Abstract
Dicers are proteins of the ribonuclease III family with the ability to process dsRNA, involved in regulation of gene expression at the post-transcriptional level. Dicers are conserved from basal metazoans to higher metazoans and contain a number of functional domains that interact with dsRNA. The completed genome sequences of over 34 invertebrate species allowed us to systematically investigate Dicer genes over a diverse range of phyla. The majority of invertebrate Dicers clearly fell into the Dicer1 or Dicer2 subfamilies. Most nematodes possessed only one Dicer gene, a member of the Dicer1 subfamily, whereas two Dicer genes (Dicer1 and Dicer2) were present in all platyhelminths surveyed. Analysis of the key domains showed that a 5′ pocket was conserved across members of the Dicer1 subfamily, with the exception of the nematode Bursaphelenchus xylophilus. Interestingly, Nematostella vectensis DicerB grouped into Dicer2 subfamily harbored a 5′ pocket, which is commonly present in Dicer1. Similarly, the 3′ pocket was also found to be conserved in all Dicer proteins with the exceptions of Schmidtea mediterranea Dicer2 and Trichoplax adherens Dicer A. The loss of catalytic residues in the RNase III domain was noted in platyhelminths and cnidarians, and the ‘ball’ and ‘socket’ junction between two RNase III domains in platyhelminth Dicers was different from the canonical junction, suggesting the possibility of different conformations. The present data suggest that Dicers might have duplicated and diversified independently, and have evolved for various functions in invertebrates.
Introduction
Small regulatory RNA pathways are highly conserved mechanisms present in most eukaryotic organisms and play an important role in post-transcriptional gene regulation. The gene-regulatory function of microRNAs (miRNAs) and short interfering RNAs (siRNAs) is mainly through translational repression or degradation of cytoplasmic mRNAs by an RNA-induced silencing complex (RISC). miRNA and siRNA pathways share a common RNase III processing enzyme, Dicer, and together with other proteins it constitutes RISC for gene transcriptional repression [1]. Dicer is responsible for recognizing a hairpin (in pre-miRNA) or long double-strand RNA (dsRNA), and processing them into miRNA-miRNA* duplexes or siRNA duplexes [2]. These small RNA duplexes are converted to a single-stranded form and bound to Argonaute (AGO), a key component of RISC, through a process coordinated by Dicer and other RNA-binding proteins [3]. Then small RNAs target specific mRNA sequences, leading to cleavage or translational repression of these [4].
Dicer proteins are present in many eukaryotic organisms including plants, fungi, and metazoans [5], [6]. Vertebrates and nematodes have only one Dicer gene (Dicer1), whereas insects and flatworms possess two, (Dicer1 and Dicer2). Dicers normally contain a number of functional domains: an N-terminal DEAD box, an RNA helicase domain, a Dicer dimer domain, a Piwi-Argonaute-Zwille (PAZ) domain, two RNase III domains and a dsRNA binding domain [7], [8]. The crystal structure of Dicer from Giardia intestinalis revealed that the PAZ domain was responsible for binding of the 3′ terminus of dsRNA [9]. After the 3′ end bound to the PAZ domain, pre-miRNAs or dsRNAs are cleaved by the two RNase III domains which form a single dsRNA processing center through intramolecular dimerization [10]. In Dicer1, binding of the PAZ domain to the 3′ terminus of pre-miRNA is crucial for orienting the RNase III domains for cleavage, however, recent publications have revealed that 5′ terminus recognition of pre-miRNAs is also important for mature miRNAs synthesis [11], [12].
Previous studies have focused on Dicers of plants and model organisms, little is known about Dicers of invertebrates. The recent availability of genome sequences of over 34 invertebrate species from 10 phyla, including 1 choanoflagellate, 2 cnidarians, 1 placozoan, 2 annelids, 1 mollusc, 7 platyhelminths, 7 nematodes, 10 arthropods, 1 echinoderm and 3 chordates, have allowed us to perform an extensive phylogenetic analysis of Dicers.
Materials and Methods
Acquisition of sequence
For some well-annotated genomes, Dicer sequences were directly retrieved from the databases. In addition, BLASTP and TBLASTN were performed to search against their databases using Drosophila melanogaster Dicers (NP_524453 and NP_523778), Caenorhabditis elegans Dicer (NP_498761) or Schistosoma mansoni Dicers (Smp_169750.1 and Smp_033600) as query sequences. An E-value of 1×e-10 was used as a cutoff in BLAST searches and the hits were filtered to keep only those with at least 25% identity to the query sequence. Protein functional domains were identified using Pfam database and SMART database [13], [14]. The species names, abbreviations and accession numbers are provided in Table 1.
