Abstract
Cell survival in changing environments requires appropriate regulation of gene expression, including translational control. Multiple stress signaling pathways converge on several key translation factors, such as eIF4F and eIF2, and rapidly modulate mRNA translation at both the initiation and the elongation stages. Repression of global protein synthesis is often accompanied with selective translation of mRNAs encoding proteins that are vital for cell survival and stress recovery. The past decade has seen significant progress in our understanding of translational reprogramming in part due to the development of technologies that allow the dissection of the interplay between mRNA elements and corresponding binding proteins. Recent genome-wide studies using ribosome profiling have revealed unprecedented proteome complexity and flexibility through alternative translation, raising intriguing questions about stress-induced translational reprogramming. Many surprises emerged from these studies, including wide-spread alternative translation initiation, ribosome pausing during elongation, and reversible modification of mRNAs. Elucidation of the regulatory mechanisms underlying translational reprogramming will ultimately lead to the development of novel therapeutic strategies for human diseases.
INTRODUCTION
All living organisms must detect and respond to changing growth conditions and environmental stimuli. Under acute adverse conditions, such as heat shock, hypoxia, nutrient deprivation or DNA damage, gene expression undergoes coordinated changes to ensure cell survival. The past decade has seen significant progress in our understanding of gene regulation in response to stress, including chromatin remodelling, transcriptional regulation, alternative splicing and translational control. Recent advances in next-generation sequencing allow the dissection of gene regulation in an unprecedented scale and resolution.1 Although transcriptional regulation is essential in mediating the strength of stress response, translational control often provides immediate and effective changes in protein levels.2 This swift response offers a timely adaptation for cells to maximize survival under stress.3
Translation can be divided mechanistically into three stages: initiation, elongation and termination. As the rate-limiting step in translation, initiation is a complex process involving ribosome loading, scanning, and start codon selection before elongation commitment.2 Consistent with its critical role in determining the overall rate of translation, initiation is the primary target of regulation under stress. Under various stress conditions, distinct signalling pathways converge to a few initiation regulators resulting in translational inhibition. The two best characterized mechanisms are mRNA cap recognition and ternary complex formation (see below).4 Although translational control at the initiation stage has been extensively studied,5 much less is known about the regulatory mechanisms of elongation under stress conditions. Recent development of ribosome profiling technology has reignited the research interest in the translation field.6, 7 The innovative technique enables monitoring of ribosome dynamics with unprecedented resolution at the genome-wide scale.8 With this powerful tool, surprising mechanisms at post-initiation stages of translation have been uncovered.9
Protein synthesis consumes a lion’s share of energy and cellular resources, so translation is generally repressed under most if not all types of stress conditions. However, subsets of mRNAs can bypass the general inhibition and be selectively translated. Most of these mRNAs encode stress response proteins, which protect cells from damages and facilitate the post-stress recovery.10, 11. The concept of translational reprogramming fits well into the mode of translational control in stress response, allowing selective translation of mRNAs to maintain the expression of stress proteins when general protein synthesis is compromised. Such regulation can be quantitative (all-or-none vs. graded), or qualitative (enabling a single mRNA to produce several different proteins). We argue that translational reprogramming lies at the heart of the stress response and is required for rapid cellular adaptation under stress. Mechanistic details of translational reprogramming, however, are only beginning to be unfurled. In this review, we discuss mechanisms underlying global repression of translation as well as selective translation in response to stress. Although both processes are tightly coupled during translational reprogramming, for the purpose of clarity, we review each part separately by focusing on mRNA elements as well as corresponding binding proteins. We start with an overview of well-established regulatory mechanisms through initiation and then focus on the recent progress in novel modes of regulation are important in translational reprogramming in stress response.
GLOBAL REPRESSION OF TRANSLATION DURING STRESS
Overview of Eukaryotic Translation Processes
To better illustrate mechanisms underlying translational reprogramming, it is necessary to briefly revisit what we have learned regarding translation processes in eukaryotic cells. Under normal conditions, eukaryotic cells employ a cap-dependent mechanism to initiate translation for most mRNAs.12, 13 The 5′ end of eukaryotic mRNAs is modified with a m7Gppp cap structure, which is recognized by an eukaryotic initiation factor 4E (eIF4E). eIF4E forms the eIF4F complex by binding to eIF4G (a scaffold protein) and eIF4A (a helicase).14–16 The cap recognition is the first step that determines which mRNAs are to be translated, and it is not surprising that multiple signalling pathways control this rate-limiting step. Another key step is the formation of a ternary complex, which is composed of a methionine-loaded initiator tRNA and a GTP-coupled eIF2.17 The ternary complex associates with the 40S small ribosome subunit and several other initiation factors (eIF1A, eIF3, eIF1) to form the 43S pre-initiation complex (PIC). PIC is then recruited to mRNA via the scaffold eIF4G within the cap-associated eIF4F complex, forming the 48S complex. With the help of eIF4A to unwind mRNA secondary structures, PIC scans the 5′ untranslated region (5′UTR) until it encounters an initiation codon.18, 19 The efficiency of start codon recognition can be influenced by the codon context as well as initiation factors eIF1 and eIF1A, although the precise mechanism remains elusive. The event of start codon recognition is believed to trigger conformational changes of the 48S complex followed by release of the initiation factors. With the help of eIF5 and eIF5B that induce hydrolysis of eIF2-bound GTP, a 60S large ribosome subunit joins the 40S subunit, forming a complete 80S complex ready to proceed to the elongation step.18
Translation elongation is mediated by elongation factors eEF1 and eEF2, which delivers amino acid-charged tRNA to the ribosomal A site and catalyses ribosomal translocation, respectively. During elongation, the ribosome does not move at a constant speed but rather in a stop-and-go traffic manner. Both cis sequence elements and trans regulatory factors contribute to the variations of elongation speed. However, our understanding of elongation control has lagged behind the knowledge of initiation regulation. When the ribosome decoding centre reaches a stop codon, termination occurs via the concerted action of release factors eRF1 and eRF3. Notably, peptide release, tRNA dissociation, and ribosome separation do not take place simultaneously. In some cases, the 40S subunit remains associated with mRNA and could start a second round of translation from the downstream start codon, a process called re-initiation.12 Strikingly, in a reconstituted in vitro translation system, Skabkin et al found that the post-termination ribosome could migrate bi-directionally to codons cognate to the P-site tRNA.19 Although it remains to be confirmed whether this radical event occurs in vivo, the dynamic ribosome behaviour surrounding termination provides novel mechanistic insights into translation re-initiation.
Initiation Regulators and Signalling Pathways
eIF4F-mediated 5′ cap recognition
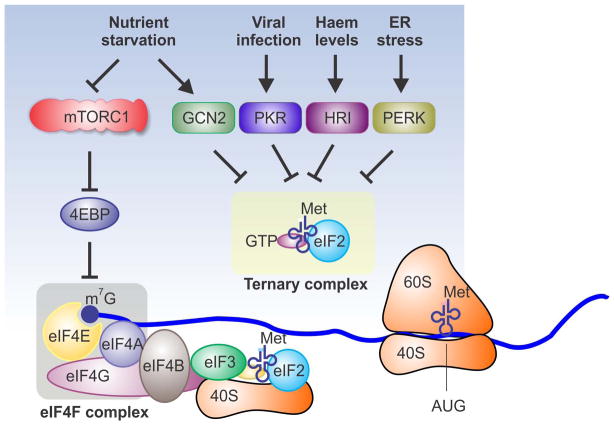
A cap-dependent mechanism accounts for the translation of the vast majority of cellular mRNAs. Under stress conditions, a diverse array of signalling pathways control the eIF4F-mediated cap recognition, thereby adjusting the rate of global protein synthesis (Figure 1). One best known regulator is the eIF4E-binding protein (4EBP), which shares a similar structure with eIF4G. By competing with eIF4G, 4EBP acts as a negative regulator of translation initiation by repressing the assembly of eIF4F complexes at the 5′ terminus of transcripts. The binding capacity of 4EBP depends on its phosphorylation status. Under normal growth conditions, 4EBP is heavily phosphorylated and has lower affinity with eIF4E.20 One major signalling pathway that mediates 4EBP phosphorylation is the mammalian target of rapamycin complex 1 (mTORC1).21, 22 mTORC1 is an evolutionarily conserved serine/threonine kinase that senses extracellular signals as well as the intracellular energy status. Nutritional stresses such as amino acid starvation inhibits global protein synthesis partially through the mTORC1 signalling pathway. mTORC1 senses amino acid levels through a sophisticated system.23 Recent studies revealed that mTORC1 activation occurs primarily at the surface of the lysosome by heterodimeric RagA/B-RagC/D GTPases.24 When amino acids are limited, Rag GTPases are inactivated, leading to GDP coupled RagA/B and GTP bound RagC/D, which are unable to recruit mTORC1 to the lysosome membrane.25 A complex named Ragulator acts as a guanine nucleotide exchange factor (GEF) for RagA and RagB, whereas another complex called GATOR1 has GTPase-activating protein (GAP) activity.26, 27 However, the direct intracellular amino acid sensor remains to be characterized. Once recruited to the lysosome surface, mTORC1 is believed to be directly activated by Ras homologue enriched in brain (Rheb).28, 29 The activated mTORC1 then phosphorylates 4EBP, leading to de-repression of eIF4F and enhanced cap-dependent translation.
