 | Surui | Japanese | Australian | 1 | 7,477 | 0.00 | 0.013 | 0.0 |
| 11 | Surui | Han | Australian | 215 | 7,261 | 0.03 | 0.013 | 2.4 |
| 12 | Surui | Dai | Australian | 169 | 7,493 | 0.02 | 0.013 | 1.7 |
| 13 | Surui | Cambodian | PNG Highlands | −195 | 64,149 | 0.00 | 0.006 | −0.5 |
| 14 | Surui | Japanese | PNG Highlands | 288 | 62,364 | 0.00 | 0.006 | 0.7 |
| 15 | Surui | Han | PNG Highlands | 393 | 60,947 | 0.01 | 0.006 | 1.0 |
| 16 | Surui | Dai | PNG Highlands | 427 | 62,925 | 0.01 | 0.006 | 1.0 |
| 17 | Surui | Cambodian | Bougainville | 319 | 64,951 | 0.00 | 0.006 | 0.8 |
| 18 | Surui | Japanese | Bougainville | 1,543 | 63,063 | 0.02 | 0.007 | 3.6 |
| 19 | Surui | Han | Bougainville | 1,577 | 62,019 | 0.03 | 0.006 | 3.9 |
| 20 | Surui | Dai | Bougainville | 1,691 | 63,585 | 0.03 | 0.006 | 4.2 |
Number of sites where a derived allele is shared between outgroup and ingroup 1 subtracted from sites where the derived allele is shared between outgroup and ingroup 2.
Number of sites where a derived allele is found in the outgroup and one of the ingroups.
D test statistics (difference divided by total).
Standard deviation (found by block jackknife).
Standardized statistics (to determine significance).
To identify which model of human dispersal best explains the data, we sequenced three Han Chinese genomes to an average depth of 23 to 24× (4) and used a test comparing the patterns of similarity between these or the Aboriginal Australian to African and European individuals (4). This test, which we call D4P, is closely related to the D test (22, 23) but is far more robust to errors and can detect subtle demographic signals in the data that may be masked by large amounts of secondary gene flow (4).
Taking those sites where the Aboriginal Australian (ABR) differs from a Han Chinese representing eastern Asia (ASN), and comparing ABR and ASN with the Centre d’Etude du Polymorphisme Humain (CEPH) European sample (CEU) representing Europe and the Yoruba representing Africa (YRI), the single-dispersal model (Fig. 1A, top) predicts an equal number of sites supporting group 1 [(YRI, ASN), (CEU, ABR)] and group 2 [(YRI, ABR), (CEU, ASN)]. In contrast, the multiple-dispersal model (Fig. 1A, bottom) predicts an excess of group 2. Indeed, we found a statistically significant excess of sites (51.4%) grouping the Yoruba and Australian genomes together (group 2) relative to the Yoruba and East Asian genomes together (group 1, 48.6%, P < 0.001), consistent with a basal divergence of Aboriginal Australians in relation to East Asians and Europeans (Table 2). Another possible explanation of our findings is that gene flow between modern European and East Asian populations caused these two populations to appear more similar to each other, generating an excess of sites showing group 2, even under the single-dispersal model. However, simulations under such a model show that the amount of gene flow between Europeans and East Asians (5) cannot generate the excess of sites showing group 2 unless Aboriginal Australian, East Asian, and European ancestral populations all split from each other around the same time, with no subsequent migration between aboriginal Australasians and East Asians (4). Such a model, however, would be inconsistent with our results from D test, PCA, and discriminant analysis of principal components (DAPC) (4), given that the Aboriginal Australian is found to be genetically closer to East Asians than to Europeans (Table 1 and Fig. 1B). Thus, our findings suggest that a model in which Aboriginal Australians are directly derived from ancestral Asian populations, as proposed by the single-dispersal model, is not compatible with the genomic data. Instead, our results favor the multiple-dispersal model in which the ancestors of Aboriginal Australian and related populations split from the Eurasian population before Asian and European populations split from each other (4).
Table 2.
Results of the D4P test. The results are from NA19239 (for YRI), NA12891 (for CEU), HG00421 (for ASN), and the Aboriginal Australian genome (ABR). The two groups are patterns representing the two ways in which eligible SNPs can partition the four genomes (they have not been polarized).
| Group 1 | Group 2 | |
|---|---|---|
| YRI | 1 | 1 |
| ABR | 0 | 1 |
| CEU | 0 | 0 |
| ASN | 1 | 0 |
| Observed number* | 13,974 | 14,765 |
| Observed proportion (95% CI)† | 48.6% (47.8 to 49.4%) | 51.4% (50.6 to 52.2%) |
| Expected proportion under multiple-dispersal model 1‡ | 48.7% | 51.3% |
| Expected proportion under multiple-dispersal model 2§ | 48.0% | 52.0% |
| Expected proportion under single-dispersal model‖ | 50.3% | 49.7% |
Average number of eligible SNPs showing groups 1 and 2 across block bootstrap replicates.
95% confidence interval obtained from a block bootstrap (4). Z test rejects the null hypothesis that this value is equal to 50% (Z = 3.3, P < 0.001).