Table 1. Distribution of endoribonuclease Dicer genes in invertebrates.
| Species | Species Abbreviation | Gene Symbol | Accession Number | Lengtha | Data originb |
| Choanoflagellatea | |||||
| Monosiga brevicollis | Mbrev | / | / | / | / |
| Cnidaria | |||||
| Nematostella vectensis | NvecA | Sca_186 | ABZ10549c | 1099 | NCBI JGI |
| NvecB | 167046039 | ABZ10551c | 1469 | ||
| Hydra magnipapillata | HmagA | HYDRA_212274 | Hma2.212274 | 835 | Metazome |
| HmagB | HYDRA_205202 | Hma2.205202 | 769 | ||
| HmagC | HYDRA_222700 | Hma2.222700 | 565 | ||
| Placozoa | |||||
| Trichoplax adhaerens | TadhA | TRIADDRAFT_18215 | XP_002108754 | 1018 | NCBI JGI |
| TadhB | TRIADDRAFT_51673 | XP_002107798 | 1418 | ||
| TadhC | TRIADDRAFT_51985 | XP_002107959 | 685 | ||
| TadhD | TRIADDRAFT_52058 | XP_002108012 | 915 | ||
| TadhE | DCL-E | ABZ10547c | 707 | ||
| Annelida | |||||
| Capitella teleta | Ctel | CAPTEDRAFT_223153 | ELU12939 | 1651 | NCBI |
| Helobdella robusta | Hrob | estExt_Genewise1.C_710069 | 103772 | 1316 | JGI |
| Mollusca | |||||
| Lottia gigantea | Lgig | gw1.12.81.1 | 61365 | 1800 | JGI |
| Platyhelminthes | |||||
| Schmidtea mediterranea | Smed1 | Dicer-1 | ASA.00018.01 | 1328 | SmedGD |
| Smed2 | Dicer-2 | mk4.000125.07.01 | 1589 | ||
| Schistosoma mansoni | Sman1 | Dicer-1 | Smp_169750.1 | 2319 | Sanger |
| Sman2 | Dicer-2 | Smp_033600 | 928 | ||
| Schistosoma japonicum | Sjap1 | Dicer-1 | Sjp_0069770 | 2480 | Sanger |
| Sjap2 | Dicer-2 | Sjp_0043700 | 923 | ||
| Echinococcus granulosus | Egra1 | Dicer-1 | EgrG_000085200 | 1904 | Sanger |
| Egra2 | Dicer-2 | EgrG_000181800 | 1924 | ||
| Echinococcus multilocularis | Emul1 | Dicer-1 | EmuJ_000085200 | 1906 | Sanger |
| Emul2 | RNC3.1 | EmuJ_000180900 | 1812 | ||
| Emul2 | RNC3.2 | EmuJ_000181800 | 1929 | ||
| Hymenolepis microstoma | Hmic1 | Dicer-1 | HmN_000252400 | 1897 | Sanger |
| Hmic2 | Endoribonuclease Dicer | HmN_000200100 | 1183 | ||
| Taenia solium | Tsol1 | Dicer-1 | TsM_000872800 | 1890 | Sanger |
| Tsol2 | Endoribonuclease Dicer | TsM_000756400 | 829 | ||
| Nematoda | |||||
| Caenorhabditis elegans | Cele | Dicer-1 | NP_498761 | 1910 | NCBI |
| Bursaphelenchus xylophilus | Bxyl | Dicer-1 | BUX_s00116.153 | 1040 | Sanger |
| Pristionchus pacificus | Ppac | Dicer-1 | WBGene00096444 | 2769 | WormBase |
| Strongyloides ratti | Srat | Dicer-1 | g5271 | 1824 | WormBase |
| Brugia malayi | Bmal | Dicer-1/Bm5026 | WBGene00225287 | 1930 | WormBase |
| Trichinella spiralis | TspiA | Tsp_01223 | XP_003377020 | 2029 | NCBI |
| TspiB | Tsp_00125 | XP_003375890 | 1903 | NCBI | |
| Loa loa filariasis | Lloa | LOAG_02227 | XP_003137813 | 1928 | NCBI |
| Arthropoda | |||||
| Daphnia pulex | DpulA | DAPPUDRAFT_308316 | EFX72380 | 1979 | NCBI JGI |
| DpulB | DAPPUDRAFT_329028 | EFX69538 | 1459 | ||
| DpulC | DAPPUDRAFT_309030 | EFX86072 | 1604 | ||
| Pediculus humanus corporis | Phum | Dcier-1 | XP_002429494 | 2179 | NCBI |
| Tribolium castaneum | Tcas1 | Dicer-1 | XP_968993 | 1865 | NCBI |
| Tcas2 | Dicer-2 | NP_001107840 | 1623 | ||
