Abstract
The excitatory neurotransmitter glutamate induces modulatory actions via the metabotropic glutamate receptors (mGlus), which are class C G protein-coupled receptors (GPCRs). We determined the 2.8 Å resolution structure of the human mGlu1 receptor seven-transmembrane (7TM) domain bound to a negative allosteric modulator FITM. The modulator binding site partially overlaps with the orthosteric binding sites of class A GPCRs, but is more restricted compared to most other GPCRs. We observed a parallel 7TM dimer, mediated by cholesterols, suggesting that signaling initiated by glutamate’s interaction with the extracellular domain might be mediated via 7TM interactions within the full-length receptor dimer. A combination of crystallography, structure-activity relationships, mutagenesis, and full-length dimer modeling provides insights on the allosteric modulation and activation mechanism of class C GPCRs.
The human G protein-coupled receptor (GPCR) superfamily is composed of over 800 seven transmembrane (7TM) receptors that can be divided into four classes based on their sequence homology: class A, B, C, and Frizzled (F) (1). Class C GPCRs play important roles in many physiological processes such as synaptic transmission, taste sensation and calcium homeostasis, and include metabotropic glutamate receptors (mGlu), γ-aminobutyric acid B receptors (GABAB), calcium sensing receptor (CaS), taste 1 receptors (TAS1), as well as a few orphan receptors. A distinguishing feature of class C GPCRs is constitutive homo- or hetero-dimerization mediated by a large N-terminal extracellular domain (ECD) (Fig. 1A). The ECDs within homodimeric receptors (mGlu and CaS) are cross-linked via an intermolecular disulfide bond. The heterodimeric receptors (GABAB and TAS1) are not covalently linked, but their heterodimerization is required for trafficking to the cell surface and signaling (2). The ECD of class C GPCRs consists of a Venus flytrap domain (VFD), which contains the orthosteric binding site for native ligands (Fig. 1A), and a cysteine-rich domain (CRD), except for GABAB receptors. The CRD, which mediates the communication between ECD and 7TM domains, is stabilized by disulfide bridges, one of which connects the CRD and VFD (3).
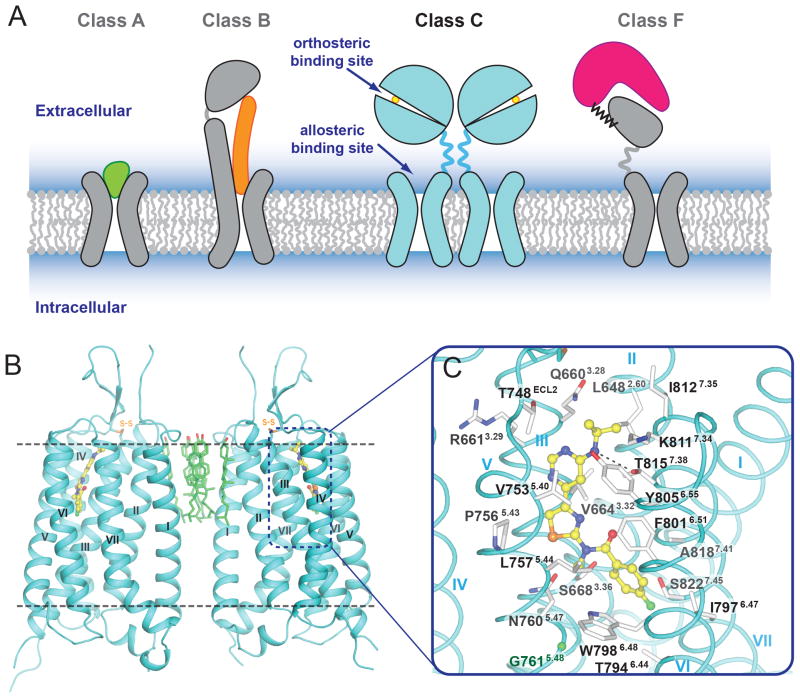
Fig. 1.
Overall structure of the mGlu1 TM domain. (A) Cartoon models for structure and endogenous ligand recognition in different GPCR classes. For class A, in most cases, the endogenous ligand (shown in green) is recognized by an orthosteric site in the 7TM domain. For class B, the endogenous peptide ligand (shown in orange) binds to both ECD and 7TM domains. For class C, the endogenous small molecule ligands (shown in yellow circle) are recognized by orthosteric sites in the VFDs. For class F, lipoprotein WNT (shown in magenta) binds the CRD domain of Frizzled receptors. (B) The mGlu1 7TM domain that crystallized as a parallel dimer is shown in cyan cartoon. Cholesterols mediating the dimer interface are shown as green carbons. (C) Side chains of the FITM binding pocket residues are shown as white carbons. Hydrogen bond interaction between the NAM and T8157.38 is shown as a dashed line. The Cα carbon of G7615.48 is shown as a green ball. In (B) and (C), the ligand FITM is shown as yellow carbons.
The mGlu family was the first group of class C GPCRs to be cloned (4, 5). Comprised of eight members, the mGlu family can be separated into three subgroups (6), termed groups I (mGlu1 and mGlu5), II (mGlu2 and mGlu3) and III (mGlu4,6,7,8), based on their sequence homology, G protein coupling profile, and pharmacology (7). Group I mGlus are predominantly coupled to Gq/11 and activate phospholipase Cβ, which hydrolyses phosphoinositides into inositol 1,4,5-trisphosphate (IP3) and diacylglycerol, inducing intracellular calcium mobilization and activating protein kinase C (PKC).
The group I mGlus, mGlu1 and mGlu5, are considered promising therapeutic targets to treat diseases including cancer, pain, schizophrenia, Alzheimer’s disease, anxiety, and autism (7, 8). However, the development of subtype-selective small molecule ligands that might serve as drug candidates for these receptors has been hampered by the conservation of the orthosteric (glutamate) binding site (Fig. 1A). This problem can be overcome by using allosteric modulators that act at alternative binding sites; these compounds bind predominantly within the 7TM domain of the class C receptors. Allosteric modulators can alter the affinity or efficacy of native ligands in positive, negative, and neutral ways, demonstrating a spectrum of activity that cannot be achieved by orthosteric ligands alone.
