Abstract
Although effective in treating an array of neurological disorders, antipsychotics are associated with deleterious metabolic side effects. Through high-throughput screening, we previously identified phenothiazine antipsychotics as modulators of the human insulin promoter. Here, we extended our initial finding to structurally diverse typical and atypical antipsychotics. We then identified the TGFβ pathway as being involved in the effect of antipsychotics on the insulin promoter, finding that antipsychotics activated SMAD3, a downstream effector of the TGFβ pathway, through a receptor distinct from the TGFβ receptor family and known neurotransmitter receptor targets of antipsychotics. Of note, antipsychotics that do not cause metabolic side effects did not activate SMAD3. In vivo relevance was demonstrated by reanalysis of gene expression data from human brains treated with antipsychotics, which showed altered expression of SMAD3 responsive genes. This work raises the possibility that antipsychotics could be designed that retain beneficial CNS activity while lacking deleterious metabolic side effects.
Keywords: Antipsychotics, TGFβ, SMAD3, Insulin Promoter
Introduction
An estimated 14.3 million Americans were taking antipsychotics in 2008, making them among the most prescribed drugs in the US1. Deleterious metabolic side effects, including obesity, insulin resistance, and diabetes, occur to some extent with almost all antipsychotics2, 3. While their therapeutic effects are thought to be related to activation of dopamine and serotonin receptors in the CNS4, it has been unclear whether the metabolic side effects of antipsychotics are due to the modulation of those receptors or whether off-target effects might also be responsible3, 5–9.
Previously, we found that phenothiazine-based antipsychotics modulated the insulin promoter10. Given the central role of insulin in metabolism and the regulation of its expression by molecules such as glucose, amino acids, and fatty acids, this raised the possibility that insights into the metabolic side effects of antipsychotics could be achieved through uncovering the mechanism by which they affected the insulin promoter.
Here, we report that antipsychotics modulate the insulin promoter through activation of SMAD3, an important downstream effector of the transforming growth factor beta (TGFβ) pathway. Activation occurred through a receptor distinct from those acted upon by TGFβ itself, and also distinct from the neurotransmitter receptors responsible for the therapeutic effects of antipsychotics on CNS function. In vivo relevance of the finding was demonstrated by bioinformatic analysis of publically available gene expression data from brains of antipsychotic-treated 11, 12 schizophrenic patients11, 12.
The TGFβ pathway and SMAD3 in particular are highly associated with obesity, insulin resistance, and diabetes13–17. Obese individuals18 and type II diabetics have higher serum levels of TGFβ than normal controls, and healthy individuals with high serum TGFβ are more likely to develop Type II diabetes13,16. Also, mice with homozygous inactivating mutations of SMAD3 show enhanced glucose tolerance and are resistant to high fat diet induced obesity and insulin resistance17, 19. The finding that antipsychotics activate SMAD3 through a mechanism distinct from that responsible for the neurological effects of those drugs therefore raises the possibility that antipsychotics could be designed that retain the beneficial neurological effects while lacking the deleterious metabolic side effects.
Results
Both typical and atypical antipsychotics modulate the insulin promoter
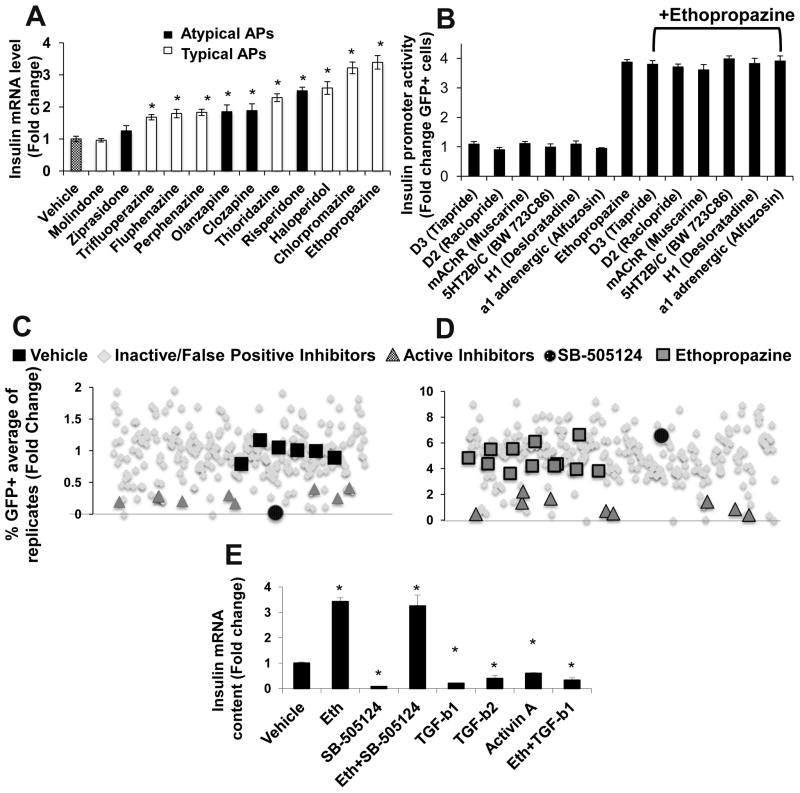
Because phenothiazine antipsychotics have been supplanted to a large degree by the structurally diverse atypical antipsychotics, we extended our analysis of antipsychotic activity beyond the phenothiazines studied previously10. Despite the structural diversity of the antipsychotics tested, almost all modulated insulin promoter activity in T6PNE (Fig. 1A, Supplemental Fig 1). There was no clear relationship between the classification of a drug as typical or atypical and its activity in the assay. Ethopropazine was the most potent, and since it is chemically stable we used it as the prototypical active antipsychotic. Also, as expected if the effects of antipsychotics on the insulin promoter in T6PNE are relevant to the metabolic side effects of the drugs, molindone and ziprasidone, which are not associated with strong metabolic side effects in patients20–22, did not affect insulin promoter activity.
Figure 1. Kinase inhibitor counterscreen revealed that antipsychotics act on the insulin promoter through a non-canonical pathway downstream of TGFβ R1.
(A) QPCR analysis of the effect of antipsychotics on endogenous insulin gene expression. T6PNE cells were treated for 48 hr with the indicated antipsychotic (n≥6). (B) Pharmacologic antagonists of neurotransmitter receptors acted on by antipsychotics were tested for activity in the T6PNE Ins-GFP insulin promoter assay in the presence and absence of ethopropazine (n=12). (C–D) The T6PNE Ins-GFP insulin promoter assay was screened against the Calbiochem 242 Compound Kinase Inhibitor Library27 in the presence (C) or absence (D) of ethopropazine (20mM). Inhibitors were added at .5 and 5uM, with each inhibitor being run in duplicate (only replicates at 5uM are shown). Ten inhibitors repressed the insulin promoter (
 ). Of the ten, one, the TGFβ R1 inhibitor SB-505124 (●), was a potent insulin promoter inhibitor in the absence but not the presence of ethopropazine. (E) RT-PCR analysis of endogenous insulin gene expression. T6PNE cells were treated for 48 hr with the indicated condition. (n=9). Error bars are SEM. * indicates p<0.05 relative to DMSO control.
). Of the ten, one, the TGFβ R1 inhibitor SB-505124 (●), was a potent insulin promoter inhibitor in the absence but not the presence of ethopropazine. (E) RT-PCR analysis of endogenous insulin gene expression. T6PNE cells were treated for 48 hr with the indicated condition. (n=9). Error bars are SEM. * indicates p<0.05 relative to DMSO control.
Neurotransmitter receptors targeted by antipsychotics are inactive in the insulin promoter assay
Since it is known that antipsychotics activate multiple neurotransmitter receptors23, 24, and many of those are expressed in islets25 where they play important roles in β-cell function26, we hypothesized that the effect of antipsychotics on the insulin promoter was through a pathway involving the known neurotransmitter receptor targets of antipsychotics. To test this, we first analyzed gene expression data (GSE18821)10 to determine which neurotransmitter receptor targets of antipsychotics were expressed in T6PNE cells, finding that many were expressed at approximately equal levels in T6PNE and primary human islets (Supplemental Fig. 2A). Specific receptor antagonists were then tested for activity on the human insulin promoter-eGFP transgene in T6PNE cells. None prevented ethopropazine from stimulating the insulin promoter, or altered insulin promoter activity in the absence of the drug (Fig. 1B), leading us to conclude that the target of the antipsychotics responsible for insulin promoter modulation was not one of the known neurotransmitter receptor targets responsible for their therapeutic benefit.