| Nasonia vitripennis | Nvit1 | Dicer-1 | XP_001605287 | 1917 | NCBI |
| Nvit2 | Dicer-2 | XP_001602524 | 1450 | ||
| Acyrthosiphon pisum | ApisA | LOC100159500 | XP_001943370 | 1626 | NCBI |
| ApisB | LOC100166428 | XP_001945890 | 1691 | ||
| Drosophila melanogaster | Dmel1 | Dicer-1 | NP_524453 | 2249 | NCBI FlyBase |
| Dmel2 | Dicer-2 | NP_523778 | 1772 | ||
| Anopheles gambiae | Agam1 | AgaP_AGAP002836 | XP_003436256 | 2336 | NCBI |
| Agam2 | AgaP_AGAP012289 | XP_320248 | 1672 | ||
| Aedes aegypti | Aaeg1 | AaeL_AAEL006794 | XP_001652212 | 1658 | NCBI |
| Aaeg2 | AaeL_AAEL001612 | XP_001659747 | 2193 | ||
| Culex pipiens quinquefasciatus | Cpip1 | CpipJ_CPIJ003169 | XP_001844757 | 2270 | NCBI |
| Cpip2 | CpipJ_CPIJ010534 | XP_001855187 | 1165 | ||
| Echinodermata | |||||
| Strongylocentrotus purpuratus | Spur | LOC586001 | XP_790894 | 1850 | Metazome |
| Chordata | |||||
| Branchiostoma floridae | Bflo | BRAFLDRAFT_202604 | XP_002610617 | 1868 | NCBI |
| Ciona intestinalis | Ciona | Endoribonuclease Dicer | ENSCINP00000017117 | 1872 | Ensemble |
| Saccoglossus kowalevskii | Skow | Sakowv30031161m.g | Sakowv30031161m | 1905 | Metazome |
Amino acid length;
JGI: Joint Genome Institute; NCBI: National Center for Biotechnology Information; SmedGD: Schmidatea mediterranea Genome Database; Sanger: Wellcome Trust Sanger Insitute; FlyBase: Drosophila database; SilkDB: silkworm database; Ensemble: Ensemble Genome Browser.
Sequence alignment and phylogenetic analysis
The data sets contained a total of 58 sequences from 34 species (in a size from 565aa to 2769aa, Text S1). The amino acid sequences of Dicer were aligned by MUSCLE [15] with default parameters and manually optimized by Jalview 2.8 [16]. The alignments were subsequently processed using Gblocks v0.91b [17] for phylogenetic reconstruction, allowing gaps in 1/2 of the sequences. ProtTest 3.2 was applied to find an appropriate model of amino acid substitution for tree building analysis [18]. A maximum likelihood tree was constructed using PhyML 3.0 program [19]. Clade support was calculated using SH-like approximate likelihood ratio test, Bayes likelihood test and bootstrap proportions (500 replicates).
Results
Identification and distribution of Dicer genes across invertebrates
The final data sets contained 58 Dicer gene sequences from two cnidarians, one placozoa, two annelids, one mollusc, seven platyhelminths, seven nematodes, eleven arthropods, one echinoderm and three chordates (Table 1). No Dicer homologues were identified in the choanoflagellate Monosiga brevicollis. Our results of genomic database searches revealed that one placozoan, two annelids, one mollusc, one echinoderm and three of the chordates investigated possessed only one Dicer1 gene. Each of nematodes had only one Dicer1 gene, except Trichinella spiralis, which expressed both Dicer1 and Dicer2 genes. Platyhelminths and arthropods possessed two Dicer genes in their genomes, with the exceptions of Daphnia pulex (three genes), Pediculus humanus corporis (one gene) and Echinococcus multilocularis (three genes).
