Abstract
Long noncoding RNAs (lncRNAs) have emerged as a major regulator of cell physiology, but many of which have no known function. CDKN1A/p21 is an important inhibitor of the cell-cycle, regulator of the DNA damage response and effector of the tumor suppressor p53, playing a crucial role in tumor development and prevention. In order to identify a regulator for tumor progression, we performed an siRNA screen of human lncRNAs required for cell proliferation, and identified a novel lncRNA, APTR, that acts in trans to repress the CDKN1A/p21 promoter independent of p53 to promote cell proliferation. APTR associates with the promoter of CDKN1A/p21 and this association requires a complementary-Alu sequence encoded in APTR. A different module of APTR associates with and recruits the Polycomb repressive complex 2 (PRC2) to epigenetically repress the p21 promoter. A decrease in APTR is necessary for the induction of p21 after heat stress and DNA damage by doxorubicin, and the levels of APTR and p21 are anti-correlated in human glioblastomas. Our data identify a new regulator of the cell-cycle inhibitor CDKN1A/p21 that acts as a proliferative factor in cancer cell lines and in glioblastomas and demonstrate that Alu elements present in lncRNAs can contribute to targeting regulatory lncRNAs to promoters.
Introduction
Long noncoding RNAs (lncRNAs), transcribed from outside (intergenic) or within protein coding regions (intragenic), are >200 nucleotides in length but do not code for proteins. Thousands of lncRNAs have been identified in mammalian cells, many with expression patterns specifically restricted by cell or tissue-type and development stage [1], [2]. The few lncRNA that have been functionally characterized often regulate gene expression, both transcriptionally and post-transcriptionally [3]. LncRNA Xist is a cis-acting RNA responsible for the initiation and spreading of inactivation of the very X chromosome that it is transcribed from, by interacting with and recruiting the Polycomb repressive complex 2 (PRC2) [4] which is key epigenetic regulator during development and tumorigenesis [5], [6] and represses gene expression at the sites where they are recruited by methylating histone H3 on lysine 27. The Xist-PRC2 complex is recruited to the X-chromosome inactivation center by DNA binding protein YY1 [7]. A larger group of trans-acting lncRNAs, like HOTAIR, regulate gene expression at sites away from where they are transcribed, by recruiting to these sites chromatin-modifying complexes such as PRC2, LSD1 and CoREST/REST [8], [9]. It remains unclear, however, how these trans-acting lncRNAs are recruited to their target gene locations.
The cyclin-dependent kinase (CDK) inhibitor p21 is expressed ubiquitously and associates with and inhibits kinases important for G1/S transition such as cyclin D/CDK4, Cyclin D/CDK6 and Cyclin E/CDK2 [10], [11]. The suppression of CDK activity allows the accumulation of hypophosphorylated Rb, which represses the E2F transcription factor to cause G1 phase cell cycle arrest [12]. p21 is a critical molecule for inhibiting cell proliferation in normal and cancer cells and is regulated at multiple levels, most notably at the transcriptional level by the tumor suppressor p53 when the latter is activated by DNA damage [13].
In this study, we identify a novel lncRNA Alu-mediated p21 transcriptional regulator (APTR) necessary for cell proliferation. APTR represses p21 transcription by recruiting the PRC2 complex to the p21 promoter. The complementary Alu (c-Alu) element embedded in APTR is required for the localization of APTR to the p21 promoter, suggesting that embedded Alu elements in lncRNAs can contribute to the functions of lncRNAs. Cellular stresses that induce p21 such as heat shock and doxorubicin treatment down-regulate APTR, and this downregulation is important for the induction of p21 independent of whether p53 is active or not. A survey of gliomas suggests that APTR and p21 levels are anti-correlated. Our results identify a new regulator of p21, an lncRNA APTR, that silences p21 epigenetically by recruiting PRC2 to the p21 promoter.
Materials and Methods
Ethics statement
Ten fresh frozen primary glioblastoma multiforme specimens and two normal brain tissue samples were obtained from patients undergoing surgical treatment at the University of Virginia Hospital following written informed consent and in accordance with a protocol approved by the University of Virginia's Institutional Review Board for Health Sciences Research. These tumors have been studied in two previous publications [14] [15]. All tumor specimens were prepared from patients who did not receive radiotherapy prior to surgery. The correlation coefficient plot with r and P value was generated using GraphPad Prism (GraphPad Software, Inc.).
Plasmid construction
The full length nucleotide sequence of APTR was obtained from the FLJ cDNA library. For MS2 pulldown assays, plasmid pUC-MS2 coat protein (MS2BP) fused to YFP (Addgene plasmid 27122) and plasmid pUC-24MS2 were used. The 24 copies of MS2 stem loops were amplified from the 24MS2 stem loop cassette (Addgene 45162) by PCR and inserted at the 3′ end of APTR in the pcDNA3-APTR plasmid (Invitrogen). For luciferase assays, various lengths (3.7 kb, 2.7 kb and 152 bp) of the p21 promoter with/without the Alu-containing region (−3389 to −2595 from the TSS) were cloned by PCR using 293T cell genomic DNA and inserted into the HindIII sites of the pGL4.20 plasmid (Promega).
Statistical analysis
In all the figures, Student's t-test was used for testing whether the differences were statistically significant, and analyzed using GraphPad Prism (GraphPad Software, Inc.). Statistical significance was defined at P<0.05.
Cell culture, synchronization and transfection
MCF10A, PC3, 293T and U87 cells were grown in DMEM, glioma cell line A172 and HCT116 cells were grown in McCoy's 5a Medium supplemented with 10% fetal bovine serum and penicillin/streptomycin. MCF10A, PC3, 293T, HCT116, A172 and U87 were purchased from the American Type Culture Collection (ATCC, Manassas, VA). HCT116 and p53−/−, p21−/− derivatives were supplied by Dr. B. Vogelstein [16]. For siRNA transfection, we used LipofectamineTM2000 or RNAiMAX (Invitrogen) according to the manufacturer's protocols. The siRNA sequences are provided in Table S1. For flow cytometry analysis, MCF10A cells were treated with BrdU (1 µM/ml) for 30 min and fixed by 70% Ethanol at −20°C. After 1 hr fixation, fixed cells were stained by BrdU-FITC antibody (BD biosciences) for 1 hr and analyzed by FACS analysis with propidium iodide (50 µg/ml). For nocodazole treatment, asynchronous 293T cells were transfected by siGL2, APTR#1 or #2. After 24 hrs of siRNA transfection, cells were treated with nocodazole (0.1 µg/ml) for 16 hrs and then harvested and analyzed by FACScaliber (BD biosciences) for DNA content.
Long noncoding RNA screening and MTT/BrdU incorporation assays
Three different siRNAs were designed against each of the 286 candidate lncRNAs to avoid off-target effects, and transfected into PC3 and MCF10A cells using three 96-well plates with three technical replicates. Two to three days after transfection, cells were incubated with 10 µM BrdU for 15 min to 1 hr, and fixed with FixDenat (Roche, Indianapolis, IN) for 30 min. Cells were blocked with 3% BSA in PBS for 1 hr, then incubated with HRP-coupled anti-BrdU antibody (Roche) diluted in 3% BSA in PBS for 1 hr. After washing three times with PBS containing 0.1% TX-100, cells were incubated with TMB substrate (Pierce, Rockford, IL) for 5–10 min and analyzed by the absorbance at 450 nm following the addition of 1 M H2SO4 to stop the reaction. For data analysis, each assay plate contained four wells of negative control (luciferase; GL2) and two wells of positive control (ORC2) siRNAs for normalization. To normalize values of BrdU incorporation, we calculated the inhibition index (%) using the following equation: Inhibition index of gene X (%) = (GL2av−X)/(GL2av−ORC2av) ×100, where X, GL2av, and ORC2av represent BrdU incorporation (absorbance at 450 nm) of gene X and average of GL2 and ORC2, respectively. Genes with a SD of inhibition indices (from 3 technical replicates) greater than a cutoff value were eliminated to select technically reproducible data.
RNA purification, RT-PCR and Quantitative PCR
Total RNA was extracted from cells using Trizol total RNA isolation reagent (Invitrogen). cDNA was synthesized with oligo (dT)18 or Random hexamer by the Superscript III First Strand Synthesis System for RT-PCR (Invitrogen) according to the manufacturer's instructions. Quantitative RT-PCR was performed by iCycler iQ™ Real time PCR Detection System (BioRad) with SYBR green Master Mix (BioRad). The Ct values were determined with the default threshold setting. Relative expressions for the target RNAs were determined by the comparative CT (2−ΔΔCt) method after normalization to GAPDH.
APTR knockdown/rescue assays
After 6hrs of siRNA transfection, 293T cells were transfected by control or APTR deletion mutant-encoding plasmids. Total RNA was analyzed by RT-PCR or Q-RT-PCR after 72 rs of siRNA transfection. The knockdown efficiency (more than 80% reduction) of endogenous APTR in each experiment was confirmed by Q-RT-PCR using the primer set 1 and 2 (Table S1).
