Abstract
The centrosome influences the shape, orientation and activity of the microtubule cytoskeleton. The pericentriolar material (PCM), determines this functionality by providing a dynamic platform for nucleating microtubules, and acts as a nexus for molecular signaling. While great strides have been made in understanding PCM activity, its diffraction-limited size and amorphous appearance under the electron microscope have limited analysis of its high-order organization. Here, we outline current knowledge of PCM architecture and assembly, emphasizing recent super-resolution imaging studies that revealed the PCM has a layered structure made of fibers and matrices conserved from flies to humans. Notably, these studies debunk the long-standing view of an amorphous PCM and provide a paradigm to dissect the supra-molecular organization of organelles in cells.
Keywords: Centrosomes, Cilia, Pericentriolar Material, Super-Resolution Microscopy, Mitosis, Cell cycle
Centrosome structure and function
The microtubule cytoskeleton is a dynamic network of filaments composed of α/β-tubulin polymers, which mediates the transport of protein complexes and segregation of genetic material through the mitotic spindle apparatus in eukaryotes. In most animal cells, the microtubule network is organized by the centrosome, a non-membrane bound organelle composed of two molecular assemblies with distinct, but integrated functions: the centriole, a 9-fold symmetric cylindrical structure encircled by microtubule blades; and the pericentriolar material (PCM), traditionally described as an amorphous, electron-dense material that surrounds the centrioles(1,2). The majority of cells have two centrosomes, each containing a mother centriole, and following duplication a daughter, whose dimensions can reach ~200 nm in diameter and typically between 200 and 400 nm in length depending on species, cell type and the cell-cycle phase. In cells with specialized functions, the size and number of centrioles can vary, reaching hundreds in number and microns in length(3). The two centrosomes, which differ in age as the centrioles, can be structurally distinguished by the presence of distal and sub-distal appendages, which confer to the older of the mother centrioles specialized microtubule anchoring capabilities(1,4,5). In most non-dividing cells, the mother centriole docks at the cell membrane and undergoes extensive structural modification to form the basal body(6), which provides a template for the formation of the microtubule-based axoneme, a structure critical for signal transduction via cilia and for cellular movement as flagella, respectively(7).
The two structural elements of centrosomes, the centrioles and the PCM, have defined and interconnected functional roles. The centrioles act as a structural scaffold to promote the organization of the PCM(8–10). Conversely, the primary functional role of the PCM is to anchor microtubules directly or through microtubule-nucleating centers (γ-tubulin ring complexes; γTuRC)(11). At the onset of mitosis, in a process termed centrosome maturation, the PCM increases in size and microtubule-nucleation capacity through an increase in the recruitment of γTuRCs from the cytosol. This leads to an increase in the nucleation of both astral and spindle microtubules, which drive spindle formation, orientation and subsequently, cytokinesis(1). The PCM also plays a role in the duplication of centrioles(12,13) and likely in cilia formation and disassembly(14,15), further underscoring the interplay between the two assemblies. In addition to its more specific roles, the PCM broadly functions as a signaling and docking station to regulate and redistribute protein assemblies through microtubule transport by motor proteins or through association with microtubule plus-ends (16,17). For example, PCM proteins have been implicated in DNA damage signaling, protein degradation, cell cycle progression and hedgehog signaling (18–23), but it remains unclear to what extent the PCM scaffold functions directly in the assembly and regulation of such complexes.
Interestingly, the centrosome can be essential, critical or dispensable for accurate division depending on the specific cell and tissue, and their reliance on oriented cell divisions. Thus eliminating the centrosome has severe, but distinct consequences for tissue organization, homeostasis and development in different species. Mice with mutations in many centrosomal proteins have dramatic developmental defects, such as microcephaly. Conversely, flies and worms do not proceed beyond early embryogenesis without centrosomes and die after a few cellular divisions(10,24–26). Interestingly, when flies survive the early embryonic stages with a parental contribution of centrosomes, they are able to develop into adults, and they eventually die only after eclosion due to lack of coordination (10). Numerical and structural centrosome aberrations are hallmarks of cells from solid tumors, and are thought to contribute to its genetic instability, cancer initiation and progression(1). Moreover, several hereditary diseases—collectively named ciliopathies—have been mapped to mutations in human genes whose products encode centrosomal proteins, with a role in cilia formation. This highlights the necessity to further comprehend how defects in centrosome organization and duplication are linked to disease states(5,26–28).
In recent years, analyses of isolated centrosomes from different cells and organisms have defined the centrosome proteome(29,30). A critical task in centrosome biology is to integrate this information into defined protein molecular complexes and understand how their structure influences activity in cells(31). Toward this end, studies by electron microscopy (EM) have provided insights into the ultrastructural organization of the centriole cartwheel(32,33) and procentrioles(34,35) or γTuRC(11). However, even with the use of electron tomography methods, which early on revealed ring-shaped proteinaceous structures embedded within PCM(36), the molecular architecture and higher-order spatial organization of the PCM has remained, until recently, murky.
Here we describe the current understanding of centrosome architecture and the molecular mechanisms that govern PCM assembly. We emphasize recent efforts to examine quantitatively the structure of the PCM using subdiffraction resolution microscopy imaging methods—three-dimensional Structured Illumination Microscopy (3DSIM), Stochastic Optical Reconstruction Microscopy (STORM)—and 3D sub-volume alignment and averaging. Recently, four independent studies examined PCM spatial organization revealing the specific contribution and organization of centrosomal proteins critical for centrosome maturation in metazoans cells. Together they showed that the PCM is composed of two major domains with distinct molecular composition and architecture(37–40). Notably, these studies establish the higher-order organization of the PCM as an evolutionary conserved property of centrosomes.
PCM architecture: historical perspective, roadblocks and recent developments
While numerous studies have revealed important structural details of the centriole and basal bodies(32–34,41), our understanding of the architecture of the PCM has remained in its infancy for over a century. The first description the PCM dates back to the origin of centrosome biology in 1900, when T. Boveri defined it as the unstructured region where microtubules appeared to originate. Decades later, convincing evidence of this initial observation was provided by Gould and Borisy, who unequivocally showed that microtubule nucleation originated from the PCM, thus defining its major functional role(42).
A central reason for this lack of progress can be attributed to the rather homogeneous electron density of the PCM under the electron microscope, which precludes detailed studies of its organization (Fig.1). Indeed, there is a striking paucity of evidence alluding to defined protein layers within the PCM. Most notable is the study of Paintrand and colleagues where averaging of EM micrographs exposed apparent features in the PCM region, which were dependent on the centrosome purification method (43). Electron microscopy studies on salt-stripped centrosomes of Drosophila and Giant Surf Clam have also revealed an insoluble protein matrix running throughout the PCM made of 12–15 nm elements(44,45). This structure is resistant to treatment with high concentrations of chaotropic salts and is sufficient to allow functional reconstitution of γTuRC–mediated microtubule nucleation. Unfortunately, the molecular identity of this matrix has remained elusive and the organization of its elements is unknown. This is a common issue in EM studies, where imaging of intact centrosomes is relatively straightforward, but labeling of specific proteins by immuno-EM suffers from antigen disruption due to sectioning and antibody penetration issues.
Figure 1. Subdiffraction fluorescence microscopy view of the PCM architecture.
a) Schematic representations of the architectural elements of interphase centrosome. The three layers of organization—centrioles, PCM fibers and matrices—are represented separately to help visualization. The PCM is organized in two major layers of proteins 1) molecular fibers composed of the elongated coiled-coil proteins pericentrin/PLP and Cep152/Asl have their C-termini near the centriole wall and their N-termini extending towards the periphery 2) A PCM matrix composed of Cep215/Cnn, γ-tubulin and Cep192/Spd-2 molecules. See Fig. 2 for model with mitotic PCM. b) Positional mapping of pericentrin domains on interphase human centrosome. Top. Position of antibody/probe positions on human pericentrin with predicted coiled coil regions indicated. Bottom. Measurements of pericentrin diameter derived from 3DSIM micrographs of human cells stained with anti-pericentrin domain specific antibodies. The PACT domain is localized at the C-terminus with a diameter of ~ 200 nm consistent with the measurements electron micrographs. The N-termini is positioned at distance > 500 nm from the center of the centriole. c) 2D projections of averages of aligned 3D volumes of human centrosomes stained with antibodies against the C- and the N-termini regions of pericentrin. Modified from (37)
Fluorescence microscopy studies based on deconvolution microscopy have also begun challenging the notion of an amorphous PCM organization. Different from EM, fluorescence microscopy methods readily provide molecular specificity with genetically encoded fluorescent tags and co-staining with multiple antibodies, while mostly preserving antigen structure and accessibility. However, this method suffers from the very small size of the centrosome. Pioneering work from Doxsey and colleagues indeed suggested that pericentrin is organized in a lattice structure made of multiple interconnected rings (46), while Rattner and colleagues suggested the existence of a PCM tube around centrioles composed of the appendage proteins ninein and centriolin (CEP110) and pericentrin(47). Yet, despite these efforts, our perception of the PCM as an amorphous electron-dense structure has populated the textbook, and has remained the dominant view until very recently.