Phylogenetic analysis of Dicers
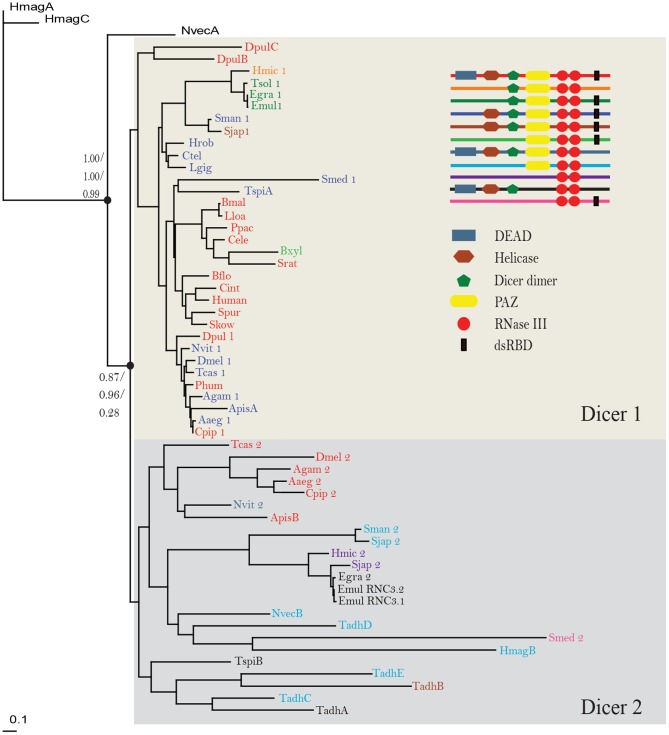
As shown in the Maximum likelihood tree (Fig. 1), Dicers of invertebrates were grouped into two lineages: Dicer1 subfamily and Dicer2 subfamily. Almost all of the arthopods and platyhelminths surveyed possessed one member of each of these subfamilies, and annelids, molluscs, nematodes, echinoderms and chordates investigated had only one Dicer gene that belongs to Dicer1 subfamily. The placozoan Trichoplax adhaerens had the most copies of Dicer genes in our investigated species; however, all of the five Dicer genes were classed into the Dicer2 subfamily. The two cnidarians N. vectensis and Hydra magnipapillata each had only one Dicer2 gene, but possessed other Dicer genes that fell outside the two subfamilies.
Figure 1. A maximum likelihood tree of invertebrate Dicers.
The tree was constructed using maximum likelihood method. Two number sets, 1.00/1.00/0.99 and 0.87/0.96/0.28, at the nodes were SH-like approximate likelihood ratio, Bayes likelihood and bootstrap values, respectively.
Organization of functional domains of Dicer family
We identified the functional domains using the Pfam database and confirmed each inferred domain using the SMART database. As shown in Fig. 1, Dicers had significant variability in domain organization. For instance, Dicers initially characterized in humans are multidomain proteins, consisting of an N-terminal DEAD box, an RNA helicase domain, a Dicer dimer domain, a PAZ domain, two RNase III domains and a dsRNA binding domain [10]. However, Taenia solium Dicer2 processed only one RNase III domain. We also observed the loss of the DEAD domain, which contains two RecA-like domains as a catalytic core and can regulate various processes involving RNA [20], in Dicer1 of mollusks, annelids, platyhelminths and most arthropods.