Cell proliferation assays
For cell proliferation assays, HCT116 p21+/+ and HCT116 p21 −/− were seeded at 1×106 cells/well in 6-cm plates and transfected by siRNA against GL2, APTR after 24 hours. Viable cells were counted every two days by Trypan Blue exclusion using auto cell counter (Invitrogen), and then replated at 1×106 cells/well. All experiments were performed on three biological replicates. P values were calculated by two-way ANNOVA analysis using GraphPad Prism (GraphPad Software, Inc.).
Northern blot analysis
Total RNA was extracted from 293T cells using TRIzol reagent (Invitrogen). Poly(A) RNA was purified from total RNA using a PolyA TRACT kit (Promega) according to the manufacturer's protocol. Northern blot analysis was performed by standard protocol. [α-32P]-dCTP-labeled DNA oligonucleotides were made by Rediprimer II DNA labeling system (GE healthcare). The probes were generated from a 300 nt fragment of APTR cDNA (651–950), amplified using the following primer sets:
Forward, 5′-TGTGGGTACAAAAGGAGAGTAACAT-3′;
Reverse, 5′-GTAGATCTGGAGCTGCAACTACAG-3′.
Heat shock and Doxorubicin treatment
For heat shock, 293T cells were treated at 37, 42 or 55°C for 30 min followed by incubation at 37°C for 60 min and then total RNA was extracted. For rescuing p21 transcription, cells were transfected with pcDNA3 or pcDNA3-APTR1-2303 for 48 hr and incubated at 37°C or 55°C for 30 min followed by recovery at 37°C for 60 min. HCT116 cells were treated with Doxorubicin (0, 10, 30 µM) for 16 hrs and total RNA was extracted. After cDNA synthesis, RNA expression was analyzed by RT-PCR. The primer sets are provided in Table S1.
Immunoprecipitation analysis
Immunoblot and immunoprecipitation analysis were performed as described previously with slight modifications [17]. Histone H1 kinase assay was performed as previously [18]. The following antibodies were used: anti- α-Tubulin (B-5-1-2), Cyclin E1 (HE111 and C-19) (Santa Cruz); p21 (CP36) (Millipore); phosphor-RB (ser807/81; D20B12) (Cell signaling). Total RB (4H1) (Cell signaling).
Crosslinking immunoprecipitation with MS2 for DNA or RNA purification
After 48 hrs of cotransfection of APTR-MS2 and MS2BP-YFP plasmids into 293T cells, cells were harvested. For MS2-CLIP assay of APTR in EZH2 or SUZ12 knockdown cells, 6 hrs following transfection of siRNAs against EZH2 or SUZ12, MS2BP-YFP and APTR-MS2 or MS2 plasmids were cotransfected into 293T cells and cells were harvested after 48 hours. Cells were then crosslinked with 1% Glutaraldehyde for 10 min and homogenized with the ChIP lysis buffer followed by sonication in the same way as the ChIP assay described below. The lysate was immunoprecipitated by ChIP grade anti-GFP antibody (Abcam, ab290) which can crossreact with the YFP protein. For MS2 pulldown/chromatin isolation, after washing and decrosslinking by heat incubation at 65°C overnight, DNA was extracted with phenol/chloroform after treatment with RNaseA at 37°C for 30 min followed by proteinase K at 55°C for 60 min. Samples were analyzed by qPCR, using specific primer sets provided in Table S1.
Chromatin immunoprecipitation assay
Chromatin immunoprecipitation assays were performed as described previously with slight modifications [17]. Briefly, 293T cells were crosslinked with 1% formaldehyde for 15 min and neutralized in the presence of 0.125 M Glycine. The cells were homogenized with ChIP lysis buffer [(50 mM Tris-HCl (pH 8.0), 10 mM EDTA, 0.1%SDS, and a proteinase inhibitor cocktail (Complete Mini) (Roche). Each sample was immunoprecipitated with pre-mixed antibody-dynabead protein G complex. The following antibodies were used: ChIP grade anti-rabbit IgG (Abcam, ab46540), anti-SUZ12 and anti-H3K27me3 (Millipore, 07-449). The immunoprecipitant was incubated at 65°C overnight to decrosslink and treated with RNaseA at 37°C for 30 min and proteinase K at 55°C for 60 min followed by phenol/chloroform treatment. Purified DNA was analyzed by qPCR analysis using the primers provided in Table S1.
RNA immunoprecipitation and biotinylated RNA pulldown assays
For RNA immunoprecipitation and biotinylated RNA pulldown, 293T cells (5×106) were fixed with 1% formaldehyde and lysed in hypertonic lysis buffer [20 mM sodium phosphate at pH 7.0, 250 mM NaCl, 0.1% NP-40, 1 mM EDTA, 1 mM DDT and a protease inhibitor cocktail (Complete Midi, Roche)] followed by sonication, and then subjected to RNA immunoprecipitation by the following antibodies; anti-EZH2 (Cell Signaling, AC22), SUZ12 (Abcam, ab12073) and rabbit polyclonal IgG antibody. For biotinylated RNA pulldown, biotinylated sense/antisense wild type or deletion mutants of APTR were generated by MEGAscript T7 kit (Life technologies) according to the manufacturer's protocol. Whole cell extracts derived from 293T cells were pulled down by Streptavidin sepharose beads-Biotinylated APTR complex after 12 hr incubation at 4°C. The precipitated RNA was subject to cDNA synthesis and analyzed by RT-PCR. The primer sets are provided in Table S1.
Dual luciferase reporter assays
293T cells were seeded on 6-well plates and co-transfected by Firefly luciferase reporter fused to various lengths of the p21 promoter (200 ng), control Renilla luciferase reporter (5 ng) and either empty vector or vector expressing wild type or deletion mutants of APTR (2 ug). After 48 hrs of transfection, the lysates were analyzed by Dual-Luciferase reporter assay system (Promega) according to the manufacturer's protocol. Firefly luciferase activity of each reporter was normalized to the Renilla luciferase activity.
Results
A targeted siRNA screen for lncRNAs required for cell proliferation identifies APTR
We previously reported a targeted RNAi (TARCOR) screen for genes required for cell proliferation, where we performed a manual, moderate-throughput siRNA transfection against a targeted gene set and measured effects on cell proliferation or viability by a quantitative comparison of BrdU incorporation in wells transfected with different siRNAs [19]. We applied the same method to identify lncRNAs required for cell proliferation and viability. The ‘Full-length Long Japan’ (FLJ) collection of sequenced human full length cDNA [20] was screened for those that did not contain an open reading frame to find 286 putative lncRNAs. We designed 3 different siRNAs against each lncRNA to guard against off-target effects, and transfected them separately into MCF10A (p53-wild type, non-transformed breast epithelial cells). BrdU incorporation was measured in each well after 72 hrs of siRNA transfection and compared to that in wells transfected with si-GL2 (negative control) and si-ORC2 (a known essential factor for cell proliferation). Knockdown of 74 lncRNAs inhibited BrdU incorporation to at least 90% of the level of inhibition seen with si-ORC2 (Table S2). Among these, we searched for lncRNAs for which there was evidence of moderate to high expression in RNA-seq data from MCF10A cells. In this paper we focus on one of these lncRNAs, Bramy2034329/SLV01230, which was renamed by us as APTR.
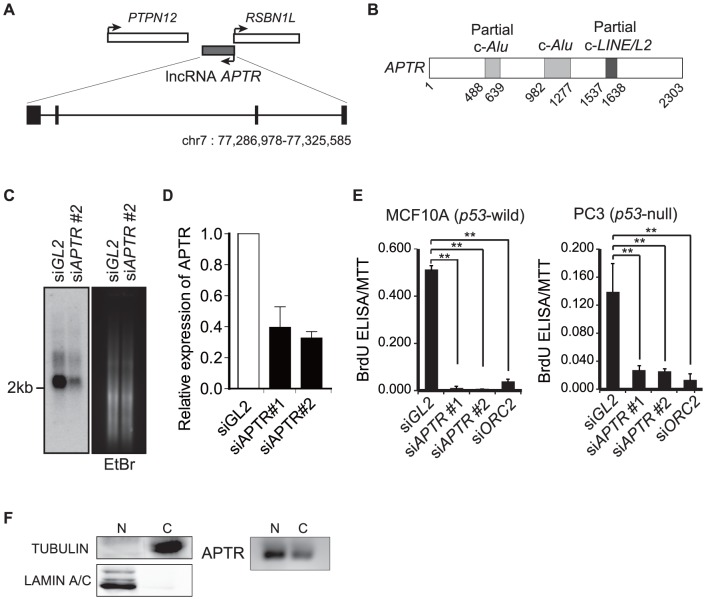
APTR is a 2303-nucleotides lncRNA expressed from the opposite strand of the intergenic region between PTPN12 and RSBN1L in chromosome 7q21 (Figure 1A, Figure S1). RepeatMasker (http://www.repeatmasker.org) found that APTR possesses two sequences complementary to SINE/Alu elements (c-Alu) as well as one sequence complementary to LINE/L2 element (Figure 1B and Figure S2). Northern blot analysis on poly(A)+RNA derived from 293T cells identified a 2.3 kb RNA that is decreased by siRNA against APTR (Figure 1C). Consistent with our lncRNA screening results, knockdown of APTR by two different siRNAs not only decreased proliferation of MCF10A, but also PC3 (prostate cancer cell with mutant p53) cells to the same extent as knockdown of ORC2 (Figure 1D and E). Since APTR is enriched by 5′ oligo capping method (Figure S1) and polyA RNA purification (Figure 1C) [20], we conclude that APTR is a capped, poly(A) tailed and spliced lncRNA expressed mainly in nucleoplasm (Figure 1F) and required for proliferation of multiple cell-lines.