Subdiffraction Resolution Fluorescence Microscopy has lately emerged as a powerful tool for the investigation of the architecture of protein complexes inside cells. Three different technologies—3DSIM, STORM, Photoactivated Localization Microscopy (PALM) and Stimulated Emission Depletion (STED)—have been reliably employed to study diverse cellular structures, by taking advantage of different principles to circumvent Abbe’s diffraction limit(48–51). 3DSIM relies on modulation of the excitation light by spatial patterning, and, in its most practical implementation, provides a resolution gain of only a factor of two (125nm in the x/y plane and 250nm in z, Box1), yet has the least stringent requirement for fluorophore selection and readily accomodates multiple labeling5,60. Conversely, PALM and STORM methods identify fluorophore position with higher resolution (~10 nm in x/y and 25 nm in z plane) by gaussian fitting methods, but necessitate more stringent control of fluorophore photoactivation and density(50,51). Lastly, STED microscopy is based on the concept of stimulated depleted emission, which uses a powerful donut-shaped laser to bring fluorophores away from the excitation focal point thus reducing the effective PSF(49). STED reaches similar resolution to PALM/STORM methods, but requires a more sophisticated optical setup, and is more challenging in 3D. All these methods improve the resolution from two- to about ten-fold compared to diffraction-limited fluorescence microscopy thus allowing a quantum leap in the structural analysis of supra-molecular complexes in cells, which are in large part beyond reach of classical structural biology methods (i.e. crystallography, cryo-EM) because of their size and need for cellular extraction and purification.
Box 1. Super Resolution Fluorescence Microscopy and 3D Volume Data Analysiss.
Super-resolution Fluorescence Microscopy experiments generate remarkable images, but, more importantly, they can provide a substantial amount of quantitative positional information, all inside the cell. The ensemble distribution of molecules, and—in many cases—domain localization and molecular orientation within individual molecules can be obtained providing an unprecedented snapshot of the organization of supra-molecular complexes in cells(37,38,88,89).
Recently, powerful image processing and analysis tools inspired by cryotomographic analyses have been implemented for the first time in super-resolution microscopy. In essence, this method aligns extracted 3D sub-volumes in real space by translating, tilting and in plane rotating the volumes to a reference by crosscorrelation of voxel intensities. The process is rendered unbiased by using an average of the aligned volume as a reference for further rounds of refinement by iterative alignment. The considerable advantage of this method is that it provides excellent quantitative metrics of the spatial distribution of centrosomal components with error in the range of a few nanometers rather than merely anecdotal examples(37). This in turn favors a precise and unambiguous protein position assignment. The sub-volume image stacks can be analyzed individually, to obtain measurement variance and structural heterogeneity, as well as through volume averages. Why the latter? The derivation of an average structure enhances the structural similarities, while de-emphasizing individual structural and/or staining heterogeneity. This type of analysis leads to the discovery of shared structural features, and also, via 3D clustering methods, could provide insights into distinct structural states of the molecular assemblies(37).
While 3D alignment analysis substantially improves measurement precision, the accuracy might be further improved by reducing the distance between the antigen and the fluorophore. Toward this end, an alternative to antibody primary-secondary complex (~15–20 nm) are directly labeled anti-tag nanobodies, which are considerably smaller probes (2–5 nm) and are commercially available (90).
An inherent limitation of all single objective microscopic imaging methods is that the derived 3D volumes have anisotropic resolution. That is, the resolution in the x and y planes is ~2.5-fold better than along the z-axis. This introduces a distortion in the observed volumes and consequently reduces the accuracy of the distance measurements. A simple way around this is to focus on those objects oriented such that the measurements are in-plane (i.e. end-on view of centrosomes)(38–40,90). A more precise and general approach is to correct for distortion of tilted volumes during the averaging by properly merging the object information such that rotated objects contribute axial resolution information largely missing in the other views. This is best done by a hybrid real space/Fourier space approach that can optimally make use of all information available from each 3D sub-volume in an unbiased manner(37).
The architecture of the PCM has been a particularly challenging problem because of its biochemical complexity (hundreds of components) including numerous large coiled-coil containing proteins—the majority of which are completely uncharacterized structurally—and its dimensions at the boundary of the diffraction limit. Just recently, several studies have started to exploit the resolving power of subdiffraction resolution microscopy to examine centrosome molecular assembly (53).
The mechanism of Pericentriolar Material assembly: scaffolding fibers and branching matrix
In recent systematic super-resolution microscopy studies, several groups demonstrated the existence of a PCM layer in the proximity of the centriole wall with a stunning, highly ordered, protein architecture in human and Drosophila cells(37–40). The PCM proximal layer—named for its vicinity to the centriole wall—is readily visible on interphase centrosomes, and is composed of several proteins involved in centrosome maturation (Table 1). These components display distinctive distributions: some are organized as apparent molecular fibers or toroids extending away from the centriole wall (pericentrin/PLP, Cep152/Asl), while others are organized in a matrix (Cep192/Spd-2, Cep215/Cnn and γ-tubulin, Fig.1).
Table 1.
Proteins implicated in centrosome maturation
| Protein name | Function in centrosome maturation | Ref. |
|---|---|---|
| TUB1G (Hu) γ-tubulin (Dm, Ce) |
A highly conserved protein required across species for the nucleation of microtubules. As part of the γTuRC complex forms a template for microtubule formation at the minus ends. | (96) (97) (98) (99) (11) |
| GCP71WD, Nedd1 (Hu) Grip71WD (Dm) |
Involved in targeting γTuRC to the spindle and to the centrosome. It copurifies in a complex with γTuRC, but also appears to have functions beyond a γTuRC targeting factor. | (100) (57) (101) (102) |
| Cep215,CDK5RAP2 (Hu) Cnn, Centrosomin (Dm) Spd5 (Ce) |
Essential protein for PCM maturation; it is the main candidate as component of the PCM matrix that provides direct binding to the majority of the γTuRCs in the PCM. | (103) (61) (95) (104) |
| Cep192 (Hu) Spd-2 (Dm) Spd2 (Ce) |
It is a key factor involved in both centriole duplication and PCM maturation in C. Elegans, flies and human cells. It has two defined populations, one at the centriole wall and one in the PCM. | (105) (65) (66) (58) |
| Pericentrin/kendrin(Hu) Pericentrin-like protein, PLP (Dm) |
Large coiled-coil protein integral to the PCM is involved in centrosome maturation across metazoan. The c-terminal region contains the PACT domain, a conserved centrosomal targeting region. | (106) (63) (93) (14) |
| Cep152 (Hu) Asterless (Dm) |
Discovered in a mutant screen for male fertility in flies, it is primarily involved in centriole duplication by defining the site of daughter centriole formation through Plk4 kinase interaction. Asl is thought to have a role in PCM stabilization/maintenance at the centriole. | (107) (68) (94) (85) (83) (84) |
| Plk1 (Hu) Polo kinase (Dm) |
A conserved serine/threonine kinase involved in several aspects of cell division. It has been shown to phoshorylate pericentrin directly to control centrosome maturation. | (108) (57) (59) (109) |
| Aurora A (Hu, Dm) Air-1 (Ce) |
Serine threonine kinase implicated in several steps of cell division including centrosome maturation. Aurora-A is required for Cnn recruitment and it phosphorylates TACC, which form a complex with Msps to stabilize mts at the centrosome. | (71) (79) (78) (110) (77) |
| CPAP (Hu) Sas-4 (Dm) |
Conserved protein involved in centriole duplication and elongation. It might be recruiting PCM proteins to the centriole during maturation as part of a large multi-protein complex. | (9) (81) (111) (112) (113) |
| Cep97 (Hu) Cep135 (Dm) CP110 (Hu, Dm) Rcd-5/6 (Dm) Myb (Hu, Dm) Calmodulin (Hu, Dm) Map205 (Dm),PP2A(Dm) |
This list contains several other proteins implicated in centrosome maturation. Since their functional role and conservation remains unclear at this stage they will not be discussed in this review. |
(60) |
A striking example of PCM molecular fibers is exemplified by pericentrin/PLP (Fig.1). Positional mapping using probes against different domains followed by 3D sub-volume alignment and averaging revealed toroids of varying diameters(37,38)Fig.1b,c, Box1). Targeting the N-terminal region of pericentrin/PLP yielded larger diameters than the C-terminal regions and probes targeting the middle region produced intermediate results. This suggested that pericentrin/PLP adopts an extended conformation and that the molecules are anchored at the centriole wall region through their C-terminal PACT domain, which had been shown previously to be sufficient for centrosomal localization(54), while the N-terminal projects outwards the periphery. Pericentrin/PLP molecules extend a remarkable distance from the centriole wall (>300 nm in human cells Fig.1b,c)(37–40). Importantly, STORM imaging shows multiple PLP molecules forming clusters in the PCM region, which might follow the nine-fold symmetry of the centriole(37)(Fig.2a). These findings suggest that pericentrin/PLP forms elongated fibers in the PCM and support the notion that centriole symmetry is not confined to its perimeter, but acts as an organizing principle that extends into the PCM.