In this study, we report the crystal structure of the human mGlu1 7TM domain bound to a negative allosteric modulator (NAM), 4-fluoro-N-(4-(6-(isopropylamino)pyrimidin-4-yl)thiazol-2-yl)-N-methylbenzamide (FITM) (9) at 2.8 Å resolution (Table S1) (10). This structure provides a 3D framework for understanding the molecular recognition and facilitating the discovery of allosteric modulators for the mGlu family and other class C GPCRs. It also complements crystallographic studies of the transmembrane domain structures of class A (11, 12), B (13, 14) and F (15) GPCRs and extends the knowledge base upon which to study the diversity and evolution of the GPCR superfamily.
Overall structure of the mGlu1 7TM domain
The human mGlu1 7TM domain (residues 581-860) (Fig. S1), complexed with FITM, was crystallized by the lipidic cubic phase method using the thermostabilized apocytochrome b562RIL (BRIL) N-terminal fusion strategy (10). A series of in vitro pharmacological studies were performed to verify that this truncated construct binds FITM and is functional in G protein coupling (Figs. S2 and S3). The structure was solved using a 4.0 Å single wavelength anomalous dispersion (SAD) dataset collected from a single crystal soaked with tantalum bromide cluster, followed by extending the resolution to 2.8 Å using native data collected from 14 crystals (Table S1 and Fig. S4).
The mGlu1 7TM domain forms a parallel dimer in each asymmetric unit with a dimer interface mediated mainly through helix I (Fig. 1B; see also Figs. S5 and S6). Interestingly, we observed six well-resolved cholesterol molecules packed against hydrophobic residues on the extracellular side of helices I and II, mediating the dimer formation. The extracellular loop (ECL) 2 adopts a β-hairpin conformation, pointing to the extracellular space, which has also been observed in many peptide class A GPCRs (16, 17). This β-hairpin is connected to the top of helix III through a disulfide bond (C657-C746) that is conserved through all classes of GPCRs. The mGlu1 NAM, FITM, binds within a pocket formed by the 7TM bundle close to the extracellular side (Fig. 1B and 1C), a region partially overlapping with the orthosteric binding sites observed for class A GPCRs (11). The intracellular loop (ICL) 1 forms an ordered helical turn, while a large part of ICL2 (residues 688-695 in molecule A and residues 689-693 in molecule B) is missing in the structure due to the long and presumably flexible nature of this loop. ICL3 is well resolved and forms a short link connecting the intracellular ends of helices V and VI. In addition, we did not observe helix VIII, reported in most class A GPCR structures, as well as in classes B and F. Instead, electron densities for C-terminal residues (844-860 in molecule A and 847-860 in molecule B) are missing in the mGlu1 structure, indicating that this region can be disordered.
Major structural differences with other GPCR classes
Superposition of 7TM domains between mGlu1 and GPCRs of different classes (Fig. S7) reveals that, despite the lack of sequence conservation (<15% identical residues) or common functional motifs (Figs. S8 and S9), the overall fold is preserved across the whole GPCR superfamily (RMSD <3.5 Å for 7TM regions). Based on the structural superposition, we generated a structure-based alignment in the 7TM domain with class A GPCRs and transplanted the class A Ballesteros & Weinstein (B&W) numbering (18) to class C GPCRs (Figs. S8 and S9) (19).
Differences, however, are observed in the 7TM helices between class C and other classes, including distinct distribution patterns of proline-induced kinks in the helical backbone. Instead of having conserved prolines in the X.50 position of helices V, VI and VII, which induce kinks as observed in class A, residues at 5.50, 6.50 and 7.50 positions in mGlu1 are all non-proline residues (Fig. S8). Notably, P8337.56 (20) at the intracellular end of helix VII in mGlu1 induces a kink, resulting in an outward orientation of the C-terminal part of this helix (Fig. S7B). In contrast, the proline conserved in the class A NP7.50xxY motif is on the opposite side of helix VII and induces an inward kink (Fig. S7B).
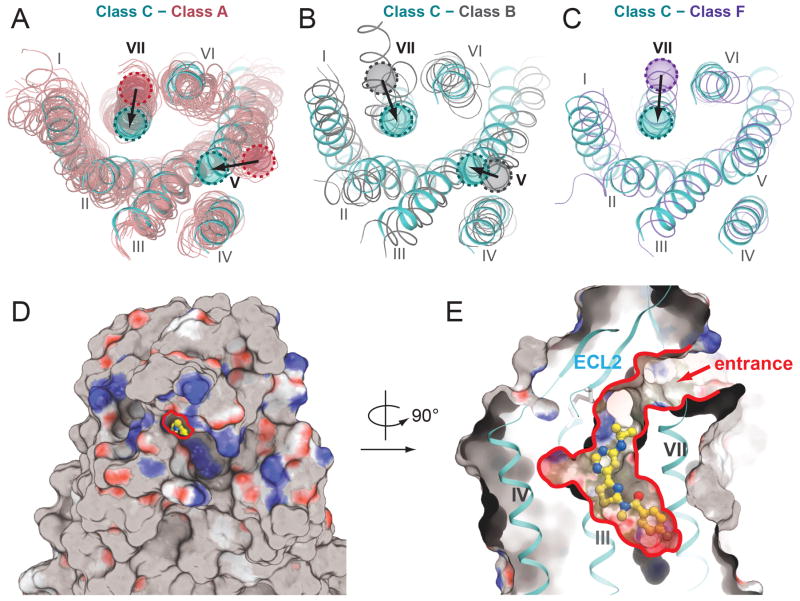
Helices I-IV of mGlu1 overlay relatively well with other GPCR structures, while helices V-VII demonstrate more obvious differences. Compared to class A and B receptors (Fig. 2, A and B), helix V of mGlu1 is shifted inward to the center of the 7TM bundle. Additionally, the extracellular end of helix VII is shifted inward compared to all other classes. These shifted helices, together with ECL2, restrict access to the NAM binding cavity (Fig. 2, D and E). The recently solved class F smoothened receptor (15) also has a narrow cavity embedded in the extracellular half of its 7TM bundle resulting partly from the inward positioning of helix V, but its ECL2 β-hairpin is located inside the 7TM bundle and forces an outward shift of helix VII as compared to mGlu1 (Fig. 2C). This more restricted 7TM cavity in class C receptors is consistent with interactions of known native ligands with the ECD rather than the 7TM domain.