Figure 1.
Multiple stress signals converge on initiation factors and inhibit global protein synthesis. Cap-dependent translation initiation requires cap binding, eIF4F complex assembly (light grey square), and ternary complex formation (light yellow square). Nutrient signalling mTORC1 controls eIF4F complex formation by phosphorylating 4EBP, which releases eIF4E for cap binding. Nutrient starvation not only inhibits the mTORC1 signalling pathway, but also triggers GCN2 kinase activity. GCN2 phosphorylates eIF2α that inhibits ternary complex formation. In addition to the GCN2 kinase, other kinases integrate many stress conditions by phosphorylating eIF2α, forming an integrated stress response targeting translation initiation.
At the lysosome surface, Rheb activity is subject to regulation by phosphoinositide 3-kinase (PI3K) pathways. Therefore, both the amino acid sensing system and the insulin signalling pathway converge on mTORC1. Rheb activity is negatively regulated by tuberous sclerosis complex (TSC) 1 and 2, in which TSC2 acts as a GAP towards Rheb.30, 31 Several stress signals integrate into mTORC1 via TSC. For instance, oxidative stress activates AMP-responsive protein kinase (AMPK) pathway, which supress mTORC1 by phosphorylating its negative regulator TSC2.32 In addition, TSC is found to be localized on the peroxisome and inhibit mTORC1 in response to endogenous reactive oxygen species.33 DNA damage could be sensed in both p53 dependent and independent pathways.34, 35 The p53-dependent pathway requires the transcriptional activation of Sestrin1 and Sestrin2. Increased Sestrin1 and 2 activate TSC2 through AMPK, eventually repressing mTORC1 activity.36 For the p53 independent recognition, DNA damage is sensed by a protein kinase ATM (ataxia telangiectasia mutated) and the signal is transduced through liver kinase B1 (LKB1)/AMPK1 to target TSC2 and inhibits mTORC1.37
As mentioned above, 4EBP is one of the direct targets of mTORC1. Under supressed mTORC1 activity during stress, the hypo-phosphorylated 4EBP sequester eIF4E from the 5′cap of mRNAs, preventing the formation of eI4F complex and the cap-dependent initiation.38 Employing ribosome profiling technique, several recent studies investigated the translational response when mTORC1 was inhibited by chemical inhibitors.39 Inhibiting mTORC1 activity by Torin significantly reduced the translation of mRNAs containing 5′ terminal oligopyrimidine (TOP) motifs or TOP-like motifs.40, 41 These mRNAs mostly encode ribosomal proteins and translation factors. In addition, several transcripts whose translation is highly regulated by mTORC1 are involved in cell proliferation, metabolism and invasion, confirming the critical role of translational control in cancer progression.41 Given the widely accepted notion that eIF4F complex formation controls the majority of cap-dependent translation, it is surprising to find that only a subset of mRNAs whose translation is influenced by mTORC1 inhibition. Indeed, in cells lacking both 4EBP1 and 4EBP2, not all mRNA translation is equally upregulated.42 Interestingly, mRNAs involving cell proliferation are preferentially subjected to translational control by 4EBP. Hence, translation of individual mRNAs has different sensitivity to the perturbation of cap-recognition.
eIF2-controlled ternary complex formation
Many stress conditions trigger the phosphorylation of eIF2α. In mammals, there are four different types of eIF2α kinases activated by different stressors: general control non-derepressible-2 (GCN2) for amino acid starvation, protein kinase RNA (PKR) for double-stranded RNAs during virus infection, PKR-like ER kinase (PERK) for unfolded proteins in ER, and heme-regulated inhibitor kinase (HRI) for heme deprivation.4 eIF2α is a subunit of eIF2 that is part of the ternary complex. As GTP is hydrolysed during translation initiation, eIF2 needs to be recharged by initiator tRNA. This recharging is accomplished by eIF2B-catalyzed GDP-GTP exchange. Under stress conditions, Ser51 of eIF2α subunit is phosphorylated by stress sensing kinases mentioned above. Phosphorylation of eIF2α inhibits the GDP-GTP exchange by reducing the dissociation rate of eIF2B.43 As a result, ternary complex formation is supressed and global translation is reduced. Therefore, different types of stress conditions converge on eIF2α, resulting in the inhibition of ternary complex formation (Figure 1). Further supporting this notion, GCN2 also responds to UV exposure and DNA damage response.44, 45 Moreover, both hypoxia and oxidative stress could activate PERK, resulting in phosphorylation of eIF2α.46
It is clear that the same type of stress could trigger multiple signalling pathways leading to global protein synthesis inhibition. For instance, amino acid starvation not only supresses eIF4-mediated cap recognition through aforementioned mTORC1 signalling pathways, but also activates GCN2 via the accumulation of uncharged tRNA.47 Consequently, both cap-recognition and ternary complex formation are supressed under nutrient starvation. It seems that both stress signalling pathways act in parallel. However, cells lacking GCN2 blunted the responsiveness of mTORC1 to amino acid deprivation.48 Much remains to be learned for the crosstalk between GCN2/eIF2α and mTORC1 signalling pathways.
Elongation Modulators and Signalling Pathways
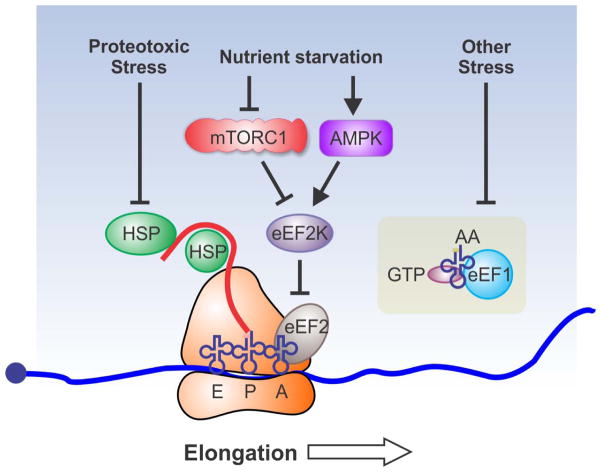
Despite the extensive regulation at the initiation stage, a growing body of evidence suggests that elongation step is subject to more rigorous regulation than is previously assumed (Figure 2).49 Like some initiation factors, one common regulatory mechanism of elongation factors is phosphorylation. For instance, elongation factor eEF2 undergoes phosphorylation at Thr56 within the GTP-binding domain in response to oxidative stress and this modification interferes with its ability to bind to the ribosome.50–53 mTORC1 negatively regulates its cognate kinase eEF2K and thereby activates eEF2.54 Thus, mTORC1 regulates protein translation at multiple stages. The activity of eEF2 can also be regulated by RNA-binding proteins. For instance, cytoplasmic polyadenylation element binding protein 2 (CPEB2) reduces the GTP hydrolysis of eEF2.55 Interestingly, CPEB2 slows down the translation of HIF1A mRNA under normal conditions by binding to the 3′UTR. When cells encounter hypoxic stress, CPEB2 dissociates from HIF-1α mRNA, leading to rapid synthesis of HIF-1α for hypoxic adaptation. Further supporting the physiological significance of eEF2, eEF2 is repressed by the activation of AMPK-eEF2K-eEF2 pathway under a series of stress conditions, including endoplasmic reticulum stress, hypoxia-induced energy stress, genotoxic stress, and nutrient deprivation.56–58 Various stress signals trigger the activation of eEF2K by AMPK-mediated phosphorylation on serine 398. Activated eEF2K phosphorylated eEF2 and induce a temporary ribosomal slowdown at the stage of elongation. During recovery stage, eEF2K is degraded by the ubiquitin-proteasome system, allowing the rapid resumption of translation elongation. Remarkably, transformed tumour cells rely on this AMPK-eEF2K axis to survive under nutrient stress conditions. Indeed, expression of eEF2K strongly correlated with overall survival in human medulloblastoma and glioblastoma multiforme.59 In addition to eEF2, eEF1A also undergoes similar regulation. One example is the role of eEF1A in epithelia-to-mesenchymal transitions (EMT) which occurs in tumour metastasis. This regulation is mediated by transforming growth factor β (TGF β) signalling pathway.60 In the absence of TGF β signalling, 3′UTRs of specific mRNAs are recognized by a RNA-binding protein heterogeneous nuclear ribonucleoprotein E1 (hnRNP E1), which blocks the translocation of ribosomes by associating with eEF1A. Active TGF β signalling phosphorylates hnRNP E1 and releases eEF1A from ribosomes, allowing the elongation to proceed on mRNAs and promoting EMT. In addition, TGF-β1 also causes dissociation of ribosomal protein RPL26 and eEF1A from p53 mRNA, thereby reducing p53 mRNA translation in response to cellular stress.61 Finally, eukaryotic initiation factor 5A (elongation factor P in prokaryotes) has been recently identified to promote elongation of polyproline motifs.62–67 In bacteria, EF-P influences the stress response of pH receptor CadC and translation of other polyproline-containing proteins, suggesting similar functions of eIF5A in eukaryotes.