Figure 1. LncRNA APTR knockdown inhibits cell proliferation in a p53-independent manner.
(A) Schematic of the APTR locus. Black boxes represent exons. (B) Schematic of lncRNA APTR. “c-“: complementary to Alu or LINE elements. (C) Northern blot of APTR on poly(A) RNA in 293T cells transfected with siGL2 or siAPTR#2, at 72 hr after siRNA transfection. Probe: APTR cDNA (651–950). (D) Relative APTR expression levels normalized to GAPDH in 293T cells transfected with siGL2 or siAPTR#1, #2, at 72 hr after siRNA transfection. (E) APTR knockdown inhibits cell proliferation. BrdU incorporation (DNA synthesis) was measured by BrdU ELISA, 72 hr after siRNA transfection, and normalized to MTT assays (number of viable cells). siOrc2 was used as a positive control known to inhibit cell proliferation. **: P<0.005. (F) Subcellular expression levels of APTR in 293T cells. α-TUBULIN and LAMIN A/C were analyzed as markers for cytoplasm or nuclear fraction (left panel). APTR expression levels in the fractions were analyzed by RT-PCR (right panel).
APTR regulates cell proliferation via p21 transcriptional suppression
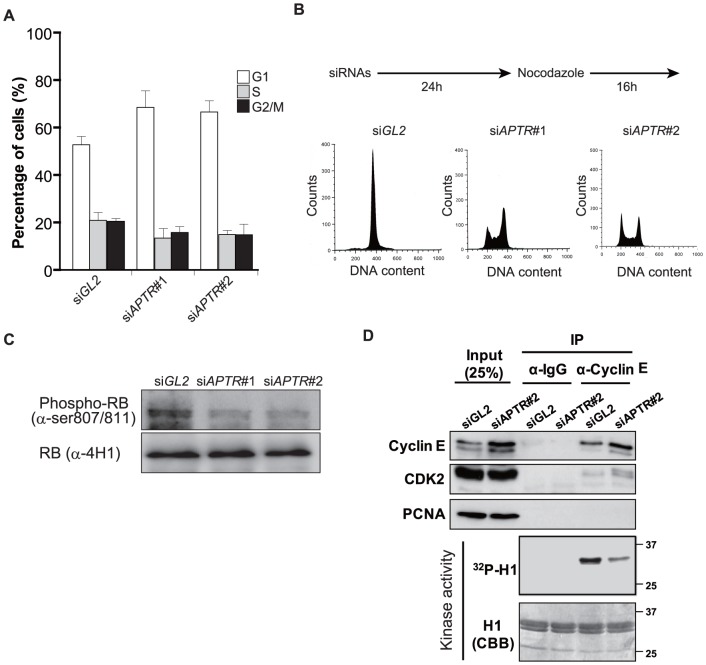
To explore the molecular mechanism for cell proliferation, we next examined whether APTR regulates cell cycle progression. Depletion of APTR by two different siRNAs in asynchromous MCF10A resulted in an increased G1 population (Figure 2A). Progression of 293T cells through the cell-cycle was also analyzed after APTR knockdown in the presence of nocodazole, a chemical that blocks cells in M phase. After 24 hrs of siAPTR#1 or #2 transfection, asynchronous 293T cells were treated with nocodazole (0.1 µg/ml) for 16 hrs, and then analyzed by FACS analysis for propidium iodide staining for DNA content. As shown in Figure 2B, while siGL2 transfected control cells accumulated in M phase, depletion of APTR led to an increase of G1 and S populations. In agreement with the increase in G1 population, depletion of APTR resulted in a decreased RB phosphorylation (Figure 2C). Cyclin E1/CDK2 kinase was decreased in activity on histone H1 in vitro (Figure 2D). These findings suggest that APTR is important for G1-S transition most likely due to a requirement for the activity of CyclinE/CDK2 kinase [12].
Figure 2. APTR depletion suppresses the G1/S phase progression.
(A) MCF10A cells transfected with siAPTR #1 or #2 accumulate in the G1 phase of the cell-cycle as measured by two color FACS for propidium-iodide and BrdU (mean ±s.e.m., n = 3). (B) 293T cells transfected with siGL2 or siAPTR #2 were analyzed by FACS for propidium-iodide in presence of Nocodazole (0.1 µg/ml). Schematic of the Nocodazole treatment procedure was presented on the top. (C) Immunoblot shows that Retinoblastoma protein is hypo-phosphorylated in 293T cells transfected with siAPTR #1 and #2. Total RB protein was analyzed as a loading control. (D) Reduced kinase activity of Cyclin E/CDK2 in 293T cells transfected with siAPTR#2. Cyclin E1 and CDK2 were analyzed by IP and immunoblot with indicated antibodies. PCNA was analyzed as a loading control. Kinase activity: autoradiogram of 32P labeled histone H1 after in vitro kinase assays with immunoprecipitates. CBB; coomassie blue staining to show equal amounts of H1 were added to all the lanes.
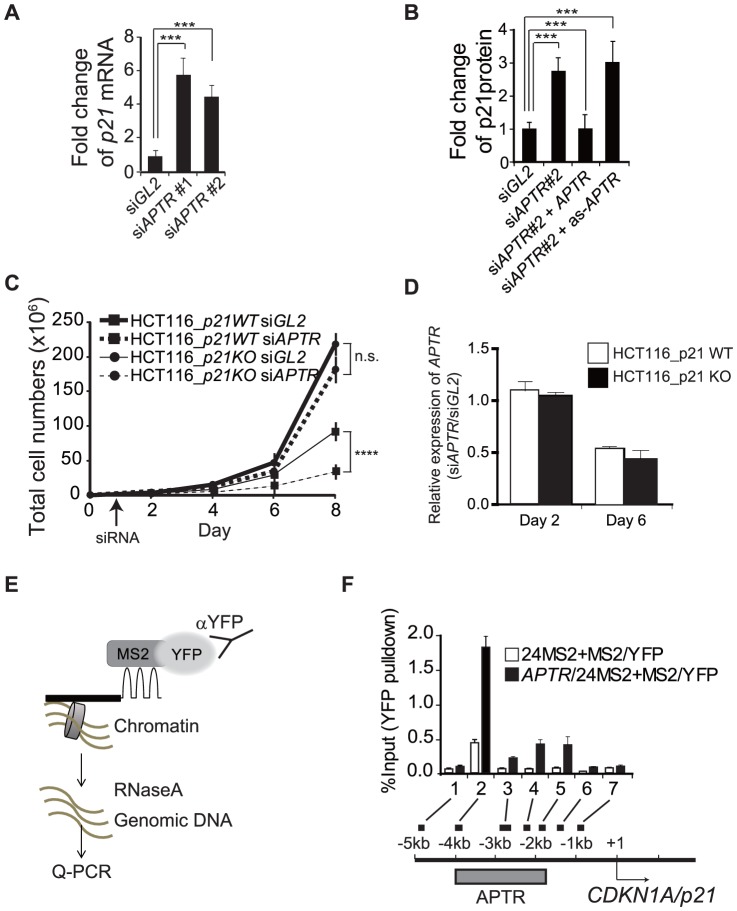
Next, we surveyed the levels of CDK inhibitors in the APTR knockdown cell lines and found that p21 mRNA and protein were increased after APTR knockdown (Figure 3A and B). Induction of p21 after APTR knockdown can be prevented by over-expression of exogenous sense-strand APTR (but not anti-sense APTR) (Figure 3B), suggesting that induction of p21 is specifically due to APTR knockdown. The exogenous APTR is expressed at a sufficiently high level that enough APTR remains in the cells even after siAPTR transfection (Figure S3). Notably, depletion of APTR induced p21 even in p53-deficient cell lines (Figure S4), indicating that APTR suppresses p21 in a p53-independent manner.
Figure 3. APTR suppresses p21 transcription.