Antipsychotics signal to the insulin promoter downstream of the TGFβR1 kinase
Given the negative results with known antipsychotic targets, we switched to an unbiased approach, screening the T6PNE insulin promoter assay with a diverse library of kinase inhibitors (Calbiochem Inhibitor) which has been used previously in high-throughput screens27. The rationale for this approach was that essentially all signaling pathways involve phosphorylation, and so we hoped to gain insight into the pathways being acted upon by antipsychotics by screening a library containing a large number of kinase inhibitors. The library was screened in the presence and absence of ethopropazine to ascertain kinase inhibitors with activity that was selectively affected by antipsychotics (Figure 1C, D).
Ten compounds repressed the insulin promoter in the absence of ethopropazine, (Fig. 1C,
 and ●); other apparent repressors were cytotoxic false-positives. One of the ten, SB-505124, an inhibitor of TGFβ type 1 receptors, of which there are seven family members (designated ALK1-7)28, completely lost activity in the presence of ethopropazine (Fig. 1D ●). This indicates that ethopropazine is epistatic to the TGFβ type 1 receptor kinase, signaling to the insulin promoter through the TGFβ pathway at a point downstream. A dose-response study of the effect of SB-505124 demonstrated potent repression of the human insulin promoter-eGFP transgene with an IC50 of 0.34uM (Supplemental Fig. 3), which was confirmed on the endogenous insulin gene in the absence, but not in the presence of ethopropazine, supporting the results with the transgene (Fig. 1E).
and ●); other apparent repressors were cytotoxic false-positives. One of the ten, SB-505124, an inhibitor of TGFβ type 1 receptors, of which there are seven family members (designated ALK1-7)28, completely lost activity in the presence of ethopropazine (Fig. 1D ●). This indicates that ethopropazine is epistatic to the TGFβ type 1 receptor kinase, signaling to the insulin promoter through the TGFβ pathway at a point downstream. A dose-response study of the effect of SB-505124 demonstrated potent repression of the human insulin promoter-eGFP transgene with an IC50 of 0.34uM (Supplemental Fig. 3), which was confirmed on the endogenous insulin gene in the absence, but not in the presence of ethopropazine, supporting the results with the transgene (Fig. 1E).
SB-505124 exhibits a strong preference for TGFβ type I receptors ALK 4/5/7 over ALK 1/2/3/6, and has no or minimal activity on a panel of 27 other protein kinases at the highest concentration used in the kinase inhibitor screen (5uM)28. We examined the expression of the TGFβ receptor family in T6PNE using gene expression microarray data, finding good concordance between the level of expression in T6PNE and primary human islets (GSE18821)10. While ALK2 was the most highly expressed, ALK4, ALK5, and ALK7 were also present, consistent with the preferential activity against ALK4/5/7 exhibited by SB-505214.
Antipsychotics activate the TGFβ pathway
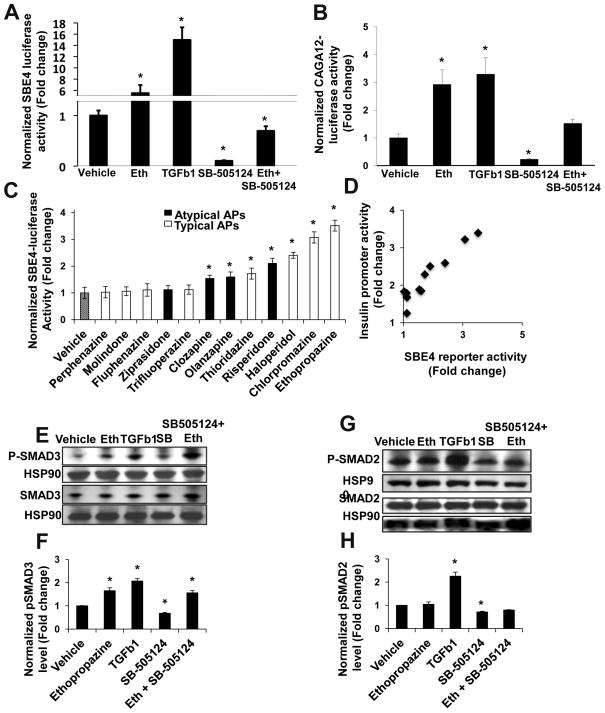
We next tested if the major downstream mediators of the TGFβ pathway, the SMAD transcription factors, were activated by antipsychotics. In the classical TGFβ signaling pathway, receptor-regulated SMADs (R-SMADs) are phosphorylated by a TGFβ type I receptor, promoting nuclear translocation and binding to the SMAD binding element (SBE) CAGAC in promoters to modulate gene expression29. However, noncanonical SMAD activation is well described30. As expected, TGFβ1 activated and SB-505124 inhibited the R-SMAD reporters SBE4-Luc31 and CAGA12-Luc32 (Fig. 2A, 2B), which contain multimerized sequence elements responsive to R-SMADs driving a luciferase reporter31. Ethopropazine activated R-SMAD reporter activity in both assays (Fig. 2A, 2B).
Figure 2. Antipsychotics activate the TGFβ pathway through enhancement of SMAD3, but not SMAD2, activity.
(A, B) Ethopropazine increases activity of the SMAD2/3 responsive SBE4-luc reporter (A) and the SMAD3 specific CAGA12-luc reporter (B). Cells were treated for 24 hr with the indicated condition before harvesting for luciferase assay. (n≥6). (C) SAR of typical and atypical antipsychotics on the SBE4-luc reporter. (n≥6). (D) SBE4 reporter activity of antipsychotics from 2C is plotted against the insulin promoter activity from Figure 1A. Each point represents a single antipsychotic (Pearson correlation coefficient = 0.93, p=0.00012). (E,G) Immunoblot analysis of phospho-SMAD levels. T6PNE cells were treated for 1hr with the indicated condition and a Western blot was performed with antibodies towards total and pSMAD3 (E) or total and pSMAD2 (G), and HSP90. (F, H) Quantification of pSMAD3 (F) and pSMAD2 (H) protein levels demonstrated that antipsychotics increased pSMAD3 but not pSMAD2. The pSMAD intensity was divided by the total SMAD intensity (n=3, blot shown is representative of 3 independent experiments). Error bars are SEM. * indicates p<0.05 relative to DMSO control.
Next, we performed a structure-activity relationship (SAR) study on the SBE4-Luc reporter with the array of typical and atypical antipsychotics used previously (Fig. 2C). Strikingly, the two SARs (insulin promoter versus SBE4-Luc reporter) were highly correlated (Pearson correlation coefficient = 0.93, p=.00012)(Fig. 2D). This high correlation coefficient strongly supports a model in which antipsychotics signal to the insulin promoter through activation of the TGFβ pathway.
Antipsychotics promote phosphorylation of SMAD3 but not SMAD2
Since it is well established that R-SMADs are activated by phosphorylation33, and antipsychotics activated two R-SMAD reporters, we tested if antipsychotics promoted increased phosphorylation of the R-SMADs responsive to TGFβ, SMAD2 and SMAD3.. As expected, TGFβ1 increased and SB-505124 decreased the levels of both phosphorylated SMADs assayed by immunoblot with phospho-specific antibodies (Fig. 2E–H). However, ethopropazine increased the level of phospho-SMAD3 (Fig. 2E, F) but not phospho-SMAD2 (Fig. 2G, H). Also, ethopropazine antagonized the effect of SB-505124 on SMAD3 (Fig. 2E, F) but not on SMAD2 (Fig. 2G, H). These experiments led us to conclude that TGFβ and antipsychotics differentially activate SMAD3 and SMAD2, and there must be distinct pathways by which antipsychotics and TGFβ promote phosphorylation of R-SMADs.
SMAD3 is required for the effect of antipsychotics on the insulin promoter
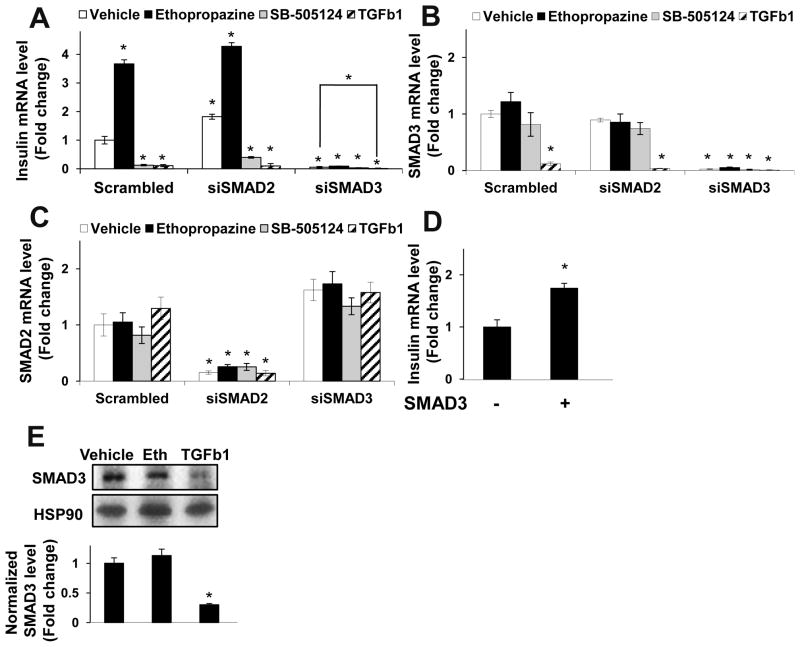
If antipsychotics signal to the insulin promoter solely through SMAD3, then downregulation of SMAD3 but not SMAD2 by siRNA should ablate the effect of antipsychotics on the insulin promoter. We found this to be true; SMAD2 siRNAs did not affect the effect of antipsychotics on the insulin promoter, while those effects were almost completely ablated by a SMAD3 siRNA (Fig. 3A). We also found that SMAD3 overexpression, confirmed by immunoblot to result in a 2.1-fold increase in SMAD3 protein, enhanced insulin transcription (Fig. 3D), while SMAD3 siRNA potently inhibited the insulin promoter (Fig. 3A).