Figure 2. Nanometer scale organization of the PCM proximal layer.
a) STORM images of S2 cell stained with antibodies against PLP, the Drosophila pericentrin orthologue. (Left) End on view. (Right) Side view. Note the quasi ninefold symmetric distribution of PLP and a ~200 nm gap, which coincides with the position of the nascent daughter centriole. b) 2D projections of 3DSIM micrographs of Drosophila S2 cells stained with antibodies against Plk4 and PLP. Note the interleaved distribution of PLP and Plk4 on interphase centrioles. The N-termini of Asl has been shown to have a similar distribution to Plk4 on interphase S2 cells and the two proteins interact directly in vitro. c) Overview of the experimental scheme used for 3D volume alignment and averaging of subdiffraction resolution images. See Box1. Modified from (37)
The coiled-coil protein Cep152/Asl has a similar orientation and distribution to pericentrin/PLP. Labeling the two termini shows that Cep152/Asl radiates outward with its C-terminus at the centriole wall and the N-terminus extending away(37,39). Subdiffraction imaging shows that PLP and Asl are mostly organized in an interleaved fashion with few areas of overlap, largely ruling out the formation of heteroligomeric structures through their coiled-coil regions(40) (Fig.2b).
Distinct from pericentrin/PLP and Cep152/Asl, molecules of Cep192/Spd-2 are distributed rather homogenously around the centriole wall; analysis with multiple antibodies against different regions of Cep192/Spd-2 showed no clear polarity, suggesting that Cep192/Spd-2 is a globular structure, organized in a tightly packed matrix. This is consistent with the lack of extensive predicted coiled-coil domains in its sequence (37–39). Cep215/Cnn and γ–Tubulin molecules also appear randomly oriented around the centriole wall and organized in a matrix which can extend to the outer region occupied by the N-termini of pericentrin/PLP(37–40). Thus in interphase centrosomes two layers of organization of the PCM are present: a proximal layer made of pericentrin/PLP and Cep152/Asl which form fibers extending up to hundreds of nanometers away from the centriole wall; and in stark contrast a matrix of interdispersed Cep215/Cnn, γ-Tubulin, and Cep192/Spd-2 molecules (Fig.1a).
In the early phases of mitosis, during centrosome maturation, the PCM proximal layer expands into a larger outer matrix to form the fully nucleating mitotic centrosome (Fig.3). While nearly the same set of proteins is present, their architecture in the outermost PCM layer changes. In particular, two components appear to adopt a different architecture. Pericentrin maintains its fiber-like organization around the mother centriole, but also expands into the PCM outer matrix in human cells to form the bulk of the PCM together with Cep215/Cnn (38,39). Cep192/Spd-2 instead goes from a tight toroidal organization around the mother centriole to a matrix (Cep192)/array (Spd-2) structure in mitosis (37,38,40). Notably, the PCM proximal layer is buried inside a large PCM matrix in humans and becomes apparent only after PCM fragmentation—upon for example depletion of augmin activity—while it remains visible in Drosophila centrosomes, likely due to the differences in PCM architecture compared to human cells(see below 55,56). 3D volume alignment and cross-correlation analysis (Box1) of the components of the outer PCM matrix shows that they largely overlap, thus suggesting the formation of a matrix of interconnecting proteins within which, specific domains of PCM components might maintain their relative spatial positioning(37,38).
Figure 3. The PCM architecture during centrosome maturation.
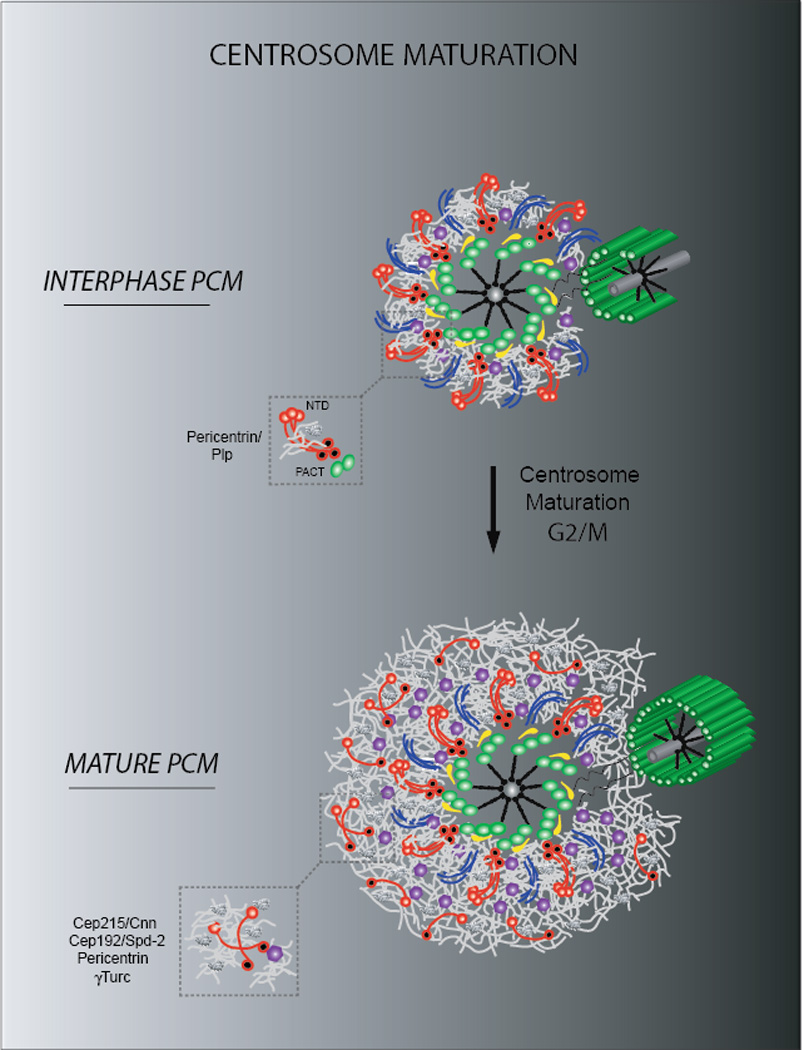
Schematic representation of a mammalian centrosome during centrosome maturation. Note the expansion of the PCM proximal layer of interphase cells during G2/M through the formation of an outer matrix of Cep215, pericentrin and Cep192 molecules. γTuRC are embedded within the PCM and promote microtubule nucleation during mitosis. In contrast, Drosophila PCM expansion does not require the pericentrin homologue PLP.
What is the functional interplay of the fibers and matrices of the proximal and outer layers of the PCM during maturation? Loss-of-function experiments suggest the existence of separate, but interdependent pathways for building the PCM(57,58). A scaffolding pathway, named after pericentrin/PLP’s elongated structures, organizes the PCM proximal layer around the centriole wall and is critical for proper 3D organization of the PCM outer layer during maturation. A second pathway is mainly involved in matrix formation, and relies on Cep192/Spd-2, Cep215/Cnn, and γ-tubulin, but does not appear to require pericentrin/PLP(14,37,38,59,60).