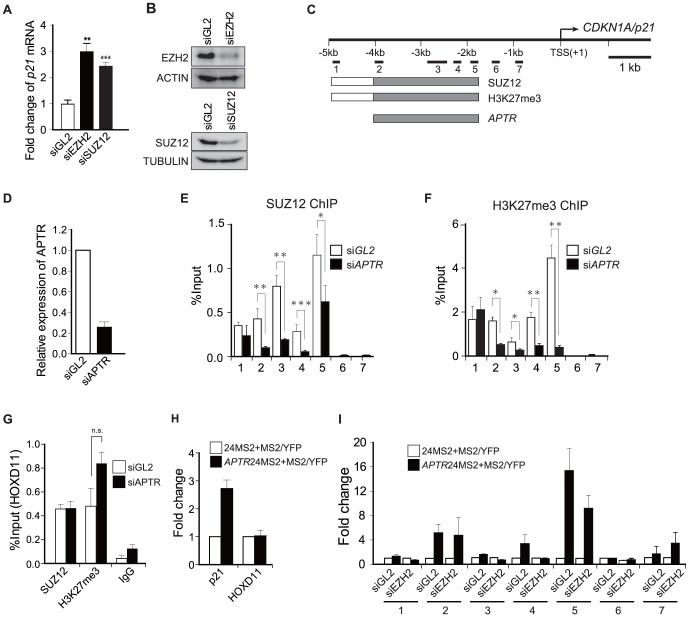
(A) Q-RT-PCR shows induction of p21 mRNA (normalized to GAPDH) after siAPTR. Fold change compared to siGL2-transfected 293T cells (mean ±s.e.m., n>6, ***: P<0.0005). (B) The induction of p21 protein in the siAPTR#2 transfected 293T cells is prevented by overexpression of sense but not antisense APTR. Fold change of p21 normalized to ACTIN, compared to siGL2-transfected cells (mean ±s.e.m., n>3, **: P<0.005). (C) Growth suppression after siAPTR is alleviated in p21 −/− HCT116 cells. Mean ±s.e.m. n = 9. Right: APTR expression levels (Normalized to GAPDH) measured by Q-RT-PCR in p21 +/+ or p21 −/− HCT116 cells at the indicated days after transfection of siRNAs (n.s.: not significant, ****: P<0.0001). (D) Q-RT-PCR shows fold change of APTR (normalized to GAPDH), compared to the siGL2-transfected cells in the two cell lines in C (mean ±.e.m., n = 3). Note that cells were transfected on Day 1, so Day 2 is 1 day after transfection and Day 6 is 5 days after transfection. Thus si-APTR does not decrease APTR on day 1 after transfection, but the APTR RNA remains low up to day 5. (E) Schematic of MS2-CLIP. The dark line is APTR RNA fused to MS2 binding sequences. (F) APTR associates with the p21 promoter. Top: The % of input DNA present in the MS2BP-YFP CLIP is shown in cells expressing MS2 alone or MS2-APTR (mean ±s.e.m, n>6). 1–7 refer to the primer pairs in the schematic. Bottom: locations of p21 promoter fragments amplified by primer pairs 1–7 in the CLIP assay. Grey bar: area where APTR binds.
Is p21 induction necessary for inhibition of cell proliferation upon APTR knockdown? To address this, we observed cell growth in p21 −/− HCT116 cell lines [21] transfected with siAPTR. Knockout and siRNA-mediated knockdown of p21 attenuated the cell growth inhibition upon APTR knockdown (Figure 3C, 3D and Figure S5), suggesting that the cell growth inhibition in siAPTR transfected HCT116 cells was dependent on p21 induction. We next sought to determine whether APTR is physically associated with the p21 promoter. MS2 tagging of RNA is widely used for following RNA localization in eukaryotic cells [22], [23]. The 24 repeats of MS2-binding stem loops of bacteriophage MS2 RNA fused to APTR (APTR/MS2) is bound by bacteriophage MS2 binding protein (MS2BP) fused to fluorescent protein YFP (MS2BP/YFP). We adapted the MS2 tagging system for MS2-crosslinking and immunoprecipitation (MS2-CLIP) to enrich the chromatin bound to MS2 tagged RNA (Figure 3E). Optimization experiments revealed that 1% glutaraldehyde fixation is needed to obtain reproducible results upon chromatin immunoprecipitation using anti-YFP antibody (data not shown). MS2-crosslinking and immunoprecipitation (MS2-CLIP) demonstrated that APTR binds to multiple DNA sites between −1.6 and −4 kb relative to the transcription start site (TSS) of p21 (Accession: NM_000389) (Figure 3F), indicating that APTR may suppress the p21 promoter directly.
APTR interacts physically with the Polycomb repressive complex2
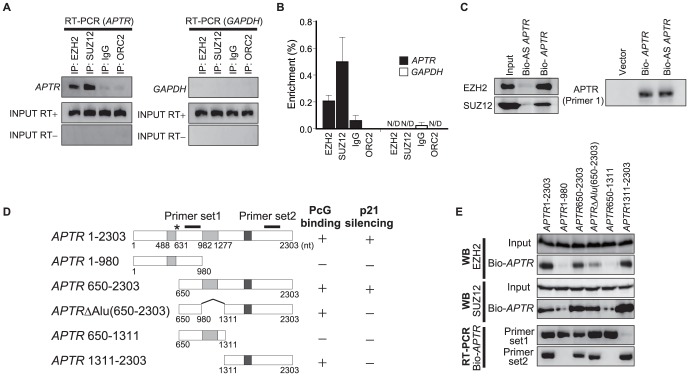
Recent studies revealed that the repressive H3K27 trimethylation mark is regulated by PRC2 on the p21 promoter and is responsible for stable p21 gene silencing [24], [25]. PRC2 contains EZH2, the enzyme that catalyzes the repressive trimethylation of histone H3 on lysine 27 (H3K27me3) [6] and SUZ12, a cofactor required for the catalytic activity of EZH2 [26]. To address the molecular mechanism by which APTR silences the p21 gene, we examined the interaction between APTR and the PRC2 complex in RNA immunoprecipitation assays with antibodies to EZH2 and SUZ12 followed by RT-PCR (Figure 4A) and Q-RT-PCR (Figure 4B). APTR is present in immunoprecipitates of EZH2 or SUZ12, but not of ORC2 (negative control protein). An unrelated RNA, GAPDH mRNA was also not precipitated with EZH2 or SUZ12 (Figure 4A, right panel). Conversely, in vitro transcribed biotinylated sense APTR associated in vitro with cellular EZH2 and SUZ12 and pulled them down on streptavidin beads (Figure 4C), but antisense APTR did not do so. Biotinylated APTR with specific deletions (Figure 4D and E) showed that the 3′ end of APTR (1311–2303) lacking any Alu elements was sufficient for binding EZH2 and SUZ12, but portions containing only the c-Alu sequences (1–980 or 650–1311) could not bind EZH2 and SUZ12 (Figure 4E). These findings indicate that the 3′ portion of APTR specifically interacts with the PRC2 complex.
Figure 4. APTR interacts with PRC2 proteins EZH2 and SUZ12.
(A) APTR interacts specifically with EZH2 and SUZ12 in vivo. Top: RNA Immunoprecipitates of 293T cell lysates probed for APTR by RT-PCR using primer set 1 in Fig. 4D. GAPDH was analyzed as a negative control in the right. IgG, ORC2: IP negative controls. Middle and bottom: input RNA analyzed with and without RT. (B) Enrichment of endogenous APTR in the RNA immunoprecipitates of endogenous EZH2 and SUZ12. RNA immunoprecipitation analysis in Figure 4A was analyzed by Q-RT-PCR. IgG and ORC2 were analyzed as a negative IP control. GAPDH was analyzed as a negative control. N/D represents not detectable. Error bars indicate Mean ±s.e.m. (n = 3). (C) Left: Immunoblot for EZH2 and SUZ12 after mixing in vitro transcribed biotinylated sense-/antisense-APTR with cell lysates and pull-down on streptavidin beads. Right: RT-PCR to show equal levels of biotinylated sense- or antisense-APTR (input RNA). (D) Schema of wt APTR (as in Fig. 1B) and deletion mutants with locations of primer sets 1 and 2 used to detect APTR. Summary of Figure 4D (PcG binding) and Figure 6A (p21 silencing) indicated on the right. (E) EZH2 and SUZ12 interact with the 3′-portion of APTR in an experiment similar to Figure 4C. Top 4 panels: Proteins input or pulled down by biotinylated APTR detected by immunoblots. Bottom 2 panels: WT and mutant APTR were pulled down at comparable levels on strepavidin beads as detected by RT-PCR with primer sets 1 and 2 (note that some deletions can be detected only by one primer pair).