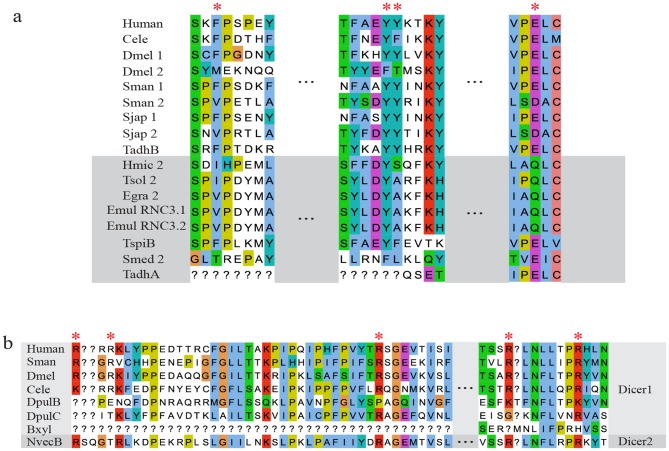
A PAZ domain is an RNA-binding module found in PPD proteins (PAZ and Piwi domain proteins) and Dicer orthologs, and anchors the 2-nucleotide 3′ overhang of dsRNA with its highly conserved binding pocket [10]. After searching annotated domains using Pfam and SMART databases, we did not find the PAZ domain in Dicer2 of the platyhelminths Schmidtea mediterranea, Hymenolepis microstoma, T. solium, Echinococcus granulosus, E. multilocularis, the placozoan T. adhaerens and the nematode T. spiralis. There are two possibilities: the sequences are too divergent to be clearly recognized or they may have lost the PAZ domain during evolution. We therefore aligned the key amino acid residues in the PAZ domain of Dicers in the above species. As shown in Fig. 2a, most of the key residues in the PAZ domain were conserved in Dicer2 sequences with the exceptions of S. mediterranea and T. adhaerens, indicating the absence of the PAZ domain in those two species. Recently studies have revealed that human Dicer anchors not only the 3′ end but also the 5′ end, and the 5′ end recognition by Dicer is important for the precise and effective biogenesis of miRNAs [12]. A previous study revealed that the 5′ binding residues (Arg778, Arg780 and Arg811 within the N-terminal extension of PAZ domain, and Arg996 and Arg1003 within PAZ domain) were conserved across invertebrate Dicer1 and absent in other Dicers [12]. However, we found that these five key residues were present in N. vectensis DicerB, which was classed into the Dicer2 family (Fig. 2b).
Figure 2. The key residues critical for recognition of 3′ and 5′ pockets.
The key residues involved in 3′ pocket (a) and 5′ pocket recognition (b) were indicated using asterisks in red. The Dicers in which a PAZ domain was not identified using Pfam and SMART are highlighted in grey (a). Gaps are filled using question marks (?).
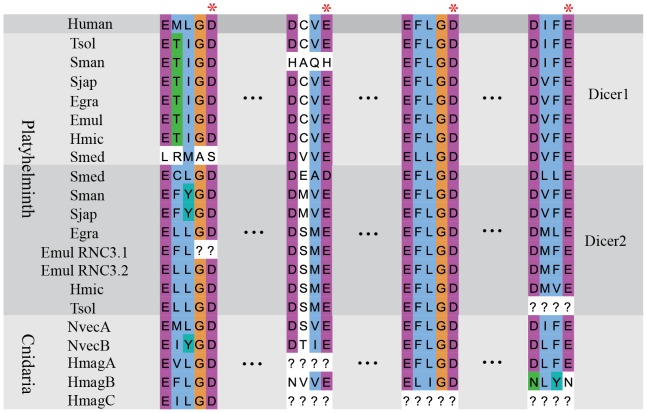
After dsRNA binding to the 5′ and 3′ pockets, two RNase III domains of Dicer cleave targeted molecules. Based on the alignment of Dicer RNase III domain, we found that the catalytic core in most invertebrates was highly conserved (Fig. 3). However, Schistosoma mansoni Dicer1, S. mediterranea Dicer1, T. solium Dicer2 and E. multilocularis Dicer RNC3.1 showed variations in this key region (Fig. 3). Compared to platyhelminths, the RNase III domain seemed to be divergent in cnidarians, and most of the key residues were altered in H. magnipapillata DicerC (Fig. 3), indicating the possibility of loss of dsRNA cleavage ability. During the cleavage of targeted molecules by sRNAs, two RNase III domains of Dicer form a tight dimer of which the subunit interface is hydrophobic [9]. The crystal structure showed that a tight dimer was formed by two Aquifex aeolicus RNase III proteins, each of which possessed only one RNase III domain. A total of 128 hydrophobic interactions (<4.0 Å) were found between the two molecules, whereas only 20 hydrogen bonds/salt bridges existed at the dimer interface. In the dimer, two identical “ball-and-socket” junctions were formed at each end of the interface. The ‘ball’ was the hydrophobic side chain of Phe41 and the ‘socket’ was a cavity formed by side chains of Val52, Val56, Leu67, Ser68, and Lys71 [9]. Subsequent studies showed that a Met1317 within the human RNase III a domain was located in the position of the ‘ball’ residue and the corresponding socket residues Thr1717, Tyr1721, Leu1732, Thr1732 and Arg1736 were located in RNaseIII b domain [21]. Interestingly, we found that the ‘ball’ residue in the RNase III a domain of platyhelminths Dicer1 was replaced by a hydrophilic amino acid-threonine, whereas the corresponding ‘socket’ residues were still conserved (Fig. 4).