Figure 3. SMAD3, but not SMAD2, is required for the effect of antipsychotics on the insulin promoter.
(A) siRNA to SMAD3 but not SMAD2 inhibited insulin gene expression in the absence and presence of antipsychotics. (B,C) SMAD3 (B) and SMAD2 (C) siRNAS demonstrated efficacy and specificity. Insulin (A), SMAD3 (B), or SMAD2 (C) mRNA content was measured by quantitative RT-PCR (n=6, data shown are representative of 2 independent experiments). (D) SMAD3 overexpression enhanced insulin promoter activity. Insulin mRNA was measured by quantitative RT-PCR (n=6). (E) SMAD3 protein levels decreased following chronic TGFβ exposure. T6PNE cells treated with the indicated condition for 48 hours were harvested for immunoblot analysis with antibodies towards SMAD3 and HSP90. (n=3, Blot shown is representative of 3 independent experiments). Error bars are SEM. * indicates p<0.05 relative to DMSO control.
TGFβ inhibits the insulin promoter through downregulation of SMAD3 transcription
The fact that SB-505124 repressed insulin gene expression in T6PNE cells implies that there is a basal level of TGFβ pathway activity in those cells, acting to stimulate promoter activity. This is consistent with the effects of SMAD3 siRNA and overexpression (Fig. 3), and with previous data on TGFβ pathway activation in β-cells15, 34, 35. However, TGFβ1, TGFβ2, and Activin A all exhibited dose-dependent inhibition of the human insulin promoter-eGFP transgene (Supplemental Fig. 4A–C) and the endogenous insulin promoter (Fig. 1E), with TGFβ1 being the most potent. Since it has been reported previously that TGFβ1 can inhibit SMAD3 gene expression36, 37, we tested by quantitative RT-PCR the effect of TGFβ1 on SMAD mRNA levels in T6PNE cells. TGFβ1 induced a dramatic decrease in SMAD3 but not SMAD2 mRNA levels, while ethopropazine had no effect on either SMAD3 or SMAD2 mRNA (Fig. 3B, C). In addition, 48 hours of treatment with TGFβ but not ethopropazine decreased SMAD3 protein levels assayed by immunoblot (Fig. 3E). Thus, in the acute 24 hour setting of a transient transfection assay, the effect of TGFβ1 on SMAD phosphorylation is dominant, while in the longer duration 48 hour assay on insulin gene expression, its effect on SMAD3 gene expression predominates.
To determine the generality of the effect of TGFβ on SMAD gene expression, we used the Pubmed Gene Expression Omnibus (GEO) database to examine previous reports on the effect of TGFβ in a variety of different cells. The vast majority of those studies found substantial effects of TGFβ on SMAD3 (average fold change = −2.15), but not SMAD2 mRNA (average fold change = −0.31)(Supplemental Table 1).
Antipsychotics signal to the insulin promoter through modulation of E-box activity downstream of SMAD3
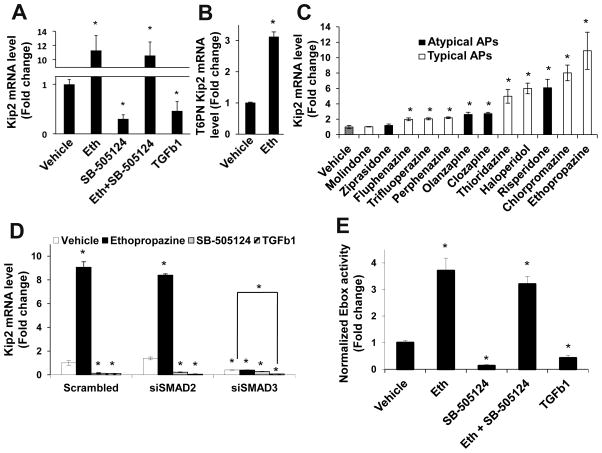
There are multiple mechanisms by which SMAD proteins could affect insulin promoter activity. While the human insulin promoter contains a SMAD binding site38, the TGFβ pathway has been shown to modulate E-box activity39, and SMAD proteins have been shown to interact directly with bHLH factors40,41, suggesting that the effect of antipsychotics on the insulin promoter could also be at E-boxes. We showed previously that CDKN1C, the gene encoding p57Kip2, is regulated in T6PNE by a specific E-box in the promoter, resulting in co-regulation with the insulin gene in those cells10. Since no SMAD binding elements are present in the Kip2 promoter, we tested the effect of antipsychotics on Kip2 gene expression to specifically detect effects on E-box activity. Ethopropazine potently activated Kip2 transcription in T6PNE cells (Fig. 4A), as well as in T6PN cells (Fig. 4B), which express PDX1 and NeuroD1 and low levels of endogenous E47, but not E47MER10, 42. T6PN was used to rule out the possibility that antipsychotics could be acting as agonists for the modified estrogen receptor (MER), leading to activation of E47 by a physiologically irrelevant mechanism. Of significance, the SAR for the antipsychotics on Kip2 expression was highly correlated with their effect on insulin expression, as one would expect if the mechanisms by which the drugs affect the two promoters were the same (Fig. 4C). Similar to their effects on the insulin promoter, SB-505124 and TGFβ1 decreased the level of Kip2 mRNA, while the effect of SB-505124 was blocked by ethopropazine (Fig. 4A). SMAD3 siRNA potently inhibited Kip2 gene expression while SMAD2 siRNA had no effect (Figure 4D), demonstrating the specificity of regulation of E-box activity by SMAD3.
Figure 4. Antipsychotics signal to the insulin promoter through modulation of E-box activity downstream of SMAD3.
(A–C) Antipsychotics enhanced, while TGFβ 1 and SB-505124 inhibited transcription of the E-box responsive gene Kip2 measured by QPCR in T6PNE cells (A,C) and T6PN cells without the E47mer construct expressed in T6PNE (B). Cells were treated for 48 hr with the indicated condition. (n≥6) (D) SMAD3 siRNA, but not SMAD2 or scrambled siRNAs, inhibited basal and antipsychotic-induced effects on Kip2 expression (n≥6). (E) Antipsychotics enhanced while TGFβ 1 and SB-505124 inhibited activity on the E-box reporter 4RTK-luc in T6PNE cells (n=9). Error bars are SEM. * indicates p<0.05 relative to DMSO control.
To confirm that the effects of antipsychotics were through the E-box directly and not any other sequence elements, we used a promoter-reporter construct, 4RTK-Luc, in which the luciferase reporter is under the control of a promoter consisting of multimerized E-box elements 5′ of thymidine kinase minimal promoter43. Antipsychotics activated 4RTK-Luc, while TGFβ1 and SB-505124 repressed reporter activity (Fig. 4E), with antipsychotics being epistatic to the effect of SB-505124. Thus, all of the results are consistent with a model in which antipsychotic effects on the insulin promoter are mediated by activation of SMAD3, which then acts through E-boxes to affect gene expression.
Brain tissue from antipsychotic-treated schizophrenic patients shows gene expression patterns consistent with activated SMAD3
To determine whether the finding that antipsychotics activated the TGFβ pathway in vitro was relevant to effects of antipsychotics in patients, we used a bioinformatic approach, taking advantage of published transcriptome data from schizophrenic patients treated with antipsychotics. In 2008, Mudge et al. used RNA-Seq technology to study the pattern of gene expression in the brains of schizophrenic patients treated with antipsychotics compared with untreated control subjects11. We utilized the web-based systems biology software NextBio to reanalyze their data with a specific focus on the effect of antipsychotics on SMAD-responsive genes.