Figure 2.
Translational regulation at the elongation stage. Nutrient starvation inhibits mTORC1 and activates eEF2K, which inhibits translation elongation by blocking the function of eEF2. Starvation also activates AMPK that promotes the activation of eEF2K, resulting in elongation inhibition. Many stressors could affect the activity of eEF1, although the underlying mechanism is not completely understood. In addition, ribosome-associated chaperones regulate translation elongation, enabling cells to modulate translational capacity in response to proteotoxic stress.
In addition to mechanisms regulating the elongation factors, elongation process itself can corporate with other stress response pathways to coordinate regulations at various levels. mRNA translation proceeds not at a constant rate but rather in a stop-and-go traffic manner.68 Variations of elongation speed may result from local stable mRNA structure, or the presence of rare codons.69–74 Interestingly, nascent chains could also induce translational pausing in a sequence-specific manner. Several recent studies have revealed the importance of elongation pausing in stress response. One example is the splicing of X-box-binding protein 1 messenger RNA (XBP1u mRNA) upon endoplasmic reticulum stress.75 An evolutionarily conserved peptide module at the carboxyl terminus is responsible for the translational pausing and required for the efficient targeting mRNA-ribosome-nascent chain (R-RNC) complex to the ER membrane and efficient splicing of the XBP1u mRNA. In addition, ribosomal stalling in the upstream ORF causes mRNA remodeling and formation of an active IRES (discuss in more details below), stimulating the translation of cat-1 Arg/Lys transporter under amino acid starvation.76 Using ribosome profiling, several recent studies discovered an early ribosome pausing under a variety of stress conditions, including heat shock, proteotoxic stress, and oxidative stress.77–79 Intriguingly, most of the ribosomes paused within the first 50 codon window of almost all coding sequences, a region corresponding to the length of nascent chains occupying the ribosomal exit tunnel. Since ribosome-associated chaperone molecules are located near the exit of the tunnel, it is postulated that translation elongation is influenced by chaperone availability. It is still unclear mechanistically how the absence of chaperones brings translation to a halt. This phenomenon nevertheless reveals that translating ribosomes, via associated factors, fine-tune the elongation rate by sensing the intracellular folding environment. The early elongation pausing may represent a co-translational stress response to maintain the intracellular protein homeostasis.
Growing ribosome profiling data has enabled computational simulation of translation process in yeast.80–82 Consistent with previous studies, initiation and ribosome availability were shown to be the rate-determining factors of translation under normal growth conditions. However, the simulative results suggest that elongation becomes the limiting step under severe amino acid starvation conditions. The authors argued that reduced initiation rate under stress might increase the free ribosome and tRNA, thereby promoting elongation. Although this hypothesis awaits experimental validation, it supports the importance of elongation regulation under stress.
Stress-Induced RNA Modification
Numerous modifications (>100) have been identified on the four canonical bases in most types of RNA. Some of the RNA modifications serve as sentinels for various stress conditions, while others directly affect the decoding process of translation.83 Emerging evidence points to a critical role for tRNA and rRNA modifications in the various cellular responses to stress (Figure 3). Using a quantitative system approach, Chan et al reported signature changes in the spectrum of tRNA modifications in S. cerevisiae upon oxidative stress.84 Interestingly, there was an increase in the proportion of tRNALeu(CAA) containing m5C at the wobble position. This modification causes selective translation of mRNA from genes enriched in the TTG codon. In addition to tRNA modifications, several recent studies reported that oxidative stress triggers endonucleolytical cleave of tRNAs around the anticodon, giving rise to small RNA species that may participate in various stress signalling pathways.85–89 The nucleases responsible for stress-induced tRNA cleavage are Rny1 in yeast and angiogenin in mammals. The oxidative-stress activated nucleases cleave within the conserved single-stranded 3′-CCA termini of all tRNAs, thereby blocking their use in translation. This CCA deactivation is reversible and repairable by the CCA-adding enzyme [ATP(CTP):tRNA nucleotidyltransferase].90 Through this mechanism the eukaryotic cell dynamically represses and reactivates translation at low metabolic costs. In non-stressed cells, these enzymes cannot gain access to cytosolic tRNAs, suggesting that stress-induced tRNA cleavage is a highly regulated process. However, not all stress conditions can trigger tRNA cleavage. Oxidative stress seems to preferentially affect tRNA biology. Interestingly, up to tenfold increase of methionine-misacylation occurs at tRNA when cells are exposed to oxidative stress.91 Likewise, virus infection, treating cells with toll-like receptor ligands or chemicals also induced tRNA mis-acylation. Physiological significance of modified translation fidelity remains unclear. It has been proposed that misincorporation of methionine into cellular proteins could possibly protect cells from reactive oxygen species (ROS)-mediated damage.91 A recent study reported that thiolation status of tRNA wobble-uridine nucleotides is correlated with the intracellular availability of sulphur amino acids methionine and cysteine.92 Interestingly, changing tRNA thiolation regulates translational reprogramming and enables cells to modulate translational capacity according to metabolic homeostasis.
Figure 3.
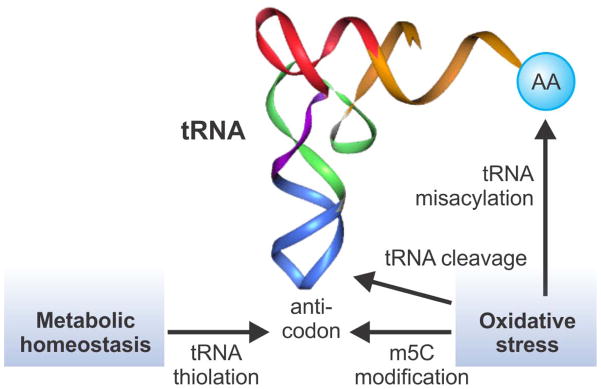
Translational regulation by tRNA modification. tRNA contains many modified nucleobases. Anti-codon modification influences decoding processes and the overall translation capacity. Oxidative stress has multiple effects on tRNA metabolism, including m5C at the wobble position, tRNA misacylation, and tRNA cleavage. In addition, metabolic homeostasis such as sulfur amino acid levels regulates tRNA thiolation at the wobble position. These tRNA modifications trigger translational reprogramming in response to stress conditions.