Fig. 2.
7TM domain comparison of mGlu1 with GPCRs of other classes. Extracellular view of superimpositions between mGlu1 shown in cyan and (A) class A GPCRs shown in salmon, (B) class B GPCRs (CRFR1 and glucagon receptor; PDB ID 4K5Y and 4L6R, respectively) shown in grey, and (C) class F GPCR, smoothened receptor (PDB ID 4JKV) shown in purple. In (A)–(C), shifts of helix V or VII in mGlu1 compared to other classes are indicated by arrow. (D) Extracellular view of the surface presentation of the structure showing the narrow entrance (highlighted by red line) to the allosteric ligand binding cavity in the center. (E) A cut-through surface side view of the ligand binding cavity. The arrow indicates the extracellular entrance to the allosteric ligand binding cavity. In the surface presentations in (D) and (E), non-polar residues are shown in gray, hydrogen bond acceptors are shown in red, and hydrogen bond donors are shown in blue. PDB ID of class A GPCRs structures used in (A): 1U19, 2RH1, 2YCW, 3RZE, 3PBL, 3UON, 4DAJ, 3EML, 3V2W, 3ODU, 4DJH, 4EA3, 4DKL, 4EJ4, 3VW7, 4GRV.
The FITM binding pocket
Analogous to the orthosteric site for many family A GPCRs, the binding pocket for the ligand FITM is defined by residues on helices II, III, V, VI, VII and ECL2 (Fig. 1C and Fig. S10). ECL2 forms a lid on the top of the ligand binding cavity, leaving a small opening through which the pocket is accessible from the extracellular side (Fig. 2, D and E). The ligand, FITM, fits tightly into the long and narrow pocket. Most of the ligand-receptor interactions are hydrophobic with the exception of the contacts of the pyrimidine-amine group with the T8157.38 side chain. Substitution of T8157.38 with methionine or alanine reduces the affinity and potency of FITM (Fig. 3B, Fig. S11F, and Table S2), as well as other mGlu1 NAMs from different scaffolds (21–23). The p-fluorophenyl moiety of the ligand points to the bottom of the pocket, making contacts with W7986.48, a residue that is conserved among mGlus as well as in many class A receptors. However, unlike the conformation of the W6.48 side chain observed in most class A GPCRs, which points into the center of the helical bundle, W7986.48 in the structure of FITM-bound mGlu1 points outward. This conformation of the bulky indole group is accommodated by G7615.48 on helix V which has no side chain (Fig. 1C). However, in other mGlus except mGlu5, residues at position 5.48 have relatively large side chains and W6.48 may adopt a different conformation.
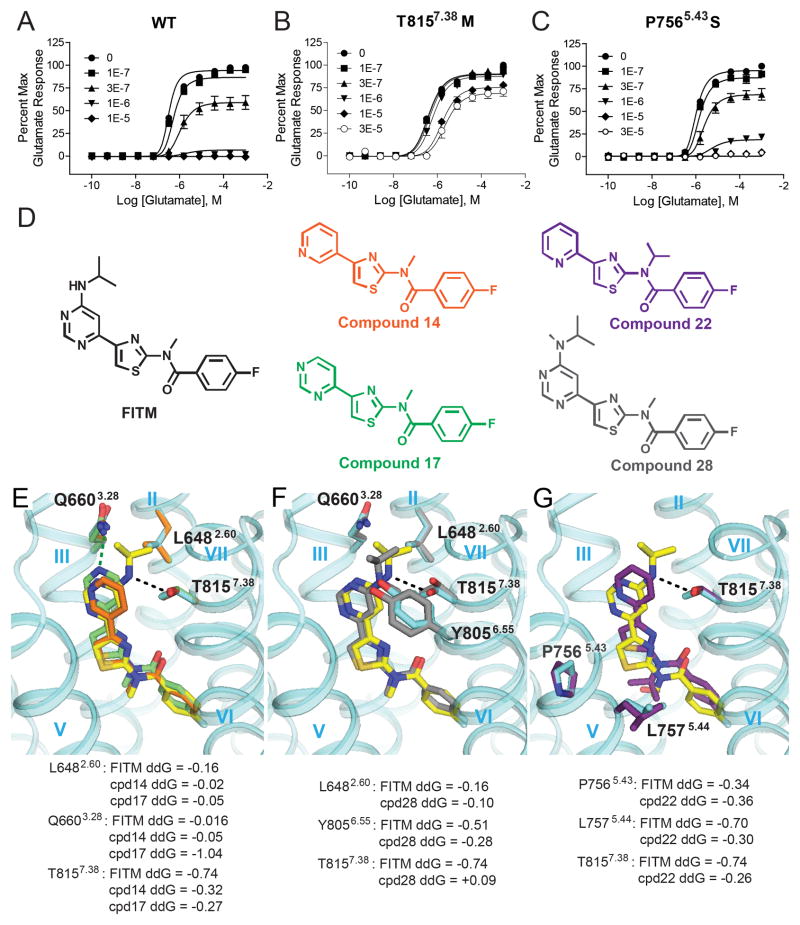
Fig. 3.
Critical FITM-receptor interactions are revealed by mutations and structure activity relationships. (A) FITM is a full NAM of the wild-type (WT) full-length human mGlu1 receptor, while the affinity of FITM and the degree of negative cooperativity with glutamate are reduced in (B) T8157.38M and (C) P7565.43S mutants. (D) Structures of FITM and FITM-related NAMs used for study. Binding pose of FITM (yellow carbons; IC50: 5 nM) in comparison with lower potency analogs, (E) compound 17 (green carbons; IC50: 10 nM) and compound 14 (orange carbons; IC50: 230 nM), (F) compound 28 (grey carbons; IC50: 77 nM) and (G) compound 22 (purple carbons; IC50: 2 μM). Per-residue binding energy ddG is predicted by Rosetta in Rosetta Energy Units (REU) (37). In (E), (F) and (G), side chain rotamers from the top 1% of key amino acids are depicted in sticks and colored corresponding to their respective docked ligand, with the exception of those from the crystal structure shown in cyan; the dashed lines indicate hydrogen bond interactions between the receptor and the ligands.