NextBio is a database comprising lists of genes sharing a common property such as possessing a particular transcription factor binding site in their promoters, or being modulated in response to a particular intervention. It compares those lists with gene lists provided by the user and generates a statistical measure of the association between the two lists, expressed as a p-value and calculated using a “running Fisher’s test” algorithm44. To validate the NextBio algorithm with the Mudge et al., dataset, we analyzed in NextBio the list of genes altered in all the antipsychotic treated patients compared to the healthy, untreated controls. Consistent with the conclusions of Mudge et al11, the Golgi apparatus and vesicle-mediated transport gene ontology (GO) lists were statistically significantly associated with the genes altered in the antipsychotic treated patients (Supplemental. Table 2, p=0.00037 and 0.0089, respectively).
Next, we extracted lists of genes affected by particular antipsychotics from the Mudge et al., dataset to determine whether they were statistically significantly similar to lists of genes containing particular transcription factor binding sites in their promoters, with a focus on genes downstream of the TGFβ pathway. Those lists came from an analysis done by Xie et al., who used a genome-wide comparative analysis of gene promoter sequences across 4 species combined with the TRANSFAC dataset of transcription factor binding sites45, to identify all genes with promoter binding sites for specific transcription factors, including SMAD1 and SMAD346 (Supplemental Table 4).
When the data from all patients were considered together, the significance of the association between the list of genes altered in antipsychotic treated patients and the list of genes with SMAD3 binding sites in their promoters was marginal (p=0.054, Supplemental Table 2). However, when we restricted the analysis to the patients taking the four antipsychotics (chlorpromazine, haloperidol, risperidone, and thioridazine) that were most active in the SMAD reporter assay (Fig. 2C), there was a highly significant association with SMAD3 responsive genes (p=0.0096, Supplemental Table 2). To test the specificity of the association between our in vitro SAR and the in vivo effects of the drugs on genes containing SMAD3 sites in their promoters, we examined the effect of antipsychotics on genes containing SMAD1 sites, finding no association regardless of whether all antipsychotics or the 4 most potent in the in vitro assays were considered (p=0.11 and 0.18 respectively, Supplemental Table 2). This is important since, while SMAD3 and SMAD1 are highly related, SMAD1 is an effector of BMP and not TGFβ signaling47.
As a further test of clinical relevance, we analyzed a second set of gene expression data from the brains of patients inflicted with schizophrenia who were treated with antipsychotics compared to healthy matched controls12. In this study, using microarrays rather than RNA-Seq, the number of altered genes declined almost to zero with increasing duration of illness. Focusing therefore on patients with short duration of illness, we found an increase in the statistical significance of the association between the genes with SMAD3 binding sites and the genes altered by antipsychotics when only the antipsychotics that were most potent in vitro were considered (0.043 vs. 0.071 for all drugs (Supplemental Table 3).
Having shown in two independent sets of gene expression data from antipsychotic-treated patients a correlation between the most active drugs in vitro with effects on SMAD3-responsive genes in vivo, we increased the granularity of the study by analyzing each antipsychotic individually, calculating the statistical association between the genes altered in each antipsychotic treated patient brain with SMAD3 or SMAD1-responsive genes. The values for each antipsychotic were averaged and plotted against the data from the SMAD reporter SAR (Fig. 2C). For SMAD3 regulated genes (Fig. 5A), the correlation of these two parameters was highly significant, with a Pearson correlation coefficient of 0.89 (p=0.0013), while for SMAD1 (Fig. 5B) there was poor correlation (correlation coefficient=0.42, p=0.24). The striking and highly significant correlation between the insulin promoter and SMAD reporter data generated in vitro in T6PNE cells and transcriptome data generated by two independent groups in samples from patients with schizophrenia provides strong evidence that our finding of TGFβ pathway activation by antipsychotics is clinically relevant.
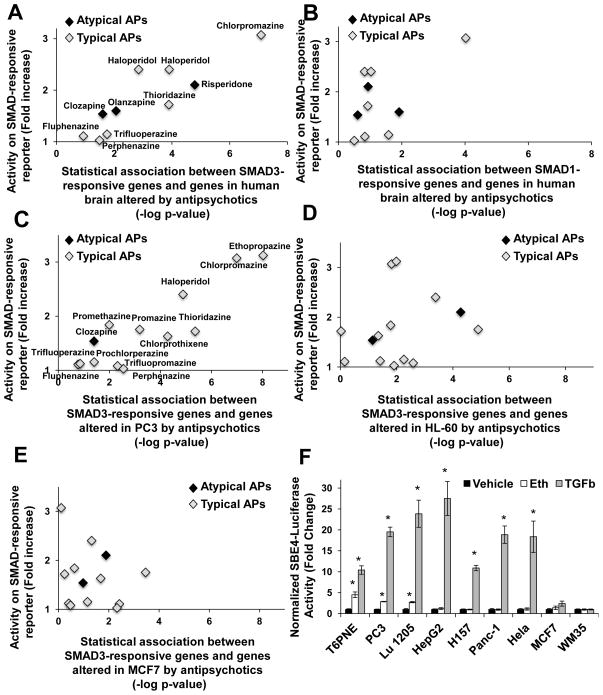
Figure 5. Antipsychotics modulated SMAD3-responsive genes in brains from antipsychotic-treated patients and a subset of cell lines.
(A,B) SAR data from the SBE4-luc SMAD reporter in T6PNE (Figure 2C) correlated with the statistical association between genes altered in human brains of antipsychotic treated patients with schizophrenia and genes containing SMAD3 (A) or SMAD1 (B) binding sites in their promoters. Data from Mudge et al.11 are in black and data from Narayan et al.12 are in green. For A, correlation coefficient=0.89 (p=0.0013) and for B, correlation coefficient=0.42 (p=0.24). (C–E) The same analysis used in A and B was performed on data from the Broad Connectivity Map (CMAP 2.0) compound database48. The SBE4 reporter SAR (Figure 5C) was correlated with SMAD3 responsiveness in PC3 cells (correlation coefficient=.88, p=0.00036)(C), but not MCF7 cells (correlation coefficient=−0.10, p=0.75) (D) or HL-60 cells (correlation coefficient=.22, p=0.47)(E). (F) Ethopropazine increased activity on the SMAD2/3 responsive SBE4-luc reporter in PC3, LU1205, and T6PNE, but not MCF7, WM35, HepG2, H157, Panc-1, and Hela cells. (n≥3), Error bars are SEM. * indicates p<0.05 relative to DMSO control.
Antipsychotics activate the TGFβ pathway in only a subset of cell lines
Since antipsychotics and TGFβ appear to act through distinct pathways that converge on SMAD3, we speculated that there might be cell lines in which antipsychotics and TGFβ differ in their ability to activate SMAD3, due to differential expression of proteins that act in the distinct pathways. If such cell lines could be found, they would be of great value in identifying pathway-specific components, facilitating the design of novel, non-diabetogenic antipsychotics. Thus, we examined published data on patterns of gene expression in cell lines treated with antipsychotics in vitro.
The Broad Connectivity Map (CMAP 2.0) is a database consisting of gene expression data from three cultured human cells treated with bioactive small molecules, including 13 antipsychotics48. Using this dataset, we performed the same Nextbio analysis used previously for the patient samples, calculating the significance of the association between genes altered by treatment with a specific antipsychotic and genes with SMAD3 binding sites in their promoters (a measure of whether antipsychotics are modulating SMAD3-responsive genes) and determining its correlation to the in vitro SMAD reporter SAR (Fig. 5C–E). No correlation was seen for the leukemic cell line HL-60 (Fig. 5D, correlation coefficient=−0.19 p=0.53) or breast cancer line MCF7 (Fig 5E, correlation coefficient=−0.16, p=0.60), but a significant correlation was seen for the prostate cancer line PC3 (Fig. 5C, correlation coefficient=0.90, p=0.00028).
The statistical analysis predicted that in PC3, but not MCF7 or HL-60 cells, antipsychotics should activate the SBE4 SMAD reporter. This proved true, ethopropazine activated the SMAD-responsive SBE4 reporter in PC3 but not in MCF7 cells (Fig. 5F). However, TGFβ exhibited the same pattern, suggesting that the defect in MCF7 was in a pathway leading to SMAD3 activation shared by antipsychotics and TGFβ. MCF7 has been reported to be unresponsive to TGFβ due to an effect of Notch449.
To determine whether cell lines with defects in a pathway specific to antipsychotic-mediated SMAD3 activation existed, we studied a panel of 7 other cell lines. Similar to T6PNE and PC3, both ethopropazine and TGFβ activated the SMAD reporter in the melanoma cell line LU-1205 (Fig. 5F). Similar to MCF7, WM35 was refractory to both ethopropazine and TGFβ (Fig. 5F). However, in HepG2, H157, Panc-1, and HeLa cells, TGFβ potently activated the SMAD reporter but antipsychotics were completely inactive, suggesting that a component unique to the pathway linking antipsychotics to SMAD3 activation is absent or defective in those cell lines (Fig. 5F).