In eukaryotic mRNA, different types of methylation modification have been documented. One abundant and conserved mRNA modification is N6-methyladenine (m6A). The abundance of m6A has been estimated to be 3–5 residues per mRNA on average in HeLa cells.83 Importantly, the m6A modification is dynamic and can be reprogrammed under different conditions. Yeast cells have low levels of m6A modification during regular mitosis growth, but appropriate 50% of mRNAs contain m6A sites during meiosis.93 It has been suggested m6A modification may regulate translation efficiency. Using m6A-specific antibodies, two recent studies revealed a wide-spread distribution of m6A across the mammalian transcriptome.94, 95 Surprisingly, the mapped m6A sites were enriched near the stop codons and in the 3′UTRs. Further supporting the dynamic feature of m6A modification, there was a tissue-specific pattern of m6A with a dramatic increase during brain development. In addition, the m6A landscape changes in response to various stimuli. Although the exact function of m6A in mRNA remains obscure, it is certain that this dynamic modification has important regulatory roles in gene expression, including translational control.
SELECTIVE TRANSLATIONAL REGULATION DURING STRESS
Repression of global protein synthesis helps reduce the cellular burden during stress conditions. However, subsets of mRNAs undergo selective translation to produce proteins that are vital for cell survival and stress recovery.4 Cells employ a variety of mechanisms to achieve selective translation, which often involves cis sequence elements on mRNAs and trans regulatory factors recognizing specific mRNA features. Most of the cis-elements reside in the untranslated region of mRNAs, including internal ribosome entry sites (IRES), upstream open reading frames (uORFs), motifs with special sequences or secondary structures, and microRNA binding sites (Figure 4). The roles of microRNA in translational regulation during stress have been comprehensively reviewed elsewhere.96 Here we will focus on other key mechanisms regulating selective translational in response to stress.
Figure 4.
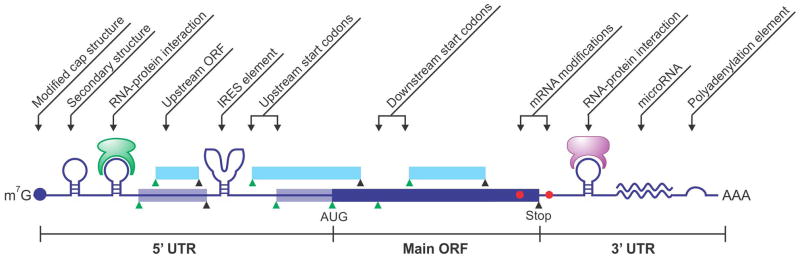
Types of cis-sequence elements that contribute to translational regulation. mRNA contains multiple start codons (green triangle) and stop codons (black triangle), generating ORFs in-frame (blue box) or out-of-frame (cyan box). Secondary structures are present in 5′UTR and/or 3′UTR, with or without interacting proteins. Reversible mRNA modification could also regulate translational reprogramming in response to stress conditions.
Cap-independent Translation Initiation
Not all the mRNAs bear the typical 5′ cap structure. The best characterized cap-independent translational mechanism is IRES.97 Originally discovered in picornavirus mRNAs, The IRES element in the 5′UTR forms complex secondary structures that directly recruit ribosome subunits without the requirement of some or even all initiation factors.98 In addition to the typical IRES elements found in viral mRNAs, a growing body of evidence suggests that certain cellular mRNAs may use the similar IRES mechanism for cap-independent translation initiation. This non-canonical translation initiation often occurs during special conditions, such as differentiation, apoptosis, and cellular stress.99 Under genotoxic stress, transcripts encoding c-Myc, p53, X-linked inhibitor of apoptosis (XIAP) and B-cell CLL/lymphoma 2 (BCL-2) are translationally upregulated and these mRNAs are believed to contain IRES at their 5′UTRs.100–102 During endoplasmic reticulum stress, the inhibitor of apoptosis protein HIAP2 undergoes IRES-mediated translational induction.103 In response to hypoxia, translational increase of vascular endothelial growth factor (VEGF) and HIF-1 is also IRES-dependent.104, 105 Additionally, translation of cold inducible RNA binding protein (CIRP) and heat shock inducible BIP, BAG-1 is also thought to be mediated through IRES.106, 107 With individual experimental validation, the list of potential IRES-containing mRNAs is expanding rapidly. Using an in vitro selection approach based on mRNA display, a recent study identified over 12,000 random genomic sequences that could act as cap-independent translation-enhancing elements (TEE).108 Interestingly, the TEE-enriched regions are overrepresented in the 5′UTR, suggesting that cap-independent translational activities might be widespread in the human genome.
Efficient IRES-mediated translation initiation requires RNA binding proteins that are known as IRES trans-acting factors (ITAFs).109 It is hypothesized that ITAFs may act as RNA chaperones to facilitate the formation of IRES secondary structures.110 However, our understanding of how IRES-ITAF interaction determines translation initiation is far from complete. For several IRES-containing transcripts mentioned above, such as p53 and BAG-1, polypyrimidine tract binding protein (PTB) functions as the ITAF.111, 112 During starvation-induced yeast differentiation, an A-rich element in the 5′UTR of some mRNAs involved in invasive growth mediates internal initiation by recruiting polyA binding protein (Pab1).113 It is likely that different IRES elements and corresponding ITAF factors interplay in distinct manners. However, functional characterization of cellular proteins serving as ITAF has lagged far behind the identification of IRES elements. It remains to be clarified whether the cellular IRES element functions in an exact same manner as the viral IRES.
The presence of both IRES and ITAF does not necessarily guarantee efficient cap-independent translation initiation. Under normal growth conditions, the limiting translation machinery prefers canonical cap-dependent translation. The functional balance between cap-dependent and cap-independent initiation underlies the central translational reprogramming in stress response. Indeed, cap-independent translation dominates only when the general cap-dependent translation is inhibited by cellular stress.114 This explains why most IRES elements are found in genes whose protein products are involved in cell survival and cell death. Further supporting the coordination between cap-dependent and cap-independent translation, overactivation of nutrient signalling pathway mTORC1 compromises the cap-independent synthesis of stress proteins like Hsp70 and consequently attenuates stress responses.115 Taken together, cap-independent translation provides an effective means for escaping the global decline in protein synthesis, while permitting the selective translation of specific mRNAs.
Alternative Translation Initiation
Proper selection of the translation initiation site on mRNAs is crucial for the production of desired protein products. In eukaryotes, ribosomal scanning is a well-accepted model for start codon selection.116 It is commonly assumed that the first AUG codon that the scanning ribosome encounters serves as the start site for translation. However, one or more potential initiation sites could exist upstream of the main start codon, forming upstream open reading frames (uORF).117 Likewise, many AUG codons downstream of the main start codon could also potentially serve as initiators. Many factors influence the start codon selection. For instance, the initiator AUG triplet is usually in an optimal context with a purine at position -3 and a guanine at position +4. The presence of mRNA secondary structure at or near the start codon also influences the recognition efficiency. In addition to these cis sequence elements, the stringency of start codon selection is also subject to regulation by trans acting factors such as eIF1 and eIF1A. Inefficient recognition of an initiator codon results in a portion of 43S PIC continuing to scan and initiating at a downstream site, in a process known as leaky scanning. Many recent studies have uncovered a surprising variety of potential translation start sites in addition to the annotated start codons. Using ribosome profiling coupled with translation inhibitors specifically targeting the initiating ribosomes, several groups have identified multiple initiation sites in almost half of the transcripts in human and mouse transcriptome.118, 119 Intriguingly, many non-AUG codons, especially CUG, act as alternative start codons for initiating uORF translation.
One expected consequence of alternative translation initiation is an expanded proteome diversity that has not been and could not be predicted by in silico analysis of AUG-mediated main ORFs. Indeed, many eukaryotic proteins exhibit a feature of NH2-terminal heterogeneity presumably due to alternative translation. Stress-triggered alternative initiation may generate isoforms with different N-terminus, leading to distinct functions or cellular localization.79 One well-characterized example is C/EBP, a family of transcription factors that regulate the expression of tissue-specific genes during differentiation. C/EBP mRNA produces protein isoforms with opposite functions according to the level of upstream hormones and signals in a tissue-specific manner.120 Alternative start codon selection could also produce functionally distinct protein isoforms. Such a strategy has been widely used by the compact genome in viruses.121 Comprehensive cataloguing of global translation initiation sites and the associated ORFs is just the beginning in unveiling the role of translational reprogramming in gene expression. The illustration of alternative translation events in response to various stress conditions represents an exciting research field to be fully exploited.