Determinants of subtype selectivity within the common allosteric site
Previous mutagenesis studies have proposed at least one common allosteric site for the mGlu family within the 7TM domain. The mGlu1 binding pocket for FITM (Fig. 1C) largely corresponds to mutagenic data for the common allosteric site in mGlus and likely extends to other class C GPCRs (see more details in Table S3). Despite the evidence that binding of various chemotypes of class C GPCR allosteric modulators involve similar residue positions, many mGlu modulators display a high degree of subtype selectivity, including FITM which shows high affinity (Ki = 2.5 nM, Fig. S2) and selectivity for mGlu1 over mGlu5 (Fig. S12) (9). Examination of the contact residues in the binding pocket reveals only four residues of mGlu1 that differ from mGlu5: V6643.32, S6683.36, T8157.38, and A8187.41, all of which have previously been implicated in subtype selectivity by mutagenesis-based studies (24–26). Therefore, we mutated these four residues to their corresponding amino acid in mGlu5 (Fig. S11 and Table S2) and compared FITM-mediated antagonism of the mutant receptors to the wide-type (WT) full-length human mGlu1 (Fig. 3A). Methionine substitution of T8157.38 (Fig. 3B) had the most profound effect, reducing FITM affinity ~6 fold and decreasing negative cooperativity with glutamate (Table S2). Thus, T8157.38 is a key selectivity determinant for FITM, consistent with the observed polar interaction between T8157.38 and the ligand in the structure.
In addition, we assessed mutations known to influence the allosteric modulation of other mGlu subtypes that had not previously been explored in mGlu1. T7946.44A and S8227.45A had no effect on FITM; while P7565.43S significantly reduced FITM affinity (~ 3 fold) as well as negative cooperativity (Fig. 3C and Table S2). Location of P7565.43 in the ligand binding pocket suggests that a P7565.43S mutation may induce conformational changes in the backbone altering the shape of the binding pocket in relation to the thiazole core of FITM. Interestingly, multiple mGlu5 modulator scaffolds are known to be sensitive to mutations of two non-conserved residues, S6.39 and A7.46 (23, 25, 27–31); neither is observed here as contributing to the FITM binding pocket. However, both of these residues contribute to a small pocket separated from the FITM pocket by the Y6723.40 side chain. Given that S3.36 in mGlu1 is replaced by P3.36 in mGlu5, it is conceivable that the proline induced kink in helix III particular to mGlu5 can significantly change the shape of the pocket, making S6.39 and A7.46 of mGlu5 accessible to ligands.
To further improve our understanding of the critical ligand-receptor interactions for FITM binding within the pocket, we docked a selection of FITM analogs (Fig. 3D-G and Fig. S13) (9) into the crystal structure. Re-docking FITM (Fig. S13, A-C) and analyzing the binding energy contribution per residue (Fig. S13D) revealed that T8157.38 forms an energetically favorable hydrogen bond with FITM (Fig. S13, C and D). Compound 17 lacks not only the hydrogen bond with T8157.38, but also a non-polar interaction with L6482.60 (Fig. 3, D and E); however, a potential hydrogen bond between Q6603.28, which is not observed in FITM binding (Fig. S11A and Table S2), may compensate for this loss and account for the retained activity at 10 nM. The 3-pyridyl analog (compound 14, Fig. 3, D and E) lacks this potential interaction with Q6603.28, accounting for its further decreased potency (230 nM). Compound 28 exhibits approximately 10 fold lower potency and differs from FITM by the introduction of a methyl group to the amine on the pyrimidine ring. Docking compound 28 reveals a major energy penalty that arises from the loss of a polar interaction and the introduction of steric clash with T8157.38 (Fig. 3, D and F). Compound 28 also lacks a polar interaction with Q6603.28 and requires movement of T8157.38 and Y8056.55 to accommodate the methyl group (Fig. 3F). Compound 22 (Fig. 3, D and G), which contains an iso-propyl group on the amide linker, requires movement of two residues in helix V (P7565.43 and L7575.44) to fit in the pocket, and also lacks hydrogen bonding capacity with either T8157.38 or Q6603.28, accounting for its reduced (micromolar) potency. Collectively, by comparing the binding of FITM with those of other less active or inactive compounds, we attribute the superior potency observed for FITM to the polar interaction between T8157.38 and the amine derivative on the 5′ position of the pyrimidine ring, as well as the perfect fit of the ligand shape within the narrow binding pocket.
NAM-bound mGlu1 is in an inactive state
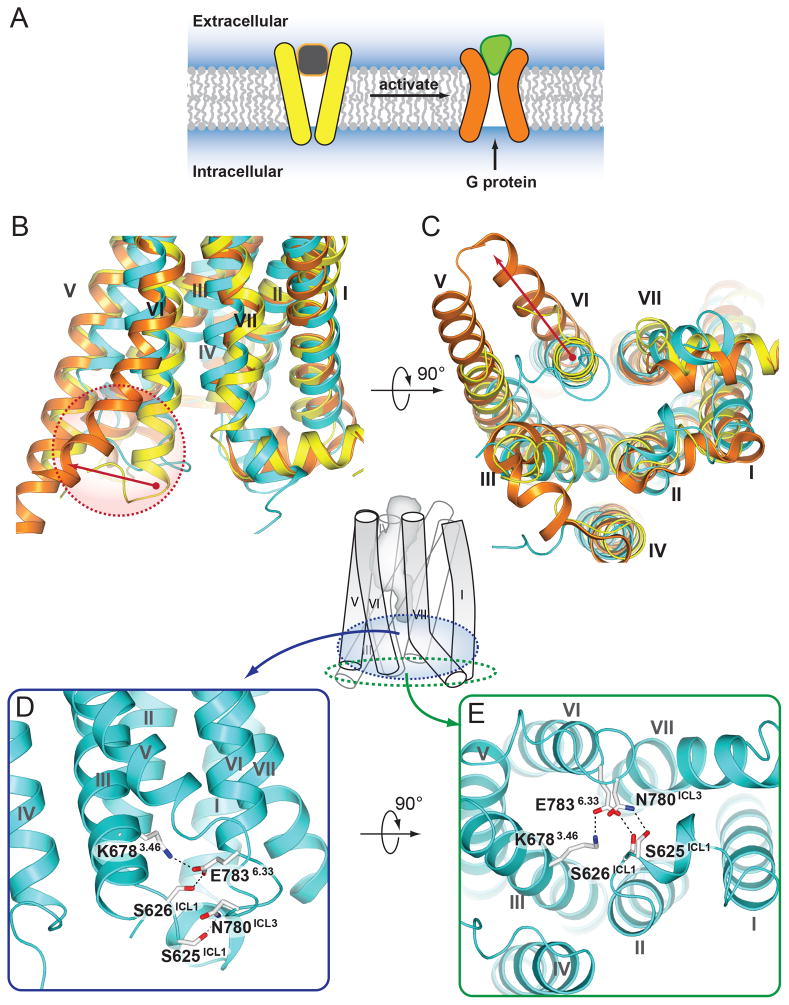
In the NAM-bound structure of mGlu1, the intracellular site responsible for G protein interaction is in a conformation similar to the inactive conformation observed in class A GPCRs (Fig. 4A–C). One of the interactions apparently stabilizing this conformation is a salt bridge between the K6783.46 side chain at the intracellular end of helix III and the E7836.33 side chain at the intracellular end of helix VI (Fig. 4, D and E); both residues are well conserved in all class C receptors. In class A GPCRs, a similar interaction called an “ionic lock” is observed between the conserved R3.50 of the D(E)R3.50Y motif and an acidic residue in the 6.30 position. The “ionic lock” plays a role in stabilizing the receptor’s inactive state, by restricting the activation-related outward movement of helix VI. In addition to the salt bridge between K6783.46 and E7836.33, S626ICL1, a residue conserved in most class C GPCRs, also participates in this interaction network. S625ICL1 and N780ICL3 form a hydrogen bond, stabilizing the interaction between ICL1 and ICL3, and further occluding the G protein binding site (Fig. 4, D and E). Thus, polar interactions within the intracellular crevice may be involved in the regulation of G protein binding and receptor activation in both class A and C GPCRs, though through distinct residue positions.