Expected proportion from a multiple-dispersal model in which aboriginal Australasians split from Eurasian populations 2500 generations ago, before the split of European and Asian populations. This split time was estimated using the Aboriginal, NA12891, and HG00421 sequences (4). These were the same individuals used for the D4P analysis.
Expected proportion from a multiple-dispersal model in which aboriginal Australasians split from Eurasian populations 2750 generations ago, before the split of European and Asian populations. This split time was estimated using the Aboriginal Australian and all Eurasian sequences (4).
To estimate the times of divergence, we developed a population genetic method for estimating demographic parameters from diploid whole-genome data. The method uses patterns of allele frequencies and linkage disequilibrium to obtain joint estimates of migration rates and divergence times between pairs of populations (4). Using this method, we estimate that aboriginal Australasians split from the ancestral Eurasian population 62,000 to 75,000 years B.P. This estimate fits well with the mtDNA-based coalescent estimates of 45,000 to 75,000 years B.P. of the non-African founder lineages (4, 15, 25, 26). Furthermore, we find that the European and Asian populations split from each other only 25,000 to 38,000 years B.P., in agreement with previous estimates (5, 6). All three populations, however, have a divergence time similar to the representative African sequence. Additionally, our estimated split time between aboriginal Australasians and the ancestral Eurasian population predicts the observed excess of sites showing group 2 discussed above (Table 2). To obtain confidence intervals and test hypotheses, we used a block bootstrap approach. In 100 bootstrap samples, we always obtained a longer divergence time between East Asians and the Aboriginal Australian than between East Asians and Europeans, showing that we can reject the null hypothesis of a trichotomy in the population phylogeny with statistical significance of approximately P < 0.01. In these analyses we have taken changes in population sizes and the effect of gene flow after divergence between populations into account. However, our models are still relatively simple, and the models we consider are only a subset of all the possible models of human demography. In addition, we have not attempted directly to model the combined effects of demography and selection. The true history of human diversification is likely to be more complex than the simple demographic models considered here.
We used two approaches to test for admixture in the genomic sequence of the Aboriginal Australian with archaic humans [Neandertals and Denisovans (22, 23)]. We asked whether previously identified high-confidence Neandertal admixture segments in Europeans and Asians (22) could also be found in the Aboriginal Australian. We found that the proportion of such segments in the Aboriginal Australian closely matched that observed in European and Asian sequences (4). In the case of the Denisovans, we used a D test (22, 23) to search for evidence of admixture within the Aboriginal Australian genome. This test compares the proportion of shared derived alleles between an outgroup sequence (Denisovan) and two ingroup sequences. This test showed a relative increase in allele sharing between the Denisovan and the Aboriginal Australian genomes, compared to other Eurasians and Africans including Andaman Islanders (4), but slightly less allele sharing than observed for Papuans. However, we found that the D test is highly sensitive to errors in the ingroup sequences (4), and shared errors are of particular concern when the comparisons involve both an ingroup and outgroup ancient DNA sequence. Although we cannot exclude these results being influenced by such errors, the latter result is consistent with the hypothesis of increased admixture between Denisovans or related groups and the ancestors of the modern inhabitants of Melanesia (23). This admixture may have occurred in Melanesia or, alternatively, in Eurasia during the early migration wave.
The degree to which a single individual is representative of the evolutionary history of Aboriginal Australians more generally is unclear. Nonetheless, we conclude that the ancestors of this Aboriginal Australian man—and possibly of all Aboriginal Australians—are as distant from Africans as are other Eurasians, and that the Aboriginal ancestors split 62,000 to 75,000 years B.P. from the gene pool that all contemporary non-African populations appear to descend from. Rather than supporting a single early human expansion into eastern Asia, our findings support the alternative model of Aboriginal Australians descending from an early Asian expansion wave some 62,000 to 75,000 years B.P. The data also fit this model’s prediction of substantial admixture and replacement of populations from the first wave by the second expansion wave, with a few populations such as Aboriginal Australians, and possibly PNG Highlands and Aeta, being remnants of the early dispersal (Fig. 2). This is compatible with mtDNA data showing that although all haplogroups observed in Australia are unique to this region, they derive from the same few founder haplogroups that are shared by all non-African populations (4). Finally, our data are in agreement with contemporary Aboriginal Australians being the direct descendants from the first humans to be found in Australia, dating to ~50,000 years B.P. (7, 8). This means that Aboriginal Australians likely have one of the oldest continuous population histories outside sub-Saharan Africa today.
Fig 2.
Reconstruction of early spread of modern humans outside Africa. The tree shows the divergence of the Aboriginal Australian (ABR) relative to the CEPH European (CEU) and the Han Chinese (HAN) with gene flow between aboriginal Australasians and Asian ancestors. Purple arrow shows early spread of the ancestors of Aboriginal Australians into eastern Asia ~62,000 to 75,000 years B.P. (ka BP), exchanging genes with Denisovans, and reaching Australia ~50,000 years B.P. Black arrow shows spread of East Asians ~25,000 to 38,000 years B.P. and admixing with remnants of the early dispersal (red arrow) some time before the split between Asians and Native American ancestors ~15,000 to 30,000 years B.P. YRI, Yoruba.