Discussion
The major finding presented here is that antipsychotics activated SMAD3, a downstream effector of TGFβ signaling, through a non-canonical pathway that involves neither the TGFβ receptor complex nor the neurotransmitter receptors that are believed to be responsible for the therapeutic effects of antipsychotics. Our data support a model in which antipsychotics and TGFβ signal to SMAD3 through independent pathways that converge downstream to activate SMAD3. Support for this comes from multiple lines of evidence. TGFβ, but not antipsychotics, activated SMAD2 and repressed SMAD3 gene expression. Cell lines exhibiting potent SMAD activation by TGFβ but that were unresponsive to antipsychotics also indicates differences in the pathways acted on by TGFβ and antipsychotics.
Previously proposed mechanisms for the metabolic effects of antipsychotics are diverse3, 5, 6, 8, 50, but many postulate that their CNS effects resulted in increased appetite and hence weight gain8, 50. However, this could not be sufficient to account for all metabolic side effects, since a direct correlation between weight gain and diabetes in antipsychotic treated patients is often not seen20, 51, 52.
The TGFβ pathway is highly associated with obesity, insulin resistance, and diabetes. Higher levels of serum TGFβ have been shown to predict the development of type II diabetes13,16, and mice with genetic deletion of SMAD3 are resistant to high fat diet induced obesity and insulin resistance17, 19. However, the effects of TGFβ signaling are complex and indicate that the level of pathway activation needs to be maintained in a tightly regulated range. It has been reported that SMAD3 both enhances and inhibits insulin gene expression15, 53. Studies with transgenic models of TGFβ inhibition in mouse β-cells suggest that TGFβ signaling is required for insulin production and proper β-cell function, but studies of TGFβ overexpression found that it led to β-cell dysfunction and hyperglycemia19, 34, 54.
In vivo relevance of SMAD activation by antipsychotics came from analysis of two independent gene expression datasets from the brains of antipsychotic-treated patients and normal controls11, 12. In both of those previous studies, antipsychotics were analyzed as a group, obscuring the effect of particular antipsychotics on the TGFβ pathway, and neither identified TGFβ signaling as being affected by antipsychotics. However, performing the analysis in light of the SAR of antipsychotic effects on the SMAD reporter produced a dramatically different picture, revealing a highly significant correlation between the effects of particular antipsychotics on SMAD3-responsive genes in patients and their effect on SMAD and insulin promoter activity in vitro.
If effects of antipsychotics on SMAD3 activation are responsible for the metabolic side effects of those drugs, one might predict that the drugs that have the greatest propensity to cause metabolic side effects would cause the greatest activation of SMAD3. In particular, first generation antipsychotics, that are often described as having a lower propensity to cause metabolic side effects55, were quite potent in their ability to activate SMAD3 in our assays. However, there are extensive inconsistencies in the literature on the relative propensity of antipsychotics to cause metabolic side effects3, 5–9, with part of the problem being that newer drugs have been more intensively studied in that regard56, 57. Also, it is well known that small perturbations of TGFβ signaling can have large physiological effects, and the TGFβ pathway has complex feedback loops34, 58, potentially confounding such a correlation, particularly with drugs that are most effective at inducing SMAD3 activation. Finally, antipsychotics are often dose-limited by side effects such as extrapyramidal symptoms and dyskinesias, which are related to dopamine D2 receptor blockade and are more common in first generation antipsychotics such as chlorpromazine and haloperidol, while our in vitro assays were done at a dose determined by activity in cell lines. The effect of non-metabolic dose-limiting side effects in patients and the lack of precise dose information for the patients used in the genomic analyses used here could significantly affect the relationship between the reported incidence of metabolic side effects for a particular drug and our determination of its propensity to activate SMAD3. However, we did observe two antipsychotics that have consistently been found to have a weak or no association with obesity and diabetes, molindone and ziprasidone20–22, were low or inactive in the insulin promoter and SMAD reporter assays. These two antipsychotics fall into a structurally distinct class, containing dihydroindolone and indolinone cores, respectively, which are not found in the phenothiazines or any of the other typical or atypical antipsychotics.
The structural specificity of antipsychotics for effects on SMAD3 activation, combined with the data on the differential responsiveness of cell lines to antipsychotics, indicates that the effects of antipsychotics are mediated through a specific target that is affected by structural features of particular antipsychotics. Based on the known binding profile of antipsychotics, we hypothesized that the direct target of antipsychotics leading to SMAD3 activation was likely to be a GPCR. Unfortunately, attempts to use screening of siRNAs to GPCRs, in combination with gene expression data from the cell lines that responded or did not respond to antipsychotics, was unsuccessful in identifying an antipsychotic target that activated SMAD3. Some intracellular proteins and processes are also targeted by antipsychotics, including calmodulin, Protein Kinase C, and clathrin-mediated endocytosis59, 60, but we found none to be involved in the signaling of antipsychotics to SMAD3.
The correlation between the propensity of particular antipsychotics to cause metabolic side effects and their ability to activate SMAD3 strongly suggests that the activity of antipsychotics on the TGFβ pathway is a significant factor in causing those side effects. The fact that the neurotransmitter receptor targets of the antipsychotics relevant to their use in treating psychosis are unrelated to their modulation of the TGFβ pathway opens the door to the possibility of developing antipsychotics that retain activity on the therapeutically beneficial neurotransmitter receptors, while not having effects on the TGFβ pathway. While such drug design efforts would be aided by the identification of the direct target of antipsychotics responsible for signaling to SMAD3, the availability of a sensitive assay for antipsychotic effects mediated by SMAD3 activation, i.e., the insulin promoter assay in T6PNE cells, allows novel antipsychotics to be rapidly screened for their propensity to activate SMAD3. This has the potential to lead to optimized antipsychotics that retain a favorable profile of activity on the neurotransmitter receptors that are responsible for their clinical benefit, while lacking effect on the target that signals to the TGFβ pathway.
Supplemental Methods
Compounds
All antipsychotics and neurotransmitter receptor antagonists were purchased from Sigma, except olanzapine (Toronto Research Chemicals). SB-505124 was purchased from Calbiochem and recombinant Activin, TGFβ1, and TGFβ2 were from Thermo Scientific. All antipsychotics were used at 10 or 20 μM, the concentration at which dose-response analysis revealed maximum activity on the human insulin promoter-eGFP transgene was evident without substantial cytotoxicity (Supplemental. Fig. 1B). SB-505124 was used at 2.5 uM. TGFβ 1, TGFβ 2, and Activin A were used at 10 ng/ml.
Transfection studies
T6PNE cells were transfected in serum free media with Lipofectamine 2000 (1.2ul per 100ul media, Invitrogen) and .5ug plasmid DNA (4RTK-luc 1, SBE4-Luc 2, CAGA-Luc 3 or SMAD3 expression plasmid4), media was changed and .6uM Tamoxifen was added at 4 hours, compound was added at 24 hours, and cells were harvested at either 48 or 72 hours with lysis buffer. For experiments using the reporter plasmids, activity was measured by Luciferase Reporter gene assay kit (Roche) and normalized to protein content determined by BCA Assay (Thermo Scientific), and divided by vehicle treated control to yield a fold change. Significance was determined with a one-tailed T-test. For the SMAD3 overexpression, cell lysates were subjected to protein purification for Western blot, or RNA purification (Qiagen) and cDNA synthesis for QPCR as described below.
siRNA studies
T6PNE cells were transfected with Lipofectamine RNAiMax Reagent (Invitrogen), OptiMEM, and 8 μMoles siRNA. 0.6uM Tamoxifen was added at 24 hours, compound was added at 48 hours, and then at 96 hours the cells were harvested for RNA isolation (Qiagen), cDNA synthesis, and QPCR as described below. siRNAs were from Applied Biosystems (Silencer Select s8402 for SMAD3 and s8398 for SMAD2). Scrambled siRNA was a mix of 48 non targeting siRNAs.
RT PCR analysis
Following RNA purification (Qiagen) and cDNA synthesis (Quanta Master Mix, MJ Research PTC-200), quantitative PCR was performed on cDNA corresponding to 100 ng of RNA using the Opticon Real-Time System (Bio-Rad) and BioPioneer QPCR Super Mix. All mRNA values were normalized to 18S rRNA and are expressed as fold change from vehicle treated control. Significance was determined with a one-tailed T-test.