Regulatory uORFs
It has been estimated that about 50% of mammalian transcripts contain at least one upstream open reading frame (uORF).122 Based on the leaky scanning model, the presence of uORFs is considered to supress the translation efficiency of main ORFs. Indeed, ribosome profiling results showed a dramatic increase of uORF occupancy under stress conditions such as starvation, oxidative stress, heat shock and proteotoxic stress.7, 77–79 Interestingly, the ribosome occupancy of uORFs also increased during yeast meiosis and mouse stem cell differentiation.119, 123 How the up-regulation of uORF translation is achieved under these conditions remains incompletely understood. Despite the inhibitory role of uORF in the translation of most main ORFs, presence of some uORFs could stimulate the translation of mRNAs encoding stress responsive proteins. The best characterized example is GCN4 in yeast or ATF4 in mammals.124, 125 In the case of ATF4, it contains two uORFs in the 5′UTR: one near the 5′ terminus and the other overlapping with the main ORF but in different reading frames. During normal growth conditions, the ternary complex is abundant and ribosome decodes the first uORF as well as the second uORF. Termination of uORF2 does not allow the initiation of the main ORF because of sequence overlapping. Under stress conditions that trigger eIF2α phosphorylation, reduced ternary complexes formation leads to longer time for the scanning ribosome to acquire a ternary complex. As a result, more ribosomes bypass the second uORF and become available to initiate from the downstream main ORF. It is perplexing to find that uORFs play either stimulating or inhibiting roles in the translation of main ORFs. This conundrum suggests that the uORF number, length, position, and other features might be critical for the overall regulatory effects. Notably, UV-induced DNA damage triggers selective translation of mRNAs containing uORFs in the 5′UTR, indicating that the ATF4-like regulatory mechanism is widely adopted by various stress conditions.126 It will be desirable to identify stress-specific genes whose mRNA translation depends on specific type of uORFs.
In addition to regulatory roles of uORF mentioned above, the de novo translational products of uORF could have direct cellular functions. For instance, small peptides generate by uORFs in fruit fly exert critical functions in development.127 Given the multiple roles of uORFs in translation control, the importance of UTR region in gene expression cannot be overemphasized. Recent technical advances in capturing 5 termini of transcriptome have uncovered an unexpected heterogeneity of leader sequences in many transcripts.128 Remarkably, yeast cells produce mRNA isofroms with distinct ends under different growth conditions based on carbon sources.129 The 5′end heterogeneity in transcripts is supposed to generate a variety of uORF configuration, further supporting the critical role of uORF in modulating gene expression.
Specialized Ribosomes
As a ribonucleoprotein particle responsible for the catalysis of peptide bond formation, the ribosome has long been considered a “molecular machine” with little intrinsic regulatory potential. A growing body of evidence suggests that ribosome heterogeneity prevails across species, under different developmental stages, and in varied tissues.130 Variation in ribosome composition, in both rRNA and ribosome proteins, provides a regulatory mechanism to the translation machinery. A clear example is illustrated in E. coli, in which a stress-induced endonuclease MazF cleaves the 16S rRNA and removes the anti-Shine-Dalgarno sequence.131 The resultant “stress ribosome” selectively translates the leaderless mRNAs, a group of transcripts also generated by MazF. Similar to the stress ribosome and transcripts generated by MazF in E. coli, eukaryotic cells might also rely on unique interactions between the distinctive component of specialized ribosomes and the cis-element on transcripts to achieve functional specificity.132 In yeast, deletion of RPS25 didn’t affect cap-dependent translation but influenced the IRES-mediated translation by hepatitis C virus (HCV) and cricket paralysis virus (CrPV).133 Whether RPS25 has similar specificity for cellular IRES remains to be elucidated. In plant, RPL24 has been shown to promote re-initiation of ribosomes after completing the uORF translation, thereby promoting the translation of main ORFs.134
There are an increasing number of observations that implicate the role of ribosome heterogeneity in selective translation, although mechanistic insight is still lacking. In S. cerevisiae, most genes encoding ribosomal proteins have paralogue duplicates and contain introns. A recent study revealed that deleting the intron from one gene copy affected the expression of the other in a nonreciprocal manner.135 As a result, removing introns within the ribosomal protein genes influenced the cell fitness and growth under stress. These results suggest that ribosomes with distinct composition might form under stress conditions. In mammals, certain ribosome proteins have been found to mediate transcript selectivity during translation. For example, RPL38 is required for translation of Homeobox mRNAs during mouse development.136 A recent study reported that chicken erythrocytic progenitors transformed by v-erbA oncogene led to the formation of specialized ribosome devoid of PRL11.137 It remains to be elucidated how specialized ribosomes achieve the selectivity of specific mRNAs. The interplay between specialized ribosomes and the cis sequence elements of transcripts adds a novel layer of translational control under stress conditions.
CONCLUSIONS
The field of translational reprogramming has made great progress over the past decade, in large part stemming from technological developments such as ribosome profiling.8, 9 The next decade should provide both a broader view of translational regulation, as huge data sets of translatome are integrated, and a vastly more detailed view, as structural studies continuously uncover actions of the translation machinery at the atomic level. The ability of cells to adapt to stress is crucial for their survival. Regulation of global protein synthesis coupled with selective translation allows cells to rapidly respond to a variety of stress conditions. Although accumulating evidence has begun to divulge multiple signalling pathways in the stress response, more questions than answers are brought up by studies of cellular adaptation strategies involving translational reprogramming. For instance, why is the translation of individual mRNA not equally affected by common effectors acting on cap recognition or ternary complex formation? What are the precise mechanisms by which subsets of mRNAs override the repression of protein synthesis? Given the fact that uORFs are frequent in genes with critical biological functions, how does evolution exploit this element for regulatory purposes? With the prevailing mRNA modifications and complex ribosome heterogeneity, how is the imposing goal of coordinating the expression of thousands of transcripts achieved in a cell? It will be exciting to watch the unveiling of answers to these questions and to see the inevitable elegant surprises that will emerge.
As we gain better insight into the mechanisms of translation it is clear that the combination of emerging technologies will paint a multifaceted picture of this paramount cellular process. Elucidating the mechanisms underlying translational reprogramming during stress will not only shed light on the fundamental principles of translation, but also provide deeper insight of the pathophysiology of human diseases.138, 139 Stress conditions are often an underlying cause of human diseases, including diabetes, neurodegenerative disorders, and cancer. In particular, cancer cells proliferate rapidly under limited nutrients and are relatively resistant to environmental stress. It is thus critical to understand how abnormal cells alter stress responsive pathways at the translational level. Interestingly, protein translation in cancer cells is coupled to transcription network centered on heat shock factor 1 (HSF1) and this link supports the anabolic malignant phenotype.140 Disrupting this linkage using translation initiation inhibitors showed great promise in supressing tumor growth. A better understanding of translational reprogramming in stress response might ultimately lead to the development of new therapeutic strategies for human diseases.
Acknowledgments
We apologize to those whose work could not be cited owing to space constraints. We’d like to thank Qian lab members for critical reading of the manuscript. B.L. is a recipient of the Genomics Scholars Award from Center for Vertebrate Genomics at Cornell. S.-B.Q. is supported by grants from National Institutes of Health (1 DP2 OD006449-01, 1R01AG042400-01A1), Ellison Medical Foundation (AG-NS-0605-09), and DOD Exploration-Hypothesis Development Award (W81XWH-11-1-0236).
Footnotes
Conflict of interest: The authors have declared no conflicts of interest for this article.
Contributor Information
Botao Liu, Graduate Field of Genetics, Genomics, and Development, Cornell University, Ithaca, NY, USA.
Shu-Bing Qian, Email: sq38@cornell.edu, Division of Nutritional Sciences, Graduate Field of Genetics, Genomics, and Development, Cornell University, Ithaca, NY, USA.