We next examined whether the PRC2 complex is recruited to the p21 promoter by APTR and whether its catalytic activity is required for the repression of the p21 gene. Depletion of EZH2 or SUZ12 by siRNA induces p21 mRNA in 293T cells (Figure 5A and B). The ChIP of endogenous SUZ12 revealed that SUZ12 is localized at −1.6 to −4.7 kb relative to the TSS of p21 (siGL2 in Figure 5E; summarized as boxes labeled SUZ12 in Figure 5C). These sites overlap with the −1.6 to −4 kb region where APTR binds to the p21 promoter (APTR box, Figure 5C and Figure 3F). Knockdown of APTR significantly decreased the recruitment of SUZ12 to the −1.6 to −4 kb region of the p21 promoter (siAPTR in Figure 5E; grey parts of the SUZ12 boxes in Figure 5C).
Figure 5. APTR regulates p21 epigenetically via the PRC2 complex.
(A) Q-RT-PCR shows induction of p21 mRNA normalized to GAPDH after EZH2 or SUZ12 knockdown on the left. Fold change compared to siGL2-transfected cells. Mean ±s.e.m, n>6, **: P = 0.006. ***: P<0.0001). (B) Endogenous EZH2 and SUZ12 were analyzed by immunoblot after EZH2 or SUZ12 knockdown as a control for Figure 5A. (C) Schematic of p21 promoter and primer pairs for CLIP PCR (as in Figure 3F). Boxes: binding of indicated proteins/RNA in control cells (Figure 3F, 5E–F). Grey areas: regions where binding is downregulated by siAPTR#2. (D) qRT-PCR shows APTR knockdown efficiency by siAPTR#2 in Figure 5E–G. (E–F) ChIP of SUZ12 or H3K27me3 on the p21 promoter in 293T cells transfected by either siGL2 or siAPTR#2. X-axes: primer-pairs 1–7 in Figure 3F. Y-axes: %Input values were presented. Mean ±s.e.m. n = 6. (G) ChIP of SUZ12 or H3K27me3 on the HOXD11 locus in 293T cells transfected by either siGL2 or siAPTR#2. X- axis: antibodies used for ChIP. Y-axis: % input of HOXD11 locus in precipitates. (n.s.: not significant, Mean ±s.e.m. n = 6) (H) CLIP of MS2-APTR or MS2 RNA alone in the p21 promoter (primer-pair 2 in Figure 3F) and the HOXD11 locus. Mean ±s.e.m. n = 6. Y-axis shows amount of specific DNA in the precipitate normalized to that in the CLIP of MS2 alone. (I) CLIP of MS2-APTR or MS2 RNA alone on the p21 promoter in 293T cells transfected by siGL2 or siEZH2. X-axes: primer-pairs 1-7 in Figure 3F. Y-axis: amount of specific DNA in the precipitate normalized to that in CLIP of MS2 alone. Mean ±s.e.m. n = 3.
Notably, the −1.6 to −4.7 kb area of the p21 promoter was also enriched for H3K27me3 (Figure 5F; H3K27me3 box in Figure 5C), and knockdown of APTR selectively decreased the repressive H3K27me3 modification at −1.6 to −4 kb relative to the TSS (Figure 5F; grey part of the H3K27me3 box in Figure 5C). The global level of H3K27me3 does not change after APTR knockdown (Figure S6).
The ChIP enrichment of SUZ12 and H3K27me3 in the p21 promoter are comparable to that seen at the known PRC2 target locus, HOXD11 [27] (siGL2 in Figure 5G). Knockdown of APTR did not decrease SUZ12 or H3K27me3 at the HOXD11 locus (siAPTR in Figure 5G). Compatible with these results, MS2-CLIP showed no enrichment of APTR in the HOXD11 locus (Figure 5H). Thus APTR is co-localized with the PRC2 complex selectively at the p21 locus and not at all PRC2-bound promoters. The increase of H3K27me3 at the HOXD11 promoter after knockdown of APTR is clearly not due to an increase in recruitment of PRC2 (SUZ12 in Fig. 5G), and not due to an increase in global levels of H3K27me3 (Fig. S6). The increase could be due to a local decrease in demethylation, which may not be directly dependent on APTR, but is beyond the scope of this paper.
Next, we asked whether the PRC2 complex is required for recruitment of APTR to the p21 promoter. Fig. 3F shows that APTR is maximally recruited to sites 2, 4 and 5 in the p21 promoter. MS2-CLIP of APTR after siGL2 and siEZH2, showed that APTR was still recruited to sites 2 and 5 after EZH2 depletion (Figure 5I), though there was a slight decrease at site 4 and increase in site 7. The results suggest that PRC2 is not required to recruit APTR to two of the three major sites of recruitment in the p21 promoter.
Collectively, these results indicate that the PRC2 complex is recruited by APTR to the p21 promoter specifically between −1.6 to −4 kb relative to the TSS, where the complex catalyzes the repressive modification of chromatin to repress the p21 promoter.
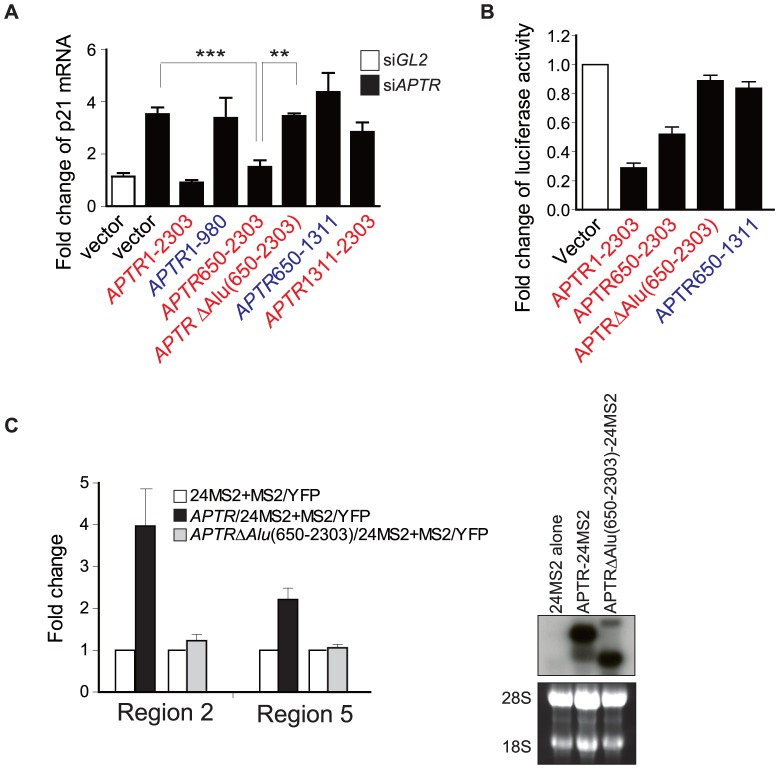
Identification of the portions of APTR required for p21 gene silencing
To perform structure-function studies on APTR, we rescued the APTR knockdown by overexpressing exogenous APTR (wt or deletion mutants). Knockdown of endogenous APTR increases p21, but this was reversed by over-expressing exogenous full length APTR (Figure 6A, first three bars). Overexpression of deletion mutants of APTR indicated which portions of APTR are required for p21 silencing (Figure 6A and summarized in Figure 4D). All the versions of exogenous APTR are expressed at comparable levels (Figure S7). APTR (650–2303), deleted of the first partial c-Alu element but retaining the second complete c-Alu, silenced p21. APTR that cannot bind PRC2 (1–980 or 650–1311) was unable to silence p21, suggesting that interaction with PRC2 is essential for silencing p21. Interestingly, APTRΔAlu(650–2303) missing all c-Alu elements did not silence the p21 promoter even though it could bind to PRC2 (Figure 6A and Figure 4D).
Figure 6. The embedded c-Alu and the 3′ end of APTR are each required for p21 suppression.
(A) Q-RT-PCR of p21 mRNA after siAPTR#2 (black bars) and transfection of empty vector or vector expressing wild-type or mutant APTR in 293T cells. X-axis: empty vector or form of APTR expressed by transfected plasmid. Red represents the mutants that bind to PRC2. Blue represents the ones that do not bind to PRC2. Y-axis: fold change of p21 normalized to GAPDH relative to siGL2-transfected cells receiving empty vector (white bar). Mean ±s.e.m; n = 6, ***: P<0.0005. **: P<0.005. (B) Firefly luciferase activity of p21 promoter in cells transfected with empty vector or vector expressing wild-type or deletion mutants of APTR. Red represents the mutants that bind to PRC2. Blue represents the ones that do not bind to PRC2. Firefly luciferase activities were normalized to Renilla luciferase from co-transfected plasmid. (C) CLIP of MS2-APTR or MS2-APTRΔAlu(650–2303) or MS2 RNA alone (negative control) on two sites in the p21 promoter (2 and 5 as defined in Figure 3F). Y-axis: amount of specific DNA in the precipitate normalized to that in CLIP of MS2 RNA alone. Mean ±s.e.m., n = 6. Northern blot on the right with APTR cDNA (651–950), shows that the APTR fusion RNAs are expressed.