Fig. 4.
The intracellular crevice in NAM-bound mGlu1 adopts a closed conformation. (A) A cartoon demonstrating agonist triggered opening of the intracellular cavity for G protein binding. (B) Side and (C) intracellular views of the superposition of mGlu1 (cyan) with inactive state of β2-adrenergic receptor shown in yellow (PDB ID 2RH1) and a fully active G protein-coupled state of β2-adrenergic receptor shown in orange (PDB ID 3SN6). Red arrows in (B) and (C) indicate movement of the intracellular end of helix VI, highlighted in red dashed circle in (B), during activation of β2-adrenergic receptor. (D) Side and (E) intracellular views of the mGlu1 receptor, the side chains of residues involved in a hydrogen bond network that stabilize the receptor in an inactive conformation are shown as white carbons.
Communication between ECD and 7TM domains
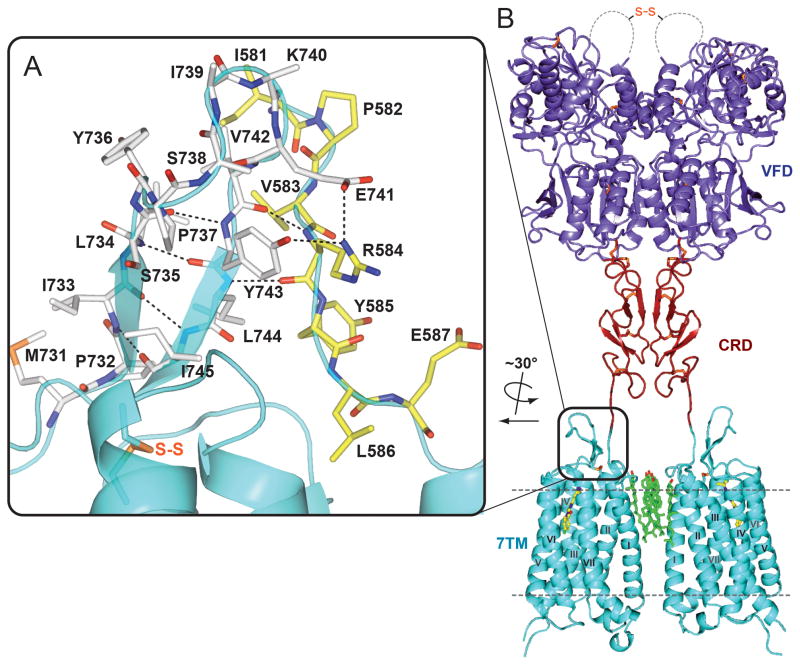
In the mGlu receptor family, as well as in other class C GPCRs, a signal is initiated by the native ligand binding to the ECD, which induces conformational changes in the ECD. In our structure, the linker region (I581-E592) between the ECD and 7TM domain is resolved. The linker forms strong interactions with the ECL2 β-sheet through main chain and side chain hydrogen bonds (Fig. 5A). ECL2 is connected by a covalent disulfide bond to the top of helix III, known to be important in triggering activation in class A GPCRs (32). This observation raises a possibility that this interaction network might contribute to the communication between the ECD and the 7TM domain during receptor activation. In addition, part of the linker residues (e.g. W588, a residue conserved in all mGlus) insert into the lipid bilayer, where they form extensive contacts with cholesterol molecules that mediate the observed dimerization of the 7TM domain (Fig. S6). These interactions suggest a potential role of dimerization and/or lipid components in the coupling between the ECD and 7TM domain during the activation process.
Fig. 5.
A full-length mGlu1 dimer model with highlighted details of interactions between ECL2 and the 7TM-to-CRD linker. (A) Shown in cyan is the extracellular part of the mGlu1 7TM. ECL2 residues (M731-I745) are shown as white carbons, while the linker region residues (I581-E587) are shown as yellow carbons. Hydrogen bond interactions between ECL2 and the linker region are shown as dashed lines. (B) Full-length model of mGlu1 with the VFD in the Acc (active closed-closed) state. VFD, CRD and 7TM domains are colored in slate, firebrick and cyan, respectively. The current model probably does not capture the specific conformation and interaction between CRD and 7TM domain, and a more tightly packed domain interaction is very likely. This model is presented to generate discussion and show the general features of the VFD, CRD, and 7TM domains.