Supplementary Material
Acknowledgments
Our work was endorsed by the Goldfields Land and Sea Council, the organization representing the Aboriginal Traditional Owners of the Goldfields region, including the cultural (and possibly the biological) descendents of the individual who provided the hair sample. See (4) for letter. Data are accessible through NCBI Sequence Read Archive SRA035301.1 or through http://dx.doi.org/10.5524/100010. We note the following additional affiliations: S.T. also works for the Australian Federal Police; J.D. is a partner in Dortch & Cuthbert Pty. Ltd.; P.F. is director of Genetic Ancestor Ltd. and Fluxus Technology Ltd.; and C.D.B. serves as an unpaid consultant for 23andMe. For author contributions and extended acknowledgements, see (4).
Footnotes
Supporting Online Material
http://www.sciencemag.org/cgi/content/full/science.1211177/DC1
Materials and Methods
SOM Text
Figs. S1 to S39
Tables S1 to S28
References
References and Notes
- 1.Ramachandran S, et al. Proc. Natl. Acad. Sci. U.S.A. 2005;102:15942. doi: 10.1073/pnas.0507611102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu H, Prugnolle F, Manica A, Balloux F. Am. J. Hum. Genet. 2006;79:230. doi: 10.1086/505436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.HUGO Pan-Asian SNP Consortium. Science. 2009;326:1541. [Google Scholar]
- 4.See supporting material on Science Online.
- 5.Gutenkunst RN, Hernandez RD, Williamson SH, Bustamante CD. PLoS Genet. 2009;5:e1000695. doi: 10.1371/journal.pgen.1000695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Keinan A, Mullikin JC, Patterson N, Reich D. Nat. Genet. 2007;39:1251. doi: 10.1038/ng2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Summerhayes GR, et al. Science. 2010;330:78. doi: 10.1126/science.1193130. [DOI] [PubMed] [Google Scholar]
- 8.O’Connell J, Allen J, Archaeol J. Sci. 2004;31:835. [Google Scholar]
- 9.Cavalli-Sforza L, Menozzi P, Piazza A. The History and Geography of Human Genes. Princeton, NJ: Princeton Univ. Press; 1994. [Google Scholar]
- 10.Lahr MM, Foley R. Evol. Anthropol. 1994;3:48. doi: 10.1002/evan.21405. [DOI] [PubMed] [Google Scholar]
- 11.Lahr MM, Foley RA. Yearb. Phys. Anthropol. 1998;41:137. doi: 10.1002/(sici)1096-8644(1998)107:27+<137::aid-ajpa6>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 12.Rasmussen M, et al. Nature. 2010;463:757. doi: 10.1038/nature08835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Binladen J, et al. Genetics. 2006;172:733. doi: 10.1534/genetics.105.049718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gilbert MTP, et al. Science. 2008;320:1787. doi: 10.1126/science.1159750. [DOI] [PubMed] [Google Scholar]
- 15.Hudjashov G, et al. Proc. Natl. Acad. Sci. U.S.A. 2007;104:8726. [Google Scholar]
- 16.Ingman M, Gyllensten U. Genome Res. 2003;13:1600. doi: 10.1101/gr.686603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Holst Pellekaan SM, Ingman M, Roberts-Thomson J, Harding RM. Am. J. Phys. Anthropol. 2006;131:282. doi: 10.1002/ajpa.20426. [DOI] [PubMed] [Google Scholar]
- 18.Karafet TM, et al. Mol. Biol. Evol. 2010;27:1833. doi: 10.1093/molbev/msq063. [DOI] [PubMed] [Google Scholar]
- 19.Lahr M. The Evolution of Modern Human Diversity: A Study of Cranial Variation. Cambridge: Cambridge Univ. Press; 1996. [Google Scholar]
- 20.Whitehouse P, Usher T, Ruhlen M, Wang WS-Y. Proc. Natl. Acad. Sci. U.S.A. 2004;101:5692. doi: 10.1073/pnas.0400233101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reich D, Thangaraj K, Patterson N, Price AL, Singh L. Nature. 2009;461:489. doi: 10.1038/nature08365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Green RE, et al. Science. 2010;328:710. doi: 10.1126/science.1188021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reich D, et al. Nature. 2010;468:1053. doi: 10.1038/nature09710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goebel T, Waters MR, O’Rourke DH. Science. 2008;319:1497. doi: 10.1126/science.1153569. [DOI] [PubMed] [Google Scholar]
- 25.Endicott P, Ho SYW, Metspalu M, Stringer C. Trends Ecol. Evol. 2009;24:515. doi: 10.1016/j.tree.2009.04.006. [DOI] [PubMed] [Google Scholar]
- 26.Soares P, et al. Am. J. Hum. Genet. 2009;84:740. doi: 10.1016/j.ajhg.2009.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schaffner SF, et al. Genome Res. 2005;15:1576. doi: 10.1101/gr.3709305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alexander DH, Novembre J, Lange K. Genome Res. 2009;19:1655. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.