In interphase centrioles, pericentrin/PLP appears to be at the top of the hierarchy of the PCM assembly of the proximal layer. pericentrin/PLP is required to recruit Cep215/Cnn(14,37,38,61), but not vice versa(61,62). In turn, pericentrin is sufficient to recruit γTuRCs in the absence of Cep215(38). These activities might be determined by pericentrin/PLP directly binding to Cep215/Cnn and γ–tubulin(46,63,64). During centrosome maturation, pericentrin and Cep215/Cnn are the first to expand into a large matrix in G2, but this is not sufficient for full centrosome maturation, which requires Plk1 phosphorylation of pericentrin. Interestingly, only a subpopulation of mitotic pericentrin located close to the centriole is phosphorylated and is required for recruitment of Cep192 and γ–tubulin(59). In addition, overexpression of pericentrin during interphase is sufficient to recruit Cep215 and build a massive PCM matrix similar to mitotic centrosomes. These observations support the notion that the pericentrin layer proximal to the centriole wall has an important role in PCM expansion under regulatory control. Conversely, in Drosophila, the pericentrin homologue PLP maintains its fibrous/toroid structure around the mother centriole, but does not expand into the outer PCM matrix during centrosome maturation. This would suggest that pericentrin might have acquired during evolution a polymerizing molecular activity much like Cep215/Cnn that Drosophila PLP lacks and explains the less dramatic effect of PLP depletion compared to pericentrin in centrosome maturation in flies(14,37,38,60).
Notably, a population of Cep215/Cnn, Cep192/Spd-2 and γ–tubulin is recruited to mitotic and interphase centrosomes in the absence of pericentrin/PLP(37,38,57,58,60,62) indicating that Cep215/Cnn can self-assemble and/or have other molecular interactions independently from pericentrin (i.e. with Cep192/Spd-2) that are sufficient to form a reticular structure. This suggests the existence of a second pathway, which appears to be mainly involved in matrix formation, and relies on Cep192/Spd-2, Cep215/Cnn, and γ–tubulin. Indeed, RNAi and genetic mutants show that Cep192/Spd-2 molecules are critical for Cep215/Cnn matrix formation, thus making Cep192/Spd-2 the main candidate to drive Cep215/Cnn oligomerization(58,65–67). Surprisingly, in flies Spd-2 does not reach the further region of the PCM where Cnn forms a matrix, but it does have a reticular structure that might play a branching role in a sub-region of the PCM(37,40). Alternatively, Cep192/Spd-2 might promote PCM matrix formation through recruitment of a regulatory factor such as Plk1(40,59,67).
The existence of two separate pathways is further supported by studies in humans and flies that show that Cep192/Spd-2 and Cep152/Asl are mostly codependent for their localization, but do not require pericentrin/PLP(58,68). Altogether, these experiments suggest that pericentrin/PLP and Cep192/Spd-2 are indirectly linked in the PCM and provide more evidence for the notion that Cep192/Spd-2 and pericentrin/PLP lie in independent pathways for PCM organization through their mutual ability to interact with Cep215/Cnn.
The experimental evidence summarized above is consistent with a combination of a template and self-organization model for PCM formation(69). The proximal interphase PCM layer, which is made of pericentrin/PLP fibers around the mother centriole, provides a molecular scaffold, which recruits Cep215/Cnn. In turn, this scaffold provides a template for the mitotic outer matrix expansion. This process is driven by more accumulation of Cep215/Cnn, Cep192/Spd-2, and in human cells pericentrin.
The dynamics of PCM assembly and the role of kinases
How is PCM recruitment regulated in space and time during centrosome maturation? As mentioned above, in human cells, robust centrosome maturation requires Plk1-dependent phosphorylation of a subset of pericentrin molecules in the vicinity of the centriole wall, and the subsequent recruitment of Cep 192 and γ–tubulin(59). Consistently, Plk1/Polo is also localized in proximity to the centriole wall (40,59). Interestingly, Plk1/Polo is also required for Cnn phosphorylation, suggestive of a direct role for these kinases in Cnn regulation during PCM expansion(40,57,59,60). FRAP experiments in fly embryos show that Cnn molecules in interphase are dynamically associated to the centrosome and are first recruited near the centriole, before spreading to the centrosome periphery(70). In contrast, FRAP studies in fly embryos indicate that the centriole associated PACT domain of centriolar PLP does not exchange rapidly(14). Since pericentrin/PLP population at the centriole wall is important for maturation, it would be interesting to examine whether PLP can also be phosphorylated by Polo (like its mammalian counterpart); if so possibly unveiling a conserved regulation mechanism in PCM recruitment, in which phosphorylated pericentrin/PLP at the centriole wall provides a structural scaffolding for PCM expansion.
The serine-threonine kinase Aurora A is also implicated in centrosome maturation(71–73). While its activity is required for mitotic entry through regulation of Cdk1 and Plk1 kinases in a feedback loop through the protein Bora(74–76), its function in centrosome maturation appears distinct from Plk-1. Aurora A and Cnn are mutually dependent for their centrosomal recruitment during maturation and this activity is mediated through a direct interaction between Aurora-A and a Cnn C-terminal region(77). Aurora-A also stabilizes microtubule growth by phosphorylation of TACC a protein in complex at the centrosome with the microtubule regulator MSPS(78,79). It will be of interest to better understand the interplay between these kinases and the full set of substrates they regulate during centrosome maturation.
Recent work suggests that some PCM components implicated in centrosome maturation (PLP, Asl, Cnn) are part of a large protein complex containing tubulin and Sas-4, which acts as an attachment factor to the centriole wall(80,81). While these complexes were shown to associate in embryonic extracts, it remains unclear whether they are assembled in the cytoplasm prior to centrosomal loading or if they are a consequence of PCM disruption during purification. While PLP, Asl and Cnn are present together on mother centrioles in Drosophila interphase cells, these PCM proteins are recruited to daughters at distinct times during mitosis, with Sas-4 recruited first to the daughter centriole, and subsequently, but not simultaneously, Asl and PLP around metaphase, followed by the outer matrix components Cnn/γ–tubulin(37). It is possible that Drosophila cells and embryos have different timing mechanisms for recruitment of these proteins, owing to the different time scales of cell division. Alternatively, it is possible that only a small subpopulation of these proteins might be forming a bona fide complex while transported together to the centrosome or when all are located at the centrosome after docking (16,82).
Platforms and gates in centriole duplication
Studies from worms to humans suggest that mother centrioles act as an assembly “platform” for the formation of daughter centrioles during their duplication. This process consists of the sequential recruitment of Cep192/Spd-2, Cep152/Asl, Plk4, Sas-6 and CPAP/Sas-4 which allows procentriole assembly(34,83–86). Subdiffraction resolution analysis together with 3D sub-volume alignment and averaging of centrosomes in G1 and G2 shows that Cep152/Asl and pericentrin/PLP undergo a pronounced change in architecture by forming a zone of exclusion on mother centrioles, which correlates with the position where the daughter emerges(37,38). These observations suggest a multistep mechanism, which involves changes in PCM molecular fiber organization. This includes the selection of the site on the mother centriole where daughter centrioles will emerge upon the recruitment of Cep152/Asl structures by Plk4 and subsequent disassembly of a sub-population of Cep152/Asl and pericentrin/PLP, to allow growth of the daughter centriole(37–39,87). This molecular gap, which is present in human and flies, is observed in Asl and PLP structures when labeled using anti-gfp nanobodies (~2nm in size), thus likely ruling out the possibility of lack of antigen accessibility to antibody binding (unpublished observations).