Figure 3. The key residues of RNase III domains.
Catalytic residues are marked by red asterisks and gaps filled using question marks (?).
Figure 4. The residues of “ball and socket” junction.

The ‘Ball’ residue within RNaseIII a domain is marked by a black asterisk, and the ‘socket’ residues within RNase III b domain are marked by red asterisks. Gaps are filled using question marks (?).
Discussion
In our study, the number of the Dicer genomic loci was variable, from one in several invertebrates to five in T. adhaerens. Dicers of invertebrates were clearly classed into two subfamilies, Dicer1 and Dicer2, except for several Dicers from cnidarians. Our results support the model of Dicer evolution in which a eukaryote Dicer may have duplicated independently. Interestingly, Dicer2 of E. multilocularis may have undergone duplication after species formation. Mature miRNAs have been identified in all the invertebrates investigated in our research with the exceptions of the choanoflagellate M. brevicollis and the placozoan T. adhaerens [22]. Similarly, we failed to find Dicer genes and other RISC proteins genes in the genome of M. brevicollis, a close known relative of metazoans. However, T. adhaerens, a simple known metazoan, possessed five Dicer proteins and all of them belonged to Dicer2 subfamily. These Dicers may constitute an immune defense mechanism against viral infection as placozoans are exposed to a high viral load [23].
Both cnidarians possessed multiple Dicers, and only N. vectensis DicerB and H. magnipapillata DicerB were classed into Dicer2 group, while the others fell outside the two recognized subfamilies. Recent analysis has revealed that cnidarians express species-specific miRNAs and share few miRNA families with bilateria [24], [25]. These distinct cnidarian Dicers may provide some clues to understanding of the biogenesis of species-specific miRNAs.
The recognition and cleavage of dsRNA by Dicer is a core step in miRNA and siRNA pathways. The 3′ pocket of Dicer is involved in 3′ end binding of dsRNA. The absence of key sites in the pocket in S. mediterranea Dicer2 and T. adhaerens DicerA could lead to loss of the binding ability. But these two Dicer2 genes may function with the help of other RNA binding protein, such as Drosha [26]. The 5′ pocket is positioned in close proximity to the 3′ pocket on the same surface of Dicer1, and the binding residues of the 5′ pocket are conserved [12]. Interestingly, we found most of the key binding residues of the 5′ pocket, which were previously found only in Dicer1, in N. vectensis DicerB that belonged to Dicer2 lineage. It suggests that N. vectensis DicerB may retain the bioactivities of Dicer1 as well.
After dsRNA recognition by the conserved pockets, the cleavage of dsRNA is conducted by two RNase III domains [27]. The loss of the catalytic residue-aspartate in E. multilocularis Dicer RNC3.1 could reduce its catalytic activity. However, Dicer RNC3.2, a paralogue of Dicer RNC3.1, possessed all the key residues, and therefore it may compensate for the reduced activity of Dicer RNC3.1. The dimerization of RNase III domains creates a catalytic valley which can accommodate a dsRNA substrate. The two “ball and socket” junctions may be responsible in part for the accurate positioning of the catalytic residues in the valley. A Previous study suggested that the ‘ball’ consisted of the hydrophobic side chain of amino acids, and the interaction between ‘ball’ and ‘socket’ was precluded by charged side chain or the absence of any side chain in the position of the ‘ball’ [9]. Interestingly, we found that the ‘ball’ within the RNase III a domain. In platyhelminths Dicer1 proteins contained a hydrophilic side chain of threonine, but the residues of the hydrophobic ‘socket’ were conserved across different species. This substitution may results in reduced affinity between the ‘ball’ and the hydrophobic ‘socket’ and altered conformation.
Supporting Information
Amino acid sequences of Dicers.