References
- 1.Shendure J, Ji H. Next-generation DNA sequencing. Nat Biotechnol. 2008;26:1135–1145. doi: 10.1038/nbt1486. [DOI] [PubMed] [Google Scholar]
- 2.Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Spriggs KA, Bushell M, Willis AE. Translational regulation of gene expression during conditions of cell stress. Mol Cell. 2010;40:228–237. doi: 10.1016/j.molcel.2010.09.028. [DOI] [PubMed] [Google Scholar]
- 4.Holcik M, Sonenberg N. Translational control in stress and apoptosis. Nat Rev Mol Cell Biol. 2005;6:318–327. doi: 10.1038/nrm1618. [DOI] [PubMed] [Google Scholar]
- 5.Aitken CE, Lorsch JR. A mechanistic overview of translation initiation in eukaryotes. Nat Struct Mol Biol. 2012;19:568–576. doi: 10.1038/nsmb.2303. [DOI] [PubMed] [Google Scholar]
- 6.Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ingolia NT, Brar GA, Rouskin S, McGeachy AM, Weissman JS. The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nat Protoc. 2012;7:1534–1550. doi: 10.1038/nprot.2012.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Michel AM, Baranov PV. Ribosome profiling: a Hi-Def monitor for protein synthesis at the genome-wide scale. Wiley Interdiscip Rev RNA. 2013 doi: 10.1002/wrna.1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kuersten S, Radek A, Vogel C, Penalva LO. Translation regulation gets its ‘omics’ moment. Wiley Interdiscip Rev RNA. 2013 doi: 10.1002/wrna.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Panniers R. Translational control during heat shock. Biochimie. 1994;76:737–747. doi: 10.1016/0300-9084(94)90078-7. [DOI] [PubMed] [Google Scholar]
- 11.Richter K, Haslbeck M, Buchner J. The heat shock response: life on the verge of death. Mol Cell. 2010;40:253–266. doi: 10.1016/j.molcel.2010.10.006. [DOI] [PubMed] [Google Scholar]
- 12.Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol. 2010;11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gebauer F, Hentze MW. Molecular mechanisms of translational control. Nat Rev Mol Cell Biol. 2004;5:827–835. doi: 10.1038/nrm1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marintchev A, Edmonds KA, Marintcheva B, Hendrickson E, Oberer M, Suzuki C, Herdy B, Sonenberg N, Wagner G. Topology and regulation of the human eIF4A/4G/4H helicase complex in translation initiation. Cell. 2009;136:447–460. doi: 10.1016/j.cell.2009.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schutz P, Bumann M, Oberholzer AE, Bieniossek C, Trachsel H, Altmann M, Baumann U. Crystal structure of the yeast eIF4A-eIF4G complex: an RNA-helicase controlled by protein-protein interactions. Proc Natl Acad Sci U S A. 2008;105:9564–9569. doi: 10.1073/pnas.0800418105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gross JD, Moerke NJ, von der Haar T, Lugovskoy AA, Sachs AB, McCarthy JE, Wagner G. Ribosome loading onto the mRNA cap is driven by conformational coupling between eIF4G and eIF4E. Cell. 2003;115:739–750. doi: 10.1016/s0092-8674(03)00975-9. [DOI] [PubMed] [Google Scholar]
- 17.Pisarev AV, Hellen CU, Pestova TV. Recycling of eukaryotic posttermination ribosomal complexes. Cell. 2007;131:286–299. doi: 10.1016/j.cell.2007.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pestova TV, Lomakin IB, Lee JH, Choi SK, Dever TE, Hellen CU. The joining of ribosomal subunits in eukaryotes requires eIF5B. Nature. 2000;403:332–335. doi: 10.1038/35002118. [DOI] [PubMed] [Google Scholar]
- 19.Skabkin MASO, Hellen CUT, PestovaSee TV. Reinitiation and Other Unconventional Posttermination Events during Eukaryotic Translation. Mol Cell. 2013 doi: 10.1016/j.molcel.2013.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pause A, Belsham GJ, Gingras AC, Donze O, Lin TA, Lawrence JC, Jr, Sonenberg N. Insulin-dependent stimulation of protein synthesis by phosphorylation of a regulator of 5′-cap function. Nature. 1994;371:762–767. doi: 10.1038/371762a0. [DOI] [PubMed] [Google Scholar]
- 21.Ma XM, Blenis J. Molecular mechanisms of mTOR-mediated translational control. Nat Rev Mol Cell Biol. 2009;10:307–318. doi: 10.1038/nrm2672. [DOI] [PubMed] [Google Scholar]
- 22.Zoncu R, Efeyan A, Sabatini DM. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol. 2011;12:21–35. doi: 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jewell JL, Russell RC, Guan KL. Amino acid signalling upstream of mTOR. Nat Rev Mol Cell Biol. 2013;14:133–139. doi: 10.1038/nrm3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L, Sabatini DM. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 2008;320:1496–1501. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim E, Goraksha-Hicks P, Li L, Neufeld TP, Guan KL. Regulation of TORC1 by Rag GTPases in nutrient response. Nat Cell Biol. 2008;10:935–945. doi: 10.1038/ncb1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bar-Peled L, Schweitzer LD, Zoncu R, Sabatini DM. Ragulator is a GEF for the rag GTPases that signal amino acid levels to mTORC1. Cell. 2012;150:1196–1208. doi: 10.1016/j.cell.2012.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bar-Peled L, Chantranupong L, Cherniack AD, Chen WW, Ottina KA, Grabiner BC, Spear ED, Carter SL, Meyerson M, Sabatini DM. A Tumor suppressor complex with GAP activity for the Rag GTPases that signal amino acid sufficiency to mTORC1. Science. 2013;340:1100–1106. doi: 10.1126/science.1232044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Inoki K, Li Y, Xu T, Guan KL. Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes Dev. 2003;17:1829–1834. doi: 10.1101/gad.1110003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Y, Gao X, Saucedo LJ, Ru B, Edgar BA, Pan D. Rheb is a direct target of the tuberous sclerosis tumour suppressor proteins. Nat Cell Biol. 2003;5:578–581. doi: 10.1038/ncb999. [DOI] [PubMed] [Google Scholar]
- 30.Inoki K, Li Y, Zhu T, Wu J, Guan KL. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat Cell Biol. 2002;4:648–657. doi: 10.1038/ncb839. [DOI] [PubMed] [Google Scholar]
- 31.Potter CJ, Pedraza LG, Xu T. Akt regulates growth by directly phosphorylating Tsc2. Nat Cell Biol. 2002;4:658–665. doi: 10.1038/ncb840. [DOI] [PubMed] [Google Scholar]
- 32.Inoki K, Zhu T, Guan KL. TSC2 mediates cellular energy response to control cell growth and survival. Cell. 2003;115:577–590. doi: 10.1016/s0092-8674(03)00929-2. [DOI] [PubMed] [Google Scholar]
- 33.Benjamin D, Hall MN. TSC on the peroxisome controls mTORC1. Nat Cell Biol. 2013;15:1135–1136. doi: 10.1038/ncb2849. [DOI] [PubMed] [Google Scholar]
- 34.Braunstein S, Badura ML, Xi Q, Formenti SC, Schneider RJ. Regulation of protein synthesis by ionizing radiation. Mol Cell Biol. 2009;29:5645–5656. doi: 10.1128/MCB.00711-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Connolly E, Braunstein S, Formenti S, Schneider RJ. Hypoxia inhibits protein synthesis through a 4E-BP1 and elongation factor 2 kinase pathway controlled by mTOR and uncoupled in breast cancer cells. Mol Cell Biol. 2006;26:3955–3965. doi: 10.1128/MCB.26.10.3955-3965.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Budanov AV, Karin M. p53 target genes sestrin1 and sestrin2 connect genotoxic stress and mTOR signaling. Cell. 2008;134:451–460. doi: 10.1016/j.cell.2008.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alexander A, Cai SL, Kim J, Nanez A, Sahin M, MacLean KH, Inoki K, Guan KL, Shen J, Person MD, et al. ATM signals to TSC2 in the cytoplasm to regulate mTORC1 in response to ROS. Proc Natl Acad Sci U S A. 2010;107:4153–4158. doi: 10.1073/pnas.0913860107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Haghighat A, Mader S, Pause A, Sonenberg N. Repression of cap-dependent translation by 4E-binding protein 1: competition with p220 for binding to eukaryotic initiation factor-4E. EMBO J. 1995;14:5701–5709. doi: 10.1002/j.1460-2075.1995.tb00257.