We confirmed these results by co-transfecting a firefly luciferase reporter driven by the p21 promoter (2.7 kb in length from TSS) with a plasmid expressing wt or mutant APTR (Figure 6B). Here again, APTR(650–2303) repressed the p21 promoter, but deletions that removed the c-Alu elements or the 3′ end of APTR abrogate this repression. These findings suggest that the 3′ terminal portion and the embedded second c-Alu of APTR are both required for p21 silencing.
The 3′ portion of APTR is required for APTR to interact with PRC2. To explore whether the embedded second c-Alu element is required for targeting APTR to the p21 promoter, we assessed the localization of APTRΔAlu(650–2303) in MS2-CLIP assay. As shown in Figure 6C, APTRΔAlu(650–2303)/MS2 was not recruited to the p21 promoter. Consistent with this and Figure 6B, over-expressing exogenous APTR/MS2 suppressed the p21 promoter luciferase activity (2.7kb in length from TSS), but APTRΔAlu(650–2303)/MS2 failed to do so (data not shown). These findings suggest that the c-Alu element of APTR is important for recruiting the APTR-PRC2 complex to the p21 promoter (schematic in Figure 7A).
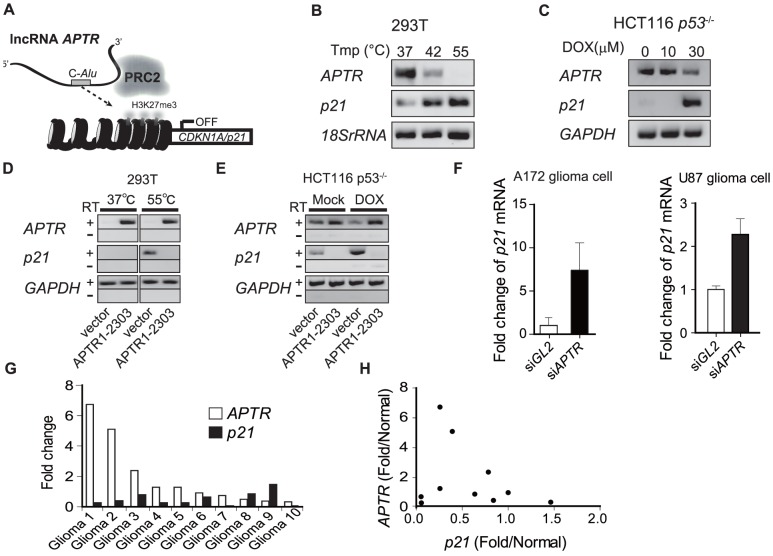
Figure 7. The implication of APTR-mediated p21 silencing in normal cell function and cancer.
(A) A schematic of the lncRNA APTR-mediated p21 gene silencing. APTR suppresses p21 gene expression by guiding the PRC2 complex to the p21 promoter. (B–C) RT-PCR of indicated transcripts in indicated cells. GAPDH and 18SrRNA are loading controls. (D–E) Overexpression of exogenous APTR (indicated at bottom) suppresses the induction of p21 after heat shock or after Doxorubicin in indicated cells. RT: reverse transcriptase. Fewer PCR cycles were done compared to Figure 7B explaining why endogenous APTR is not seen. (F) siAPTR induces p21 in human glioma cells. Q-RT-PCR of p21 mRNA normalized to GAPDH. Mean ±s.e.m; n = 3. (G) Levels of APTR and p21 mRNAs in ten GBMs. Fold change in the GBM relative to normal brain tissue (average of two normal brains). (H) Scatter plot of the data in Figure 7G to show the anti-correlation between APTR and p21 RNAs. Pearson R = −0.254. unpaired t test P = 0.004, n = 10.
Decrease of APTR correlates with p21 induction by cellular stress and in glioblastomas
Finally, we examined whether the regulation of p21 by APTR is important for normal cellular physiology or for disease. Cellular stresses that induce p21, such as heat shock (Figure 7B), or the DNA damage-inducing chemotherapy drug doxorubicin (Figure 7C), down-regulate APTR expression while up-regulating p21. Over-expression of exogenous APTR prevents induction of p21 after heat shock or doxorubicin-induced DNA damage (Figure 7D and 7E), indicating that the decrease of APTR is important for the induction of p21 following heat shock or DNA damage.
Glioblastoma multiforme (GBM), the most aggressive primary brain cancer in humans, often inactivates the p53–p21 signaling pathway [28], [29]. p21 is also critical for the radiation sensitivity and/or chemosensitivity of GBM cells [30], [31]. We wondered whether variation in APTR expression could affect p21 expression in human GBM. siRNA depletion of APTR in two GBM cells (A172 and U87) up-regulated p21 mRNA (Figure 7F) and caused cell growth inhibition (Figure S8). Notably, co-knockdown of p21 in the U87 cells alleviates the growth inhibition produced by APTR knockdown (Figure S8). In addition, quantitative RT-PCR of total RNAs derived from 12 frozen GBM samples showed anti-correlation of APTR and p21 mRNAs (Pearson r = −0.2549, P = 0.0396) in these GBM samples (Figure 7G, 7H and Figure S9). Therefore the level of expression of APTR is important for determining the expression of p21 in GBM cell lines and tumors.
Discussion
A novel lncRNA APTR is a bona fide p21 regulator for cell proliferation
We identify a new lncRNA, APTR, which is expressed from a distant site in the genome, but targets the p21 promoter in trans for epigenetic repression. Depletion of APTR resulted in the dissociation of the PRC2 complex throughout the APTR localization sites in the p21 promoter (Figure 3 and 5). Most interesting, cellular stresses that induce p21, also decrease the expression level of the repressor APTR and forced expression of APTR inhibited p21 induction in the stressed condition (Figure 7B–E). Numerous studies show that p21 gene is regulated at multiple levels, transcriptional, post-transcriptional and post-translational, in response to intra- or extracellular stress [13]. We identified an lncRNA regulator that plays an important role in the constitutive suppression of p21 and in the induction of p21 by stress, independent of p53.
Despite the number of independent factors that regulate p21, it is heartening to see that APTR and p21 levels were anti-correlated in glioblastomas, suggesting that at least in these tumors APTR is a significant repressor of p21. We do not know whether the variation in levels of APTR stem from genetic differences between the tumors or differences in the growth rate, oxygenation or other factors that affect the level of stress on the tumors. In addition, larger, well-annotated tumor sets will be examined in the future to ascertain whether APTR levels increase with glioma progression and/or whether APTR is an useful prognostic marker both for survival and for response of the tumor to radio- or chemotherapy.
The p21 gene locus expresses several cis- and trans-acting lncRNAs in normal and stressed cells. lncRNA linc-p21 and PANDA are respectively transcribed from ∼15 or 5 kb upstream to the p21 TSS, induced by p53 upon DNA damage and involved in the regulation in trans of multiple genes downstream of p53 [32], [33]. However, the transcriptional regulation of p21 itself is independent of linc-p21 and PANDA, [32], [33]. In contrast, antisense-p21 (AS-p21) epigenetically suppresses p21 transcription in cis, when transcribed from the antisense strand and overlapping with the p21 transcript [34]. APTR is clearly very different from these known lncRNAs, in that it is encoded elsewhere in the genome, and acts in trans on the p21 promoter itself.
Interaction of APTR with PRC2 is necessary for p21 silencing
One essential activity of APTR is its ability to interact with PRC2 so that the latter can be recruited to the p21 promoter. More than 20% of lncRNAs are bound by PRC2 in various cells [8], [9], and some of these lncRNAs, like XIST and HOTAIR, interact with local chromatin or DNA binding proteins to recruit the chromatin modifying machinery to specific sites in the target loci [1], [4], [7]. APTR similarly appears to recruit PRC2 to the p21 promoter, although not to all PRC2 target loci, as exemplified by HOXD11. Future studies will explore which proteins in PRC2 interact with APTR and what sequence or structural features of APTR are essential for interaction with PRC2. Clearly significant structural studies are necessary to understand exactly how these critical protein:RNA interactions are established and what determines the specificity and affinity of the various PRC2:lncRNA interactions. An interesting question is whether different lncRNAs that interact with PRC2 have similar or different affinities for the protein complex and whether they compete with each other to regulate gene expression by targeting PRC2 to certain sites in the genome and away from other sites. It will also be interesting to know in the future whether certain lncRNAs alter the conformation and enzymatic activity of PRC2 complex, or whether their activity is limited to the targeting of PRC2 to specific sites in the genome.
The embedded complementary Alu element in APTR is required to recruit the lncRNA to the p21 promoter
Importantly, the c-Alu deletion mutant of APTR could not suppress p21 transcription because the mutation prevented APTR recruitment to the p21 promoter. We do not know how APTR interacts with the p21 promoter and how the APTR-embedded Alu element is required for this interaction. For example, is there a specific “receptor” for the c-Alu element, or c-Alu-bound proteins, in the p21 promoter? Or does the c-Alu region of APTR play a structural role so that APTR can fold in the correct conformation before it can interact with the relevant targeting proteins? Regardless of the mechanism this is another example of how Alu elements encoded in RNAs are functionally important.