Primers
Insulin
FWD: CTACCTAGTGTGCGGGGAAC
REV: GCTGGTAGAGGGAGCAGATG1
Kip2
FW:GGCGATCAAGAAGCTGTCC
REV: GGGCTCTTTGGGCTCTAAAT
18S rRNA
FW:GATATGCTCATGTGGTGTTG
REV:AATCTTCTTCAGTCGCTCCA
SMAD3
FW: GAGAAATGGTGCGAGAAGGCGGTC
REV: TTCCGATGGGACACCTGCAACC
SMAD2
FW: CCTCCAATCGCCCATTCCCCTCT
REV: CAAAGGCAGCAAGCCACGCTAG
Immunoblot
Whole-cell extracts were prepared by incubation in Cell Extraction Buffer (Invitrogen) supplemented with protease/phosphatase inhibitor cocktail (Thermo Scientific). Protein (20μg) was separated on 4 20% Longlife gels (Invitrogen) and transferred to Invitrolon Membrane (Invitrogen). After overnight blocking in PBS-Tween (PBST) with 3% milk, membrane was incubated with pSMAD3 (#9502S), p-SMAD2(#310S), total SMAD3/SMAD2 (#3102) antibody (Cell Signaling), or Loading Control HSP-90 (#610410, BD), followed by secondary antibody conjugated to horseradish peroxidase (Amersham/GE, Buckinghamshire, UK). Signal was revealed by ECL (Amersham/GE) and blot Intensity quantified using Image J as described in Gassmann et al.5. All protein levels were normalized to HSP90 levels and expressed as fold change from vehicle treated control. Significance was determined with a one-tailed t-test.
Nextbio analysis
The gene expression dataset from Mudge et al. 6, was downloaded from GEO (GSE12297), and a list of differentially expressed genes was created by calculating the fold change of the average gene expression value for all 14 treated subjects, or that for those taking the 4 most potent antipsychotics, from the average value for that gene in the 6 control subjects who were not treated with antipsychotics. The two lists were then filtered with a +/−1.8X fold change cutoff, and 5 random gene lists were created of the same size as each of the two experimental gene lists. This was done using a function in the statistical software R which samples the given number of genes from the NCGR Human RefSeq 7 08 platform used in the original experiment and assigns a random fold change between −5 and +5 to each. All gene lists (random and experimental) were then loaded into Nextbio, allowing its “Running-Fisher” algorithm 7 to generate the p-values shown in Table 2 for the significance of the association between our uploaded gene lists and the Nextbio gene lists (SMAD3 Binding Sites, SMAD1 Binding Sites, Neurotransmitter Receptor Activity, Golgi Apparatus, Vesicle-Mediated Transport). The Golgi apparatus and vesicle-mediated transport gene lists in Nextbio were derived from the GO database8 and the SMAD1/3 gene lists came from Xie et al.9, who performed applied a comparative genomic approach, applying data from the TRANSFAC database10 to identify genes containing particular transcription factor binding sites in their promoters, including lists of genes with SMAD1/SMAD3 sites. In all instances, the p-values used from Nextbio were the minimum p-value across that calculated for the up-regulated genes, down-regulated genes and both lists considered together. The p-values listed for the random gene lists in Table 2 are the average of the p-values across the 5 random gene lists used for each experimental list.
The data plotted on the x-axis in figure 5 were produced by calculating the fold change of the gene expression data from each of the 14 patient samples considered individually from the average of the 6 control samples, creating 14 lists of differentially expressed genes. These lists were then filtered with a +/−1.8X fold cutoff and 5 random gene lists of the same size as each of the 14 experimental gene lists were created. All gene lists (random and experimental) were then analyzed by the Nextbio algorithm, generating a p-value for each individual gene list. The p-value was -log transformed to normally distribute and equalize the variance between the treatment groups and then normalized to the average of the log transformed p-values for the 5 random gene list associated with that individual. This value was then averaged across each group of individuals taking the same antipsychotic.
For Narayan et al.11, data were downloaded from GEO (GSE21138), and analyzed with the RMA algorithm (quantile normalization) to generate a log2 expression value for each array. The hybridization quality controls for each array were assessed by the percent present calls and GAPDH 3′/5′ ratios. Hierarchical clustering methods were next performed based on the gene expression patterns for those patients taking the same antipsychotic and the results, combined with the QC results, led to the removal of two outliers and a batch effect adjustment, which is consistent with the data processing conducted in Narayan et al.11. Of the remaining 57 samples, only the short duration of illness samples were used because the low number of differentially expressed genes among the individuals with longer durations of illness precluded analysis. This left 8 patients treated with antipsychotics and 8 age and sex matched healthy, untreated controls. These were then analyzed identically to the data from Mudge et al. except fold change was calculated for each patient compared to its matched control, not the average across all controls.
Supplementary Material
Acknowledgments
We thank Drs. Joaquim Teixeira and Mark Mercola for the SMAD reporter constructs and for helpful discussion. Drs. Susanne Heynen, Eduard Sergienko, and Fu-Yue Zeng at the SBMRI Conrad Prebys Center for Chemical Genomics provided the kinase inhibitor library and expertise in high-throughput screening. Drs. Alexey Eroshkin and Xiayu Huang at the SBMRI Bioinformatics Core helped with processing gene expression data from GEO. Dr. Tony Pinkerton was helpful in providing insights into structural aspects of the antipsychotics. Dr. Elizabeth Thomas of the Scripps Research Institute provided access to additional data from her published microarray experiments.
Funding: This work was funded by the Sanford Children’s Health Research Center and the UCSD Biomedical Sciences Genetics Training Grant.
References
- 1.Alexander GCGS, Mascola A, Moloney RM, Stafford RS. Increasing off-label use of antipsychotic medications in the United States, 1995–2008. Pharmacoepidemiol Drug Saf. 2011;20(2):177–184. doi: 10.1002/pds.2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ostbye T, Curtis LH, Masselink LE, Hutchison S, Wright A, Dans PE, et al. Atypical antipsychotic drugs and diabetes mellitus in a large outpatient population: a retrospective cohort study. Pharmacoepidemiol Drug Saf. 2005;14 (6):407–415. doi: 10.1002/pds.1016. [DOI] [PubMed] [Google Scholar]
- 3.Citrome LL, Holt RI, Zachry WM, Clewell JD, Orth PA, Karagianis JL, et al. Risk of treatment-emergent diabetes mellitus in patients receiving antipsychotics. Ann Pharmacother. 2007;41(10):1593–1603. doi: 10.1345/aph.1K141. [DOI] [PubMed] [Google Scholar]
- 4.Nasrallah H. Atypical antipsychotic-induced metabolic side effects: insights from receptor-binding profiles. Mol Psychiatry. 2008;13(1):27–35. doi: 10.1038/sj.mp.4002066. [DOI] [PubMed] [Google Scholar]
- 5.Tonnard-Neumann E. Phenothiazines and diabetes in hospitalized women. Am J Pychiatry. 1968;124:978–982. doi: 10.1176/ajp.124.7.978. [DOI] [PubMed] [Google Scholar]
- 6.Dixon L, Weiden P, Delahanty J, Goldberg R, Postrado L, Lucksted A, et al. Prevalence and correlates of diabetes in national schizophrenia samples. Schizophr Bull. 2000;26(4):903–912. doi: 10.1093/oxfordjournals.schbul.a033504. [DOI] [PubMed] [Google Scholar]
- 7.Baptista T, de Baptista EA, Lalonde J, Plamondon J, Kin NM, Beaulieu S, et al. Comparative effects of the antipsychotics sulpiride and risperidone in female rats on energy balance, body composition, fat morphology and macronutrient selection. Prog Neuropsychopharmacol Biol Psychiatry. 2004;28(8):1305–1311. doi: 10.1016/j.pnpbp.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 8.Starrenburg FC, Bogers JP. How can antipsychotics cause Diabetes Mellitus? Insights based on receptor-binding profiles, humoral factors and transporter proteins. Eur Psychiatry. 2009;24(3):164–170. doi: 10.1016/j.eurpsy.2009.01.001. [DOI] [PubMed] [Google Scholar]