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Larsson O, Morita M, Topisirovic I, Alain T, Blouin MJ, Pollak M, Sonenberg N. Distinct perturbation of the translatome by the antidiabetic drug metformin. Proc Natl Acad Sci U S A. 2012;109:8977–8982. doi: 10.1073/pnas.1201689109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thoreen CC, Chantranupong L, Keys HR, Wang T, Gray NS, Sabatini DM. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature. 2012;485:109–113. doi: 10.1038/nature11083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hsieh AC, Liu Y, Edlind MP, Ingolia NT, Janes MR, Sher A, Shi EY, Stumpf CR, Christensen C, Bonham MJ, et al. The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature. 2012;485:55–61. doi: 10.1038/nature10912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dowling RJ, Topisirovic I, Alain T, Bidinosti M, Fonseca BD, Petroulakis E, Wang X, Larsson O, Selvaraj A, Liu Y, et al. mTORC1-mediated cell proliferation, but not cell growth, controlled by the 4E-BPs. Science. 2010;328:1172–1176. doi: 10.1126/science.1187532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Clemens MJ. Targets and mechanisms for the regulation of translation in malignant transformation. Oncogene. 2004;23:3180–3188. doi: 10.1038/sj.onc.1207544. [DOI] [PubMed] [Google Scholar]
- 44.Deng J, Harding HP, Raught B, Gingras AC, Berlanga JJ, Scheuner D, Kaufman RJ, Ron D, Sonenberg N. Activation of GCN2 in UV-irradiated cells inhibits translation. Curr Biol. 2002;12:1279–1286. doi: 10.1016/s0960-9822(02)01037-0. [DOI] [PubMed] [Google Scholar]
- 45.Jiang HY, Wek RC. GCN2 phosphorylation of eIF2alpha activates NF-kappaB in response to UV irradiation. Biochem J. 2005;385:371–380. doi: 10.1042/BJ20041164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu L, Wise DR, Diehl JA, Simon MC. Hypoxic reactive oxygen species regulate the integrated stress response and cell survival. J Biol Chem. 2008;283:31153–31162. doi: 10.1074/jbc.M805056200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang P, McGrath BC, Reinert J, Olsen DS, Lei L, Gill S, Wek SA, Vattem KM, Wek RC, Kimball SR, et al. The GCN2 eIF2alpha kinase is required for adaptation to amino acid deprivation in mice. Mol Cell Biol. 2002;22:6681–6688. doi: 10.1128/MCB.22.19.6681-6688.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Anthony TG, McDaniel BJ, Byerley RL, McGrath BC, Cavener DR, McNurlan MA, Wek RC. Preservation of liver protein synthesis during dietary leucine deprivation occurs at the expense of skeletal muscle mass in mice deleted for eIF2 kinase GCN2. J Biol Chem. 2004;279:36553–36561. doi: 10.1074/jbc.M404559200. [DOI] [PubMed] [Google Scholar]
- 49.Kong J, Lasko P. Translational control in cellular and developmental processes. Nat Rev Genet. 2012;13:383–394. doi: 10.1038/nrg3184. [DOI] [PubMed] [Google Scholar]
- 50.Ayala A, Parrado J, Bougria M, Machado A. Effect of oxidative stress, produced by cumene hydroperoxide, on the various steps of protein synthesis. Modifications of elongation factor- 2. J Biol Chem. 1996;271:23105–23110. doi: 10.1074/jbc.271.38.23105. [DOI] [PubMed] [Google Scholar]
- 51.Kang KR, Lee SY. Effect of serum and hydrogen peroxide on the Ca2+/calmodulin-dependent phosphorylation of eukaryotic elongation factor 2(eEF-2) in Chinese hamster ovary cells. Exp Mol Med. 2001;33:198–204. doi: 10.1038/emm.2001.33. [DOI] [PubMed] [Google Scholar]
- 52.Piwien-Pilipuk G, Ayala A, Machado A, Galigniana MD. Impairment of mineralocorticoid receptor (MR)-dependent biological response by oxidative stress and aging: correlation with post-translational modification of MR and decreased ADP-ribosylatable level of elongating factor 2 in kidney cells. J Biol Chem. 2002;277:11896–11903. doi: 10.1074/jbc.M109530200. [DOI] [PubMed] [Google Scholar]
- 53.Shenton D, Smirnova JB, Selley JN, Carroll K, Hubbard SJ, Pavitt GD, Ashe MP, Grant CM. Global translational responses to oxidative stress impact upon multiple levels of protein synthesis. J Biol Chem. 2006;281:29011–29021. doi: 10.1074/jbc.M601545200. [DOI] [PubMed] [Google Scholar]
- 54.Ryazanov AG, Davydova EK. Mechanism of elongation factor 2 (EF-2) inactivation upon phosphorylation. Phosphorylated EF-2 is unable to catalyze translocation. FEBS Lett. 1989;251:187–190. doi: 10.1016/0014-5793(89)81452-8. [DOI] [PubMed] [Google Scholar]
- 55.Chen PJ, Huang YS. CPEB2-eEF2 interaction impedes HIF-1alpha RNA translation. EMBO J. 2012;31:959–971. doi: 10.1038/emboj.2011.448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Terai K, Hiramoto Y, Masaki M, Sugiyama S, Kuroda T, Hori M, Kawase I, Hirota H. AMP-activated protein kinase protects cardiomyocytes against hypoxic injury through attenuation of endoplasmic reticulum stress. Mol Cell Biol. 2005;25:9554–9575. doi: 10.1128/MCB.25.21.9554-9575.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu L, Cash TP, Jones RG, Keith B, Thompson CB, Simon MC. Hypoxia-induced energy stress regulates mRNA translation and cell growth. Mol Cell. 2006;21:521–531. doi: 10.1016/j.molcel.2006.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kruiswijk F, Yuniati L, Magliozzi R, Low TY, Lim R, Bolder R, Mohammed S, Proud CG, Heck AJ, Pagano M, et al. Coupled activation and degradation of eEF2K regulates protein synthesis in response to genotoxic stress. Sci Signal. 2012;5:ra40. doi: 10.1126/scisignal.2002718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Leprivier G, Remke M, Rotblat B, Dubuc A, Mateo AR, Kool M, Agnihotri S, El-Naggar A, Yu B, Prakash Somasekharan S, et al. The eEF2 Kinase Confers Resistance to Nutrient Deprivation by Blocking Translation Elongation. Cell. 2013;153:1064–1079. doi: 10.1016/j.cell.2013.04.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hussey GS, Chaudhury A, Dawson AE, Lindner DJ, Knudsen CR, Wilce MC, Merrick WC, Howe PH. Identification of an mRNP complex regulating tumorigenesis at the translational elongation step. Mol Cell. 2011;41:419–431. doi: 10.1016/j.molcel.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lopez-Diaz FJ, Gascard P, Balakrishnan SK, Zhao J, Del Rincon SV, Spruck C, Tlsty TD, Emerson BM. Coordinate transcriptional and translational repression of p53 by TGF-beta1 impairs the stress response. Mol Cell. 2013;50:552–564. doi: 10.1016/j.molcel.2013.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Saini P, Eyler DE, Green R, Dever TE. Hypusine-containing protein eIF5A promotes translation elongation. Nature. 2009;459:118–121. doi: 10.1038/nature08034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gutierrez E, Shin BS, Woolstenhulme CJ, Kim JR, Saini P, Buskirk AR, Dever TE. eIF5A promotes translation of polyproline motifs. Mol Cell. 2013;51:35–45. doi: 10.1016/j.molcel.2013.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ude S, Lassak J, Starosta AL, Kraxenberger T, Wilson DN, Jung K. Translation elongation factor EF-P alleviates ribosome stalling at polyproline stretches. Science. 2013;339:82–85. doi: 10.1126/science.1228985. [DOI] [PubMed] [Google Scholar]
- 65.Doerfel LK, Wohlgemuth I, Kothe C, Peske F, Urlaub H, Rodnina MV. EF-P is essential for rapid synthesis of proteins containing consecutive proline residues. Science. 2013;339:85–88. doi: 10.1126/science.1229017. [DOI] [PubMed] [Google Scholar]
- 66.Scuoppo C, Miething C, Lindqvist L, Reyes J, Ruse C, Appelmann I, Yoon S, Krasnitz A, Teruya-Feldstein J, Pappin D, et al. A tumour suppressor network relying on the polyamine-hypusine axis. Nature. 2012;487:244–248. doi: 10.1038/nature11126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kaiser A. Translational control of eIF5A in various diseases. Amino Acids. 2012;42:679–684. doi: 10.1007/s00726-011-1042-8. [DOI] [PubMed] [Google Scholar]