Concluding remarks
Our understanding of PCM architecture has changed substantially through the recent advancements of subdiffraction imaging. While the existence of an organized layer of proteins around centrioles with a defined polarity (PCM tube (4,15,47) had been previously hypothesized, the findings highlighted in this review reveal the existence of multiple layers in the PCM. A systematic, quantitative manner with nanometer precision established the position and orientation of a large number of proteins critical for various facets of centrosome biogenesis. These studies further explain the structural heterogeneity of the components closely associated to the centriole wall during the cell cycle by showing that their state (open vs closed) is correlated with daughter centriole formation. More precisely, they expanded the view of pericentrin architecture and function —previously described as a lattice-like structure (46)—by revealing the dual nature of pericentrin distribution (mitotic matrix-like vs interphase fibers), as well as the orientation of its molecules within the PCM. Lastly and critically, they provide a conceptual framework that will greatly facilitate understanding centrosome maturation at the structural level. The future challenge is to integrate the compositional, functional and structural data into a coherent view of PCM assembly—see Box 2 for analysis of outstanding questions. A major task is to bridge the nanometer resolution information obtained from super-resolution microscopy with the angstrom resolution maps derived from classical structural methods. Integral to this task is the generation of probes targeting various regions of all centrosomal proteins, and the isolation or reconstitution of protein complexes from purified components or extracts. In addition, the molecular role of individual components in the kinetics of PCM assembly will be clearly defined via in vitro reconstitution, enabling the basic principles of PCM formation to be elucidated. These studies should be guided by parallel efforts in cells aimed at defining a high-resolution map of the dynamics of PCM proteins and a comprehensive analysis of their regulatory networks.
Box 2. Outstanding Questions.
Several fundamentally important but unanswered questions remain. Is Cep215/Cnn capable of self-assembly? What are the precise molecular roles of the other PCM components like pericentrin/PLP and Cep192/Spd-2 in building the PCM matrix around interphase centrioles, how are these relationships modulated during centrosome maturation and how are they regulated by the cell cycle machinery? Are Cep192/Spd-2 and pericentrin/PLP required to alter the connectivity, the orientation and/or the kinetics of Cep215/Cnn assembly? To answer these questions it will be important to determine if the set of molecular interactions underpinning PCM organization during interphase is different than in mitosis. While pericentrin/PLP and Cep215/Cnn interactions appear to be maintained throughout the cell cycle, evidence suggests that Spd-2 might not play a major role in PCM assembly during mitosis but not interphase(38). Recent work suggests that Cep192 participates in the phospho-regulation of NEDD1 and controls its ability to associate with and recruit γ-tubulin to mitotic centrosomes raising the tantalizing possibility that Cep192 contributes to PCM activation at the onset of mitosis(91). A systematic biochemical analysis of the interactions with recombinant material or sub-complexes purified from cell extracts at various stages of centrosome assembly will be necessary to answer these questions directly. Considering the importance of microtubule organization and anchoring in specialized cellular processes including stem cell division, it would be important to analyze the similarities and differences in the PCM organization during interphase in cells about to undergo symmetric vs asymmetric divisions(5,92).
During metazoan evolution, pericentrin/PLP has acquired the ability to be recruited not only at the centriole wall as a fiber (in flies) but also as a matrix component in the outer PCM during maturation (in human cells). This more recently acquired property of pericentrin is under regulatory control. Indeed, overexpression of pericentrin in human cells can lead to matrix formation in interphase centrosomes, which in normal conditions have only a fiber/toroid distribution. Interestingly, this function is not required to make the outer matrix in Drosophila—or in C. elegans that seems to lack a pericentrin homologue altogether. This would suggest that the original primary role of pericentrin/PLP is to provide an interphase scaffold for PCM recruitment, and later in evolution gained additional functionality to facilitate PCM expansion in the outer matrix.
Pericentrin/PLP is expressed in metazoans in many isoforms and splicing variants(14,93). Super resolution imaging seems unable to resolve the individual isoforms, suggesting that the different pericentrin/PLP species are arranged in close proximity or bundle together into a multi-molecular fiber at the centrosome with a symmetry likely dictated by the centriole nine-fold structure (Fig.2d)(37). In vitro biochemical and structural analysis will be critical to demonstrate the existence of a pericentrin/PLP multimeric structure and to define the biochemical determinants of its formation and the role of the different isoforms in higher-order PCM matrix organization.
Interestingly, Cep152/Asl on interphase centrosomes is located in the same PCM domain and shares a similar elongated orientation to pericentrin/PLP, but neither pericentrin/PLP nor Cep152/Asl is required for the recruitment of the other implying independent anchoring to the centriole wall (37,68,87). Do the Cep152/Asl elongated structures have a role in organizing the PCM or only in providing a platform for duplication? Mutant analysis of Drosophila spermatocytes suggests that Asl is indeed necessary for PCM stabilization(68,94), while RNAi studies suggest that it is not(40). One possibility is that Asl has an indirect role in PCM organization by recruiting Spd-2 at the centriole wall, which might affect Spd-2 matrix organization. Further studies are required to address this issue.
How do pericentrin/PLP and Cep152/Asl fibers bind to the centriole wall? While Asl has been shown to bind Sas-4 directly(83), it is still unclear how PLP is anchored to the centriole wall. Since pericentrin/PLP appears to be distributed symmetrically around the centriole it is likely to take advantage of the microtubule doublets/triplets, directly or through an attachment factor, or of the space in between(37,38). Interestingly, pull down interaction experiments suggested that Sas-4 N-termini might link PLP to the centriole wall(81).
Recent discoveries defined Cep215 as the major interaction partner and regulator of the γTuRC activity in the PCM(61,95). In addition, the N-Terminal Domain (NTD) of pericentrin/PLP has been shown to bind directly γTuRC, albeit weakly through GCP2-3(46,64), where it may help activate γTuRCs for microtubule nucleation. This putative globular domain does not seem to be necessary for PCM maturation, and its precise role is still unclear(37). Further studies will be required to assess if pericentrin and γTuRC are indeed part of a bona fide complex, and if so, what their functional role is in centrosome maturation and PCM organization.
In conclusion, recent studies highlighted here demonstrate the power of combining multimodal subdiffraction resolution microscopy with quantitative image analysis to reveal molecular-scale details of organelle architecture. Importantly, they defined a fundamental map of PCM organization, and revealed a functional framework to dissect how its organization is achieved. In depth understanding of the centrosome architecture and function is crucial for the discovery and validation of protein targets as determinants of disease.
Highlights.