- 9.Baptista TDMS, Beaulieu S, Bermudez A, Martinez M. The metabolic syndrome during atypical antipsychotic drug treatment: mechanisms and management. Metab Syndr Relat Disord. 2004;2(4):290–307. doi: 10.1089/met.2004.2.290. [DOI] [PubMed] [Google Scholar]
- 10.Kiselyuk A, Farber-Katz S, Cohen T, Lee SH, Geron I, Azimi B, et al. Phenothiazine neuroleptics signal to the human insulin promoter as revealed by a novel high-throughput screen. J Biomol Screen. 2010;15(6):663–670. doi: 10.1177/1087057110372257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mudge J, Miller NA, Khrebtukova I, Lindquist IE, May GD, Huntley JJ, et al. Genomic convergence analysis of schizophrenia: mRNA sequencing reveals altered synaptic vesicular transport in post-mortem cerebellum. PLoS One. 2008;3(11):e3625. doi: 10.1371/journal.pone.0003625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Narayan S, Tang B, Head SR, Gilmartin TJ, Sutcliffe JG, Dean B, et al. Molecular profiles of schizophrenia in the CNS at different stages of illness. Brain Res. 2008;1239:235–248. doi: 10.1016/j.brainres.2008.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Azar STSI, Zantout MS, Major S. Alterations in plasma transforming growth factor beta in normoalbuminuric type 1 and type 2 diabetic patients. J Clin Endocrinol Metab. 2000;85(12):4680–4682. doi: 10.1210/jcem.85.12.7073. [DOI] [PubMed] [Google Scholar]
- 14.Smart NGAA, Gu X, Harmon EB, Topper JN, MacDonald RJ, Kim SK. Conditional expression of Smad7 in pancreatic beta cells disrupts TGF-beta signaling and induces reversible diabetes mellitus. PLoS Biol. 2006;4(2):e39. doi: 10.1371/journal.pbio.0040039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Watanabe RSZ, Tsuda K, Yamada Y. Insulin gene is a target in activin receptor-like kinase 7 signaling pathway in pancreatic beta-cells. Biochem Biophys Res Commun. 2008;377(3):867–872. doi: 10.1016/j.bbrc.2008.10.074. [DOI] [PubMed] [Google Scholar]
- 16.Herder C, Zierer A, Koenig W, Roden M, Meisinger C, Thorand B. Transforming growth factor-beta1 and incident type 2 diabetes: results from the MONICA/KORA case-cohort study, 1984–2002. Diabetes Care. 2009;32 (10):1921–1923. doi: 10.2337/dc09-0476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tan CK, Leuenberger N, Tan MJ, Yan YW, Chen Y, Kambadur R, et al. Smad3 deficiency in mice protects against insulin resistance and obesity induced by a high-fat diet. Diabetes. 2011;60(2):464–476. doi: 10.2337/db10-0801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin YNK, Ito Y, Kikuchi S, Tamakoshi A, Yagyu K, Watanabe Y, Inaba Y, Tajima Kazuo Jacc Study Group. Variations in serum transforming growth factor-beta1 levels with gender, age and lifestyle factors of healthy Japanese adults. Dis Markers. 2009;27(1):23–28. doi: 10.3233/DMA-2009-0643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin HM, Lee JH, Yadav H, Kamaraju AK, Liu E, Zhigang D, et al. Transforming growth factor-beta/Smad3 signaling regulates insulin gene transcription and pancreatic islet beta-cell function. J Biol Chem. 2009;284(18):12246–12257. doi: 10.1074/jbc.M805379200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Newcomer J. Second-generation (atypical) antipsychotics and metabolic effects: a comprehensive literature review. CNS Drugs. 2005;19 (Suppl 1):1–93. doi: 10.2165/00023210-200519001-00001. [DOI] [PubMed] [Google Scholar]
- 21.Allison DBMJ, Heo M, Chandler LP, Cappelleri JC, Infante MC, Weiden PJ. Antipsychotic-induced weight gain: a comprehensive research synthesis. Am J Psychiatry. 1999;156(11):1686–1696. doi: 10.1176/ajp.156.11.1686. [DOI] [PubMed] [Google Scholar]
- 22.Stimmel GLGM, Lee V. Ziprasidone: an atypical antipsychotic drug for the treatment of schizophrenia. Clin Ther. 2002;24(1):21–37. doi: 10.1016/s0149-2918(02)85003-2. [DOI] [PubMed] [Google Scholar]
- 23.Correll C. From receptor pharmacology to improved outcomes: individualising the selection, dosing, and switching of antipsychotics. Eur Psychiatry. 2010;25 (Suppl 2):S12–21. doi: 10.1016/S0924-9338(10)71701-6. [DOI] [PubMed] [Google Scholar]
- 24.Horacek JB-VV, Kopecek M, Palenicek T, Dockery C, Mohr P, Höschl C. Mechanism of action of atypical antipsychotic drugs and the neurobiology of schizophrenia. CNS Drugs. 2006;20(5):389–409. doi: 10.2165/00023210-200620050-00004. [DOI] [PubMed] [Google Scholar]
- 25.Satin LS, Kinard TA. Neurotransmitters and their receptors in the islets of Langerhans of the pancreas: what messages do acetylcholine, glutamate, and GABA transmit? Endocrine. 1998;8(3):213–223. doi: 10.1385/ENDO:8:3:213. [DOI] [PubMed] [Google Scholar]
- 26.Kim H, Toyofuku Y, Lynn FC, Chak E, Uchida T, Mizukami H, et al. Serotonin regulates pancreatic beta cell mass during pregnancy. Nat Med. 2010;16(7):804–808. doi: 10.1038/nm.2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Farkas T, Hoyer-Hansen M, Jaattela M. Identification of novel autophagy regulators by a luciferase-based assay for the kinetics of autophagic flux. Autophagy. 2009;5(7):1018–1025. doi: 10.4161/auto.5.7.9443. [DOI] [PubMed] [Google Scholar]
- 28.DaCosta Byfield S, Major C, Laping NJ, Roberts AB. SB-505124 is a selective inhibitor of transforming growth factor-beta type I receptors ALK4, ALK5, and ALK7. Mol Pharmacol. 2004;65(3):744–752. doi: 10.1124/mol.65.3.744. [DOI] [PubMed] [Google Scholar]
- 29.Moustakas A, Heldin CH. The regulation of TGFbeta signal transduction. Development. 2009;136(22):3699–3714. doi: 10.1242/dev.030338. [DOI] [PubMed] [Google Scholar]
- 30.Hoover LL, Kubalak SW. Holding their own: the noncanonical roles of Smad proteins. Sci Signal. 2008;1(46): pe48. doi: 10.1126/scisignal.146pe48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zawel L, Dai JL, Buckhaults P, Zhou S, Kinzler KW, Vogelstein B, et al. Human Smad3 and Smad4 are sequence-specific transcription activators. Mol Cell. 1998;1(4):611–617. doi: 10.1016/s1097-2765(00)80061-1. [DOI] [PubMed] [Google Scholar]
- 32.Dennler S, Itoh S, Vivien D, ten Dijke P, Huet S, Gauthier JM. Direct binding of Smad3 and Smad4 to critical TGF beta-inducible elements in the promoter of human plasminogen activator inhibitor-type 1 gene. EMBO J. 1998;17(11):3091–3100. doi: 10.1093/emboj/17.11.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.ten Dijke PHC. New insights into TGF-beta-Smad signalling. Trends Biochem Sci. 2004;29(5):265–273. doi: 10.1016/j.tibs.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 34.Brown ML, Schneyer AL. Emerging roles for the TGFbeta family in pancreatic beta-cell homeostasis. Trends Endocrinol Metab. 2010;21(7):441–448. doi: 10.1016/j.tem.2010.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sayo Y, Hosokawa H, Imachi H, Murao K, Sato M, Wong NC, et al. Transforming growth factor beta induction of insulin gene expression is mediated by pancreatic and duodenal homeobox gene-1 in rat insulinoma cells. Eur J Biochem. 2000;267(4):971–978. doi: 10.1046/j.1432-1327.2000.01080.x. [DOI] [PubMed] [Google Scholar]
- 36.Kirwan RPLM, Murphy M, Clark AF, O’Brien CJ. Transforming growth factor-beta-regulated gene transcription and protein expression in human GFAP-negative lamina cribrosa cells. Glia. 2005;52(4):309–324. doi: 10.1002/glia.20247. [DOI] [PubMed] [Google Scholar]
- 37.Poncelet AC, Schnaper HW, Tan R, Liu Y, Runyan CE. Cell phenotype-specific down-regulation of Smad3 involves decreased gene activation as well as protein degradation. J Biol Chem. 2007;282(21):15534–15540. doi: 10.1074/jbc.M701991200. [DOI] [PubMed] [Google Scholar]
- 38.Watanabe H, Saito H, Nishimura H, Ueda J, Evers BM. Activation of phosphatidylinositol-3 kinase regulates pancreatic duodenal homeobox-1 in duct cells during pancreatic regeneration. Pancreas. 2008;36(2):153–159. doi: 10.1097/MPA.0b013e318157753e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Allen RRQL, Higgins PJ. Upstream stimulatory factor regulates E box-dependent PAI-1 transcription in human epidermal keratinocytes. J Cell Physiol. 2005;203(1):156–165. doi: 10.1002/jcp.20211. [DOI] [PubMed] [Google Scholar]