Interestingly, the APTR localization sites in the p21 promoter (−1.6 kb to −4 from the TSS) contains tandem inverted partial and full Alu elements (Figure S10). Furthermore, we observed at least two uncharacterized promoter lncRNAs transcribed across the Alu element from both strands of this site at very low levels (our unpublished data). Thus, we tested the possibility that Alu: c-Alu base-pairing interactions between cis-lncRNAs from the p21 promoter and APTR tether the latter to the p21 promoter. Luciferase assays on p21 promoter with or without the Alu region (3.7 kb or 2.7 kb of the promoter upstream from the TSS, respectively), however, clearly showed that APTR suppressed the p21 promoter without the Alu region (Figure S10). Thus a simple Alu:c-Alu base-pairing interaction between the p21 promoter and APTR is not the explanation for how APTR is recruited to the promoter.
An alternative possibility is that RNA binding proteins interact with the c-Alu sequence of APTR to recruit the latter to the p21 promoter. Past studies on SINE/Alu revealed that some proteins bind physically to Alu DNA or RNA [35], [36]. Examples include signal recognition particle SRP9/14 heterodimer interacting with Alu element containing RNAs in the cytoplasm [37] and double strand RNA binding protein Staufen 1 (STAU1) recognizing intermolecular Alu: c-Alu element pairs and flanking sequences [38]. Thus another possibility is that similar factor(s) bind to the c-Alu element in APTR to target it to the p21 promoter.
Clearly much more needs to be done to understand what decides target specificity of APTR. Genome-wide ChIP-seq studies of APTR-binding sites on the genome and proteomic studies of proteins that bind to APTR will be helpful in this regard. However, our data raise the intriguing possibility that embedded repetitive elements in an lncRNA could contribute to the targeting of an lncRNA to the genome.
Supporting Information
The nucleotide sequence of long noncoding RNA APTR . Shaded boxes represent sequences in the RNA complementary to SINE/Alu elements and open box represents sequence in the RNA complementary to LINE/L2 elements.
(EPS)
Alignment of sequences from the repetitive elements in APTR with the retrotransposon SINE/Alu or LINE/L2 .
(EPS)
Enough overexpressed APTR persists after si APTR transfection to exceed the amount of endogenous APTR present normally. APTR expression levels normalized to GAPDH in cells transfected either siGL2 or siAPTR#2 as analyzed by Q-RT-PCR. Residual expression level of the siRNA-sensitive APTR after knockdown is higher than that in wild type cells transfected with siGL2 and empty vector. Percentages represent extent of reduction of APTR in siAPTR-transfected cells compared to that in siGL2-transfected cells.
(EPS)
APTR knockdown results in induction of p21 in multiple cancer cells without wild type p53. (A) RT-PCR shows APTR knockdown efficiency by siAPTR#2 in figure S4B. (B) Induction of p21 protein in indicated cells transfected with siGL2 or siAPTR#2. PC3 and H1299 have mutant, and 293T has inactivated p53.
(EPS)
Knockdown of p21 alleviates cell growth inhibition upon APTR knockdown. (A) Growth suppression after siAPTR is alleviated in p21 knockdown HCT116 cells. The indicated siRNAs were transfected at Day1. Data represents total cell number at Day 0 and Day 4 (after 3 days of transfection) with mean ±s.e.m., n = 3, *:P<0.05. (B) The expression levels of APTR and p21 relative to siGL2-transfected cells were measured by Q-RT-PCR at Day 6 (after 5 days of transfection).
(EPS)
Expression level of Histone H3K27me3 does not change after APTR knockdown. Immunoblot of H3K27me3 in 293T cells transfected with siGL2 or siAPTR#2.
(EPS)
Expression of exogenous APTR RNA in APTR knockdown- APTR rescue assays. (A) All cells express the expected exogenous APTR RNA in Figure 6A. RT-PCR with primer sets shown in Figure 4C. RT: reverse transcriptase. (B) RNA expression change of the siRNA-sensitive APTR WT and mutants (1–980) after transfection of siAPTR#2. Percentages represent reduction rate of APTR in siAPTR-transfected cells compared to that in siGL2-transfected cells. The residual level of APTR1-980 after siAPTR is still significantly higher than endogenous APTR levels.
(EPS)
Knockdown of p21 alleviates cell growth inhibition in APTR knockdown glioma cells. (A) MTT assays (after 48 hrs of siRNA transfection) show that growth suppression after siAPTR is alleviated in p21 knockdown U87 glioma cells. Data represents mean ±s.e.m., n = 3, **:P<0.005. (B) Q-RT-PCR shows that the expression levels of APTR and p21 relative to siGL2-transfected cells in Fig. S8A.
(EPS)
Anti-correlation between APTR and p21 RNAs in human glioma cells. The two technical replicates of the experiment in Figure 7G.
(EPS)
The tandem inverted Alu elements in the APTR regulatory p21 promoter is not involved in the APTR -mediating p21 silencing. (A) Schema represents firefly luciferase plasmid construction with various length of p21 promoter. (B) Firefly luciferase activity of various p21 promoters in cells transfected with empty vector or vector expressing wild-type APTR. Firefly luciferase activities were normalized to Renilla luciferase from co-transfected plasmid (mean±s.e.m; n = 3).
(EPS)
Sequences of primers and siRNAs used for RT-PCR, Q-RT-PCR, ChIP/CLIP assays and siRNA interference in this study.
(EPS)
siRNA based lncRNA functional screening in MCF10A cells. F-BRAMY2034329_1 was renamed as APTR. The positive control siORC2 and negative control siGL2 were transfected in six to eight wells in each biological replicate. The inhibition of BrdU incorporation by each siRNA is expressed as a % of the extent of inhibition by siORC2. For each lncRNA, we showed the % inhibition for each of three siRNAs and the average and standard deviation of the inhibition. Each siRNA was tested in triplicate. The average inhibition was calculated from all three siRNAs, unless the SD>0.2 x mean, in which case the outlier siRNA was discarded when calculating the average.
(XLSX)
Supplementary Method.
(DOC)
Acknowledgments
We thank members of the Dutta laboratory for advice and helpful discussions.