The PCM is not an amorphous structure
The PCM is composed of molecular fibers and components organized in a matrix
During centrosome maturation the PCM proximal layer acts as a scaffold for PCM expansion
3D volume averaging provide protein position with error of few nanometers
Multimodal super-resolution microscopy with quantitative analysis reveals organelle architecture
Acknowledgements
We would like to sincerely apologize to the authors whose studies we could not include in our reference list for space restrains.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Nigg E, Stearns T. The centrosome cycle: Centriole biogenesis, duplication and inherent asymmetries. Nat. Cell Biol. 2011;13(10):1154–1160. doi: 10.1038/ncb2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bornens M. The centrosome in cells and organisms. Science. 2012;335(6067):422–426. doi: 10.1126/science.1209037. [DOI] [PubMed] [Google Scholar]
- 3.Cunha-Ferreira I, et al. From zero to many: control of centriole number in development and disease. Traffic. 2009;10(5):482–498. doi: 10.1111/j.1600-0854.2009.00905.x. [DOI] [PubMed] [Google Scholar]
- 4.Bornens M. Centrosome composition and microtubule anchoring mechanisms. Curr. Opin. Cell Biol. 2002 Feb;14(1):25–34. doi: 10.1016/s0955-0674(01)00290-3. [DOI] [PubMed] [Google Scholar]
- 5.Pelletier L, Yamashita YM. Centrosome asymmetry and inheritance during animal development. Curr. Opin. Cell Biol. 2012;24(4):541–546. doi: 10.1016/j.ceb.2012.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kobayashi T, Dynlacht BD. Regulating the transition from centriole to basal body. J. Cell Biol. 2011 May 2;193(3):435–444. doi: 10.1083/jcb.201101005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ishikawa H, Marshall WF. Ciliogenesis: building the cell’s antenna. Nat. Rev. Mol. Cell Biol. 2011 May;12(4):222–234. doi: 10.1038/nrm3085. [DOI] [PubMed] [Google Scholar]
- 8.Abal M, et al. Centrioles resist forces applied on centrosomes during G2/M transition. Biol. Cell. 2005 Jun;97(6):425–434. doi: 10.1042/BC20040112. [DOI] [PubMed] [Google Scholar]
- 9.Kirkham M, et al. SAS-4 is a C. elegans centriolar protein that controls centrosome size. Cell. 2003 Feb 21;112(4):575–587. doi: 10.1016/s0092-8674(03)00117-x. [DOI] [PubMed] [Google Scholar]
- 10.Basto R, et al. Flies without centrioles. Cell. 2006 Jun 30;125(7):1375–1386. doi: 10.1016/j.cell.2006.05.025. [DOI] [PubMed] [Google Scholar]
- 11.Kollman JM, et al. Microtubule nucleation by γ-tubulin complexes. Nat. Rev. Mol. Cell Biol. 2011 Nov;12(11):709–721. doi: 10.1038/nrm3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dammermann A, et al. Centriole assembly requires both centriolar and pericentriolar material proteins. Dev. Cell. 2004 Dec;7(6):815–829. doi: 10.1016/j.devcel.2004.10.015. [DOI] [PubMed] [Google Scholar]
- 13.Loncarek J, et al. Control of daughter centriole formation by the pericentriolar material. Nat. Cell Biol. 2008 Mar;10(3):322–328. doi: 10.1038/ncb1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martinez-Campos M, et al. The Drosophila pericentrin-like protein is essential for cilia/flagella function, but appears to be dispensable for mitosis. J. Cell Biol. 2004 Jun 7;165(5):673–683. doi: 10.1083/jcb.200402130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moser JJ, et al. The PCM-basal body/primary cilium coalition. Semin. Cell Dev. Biol. 2010 Apr;21(2):148–155. doi: 10.1016/j.semcdb.2009.06.006. [DOI] [PubMed] [Google Scholar]
- 16.Zimmerman W, Doxsey SJ. Construction of centrosomes and spindle poles by molecular motor-driven assembly of protein particles. Traffic. 2000 Dec;1(12):927–934. [PubMed] [Google Scholar]
- 17.Kumar P, Wittmann T. +TIPs: SxIPping along microtubule ends. Trends Cell Biol. 2012 Aug;22(8):418–428. doi: 10.1016/j.tcb.2012.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takada S, et al. Drosophila checkpoint kinase 2 couples centrosome function and spindle assembly to genomic integrity. Cell. 2003 Apr 4;113(1):87–99. doi: 10.1016/s0092-8674(03)00202-2. [DOI] [PubMed] [Google Scholar]
- 19.Basto R, et al. Hsp90 is required to localise cyclin B and Msps/ch-TOG to the mitotic spindle in Drosophila and humans. J. Cell Sci. 2007 Apr 1;120(Pt 7):1278–1287. doi: 10.1242/jcs.000604. [DOI] [PubMed] [Google Scholar]
- 20.Atwood SX, et al. GLI activation by atypical protein kinase C ι/λ regulates the growth of basal cell carcinomas. Nature. 2013 Feb 28;494(7438):484–488. doi: 10.1038/nature11889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Griffith E, et al. Mutations in pericentrin cause Seckel syndrome with defective ATR-dependent DNA damage signaling. Nat. Genet. 2008 Feb;40(2):232–236. doi: 10.1038/ng.2007.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Doxsey S, et al. Centrosome control of the cell cycle. Trends Cell Biol. 2005 Jun;15(6):303–311. doi: 10.1016/j.tcb.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 23.D’Angiolella V, et al. SCF(Cyclin F) controls centrosome homeostasis and mitotic fidelity through CP110 degradation. Nature. 2010 Jul 1;466(7302):138–142. doi: 10.1038/nature09140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nigg E, Raff JW. Centrioles, centrosomes, and cilia in health and disease. Cell. 2009 Dec 13;139(4):663–678. doi: 10.1016/j.cell.2009.10.036. [DOI] [PubMed] [Google Scholar]
- 25.Megraw TL, et al. Cdk5rap2 exposes the centrosomal root of microcephaly syndromes. Trends Cell Biol. 2011 Aug;21(8):470–480. doi: 10.1016/j.tcb.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bettencourt-Dias M, et al. Centrosomes and cilia in human disease. Trends Genet. 2011 Aug;27(8):307–315. doi: 10.1016/j.tig.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vitre BD, Cleveland DW. Centrosomes, chromosome instability (CIN) and aneuploidy. Curr. Opin. Cell Biol. 2012 Dec;24(6):809–815. doi: 10.1016/j.ceb.2012.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Davis EE, Katsanis N. The ciliopathies: a transitional model into systems biology of human genetic disease. Curr. Opin. Genet. Dev. 2012 Jun;22(3):290–303. doi: 10.1016/j.gde.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Andersen JS, et al. Proteomic characterization of the human centrosome by protein correlation profiling. Nature. 2003 Dec;426(6996):570–574. doi: 10.1038/nature02166. [DOI] [PubMed] [Google Scholar]
- 30.Müller H, et al. Proteomic and functional analysis of the mitotic Drosophila centrosome. EMBO J. 2010 Oct 6;29(19):3344–3357. doi: 10.1038/emboj.2010.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Habermann K, Lange BM. New insights into subcomplex assembly and modifications of centrosomal proteins. Cell Div. Cell Division. 2012 Jan;7(1):17. doi: 10.1186/1747-1028-7-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kitagawa D, et al. Structural basis of the 9-fold symmetry of centrioles. Cell. 2011 Feb 4;144(3):364–375. doi: 10.1016/j.cell.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Van Breugel M, et al. Structures of SAS-6 suggest its organization in centrioles. Science. 2011 Mar 4;331(6021):1196–1199. doi: 10.1126/science.1199325. [DOI] [PubMed] [Google Scholar]
- 34.Pelletier L, et al. Centriole assembly in Caenorhabditis elegans. Nature. 2006 Nov 30;444(7119):619–623. doi: 10.1038/nature05318. [DOI] [PubMed] [Google Scholar]
- 35.Guichard P, et al. Procentriole assembly revealed by cryo-electron tomography. EMBO J. 2010 May 5;29(9):1565–1572. doi: 10.1038/emboj.2010.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moritz M, et al. Three-Dimensional Structural Characterization of Centrosomes from Early Drosophila Embryos. J. Cell Biol. 1995 Sep;130(5):1149–1159. doi: 10.1083/jcb.130.5.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mennella V, et al. Subdiffraction-resolution fluorescence microscopy reveals a domain of the centrosome critical for pericentriolar material organization. Nat. Cell Biol. 2012 Nov;14(11):1159–1168. doi: 10.1038/ncb2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lawo S, et al. Subdiffraction imaging of centrosomes reveals higher-order organizational features of pericentriolar material. Nat. Cell Biol. 2012 Nov;14(11):1148–1158. doi: 10.1038/ncb2591. [DOI] [PubMed] [Google Scholar]