(TXT)
Funding Statement
This work was financially sponsored by Gansu Natural Science Foundation (0710RJZA081), State Public-interest Institution Basal Research Fund and by Special Fund for Agro-scientific Research in the Public Interest (201103008). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Lamontagne B, Larose S, Boulanger J, Elela SA (2001) The RNase III family: a conserved structure and expanding functions in eukaryotic dsRNA metabolism. Curr Issues Mol Biol 3: 71–78. [PubMed] [Google Scholar]
- 2. He L, Hannon GJ (2004) MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet 5: 522–531. [DOI] [PubMed] [Google Scholar]
- 3. Hammond SM, Boettcher S, Caudy AA, Kobayashi R, Hannon GJ (2001) Argonaute2, a link between genetic and biochemical analyses of RNAi. Science 293: 1146–1150. [DOI] [PubMed] [Google Scholar]
- 4. Wang Y, Juranek S, Li H, Sheng G, Wardle GS, et al. (2009) Nucleation, propagation and cleavage of target RNAs in Ago silencing complexes. Nature 461: 754–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cerutti H, Casas-Mollano JA (2006) On the origin and functions of RNA-mediated silencing: from protists to man. Curr Genet 50: 81–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Shabalina SA, Koonin EV (2008) Origins and evolution of eukaryotic RNA interference. Trends Ecol Evol 23: 578–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bernstein E, Caudy AA, Hammond SM, Hannon GJ (2001) Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409: 363–366. [DOI] [PubMed] [Google Scholar]
- 8. Margis R, Fusaro AF, Smith NA, Curtin SJ, Watson JM, et al. (2006) The evolution and diversification of Dicers in plants. FEBS Lett 580: 2442–2450. [DOI] [PubMed] [Google Scholar]
- 9. Blaszczyk J, Tropea JE, Bubunenko M, Routzahn KM, Waugh DS, et al. (2001) Crystallographic and modeling studies of RNase III suggest a mechanism for double-stranded RNA cleavage. Structure 9: 1225–1236. [DOI] [PubMed] [Google Scholar]
- 10. Zhang H, Kolb FA, Jaskiewicz L, Westhof E, Filipowicz W (2004) Single processing center models for human Dicer and bacterial RNase III. Cell 118: 57–68. [DOI] [PubMed] [Google Scholar]
- 11. MacRae IJ, Zhou K, Doudna JA (2007) Structural determinants of RNA recognition and cleavage by Dicer. Nat Struct Mol Biol 14: 934–940. [DOI] [PubMed] [Google Scholar]
- 12. Park JE, Heo I, Tian Y, Simanshu DK, Chang H, et al. (2011) Dicer recognizes the 5' end of RNA for efficient and accurate processing. Nature 475: 201–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, et al. (2012) The Pfam protein families database. Nucleic Acids Res 40: D290–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Letunic I, Doerks T, Bork P (2012) SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res 40: D302–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ (2009) Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25: 1189–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Castresana J (2000) Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Molecular Biology and Evolution 17: 540–552. [DOI] [PubMed] [Google Scholar]
- 18. Darriba D, Taboada GL, Doallo R, Posada D (2011) ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics 27: 1164–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, et al. (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59: 307–321. [DOI] [PubMed] [Google Scholar]
- 20. Kurimoto K, Muto Y, Obayashi N, Terada T, Shirouzu M, et al. (2005) Crystal structure of the N-terminal RecA-like domain of a DEAD-box RNA helicase, the Dugesia japonica vasa-like gene B protein. J Struct Biol 150: 58–68. [DOI] [PubMed] [Google Scholar]
- 21. Ji X (2006) Structural basis for non-catalytic and catalytic activities of ribonuclease III. Acta Crystallogr D Biol Crystallogr 62: 933–940. [DOI] [PubMed] [Google Scholar]
- 22. Grimson A, Srivastava M, Fahey B, Woodcroft BJ, Chiang HR, et al. (2008) Early origins and evolution of microRNAs and Piwi-interacting RNAs in animals. Nature 455: 1193–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. de Jong D, Eitel M, Jakob W, Osigus HJ, Hadrys H, et al. (2009) Multiple dicer genes in the early-diverging metazoa. Molecular Biology and Evolution 26: 1333–1340. [DOI] [PubMed] [Google Scholar]
- 24. Krishna S, Nair A, Cheedipudi S, Poduval D, Dhawan J, et al. (2013) Deep sequencing reveals unique small RNA repertoire that is regulated during head regeneration in Hydra magnipapillata. Nucleic Acids Research 41: 599–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Moran Y, Praher D, Fredman D, Technau U (2013) The Evolution of MicroRNA Pathway Protein Components in Cnidaria. Molecular Biology and Evolution 30: 2541–2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Han J, Lee Y, Yeom KH, Nam JW, Heo I, et al. (2006) Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell 125: 887–901. [DOI] [PubMed] [Google Scholar]
- 27. Ji XH (2008) The mechanism of RNase III action: How Dicer dices. Rna Interference 320: 99–116. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Amino acid sequences of Dicers.
(TXT)