Funding Statement
MN was partly supported by the UVA Cancer Center through the Farrow Fellowship Fund and the NCI Cancer Center Support Grant, P30 CA44579. The work was supported by P01 CA104106 and R01 GM 084465 to AD. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, et al. (2007) Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129: 1311–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Loewer S, Cabili MN, Guttman M, Loh YH, Thomas K, et al. (2010) Large intergenic non-coding RNA-RoR modulates reprogramming of human induced pluripotent stem cells. Nat Genet 42: 1113–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wang KC, Chang HY (2011) Molecular mechanisms of long noncoding RNAs. Mol Cell 43: 904–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT (2008) Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science 322: 750–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bracken AP, Helin K (2009) Polycomb group proteins: navigators of lineage pathways led astray in cancer. Nat Rev Cancer 9: 773–784. [DOI] [PubMed] [Google Scholar]
- 6. Margueron R, Reinberg D (2011) The Polycomb complex PRC2 and its mark in life. Nature 469: 343–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jeon Y, Lee JT (2011) YY1 tethers Xist RNA to the inactive X nucleation center. Cell 146: 119–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao J, Ohsumi TK, Kung JT, Ogawa Y, Grau DJ, et al.. (2010) Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol Cell. 2010/12/22 ed. pp. 939–953. [DOI] [PMC free article] [PubMed]
- 9. Khalil AM, Guttman M, Huarte M, Garber M, Raj A, et al. (2009) Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A 106: 11667–11672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sherr CJ, Roberts JM (1995) Inhibitors of mammalian G1 cyclin-dependent kinases. Genes Dev 9: 1149–1163. [DOI] [PubMed] [Google Scholar]
- 11. Sherr CJ, Roberts JM (1999) CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev 13: 1501–1512. [DOI] [PubMed] [Google Scholar]
- 12. Bartek J, Lukas J (2001) Mammalian G1- and S-phase checkpoints in response to DNA damage. Curr Opin Cell Biol 13: 738–747. [DOI] [PubMed] [Google Scholar]
- 13. Abbas T, Dutta A (2009) p21 in cancer: intricate networks and multiple activities. Nat Rev Cancer 9: 400–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim J, Zhang Y, Skalski M, Hayes J, Kefas B, et al.. (2014) microRNA-148a Is a Prognostic oncomiR That Targets MIG6 and BIM to Regulate EGFR and Apoptosis in Glioblastoma. Cancer Res Cancer Research, 2014 Jan 14 [Epub ahead of print]. PMID:24425048. [DOI] [PMC free article] [PubMed]
- 15. Li Y, Guessous F, Zhang Y, Dipierro C, Kefas B, et al. (2009) MicroRNA-34a inhibits glioblastoma growth by targeting multiple oncogenes. Cancer Res 69: 7569–7576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bunz F, Dutriaux A, Lengauer C, Waldman T, Zhou S, et al. (1998) Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science 282: 1497–1501. [DOI] [PubMed] [Google Scholar]
- 17. Negishi M, Saraya A, Mochizuki S, Helin K, Koseki H, et al. (2010) A novel zinc finger protein Zfp277 mediates transcriptional repression of the Ink4a/arf locus through polycomb repressive complex 1. PLoS One 5: e12373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lee KY, Bang SW, Yoon SW, Lee SH, Yoon JB, et al. (2012) Phosphorylation of ORC2 protein dissociates origin recognition complex from chromatin and replication origins. J Biol Chem 287: 11891–11898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Machida YJ, Chen Y, Machida Y, Malhotra A, Sarkar S, et al. (2006) Targeted comparative RNA interference analysis reveals differential requirement of genes essential for cell proliferation. Mol Biol Cell 17: 4837–4845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ota T, Suzuki Y, Nishikawa T, Otsuki T, Sugiyama T, et al. (2004) Complete sequencing and characterization of 21,243 full-length human cDNAs. Nat Genet 36: 40–45. [DOI] [PubMed] [Google Scholar]
- 21. Waldman T, Kinzler KW, Vogelstein B (1995) p21 is necessary for the p53-mediated G1 arrest in human cancer cells. Cancer Res 55: 5187–5190. [PubMed] [Google Scholar]
- 22. Bertrand E, Chartrand P, Schaefer M, Shenoy SM, Singer RH, et al. (1998) Localization of ASH1 mRNA particles in living yeast. Mol Cell 2: 437–445. [DOI] [PubMed] [Google Scholar]
- 23. Tyagi S (2009) Imaging intracellular RNA distribution and dynamics in living cells. Nat Methods 6: 331–338. [DOI] [PubMed] [Google Scholar]
- 24. Velichutina I, Shaknovich R, Geng H, Johnson NA, Gascoyne RD, et al. (2010) EZH2-mediated epigenetic silencing in germinal center B cells contributes to proliferation and lymphomagenesis. Blood 116: 5247–5255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fan T, Jiang S, Chung N, Alikhan A, Ni C, et al. (2011) EZH2-dependent suppression of a cellular senescence phenotype in melanoma cells by inhibition of p21/CDKN1A expression. Mol Cancer Res 9: 418–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pasini D, Bracken AP, Jensen MR, Lazzerini Denchi E, Helin K (2004) Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. EMBO J 23: 4061–4071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lan F, Bayliss PE, Rinn JL, Whetstine JR, Wang JK, et al. (2007) A histone H3 lysine 27 demethylase regulates animal posterior development. Nature 449: 689–694. [DOI] [PubMed] [Google Scholar]
- 28. Network TCGAR (2008) Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 455: 1061–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, et al. (2008) An integrated genomic analysis of human glioblastoma multiforme. Science 321: 1807–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Russo T, Zambrano N, Esposito F, Ammendola R, Cimino F, et al. (1995) A p53-independent pathway for activation of WAF1/CIP1 expression following oxidative stress. J Biol Chem 270: 29386–29391. [DOI] [PubMed] [Google Scholar]
- 31. Kraus A, Gross MW, Knuechel R, Munkel K, Neff F, et al. (2000) Aberrant p21 regulation in radioresistant primary glioblastoma multiforme cells bearing wild-type p53. J Neurosurg 93: 863–872. [DOI] [PubMed] [Google Scholar]
- 32. Hung T, Wang Y, Lin MF, Koegel AK, Kotake Y, et al. (2011) Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat Genet 43: 621–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, et al. (2010) A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell 142: 409–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Morris KV, Santoso S, Turner AM, Pastori C, Hawkins PG (2008) Bidirectional transcription directs both transcriptional gene activation and suppression in human cells. PLoS Genet 4: e1000258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lukyanov DV, Urusova ME, Shcherba KM, Podgornaya OI (2000) Alu-DNA repeat-binding protein p68 is a part of Alu-RNA containing alpha-RNP. Eur J Biochem 267: 2362–2371. [DOI] [PubMed] [Google Scholar]
- 36. Hsu K, Chang DY, Maraia RJ (1995) Human signal recognition particle (SRP) Alu-associated protein also binds Alu interspersed repeat sequence RNAs. Characterization of human SRP9. J Biol Chem 270: 10179–10186. [DOI] [PubMed] [Google Scholar]
- 37. Hasler J, Strub K (2006) Alu RNP and Alu RNA regulate translation initiation in vitro. Nucleic Acids Res 34: 2374–2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gong C, Maquat LE (2011) lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3′ UTRs via Alu elements. Nature 470: 284–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The nucleotide sequence of long noncoding RNA APTR . Shaded boxes represent sequences in the RNA complementary to SINE/Alu elements and open box represents sequence in the RNA complementary to LINE/L2 elements.
(EPS)
Alignment of sequences from the repetitive elements in APTR with the retrotransposon SINE/Alu or LINE/L2 .
(EPS)
Enough overexpressed APTR persists after si APTR transfection to exceed the amount of endogenous APTR present normally. APTR expression levels normalized to GAPDH in cells transfected either siGL2 or siAPTR#2 as analyzed by Q-RT-PCR. Residual expression level of the siRNA-sensitive APTR after knockdown is higher than that in wild type cells transfected with siGL2 and empty vector. Percentages represent extent of reduction of APTR in siAPTR-transfected cells compared to that in siGL2-transfected cells.
(EPS)
APTR knockdown results in induction of p21 in multiple cancer cells without wild type p53. (A) RT-PCR shows APTR knockdown efficiency by siAPTR#2 in figure S4B. (B) Induction of p21 protein in indicated cells transfected with siGL2 or siAPTR#2. PC3 and H1299 have mutant, and 293T has inactivated p53.
(EPS)
Knockdown of p21 alleviates cell growth inhibition upon APTR knockdown. (A) Growth suppression after siAPTR is alleviated in p21 knockdown HCT116 cells. The indicated siRNAs were transfected at Day1. Data represents total cell number at Day 0 and Day 4 (after 3 days of transfection) with mean ±s.e.m., n = 3, *:P<0.05. (B) The expression levels of APTR and p21 relative to siGL2-transfected cells were measured by Q-RT-PCR at Day 6 (after 5 days of transfection).
(EPS)
Expression level of Histone H3K27me3 does not change after APTR knockdown. Immunoblot of H3K27me3 in 293T cells transfected with siGL2 or siAPTR#2.
(EPS)
Expression of exogenous APTR RNA in APTR knockdown- APTR rescue assays. (A) All cells express the expected exogenous APTR RNA in Figure 6A. RT-PCR with primer sets shown in Figure 4C. RT: reverse transcriptase. (B) RNA expression change of the siRNA-sensitive APTR WT and mutants (1–980) after transfection of siAPTR#2. Percentages represent reduction rate of APTR in siAPTR-transfected cells compared to that in siGL2-transfected cells. The residual level of APTR1-980 after siAPTR is still significantly higher than endogenous APTR levels.
(EPS)
Knockdown of p21 alleviates cell growth inhibition in APTR knockdown glioma cells. (A) MTT assays (after 48 hrs of siRNA transfection) show that growth suppression after siAPTR is alleviated in p21 knockdown U87 glioma cells. Data represents mean ±s.e.m., n = 3, **:P<0.005. (B) Q-RT-PCR shows that the expression levels of APTR and p21 relative to siGL2-transfected cells in Fig. S8A.
(EPS)
Anti-correlation between APTR and p21 RNAs in human glioma cells. The two technical replicates of the experiment in Figure 7G.
(EPS)
The tandem inverted Alu elements in the APTR regulatory p21 promoter is not involved in the APTR -mediating p21 silencing. (A) Schema represents firefly luciferase plasmid construction with various length of p21 promoter. (B) Firefly luciferase activity of various p21 promoters in cells transfected with empty vector or vector expressing wild-type APTR. Firefly luciferase activities were normalized to Renilla luciferase from co-transfected plasmid (mean±s.e.m; n = 3).
(EPS)
Sequences of primers and siRNAs used for RT-PCR, Q-RT-PCR, ChIP/CLIP assays and siRNA interference in this study.
(EPS)
siRNA based lncRNA functional screening in MCF10A cells. F-BRAMY2034329_1 was renamed as APTR. The positive control siORC2 and negative control siGL2 were transfected in six to eight wells in each biological replicate. The inhibition of BrdU incorporation by each siRNA is expressed as a % of the extent of inhibition by siORC2. For each lncRNA, we showed the % inhibition for each of three siRNAs and the average and standard deviation of the inhibition. Each siRNA was tested in triplicate. The average inhibition was calculated from all three siRNAs, unless the SD>0.2 x mean, in which case the outlier siRNA was discarded when calculating the average.
(XLSX)
Supplementary Method.
(DOC)