- 40.Liu DBB, Derynck R. TGF-beta inhibits muscle differentiation through functional repression of myogenic transcription factors by Smad3. Genes Dev. 2001;15(22):2950–2966. doi: 10.1101/gad.925901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu DKJ, Derynck R. 2004 TGF-beta-activated Smad3 represses MEF2-dependent transcription in myogenic differentiation. EMBO J. 2004;23(7):1557–1566. doi: 10.1038/sj.emboj.7600179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Itkin-Ansari P, Marcora E, Geron I, Tyrberg B, Demeterco C, Hao E, et al. NeuroD1 in the endocrine pancreas: Localization and dual function as an activator and repressor. Dev Dyn. 2005;233(3):946–953. doi: 10.1002/dvdy.20443. [DOI] [PubMed] [Google Scholar]
- 43.Weintraub H, Davis R, Lockshon D, Lassar A. MyoD binds cooperatively to two sites in a target enhancer sequence: occupancy of two sites is required for activation. Proc Natl Acad Sci U S A. 1990;87(15):5623–5627. doi: 10.1073/pnas.87.15.5623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kupershmidt I, Su QJ, Grewal A, Sundaresh S, Halperin I, Flynn J, et al. Ontology-based meta-analysis of global collections of high-throughput public data. PLoS One. 2010;5(9) doi: 10.1371/journal.pone.0013066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Matys V, Fricke E, Geffers R, Gossling E, Haubrock M, Hehl R, et al. TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 2003;31 (1):374–378. doi: 10.1093/nar/gkg108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xie X, Lu J, Kulbokas EJ, Golub TR, Mootha V, Lindblad-Toh K, et al. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature. 2005;434(7031):338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen D, Zhao M, Harris SE, Mi Z. Signal transduction and biological functions of bone morphogenetic proteins. Front Biosci. 2004;9:349–358. doi: 10.2741/1090. [DOI] [PubMed] [Google Scholar]
- 48.Lamb JCE, Peck D, Modell JW, Blat IC, Wrobel MJ, Lerner J, Brunet JP, Subramanian A, Ross KN, Reich M, Hieronymus H, Wei G, Armstrong SA, Haggarty SJ, Clemons PA, Wei R, Carr SA, Lander ES, Golub TR. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313(5795):1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 49.Sun YLW, Kato K, Bianco C, Kenney N, Strizzi L, Raafat D, Hirota M, Khan NI, Bargo S, Jones B, Salomon D, Callahan R. Notch4 intracellular domain binding to Smad3 and inhibition of the TGF-beta signaling. Oncogene. 2005;24 (34):5365–5374. doi: 10.1038/sj.onc.1208528. [DOI] [PubMed] [Google Scholar]
- 50.Coccurello R, Moles A. A murine model of atypical antipsychotic-induced weight gain and metabolic dysregulation. Curr Protoc Neurosci. 2010;Chapter 9(Unit9 33) doi: 10.1002/0471142301.ns0933s52. [DOI] [PubMed] [Google Scholar]
- 51.Lambert MTCL, Sampson N, Duffy SA. New-onset type-2 diabetes associated with atypical antipsychotic medications. Prog Neuropsychopharmacol Biol Psychiatry. 2006;30(5):919–923. doi: 10.1016/j.pnpbp.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 52.Stahl SMML, Meyer JM. Which comes first: atypical antipsychotic treatment or cardiometabolic risk? Acta Psychiatr Scand. 2009;119(3):171–179. doi: 10.1111/j.1600-0447.2008.01334.x. [DOI] [PubMed] [Google Scholar]
- 53.Sayo YHH, Imachi H, Murao K, Sato M, Wong NC, Ishida T, Takahara J. Transforming growth factor beta induction of insulin gene expression is mediated by pancreatic and duodenal homeobox gene-1 in rat insulinoma cells. Eur J Biochem. 2000;267(4):971–978. doi: 10.1046/j.1432-1327.2000.01080.x. [DOI] [PubMed] [Google Scholar]
- 54.Smart NG, Apelqvist AA, Gu X, Harmon EB, Topper JN, MacDonald RJ, et al. Conditional expression of Smad7 in pancreatic beta cells disrupts TGF-beta signaling and induces reversible diabetes mellitus. PLoS Biol. 2006;4(2):e39. doi: 10.1371/journal.pbio.0040039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Smith M, Hopkins D, Peveler RC, Holt RI, Woodward M, Ismail K. First- v. second-generation antipsychotics and risk for diabetes in schizophrenia: systematic review and meta-analysis. Br J Psychiatry. 2008;192(6):406–411. doi: 10.1192/bjp.bp.107.037184. [DOI] [PubMed] [Google Scholar]
- 56.Baker RA, Pikalov A, Tran QV, Kremenets T, Arani RB, Doraiswamy PM. Atypical Antipsychotic Drugs and Diabetes Mellitus in the US Food and Drug Administration Adverse Event Database: A Systematic Bayesian Signal Detection Analysis. Psychopharmacol Bull. 2009;42(1):11–31. [PubMed] [Google Scholar]
- 57.Kohen D. Diabetes mellitus and schizophrenia: historical perspective. Br J Psychiatry Suppl. 2004;47:S64–66. doi: 10.1192/bjp.184.47.s64. [DOI] [PubMed] [Google Scholar]
- 58.Sekine N, Yamashita N, Kojima I, Miyazaki J, Ogata E. Bimodal effect of transforming growth factor-beta on insulin secretion in MIN6 cells. Diabetes Res Clin Pract. 1994;26(1):7–14. doi: 10.1016/0168-8227(94)90133-3. [DOI] [PubMed] [Google Scholar]
- 59.Feldkamp MD, O’Donnell SE, Yu L, Shea MA. Allosteric effects of the antipsychotic drug trifluoperazine on the energetics of calcium binding by calmodulin. Proteins. 2010;78(10):2265–2282. doi: 10.1002/prot.22739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen CL, Hou WH, Liu IH, Hsiao G, Huang SS, Huang JS. Inhibitors of clathrin-dependent endocytosis enhance TGFbeta signaling and responses. J Cell Sci. 2009;122(Pt 11):1863–1871. doi: 10.1242/jcs.038729. [DOI] [PMC free article] [PubMed] [Google Scholar]
Supplemental References
- 1.Weintraub H, Davis R, Lockshon D, Lassar A. MyoD binds cooperatively to two sites in a target enhancer sequence: occupancy of two sites is required for activation. Proc Natl Acad Sci U S A. 1990;87(15):5623–5627. doi: 10.1073/pnas.87.15.5623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zawel L, Dai JL, Buckhaults P, Zhou S, Kinzler KW, Vogelstein B, et al. Human Smad3 and Smad4 are sequence-specific transcription activators. Mol Cell. 1998;1(4):611–617. doi: 10.1016/s1097-2765(00)80061-1. [DOI] [PubMed] [Google Scholar]
- 3.Dennler S, Itoh S, Vivien D, ten Dijke P, Huet S, Gauthier JM. Direct binding of Smad3 and Smad4 to critical TGF beta-inducible elements in the promoter of human plasminogen activator inhibitor-type 1 gene. EMBO J. 1998;17(11):3091–3100. doi: 10.1093/emboj/17.11.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Matsuura IDN, Wang G, He D, Long J, Liu F. Cyclin-dependent kinases regulate the antiproliferative function of Smads. Nature. 2004;430(6996):226–231. doi: 10.1038/nature02650. [DOI] [PubMed] [Google Scholar]
- 5.Gassmann M, Grenacher B, Rohde B, Vogel J. Quantifying western blots: pitfalls of densitometry. Electrophoresis. 2009;30(11):1845–1855. doi: 10.1002/elps.200800720. [DOI] [PubMed] [Google Scholar]
- 6.Mudge J, Miller NA, Khrebtukova I, Lindquist IE, May GD, Huntley JJ, et al. Genomic convergence analysis of schizophrenia: mRNA sequencing reveals altered synaptic vesicular transport in post-mortem cerebellum. PLoS One. 2008;3(11):e3625. doi: 10.1371/journal.pone.0003625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kupershmidt I, Su QJ, Grewal A, Sundaresh S, Halperin I, Flynn J, et al. Ontology-based meta-analysis of global collections of high-throughput public data. PLoS One. 2010;5(9) doi: 10.1371/journal.pone.0013066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xie X, Lu J, Kulbokas EJ, Golub TR, Mootha V, Lindblad-Toh K, et al. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature. 2005;434(7031):338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matys V, Fricke E, Geffers R, Gossling E, Haubrock M, Hehl R, et al. TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 2003;31(1):374–378. doi: 10.1093/nar/gkg108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Narayan S, Tang B, Head SR, Gilmartin TJ, Sutcliffe JG, Dean B, et al. Molecular profiles of schizophrenia in the CNS at different stages of illness. Brain Res. 2008;1239:235–248. doi: 10.1016/j.brainres.2008.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.