- 39.Sonnen KF, et al. 3D–structured illumination microscopy provides novel insight into architecture of human centrosomes. Biol. Open. 2013 Oct 15;1(10):965–976. doi: 10.1242/bio.20122337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fu J, Glover DM. Structured illumination of the interface between centriole and peri-centriolar material. Open Biol. 2012 Aug;2(8):120104. doi: 10.1098/rsob.120104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li S, et al. Three-dimensional structure of basal body triplet revealed by electron cryo-tomography. EMBO J. 2012 Feb 1;31(3):552–562. doi: 10.1038/emboj.2011.460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gould RR, Borisy GG. The pericentriolar material in Chinese hamster ovary cells nucleates microtubule formation. J. of Cell Biol. 1977;73:601–615. doi: 10.1083/jcb.73.3.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Paintrand M, et al. Centrosome Organization Their Sensitivity and Centriole Architecture to Divalent Cations. J. of Struct. Biol. 1992;108:107–128. doi: 10.1016/1047-8477(92)90011-x. [DOI] [PubMed] [Google Scholar]
- 44.Moritz M, et al. Recruitment of the γ-Tubulin Ring Complex to Drosophila salt-stripped centrosome scaffolds. J. Cell Biol. 1998 Aug;142(3):775–786. doi: 10.1083/jcb.142.3.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schnackenberg BJ, et al. Reconstitution of microtubule nucleation potential in centrosomes isolated from Spisula solidissima oocytes. J. of Cell Sci. 2000 Mar;113:943–953. doi: 10.1242/jcs.113.6.943. [DOI] [PubMed] [Google Scholar]
- 46.Dictenberg JB, et al. Pericentrin and gamma-tubulin form a protein complex and are organized into a novel lattice at the centrosome. J. Cell Biol. 1998 Apr 6;141(1):163–174. doi: 10.1083/jcb.141.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ou YY, et al. Higher order structure of the PCM adjacent to the centriole. Cell Motil. Cytoskeleton. 2003 Jun;55(2):125–133. doi: 10.1002/cm.10115. [DOI] [PubMed] [Google Scholar]
- 48.Gustafsson MGL, et al. Three-dimensional resolution doubling in wide-field fluorescence microscopy by structured illumination. Biophys. J. 2008 Jun;94(12):4957–4970. doi: 10.1529/biophysj.107.120345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Klar T, et al. Fluorescence microscopy with diffraction resolution barrier broken by stimulated emission. Proc. Natl. Acad. Sci. U. S. A. 2000 Jul 18;97(15):8206–8210. doi: 10.1073/pnas.97.15.8206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Betzig E, et al. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006 Sep 15;313(5793):1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- 51.Rust MJ, et al. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM) Nature Methods. 2006 Oct;3(10):793–795. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schermelleh L, et al. Subdiffraction multicolor imaging of the nuclear periphery with 3D structured illumination microscopy. Science. 2008 Jun 6;320(5881):1332–1336. doi: 10.1126/science.1156947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lau L, et al. STED microscopy with optimized labeling density reveals 9-fold arrangement of a centriole protein. Biophys. J. 2012 Jun 20;102(12):2926–2935. doi: 10.1016/j.bpj.2012.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gillingham K, Munro S. The PACT domain, a conserved centrosomal targeting motif in the coiled-coil proteins AKAP450 and pericentrin. EMBO Rep. 2000 Dec;1(6):524–529. doi: 10.1093/embo-reports/kvd105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lawo S, et al. HAUS, the 8-subunit human Augmin complex, regulates centrosome and spindle integrity. Curr. Biol. 2009 May 26;19(10):816–826. doi: 10.1016/j.cub.2009.04.033. [DOI] [PubMed] [Google Scholar]
- 56.Uehara R, et al. The augmin complex plays a critical role in spindle microtubule generation for mitotic progression and cytokinesis in human cells. Proc. Natl. Acad. Sci. U. S. A. 2009 Apr 28;106(17):6998–7003. doi: 10.1073/pnas.0901587106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Haren L, et al. Plk1-dependent recruitment of gamma-tubulin complexes to mitotic centrosomes involves multiple PCM components. PLoS One. 2009 Jan;4(6):e5976. doi: 10.1371/journal.pone.0005976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gomez-Ferreria MA, et al. Human Cep192 is required for mitotic centrosome and spindle assembly. Curr. Biol. 2007 Nov 20;17(22):1960–1966. doi: 10.1016/j.cub.2007.10.019. [DOI] [PubMed] [Google Scholar]
- 59.Lee K, Rhee K. PLK1 phosphorylation of pericentrin initiates centrosome maturation at the onset of mitosis. J. Cell Biol. 2011 Dec 26;195(7):1093–1101. doi: 10.1083/jcb.201106093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dobbelaere J, et al. A genome-wide RNAi screen to dissect centriole duplication and centrosome maturation in Drosophila. PLoS Biol. 2008 Sep 16;6(9):e224. doi: 10.1371/journal.pbio.0060224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fong K, et al. CDK5RAP2 Is a Pericentriolar Protein That Functions in Centrosomal Attachment of theγ-Tubulin Ring Complex. 2008;19(January):115–125. doi: 10.1091/mbc.E07-04-0371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Graser S, et al. Cep68 and Cep215 (Cdk5rap2) are required for centrosome cohesion. J. Cell Sci. 2007 Dec 15;120(Pt 24):4321–4331. doi: 10.1242/jcs.020248. [DOI] [PubMed] [Google Scholar]
- 63.Kawaguchi S, Zheng Y. Characterization of a Drosophila Centrosome Protein CP309 That Shares Homology with Kendrin and CG-NAP. 2004 Jan 15;:37–45. doi: 10.1091/mbc.E03-03-0191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Takahashi M, et al. Centrosomal Proteins CG-NAP and Kendrin Provide Microtubule Nucleation Sites by Anchoring γ-Tubulin Ring Complex. Mol Biol. Cell. 2002 Sep;13:3235–3245. doi: 10.1091/mbc.E02-02-0112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pelletier L, et al. The Caenorhabditis elegans Centrosomal Protein SPD-2 Is Required for both Pericentriolar Material Recruitment and Centriole Duplication. 2004;14:863–873. doi: 10.1016/j.cub.2004.04.012. [DOI] [PubMed] [Google Scholar]
- 66.Giansanti MG, et al. Drosophila SPD-2 is an essential centriole component required for PCM recruitment and astral-microtubule nucleation. Curr. Biol. 2008 Feb 26;18(4):303–309. doi: 10.1016/j.cub.2008.01.058. [DOI] [PubMed] [Google Scholar]
- 67.Decker M, et al. Limiting amounts of centrosome material set centrosome size in C. elegans embryos. Curr. Biol. 2011 Aug 9;21(15):1259–1267. doi: 10.1016/j.cub.2011.06.002. [DOI] [PubMed] [Google Scholar]
- 68.Varmark H, et al. Asterless is a centriolar protein required for centrosome function and embryo development in Drosophila. Curr. Biol. 2007 Oct 23;17(20):1735–1745. doi: 10.1016/j.cub.2007.09.031. [DOI] [PubMed] [Google Scholar]
- 69.Mahen R, Venkitaraman AR. Pattern formation in centrosome assembly. Curr. Opin. Cell Biol. 2012 Feb;24(1):14–23. doi: 10.1016/j.ceb.2011.12.012. [DOI] [PubMed] [Google Scholar]
- 70.Conduit PT, et al. Centrioles regulate centrosome size by controlling the rate of Cnn incorporation into the PCM. Curr. Biol. 2010 Dec 21;20(24):2178–2186. doi: 10.1016/j.cub.2010.11.011. [DOI] [PubMed] [Google Scholar]
- 71.Hannak E, et al. Aurora-A kinase is required for centrosome maturation in Caenorhabditis elegans. J. Cell Biol. 2001 Dec 24;155(7):1109–1116. doi: 10.1083/jcb.200108051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Berdnik D, Knoblich J. Drosophila Aurora-A is required for centrosome maturation and actin-dependent asymmetric protein localization during mitosis. Curr. Biol. 2002 Apr 16;12(8):640–647. doi: 10.1016/s0960-9822(02)00766-2. [DOI] [PubMed] [Google Scholar]
- 73.Hirota T, et al. Aurora-A and an interacting activator, the LIM protein Ajuba, are required for mitotic commitment in human cells. Cell. 2003 Sep 5;114(5):585–598. doi: 10.1016/s0092-8674(03)00642-1. [DOI] [PubMed] [Google Scholar]
- 74.Seki A, et al. Bora and the kinase Aurora a cooperatively activate the kinase Plk1 and control mitotic entry. Science. 2008 Jun 20;320(5883):1655–1658. doi: 10.1126/science.1157425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hutterer A, et al. Mitotic Activation of the Kinase Aurora-A Requires Its Binding Partner Bora. Dev. Cell. 2006 Aug;11(2):147–157. doi: 10.1016/j.devcel.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 76.Macůrek L, et al. Polo-like kinase-1 is activated by aurora A to promote checkpoint recovery. Nature. 2008 Sep 4;455(7209):119–123. doi: 10.1038/nature07185. [DOI] [PubMed] [Google Scholar]