ECDs of class C GPCRs mediate receptor homo- and hetero-dimerization (2). Several dimeric structures of mGlu receptor VFDs have been solved in different conformations: putative active (A) or resting (R) state defined by the relative orientation between the VFD protomers as well as closed (c) or open (o) states defined by the conformation of each VFD (33). Comparing different conformations, the distance between the C-terminal ends of the ECDs within a dimer changes dramatically (3). In our crystal structure of the 7TM domain, we observed a parallel dimer mediated by interactions of helix I and cholesterols. In this dimer conformation, the distance between the N-terminal linkers that are attached to the C-termini of ECDs is ~20 Å. If this is a conformation that can be adopted by the full-length receptor dimer, the CRDs of each protomer should also be in close proximity. Disulfide bond crosslinking experiments suggested that the CRDs of each protomer may form close contact in an activated receptor dimer (34). Although our structure is solved in complex with a NAM and the 7TM domain appears to be in an inactive state, there is evidence supporting the existence of a glutamate-bound, but signaling incapable state, in the full-length mGlu dimer (35). Moreover, there is evidence that cholesterol can positively modulate glutamate responses by recruiting mGlus to lipid rafts (31, 36), consistent with the observation that the close proximity of the N-terminus of the 7TM domain results from a dimer conformation mediated by multiple cholesterol molecules. To test the possibility of fitting the existing ECD structures into our observed 7TM dimer conformation, we created a full-length dimer model in which the VFD adopts an Acc (active closed-closed) conformation as this conformation has the closest distance (~ 50 Å) between the C-termini of the ECDs (3) (Fig. 5B). A 20° rotation was applied to the CRD coupled with a conformational change in the Q513-V523 loop region that reduces this distance to 20 Å, fulfilling the CRD interface proposed in the cysteine mutant study (34), as well as matching the distance of the 7TM domain N-termini observed in the crystallographic dimer. This model might represent a glutamate bound, but signaling incapable, conformation of mGlu1. While consistent with the currently available experimental data, we acknowledge that this model is only one of the several possible explanations for the biological role of the 7TM domain dimer we observed, and needs to be tested in future studies. We further acknowledge that the 7TM domain dimer conformation might vary in different states of the receptor and may be modulated by several factors in biological systems, such as membrane lipid content or other protein-protein interactions.
The mGlu1 7TM structure presented here uncovers atomic details of the class C GPCR transmembrane domain, providing a missing link in our structural understanding of the GPCR superfamily. As noted for the recently solved class B and F GPCR structures, and now for class C, despite a lack of sequence and motif conservation, the architecture of the 7TM bundle is generally preserved. Furthermore, while class C GPCRs are known to form obligate dimers via the ECDs, the observed 7TM dimer suggests additional points of communication between protomers, mediated by multiple cholesterol molecules and direct protein-protein interactions. Moreover, as a robust structural template, the mGlu1 7TM domain structure will likely provide insights into pharmacology of small molecule allosteric modulators for class C GPCRs.
Supplementary Material
Acknowledgments
This work was supported by the NIGMS PSI:Biology grant U54 GM094618 for biological studies and structure production (target GPCR-68) (V.K., V.C. and R.C.S.), NIH Common Fund in Structural Biology grant P50 GM073197 for technology development (V.C. and R.C.S.); R01 NS031373 (P.J.C.), R01 MH062646 (P.J.C.), R21 NS078262 (C.M.N.), a Basic Research Grant from the International Rett Syndrome Foundation (C.M.N.), an NHMRC (Australia) Overseas Biomedical Postdoctoral fellowship (K.J.G.) and a NARSAD Maltz Young Investigator Award (K.J.G.). Work in the Meiler laboratory on computational modeling of membrane proteins and their ligand interactions is supported through NIH (R01 MH090192, R01 GM080403, R01 GM099842, R01 DK097376) and NSF (CHE 1305874). We would like to thank Dr. K. Emmitte and P. Garcia for the synthesis of RO0711401 and FITM; J. Velasquez for help on molecular biology; T. Trinh and M. Chu for help on baculovirus expression; K. Kadyshevskaya for assistance with figure preparation; A. Walker for assistance with manuscript preparation; J. Smith, R. Fischetti, and N. Sanishvili for assistance in development and use of the minibeam and beamtime at GM/CA-CAT beamline 23-ID at the Advanced Photon Source, which is supported by National Cancer Institute grant Y1-CO-1020 and National Institute of General Medical Sciences grant Y1-GM-1104. The coordinates and the structure factors have been deposited in the Protein Data Bank (PDB) under accession codes 4OR2.
Footnotes
“This manuscript has been accepted for publication in Science. This version has not undergone final editing. Please refer to the complete version of record at http://www.sciencemag.org/. The manuscript may not be reproduced or used in any manner that does not fall within the fair use provisions of the Copyright Act without the prior, written permission of AAAS.”