- 77.Terada Y, et al. Interaction of Aurora-A and centrosomin at the microtubule-nucleating site in Drosophila and mammalian cells. J. Cell Biol. 2003 Sep 1;162(5):757–763. doi: 10.1083/jcb.200305048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Barros TP, et al. Aurora A activates D-TACC-Msps complexes exclusively at centrosomes to stabilize centrosomal microtubules. J. Cell Biol. 2005 Sep 26;170(7):1039–1046. doi: 10.1083/jcb.200504097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Giet R, et al. Drosophila Aurora A kinase is required to localize D-TACC to centrosomes and to regulate astral microtubules. J. Cell Biol. 2002 Feb 4;156(3):437–451. doi: 10.1083/jcb.200108135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gopalakrishnan J, et al. Tubulin nucleotide status controls Sas-4-dependent pericentriolar material recruitment. Nat. Cell Biol. 2012 Aug;14(8):865–873. doi: 10.1038/ncb2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gopalakrishnan J, et al. Sas-4 provides a scaffold for cytoplasmic complexes and tethers them in a centrosome. Nat. Commun. 2011 Jun 21;2:359. doi: 10.1038/ncomms1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Li Q, et al. Kendrin/pericentrin-B, a centrosome protein with homology to pericentrin that complexes with PCM-1. J. Cell Sci. 2001 Feb;114(Pt 4):797–809. doi: 10.1242/jcs.114.4.797. [DOI] [PubMed] [Google Scholar]
- 83.Dzhindzhev NS, et al. Asterless is a scaffold for the onset of centriole assembly. Nature. 2010 Oct 7;467(7316):714–718. doi: 10.1038/nature09445. [DOI] [PubMed] [Google Scholar]
- 84.Cizmecioglu O, et al. Cep152 acts as a scaffold for recruitment of Plk4 and CPAP to the centrosome. J. Cell Biol. 2010 Nov 15;191(4):731–739. doi: 10.1083/jcb.201007107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hatch EM, et al. Cep152 interacts with Plk4 and is required for centriole duplication. J. Cell Biol. 2010 Nov 15;191(4):721–729. doi: 10.1083/jcb.201006049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Delattre M, et al. Sequential protein recruitment in C. elegans centriole formation. Curr. Biol. 2006 Sep 19;16(18):1844–1849. doi: 10.1016/j.cub.2006.07.059. [DOI] [PubMed] [Google Scholar]
- 87.Sonnen KF, et al. Human Cep192 and Cep152 cooperate in Plk4 recruitment and centriole duplication. J. Cell Sci. 2013 Jul 15;126(Pt 14):3223–3233. doi: 10.1242/jcs.129502. [DOI] [PubMed] [Google Scholar]
- 88.Kanchanawong P, et al. Nanoscale architecture of integrin-based cell adhesions. Nature. 2010 Nov 25;468(7323):580–584. doi: 10.1038/nature09621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Dani A, et al. Superresolution imaging of chemical synapses in the brain. Neuron. 2010 Dec 9;68(5):843–856. doi: 10.1016/j.neuron.2010.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Szymborska A, et al. Nuclear Pore Scaffold Structure Analyzed by Super-Resolution Microscopy and Particle Averaging. Science. 2013 Aug 9;341(6146):655–658. doi: 10.1126/science.1240672. [DOI] [PubMed] [Google Scholar]
- 91.Gomez-Ferreria MA, et al. Novel NEDD1 phosphorylation sites regulate γ-tubulin binding and mitotic spindle assembly. J. Cell Sci. 2012 Aug 15;125(Pt 16):3745–3751. doi: 10.1242/jcs.105130. [DOI] [PubMed] [Google Scholar]
- 92.Lerit D, et al. Organelle asymmetry for proper fitness, function, and fate. Chromosome Res. 2013 May;21(3):271–286. doi: 10.1007/s10577-013-9350-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Flory MR, Davis TN. The centrosomal proteins pericentrin and kendrin are encoded by alternatively spliced products of one gene. Genomics. 2003 Sep;82(3):401–405. doi: 10.1016/s0888-7543(03)00119-8. [DOI] [PubMed] [Google Scholar]
- 94.Blachon S, et al. Drosophila asterless and vertebrate Cep152 Are orthologs essential for centriole duplication. Genetics. 2008 Dec;180(4):2081–2094. doi: 10.1534/genetics.108.095141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Choi Y-K, et al. CDK5RAP2 stimulates microtubule nucleation by the gamma-tubulin ring complex. J. Cell Biol. 2010 Dec 13;191(6):1089–1095. doi: 10.1083/jcb.201007030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Moritz M, et al. Structure of the γ-tubulin ring complex : a template for microtubule nucleation. Nat Cell Biol. 2000 Jun;2(6):365–370. doi: 10.1038/35014058. [DOI] [PubMed] [Google Scholar]
- 97.Wiese C, Zheng Y. A new function for the γ-tubulin ring complex as a microtubule minus-end cap. Nat Cell Biol. 2000 Jun;2(6):358–364. doi: 10.1038/35014051. [DOI] [PubMed] [Google Scholar]
- 98.Keating TJ, Borisy GG. Immunostructural evidence for the template mechanism of microtubule nucleation. Nat Cell Biol. 2000 Jun;2(6):352–357. doi: 10.1038/35014045. [DOI] [PubMed] [Google Scholar]
- 99.Aldaz H. Insights into microtubule nucleation from the crystal structure of human gamma-tubulin. Nature. 2005 May 26;435(7041):523–527. doi: 10.1038/nature03586. [DOI] [PubMed] [Google Scholar]
- 100.Gunawardane RN, et al. Characterization of a New γ-TuRC Subunit with WD Repeats. 2003;14(March):1017–1026. doi: 10.1091/mbc.E02-01-0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lüders J, et al. GCP-WD is a gamma-tubulin targeting factor required for centrosomal and chromatin-mediated microtubule nucleation. Nat. Cell Biol. 2006 Feb;8(2):137–147. doi: 10.1038/ncb1349. [DOI] [PubMed] [Google Scholar]
- 102.Reschen RF, et al. Dgp71WD is required for the assembly of the acentrosomal Meiosis I spindle, and is not a general targeting factor for the γ-TuRC. Biol. Open. 2012 May 15;1(5):422–429. doi: 10.1242/bio.2012596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Megraw TL, et al. The centrosomin protein is required for centrosome assembly and function during cleavage in Drosophila. Development. 1999 Jul;126(13):2829–2839. doi: 10.1242/dev.126.13.2829. [DOI] [PubMed] [Google Scholar]
- 104.Hamill DR, et al. Centrosome Maturation and Mitotic Spindle Assembly in C . elegans Require SPD-5 , a Protein with Multiple Coiled-Coil Domains. Dev. Cell. 2002 Nov;3(5):673–684. doi: 10.1016/s1534-5807(02)00327-1. [DOI] [PubMed] [Google Scholar]
- 105.Kemp CA, et al. Centrosome Maturation and Duplication in C . elegans Require the Coiled-Coil Protein SPD-2. Dev Cell. 2004 Apr;6(4):511–523. doi: 10.1016/s1534-5807(04)00066-8. [DOI] [PubMed] [Google Scholar]
- 106.Doxsey SJ, et al. Pericentrin, a Hiqhly Conserved Centrosome Protein Involved in Microtubule Organization. Cell. 1994 Feb 25;76:639–650. doi: 10.1016/0092-8674(94)90504-5. [DOI] [PubMed] [Google Scholar]
- 107.Bonaccorsi S, et al. Spindle Self-organization and Cytokinesis During Male Meiosis in asterless mutants of Drosophila Melanogaster. J. of Cell Biology. 1998 Aug 10;142(3):751–761. doi: 10.1083/jcb.142.3.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lane HA, Nigg EA. Antibody Microinjection Reveals an Essential Role for Human Polo-like Kinase 1 (Plk1) in the Functional Maturation of Mitotic Centrosomes. J. of Cell Biology. 1996 Dec;1(6):1701–1713. doi: 10.1083/jcb.135.6.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sunkel CE, Glover DM. Polo, a mitotic mutant of Drosophila displaying abnormal spindle poles. J. Cell Sci. 1988 Jan;89(Pt 1):25–38. doi: 10.1242/jcs.89.1.25. [DOI] [PubMed] [Google Scholar]
- 110.Peset I, et al. Function and regulation of Maskin, a TACC family protein, in microtubule growth during mitosis. J. Cell Biol. 2005 Sep 26;170(7):1057–1066. doi: 10.1083/jcb.200504037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Leidel S, Gonczy P. SAS-4 Is Essential for Centrosome Duplication in C . elegans and Is Recruited to Daughter Centrioles Once per Cell Cycle. Dev Cell. 2003;4(3):431–439. doi: 10.1016/s1534-5807(03)00062-5. [DOI] [PubMed] [Google Scholar]
- 112.Tang C-JC, et al. CPAP is a cell-cycle regulated protein that controls centriole length. Nat. Cell Biol. 2009 Jul;11(7):825–831. doi: 10.1038/ncb1889. [DOI] [PubMed] [Google Scholar]
- 113.Schmidt TI, et al. Control of centriole length by CPAP and CP110. Curr. Biol. 2009 Jun 23;19(12):1005–1011. doi: 10.1016/j.cub.2009.05.016. [DOI] [PubMed] [Google Scholar]