Materials and Methods
References (38–71)
References and Notes
- 1.Lagerstrom MC, Schioth HB. Structural diversity of G protein-coupled receptors and significance for drug discovery. Nat Rev Drug Discov. 2008;7:339–357. doi: 10.1038/nrd2518. published online Epub Apr. nrd2518 [pii] [DOI] [PubMed] [Google Scholar]
- 2.Kniazeff J, Prezeau L, Rondard P, Pin JP, Goudet C. Dimers and beyond: The functional puzzles of class C GPCRs. Pharmacol Ther. 2011;130:9–25. doi: 10.1016/j.pharmthera.2011.01.006. published online Epub Apr. [DOI] [PubMed] [Google Scholar]
- 3.Muto T, Tsuchiya D, Morikawa K, Jingami H. Structures of the extracellular regions of the group II/III metabotropic glutamate receptors. Proc Natl Acad Sci U S A. 2007;104:3759–3764. doi: 10.1073/pnas.0611577104. published online Epub Mar 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Houamed KM, Kuijper JL, Gilbert TL, Haldeman BA, O’Hara PJ, Mulvihill ER, Almers W, Hagen FS. Cloning, expression, and gene structure of a G protein-coupled glutamate receptor from rat brain. Science. 1991;252:1318–1321. doi: 10.1126/science.1656524. published online Epub May 31 ( [DOI] [PubMed] [Google Scholar]
- 5.Masu M, Tanabe Y, Tsuchida K, Shigemoto R, Nakanishi S. Sequence and expression of a metabotropic glutamate receptor. Nature. 1991;349:760–765. doi: 10.1038/349760a0. published online Epub Feb 28. [DOI] [PubMed] [Google Scholar]
- 6.Nakanishi S. Molecular diversity of glutamate receptors and implications for brain function. Science. 1992;258:597–603. doi: 10.1126/science.1329206. published online Epub Oct 23 ( [DOI] [PubMed] [Google Scholar]
- 7.Niswender CM, Conn PJ. Metabotropic glutamate receptors: physiology, pharmacology, and disease. Annu Rev Pharmacol Toxicol. 2010;50:295–322. doi: 10.1146/annurev.pharmtox.011008.145533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dolen G, Carpenter RL, Ocain TD, Bear MF. Mechanism-based approaches to treating fragile X. Pharmacol Ther. 2010;127:78–93. doi: 10.1016/j.pharmthera.2010.02.008.S0163-7258(10)00058-6. published online Epub Jul. [pii] [DOI] [PubMed] [Google Scholar]
- 9.Satoh A, Nagatomi Y, Hirata Y, Ito S, Suzuki G, Kimura T, Maehara S, Hikichi H, Satow A, Hata M, Ohta H, Kawamoto H. Discovery and in vitro and in vivo profiles of 4-fluoro-N-[4-[6-(isopropylamino)pyrimidin-4-yl]-1,3-thiazol-2-yl]-N-methylbenzam ide as novel class of an orally active metabotropic glutamate receptor 1 (mGluR1) antagonist. Bioorg Med Chem Lett. 2009;19:5464–5468. doi: 10.1016/j.bmcl.2009.07.097.S0960-894X(09)01057-9. published online Epub Sep 15. [pii] [DOI] [PubMed] [Google Scholar]
- 10.Materials and methods are available as supplementary materials on Science Online.
- 11.Katritch V, Cherezov V, Stevens RC. Structure-function of the G protein-coupled receptor superfamily. Annu Rev Pharmacol Toxicol. 2013;53:531–556. doi: 10.1146/annurev-pharmtox-032112-135923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H, Fox BA, Le Trong I, Teller DC, Okada T, Stenkamp RE, Yamamoto M, Miyano M. Crystal structure of rhodopsin: A G protein-coupled receptor. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. published online Epub Aug 4 (8725 [pii]) [DOI] [PubMed] [Google Scholar]
- 13.Siu FY, He M, de Graaf C, Han GW, Yang D, Zhang Z, Zhou C, Xu Q, Wacker D, Joseph JS, Liu W, Lau J, Cherezov V, Katritch V, Wang MW, Stevens RC. Structure of the human glucagon class B G-protein-coupled receptor. Nature. 2013;499:444–449. doi: 10.1038/nature12393. published online Epub Jul 25. nature12393. [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hollenstein K, Kean J, Bortolato A, Cheng RK, Dore AS, Jazayeri A, Cooke RM, Weir M, Marshall FH. Structure of class B GPCR corticotropin-releasing factor receptor 1. Nature. 2013;499:438–443. doi: 10.1038/nature12357. published online Epub Jul 25. nature12357 [pii] [DOI] [PubMed] [Google Scholar]
- 15.Wang C, Wu H, Katritch V, Han GW, Huang XP, Liu W, Siu FY, Roth BL, Cherezov V, Stevens RC. Structure of the human smoothened receptor bound to an antitumour agent. Nature. 2013;497:338–343. doi: 10.1038/nature12167. published online Epub May 16. nature12167 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu H, Wacker D, Mileni M, Katritch V, Han GW, Vardy E, Liu W, Thompson AA, Huang XP, Carroll FI, Mascarella SW, Westkaemper RB, Mosier PD, Roth BL, Cherezov V, Stevens RC. Structure of the human kappa-opioid receptor in complex with JDTic. Nature. 2012;485:327–332. doi: 10.1038/nature10939. published online Epub May 17. nature10939 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu B, Chien EY, Mol CD, Fenalti G, Liu W, Katritch V, Abagyan R, Brooun A, Wells P, Bi FC, Hamel DJ, Kuhn P, Handel TM, Cherezov V, Stevens RC. Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists. Science. 2010;330:1066–1071. doi: 10.1126/science.1194396. published online Epub Nov 19. science.1194396 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ballesteros JA, Weinstein H. Integrated methods for the construction of three dimensional models and computational probing of structure–function relations in G-protein coupled receptors. Methods Neurosci. 1995;25:366–428. [Google Scholar]
- 19.In each helix, the following residues are assigned number 50: T6071.50, I6382.50, I6823.50, I7144.50, L7635.50, A8006.50, and L8277.50. The numbering of other residues in each helix is counted relative to the X.50 position according to the B&W numbering system
- 20.Superscripts refer to the B&W numbering for class A GPCRs and that transplanted to class C GPCRs
- 21.Suzuki G, Kimura T, Satow A, Kaneko N, Fukuda J, Hikichi H, Sakai N, Maehara S, Kawagoe-Takaki H, Hata M, Azuma T, Ito S, Kawamoto H, Ohta H. Pharmacological characterization of a new, orally active and potent allosteric metabotropic glutamate receptor 1 antagonist, 4-[1-(2-fluoropyridin-3-yl)-5-methyl-1H-1,2,3-triazol-4-yl]-N-isopropyl-N-methyl- 3,6-dihydropyridine-1(2H)-carboxamide (FTIDC) J Pharmacol Exp Ther. 2007;321:1144–1153. doi: 10.1124/jpet.106.116574. published online Epub Jun. jpet.106.116574 [pii] [DOI] [PubMed] [Google Scholar]
- 22.Fukuda J, Suzuki G, Kimura T, Nagatomi Y, Ito S, Kawamoto H, Ozaki S, Ohta H. Identification of a novel transmembrane domain involved in the negative modulation of mGluR1 using a newly discovered allosteric mGluR1 antagonist, 3-cyclohexyl-5-fluoro-6-methyl-7-(2-morpholin-4-ylethoxy)-4H-chromen-4-one. Neuropharmacology. 2009;57:438–445. doi: 10.1016/j.neuropharm.2009.06.017.S0028-3908(09)00173-7. published online Epub Sep. [pii] [DOI] [PubMed] [Google Scholar]
- 23.Malherbe P, Kratochwil N, Zenner MT, Piussi J, Diener C, Kratzeisen C, Fischer C, Porter RH. Mutational analysis and molecular modeling of the binding pocket of the metabotropic glutamate 5 receptor negative modulator 2-methyl-6-(phenylethynyl)-pyridine. Mol Pharmacol. 2003;64:823–832. doi: 10.1124/mol.64.4.823. published online Epub Oct. 64/4/823 [pii] [DOI] [PubMed] [Google Scholar]
- 24.Litschig S, Gasparini F, Rueegg D, Stoehr N, Flor PJ, Vranesic I, Prezeau L, Pin JP, Thomsen C, Kuhn R. CPCCOEt, a noncompetitive metabotropic glutamate receptor 1 antagonist, inhibits receptor signaling without affecting glutamate binding. Mol Pharmacol. 1999;55:453–461. published online Epub Mar ( [PubMed] [Google Scholar]
- 25.Pagano A, Ruegg D, Litschig S, Stoehr N, Stierlin C, Heinrich M, Floersheim P, Prezeau L, Carroll F, Pin JP, Cambria A, Vranesic I, Flor PJ, Gasparini F, Kuhn R. The non-competitive antagonists 2-methyl-6-(phenylethynyl)pyridine and 7-hydroxyiminocyclopropan[b]chromen-1a-carboxylic acid ethyl ester interact with overlapping binding pockets in the transmembrane region of group I metabotropic glutamate receptors. J Biol Chem. 2000;275:33750–33758. doi: 10.1074/jbc.M006230200. published online Epub Oct 27 ( [DOI] [PubMed] [Google Scholar]
- 26.Surin A, Pshenichkin S, Grajkowska E, Surina E, Wroblewski JT. Cyclothiazide selectively inhibits mGluR1 receptors interacting with a common allosteric site for non-competitive antagonists. Neuropharmacology. 2007;52:744–754. doi: 10.1016/j.neuropharm.2006.09.018. published online Epub Mar. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Malherbe P, Kratochwil N, Muhlemann A, Zenner MT, Fischer C, Stahl M, Gerber PR, Jaeschke G, Porter RHP. Comparison of the binding pockets of two chemically unrelated allosteric antagonists of the mGlu5 receptor and identification of crucial residues involved in the inverse agonism of MPEP. Journal of Neurochemistry. 2006;98:601–615. doi: 10.1111/j.1471-4159.2006.03886.x. [DOI] [PubMed] [Google Scholar]
- 28.Chen Y, Goudet C, Pin JP, Conn PJ. N-{4-Chloro-2-[(1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)methyl]phenyl}-2-hy droxybenzamide (CPPHA) acts through a novel site as a positive allosteric modulator of group 1 metabotropic glutamate receptors. Mol Pharmacol. 2008;73:909–918. doi: 10.1124/mol.107.040097. published online Epub Mar. mol.107.040097 [pii] [DOI] [PubMed] [Google Scholar]
- 29.Molck C, Harpsoe K, Gloriam DE, Clausen RP, Madsen U, Pedersen LO, Jimenez HN, Nielsen SM, Mathiesen JM, Brauner-Osborne H. Pharmacological Characterization and Modeling of the Binding Sites of Novel 1,3-bis(pyridinylethynyl)benzenes as Metabotropic Glutamate Receptor 5-selective Negative Allosteric Modulators. Molecular pharmacology. 2012 doi: 10.1124/mol.112.078808. published online Epub Aug 16. [DOI] [PubMed] [Google Scholar]
- 30.Gregory KJ, Noetzel MJ, Rook JM, Vinson PN, Stauffer SR, Rodriguez AL, Emmitte KA, Zhou Y, Chun AC, Felts AS, Chauder BA, Lindsley CW, Niswender CM, Conn PJ. Investigating metabotropic glutamate receptor 5 allosteric modulator cooperativity, affinity, and agonism: enriching structure-function studies and structure-activity relationships. Molecular pharmacology. 2012;82:860–875. doi: 10.1124/mol.112.080531. published online Epub Nov. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gregory KJ, Nguyen ED, Reiff SD, Squire EF, Stauffer SR, Lindsley CW, Meiler J, Conn PJ. Probing the metabotropic glutamate receptor 5 (mGlu(5)) positive allosteric modulator (PAM) binding pocket: discovery of point mutations that engender a “molecular switch” in PAM pharmacology. Mol Pharmacol. 2013;83:991–1006. doi: 10.1124/mol.112.083949. published online Epub May. mol.112.083949 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu F, Wu H, Katritch V, Han GW, Jacobson KA, Gao ZG, Cherezov V, Stevens RC. Structure of an agonist-bound human A2A adenosine receptor. Science. 2011;332:322–327. doi: 10.1126/science.1202793. published online Epub Apr 15. science.1202793 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kunishima N, Shimada Y, Tsuji Y, Sato T, Yamamoto M, Kumasaka T, Nakanishi S, Jingami H, Morikawa K. Structural basis of glutamate recognition by a dimeric metabotropic glutamate receptor. Nature. 2000;407:971–977. doi: 10.1038/35039564. published online Epub Oct 26. [DOI] [PubMed] [Google Scholar]
- 34.Huang S, Cao J, Jiang M, Labesse G, Liu J, Pin JP, Rondard P. Interdomain movements in metabotropic glutamate receptor activation. Proc Natl Acad Sci U S A. 2011;108:15480–15485. doi: 10.1073/pnas.1107775108. published online Epub Sep 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Doumazane E, Scholler P, Fabre L, Zwier JM, Trinquet E, Pin JP, Rondard P. Illuminating the activation mechanisms and allosteric properties of metabotropic glutamate receptors. Proc Natl Acad Sci U S A. 2013;110:E1416–1425. doi: 10.1073/pnas.1215615110. published online Epub Apr 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eroglu C, Brugger B, Wieland F, Sinning I. Glutamate-binding affinity of Drosophila metabotropic glutamate receptor is modulated by association with lipid rafts. Proc Natl Acad Sci U S A. 2003;100:10219–10224. doi: 10.1073/pnas.1737042100. published online Epub Sep 2. 1737042100 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meiler J, Baker D. ROSETTALIGAND: protein-small molecule docking with full side-chain flexibility. Proteins. 2006;65:538–548. doi: 10.1002/prot.21086. published online Epub Nov 15. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.