- 68.Wolin SL, Walter P. Ribosome pausing and stacking during translation of a eukaryotic mRNA. EMBO J. 1988;7:3559–3569. doi: 10.1002/j.1460-2075.1988.tb03233.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fredrick K, Ibba M. How the sequence of a gene can tune its translation. Cell. 2010;141:227–229. doi: 10.1016/j.cell.2010.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gray NK, Hentze MW. Regulation of protein synthesis by mRNA structure. Mol Biol Rep. 1994;19:195–200. doi: 10.1007/BF00986961. [DOI] [PubMed] [Google Scholar]
- 71.Elf J, Nilsson D, Tenson T, Ehrenberg M. Selective charging of tRNA isoacceptors explains patterns of codon usage. Science. 2003;300:1718–1722. doi: 10.1126/science.1083811. [DOI] [PubMed] [Google Scholar]
- 72.Lavner Y, Kotlar D. Codon bias as a factor in regulating expression via translation rate in the human genome. Gene. 2005;345:127–138. doi: 10.1016/j.gene.2004.11.035. [DOI] [PubMed] [Google Scholar]
- 73.Kramer G, Boehringer D, Ban N, Bukau B. The ribosome as a platform for co-translational processing, folding and targeting of newly synthesized proteins. Nat Struct Mol Biol. 2009;16:589–597. doi: 10.1038/nsmb.1614. [DOI] [PubMed] [Google Scholar]
- 74.Novoa EM, Ribas de Pouplana L. Speeding with control: codon usage, tRNAs, and ribosomes. Trends Genet. 2012;28:574–581. doi: 10.1016/j.tig.2012.07.006. [DOI] [PubMed] [Google Scholar]
- 75.Yanagitani K, Kimata Y, Kadokura H, Kohno K. Translational pausing ensures membrane targeting and cytoplasmic splicing of XBP1u mRNA. Science. 2011;331:586–589. doi: 10.1126/science.1197142. [DOI] [PubMed] [Google Scholar]
- 76.Fernandez J, Yaman I, Huang C, Liu H, Lopez AB, Komar AA, Caprara MG, Merrick WC, Snider MD, Kaufman RJ, et al. Ribosome stalling regulates IRES-mediated translation in eukaryotes, a parallel to prokaryotic attenuation. Mol Cell. 2005;17:405–416. doi: 10.1016/j.molcel.2004.12.024. [DOI] [PubMed] [Google Scholar]
- 77.Liu B, Han Y, Qian SB. Cotranslational response to proteotoxic stress by elongation pausing of ribosomes. Mol Cell. 2013;49:453–463. doi: 10.1016/j.molcel.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shalgi R, Hurt JA, Krykbaeva I, Taipale M, Lindquist S, Burge CB. Widespread regulation of translation by elongation pausing in heat shock. Mol Cell. 2013;49:439–452. doi: 10.1016/j.molcel.2012.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gerashchenko MV, Lobanov AV, Gladyshev VN. Genome-wide ribosome profiling reveals complex translational regulation in response to oxidative stress. Proc Natl Acad Sci U S A. 2012;109:17394–17399. doi: 10.1073/pnas.1120799109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shah P, Ding Y, Niemczyk M, Kudla G, Plotkin JB. Rate-limiting steps in yeast protein translation. Cell. 2013;153:1589–1601. doi: 10.1016/j.cell.2013.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Racle J, Picard F, Girbal L, Cocaign-Bousquet M, Hatzimanikatis V. A Genome-Scale Integration and Analysis of Lactococcus lactis Translation Data. PLoS Comput Biol. 2013;9:e1003240. doi: 10.1371/journal.pcbi.1003240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Firczuk H, Kannambath S, Pahle J, Claydon A, Beynon R, Duncan J, Westerhoff H, Mendes P, McCarthy JE. An in vivo control map for the eukaryotic mRNA translation machinery. Mol Syst Biol. 2013;9:635. doi: 10.1038/msb.2012.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yi C, Pan T. Cellular dynamics of RNA modification. Acc Chem Res. 2011;44:1380–1388. doi: 10.1021/ar200057m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chan CT, Pang YL, Deng W, Babu IR, Dyavaiah M, Begley TJ, Dedon PC. Reprogramming of tRNA modifications controls the oxidative stress response by codon-biased translation of proteins. Nat Commun. 2012;3:937. doi: 10.1038/ncomms1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Thompson DM, Lu C, Green PJ, Parker R. tRNA cleavage is a conserved response to oxidative stress in eukaryotes. RNA. 2008;14:2095–2103. doi: 10.1261/rna.1232808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Saikia M, Krokowski D, Guan BJ, Ivanov P, Parisien M, Hu GF, Anderson P, Pan T, Hatzoglou M. Genome-wide identification and quantitative analysis of cleaved tRNA fragments induced by cellular stress. J Biol Chem. 2012;287:42708–42725. doi: 10.1074/jbc.M112.371799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Thompson DM, Parker R. Stressing out over tRNA cleavage. Cell. 2009;138:215–219. doi: 10.1016/j.cell.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 88.Yamasaki S, Ivanov P, Hu GF, Anderson P. Angiogenin cleaves tRNA and promotes stress-induced translational repression. J Cell Biol. 2009;185:35–42. doi: 10.1083/jcb.200811106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ivanov P, Emara MM, Villen J, Gygi SP, Anderson P. Angiogenin-induced tRNA fragments inhibit translation initiation. Mol Cell. 2011;43:613–623. doi: 10.1016/j.molcel.2011.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Czech A, Wende S, Morl M, Pan T, Ignatova Z. Reversible and rapid transfer-RNA deactivation as a mechanism of translational repression in stress. PLoS Genet. 2013;9:e1003767. doi: 10.1371/journal.pgen.1003767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Netzer N, Goodenbour JM, David A, Dittmar KA, Jones RB, Schneider JR, Boone D, Eves EM, Rosner MR, Gibbs JS, et al. Innate immune and chemically triggered oxidative stress modifies translational fidelity. Nature. 2009;462:522–526. doi: 10.1038/nature08576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Laxman S, Sutter BM, Wu X, Kumar S, Guo X, Trudgian DC, Mirzaei H, Tu BP. Sulfur Amino Acids Regulate Translational Capacity and Metabolic Homeostasis through Modulation of tRNA Thiolation. Cell. 2013;154:416–429. doi: 10.1016/j.cell.2013.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bodi Z, Button JD, Grierson D, Fray RG. Yeast targets for mRNA methylation. Nucleic Acids Res. 2010;38:5327–5335. doi: 10.1093/nar/gkq266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell. 2012;149:1635–1646. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K, Jacob-Hirsch J, Amariglio N, Kupiec M, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- 96.Leung AK, Sharp PA. MicroRNA functions in stress responses. Mol Cell. 2010;40:205–215. doi: 10.1016/j.molcel.2010.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pelletier J, Sonenberg N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1988;334:320–325. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- 98.Gilbert WV. Alternative ways to think about cellular internal ribosome entry. J Biol Chem. 2010;285:29033–29038. doi: 10.1074/jbc.R110.150532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hellen CU, Sarnow P. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev. 2001;15:1593–1612. doi: 10.1101/gad.891101. [DOI] [PubMed] [Google Scholar]
- 100.Dobbyn HC, Hill K, Hamilton TL, Spriggs KA, Pickering BM, Coldwell MJ, de Moor CH, Bushell M, Willis AE. Regulation of BAG-1 IRES-mediated translation following chemotoxic stress. Oncogene. 2008;27:1167–1174. doi: 10.1038/sj.onc.1210723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sherrill KW, Byrd MP, Van Eden ME, Lloyd RE. BCL-2 translation is mediated via internal ribosome entry during cell stress. J Biol Chem. 2004;279:29066–29074. doi: 10.1074/jbc.M402727200. [DOI] [PubMed] [Google Scholar]
- 102.Yang DQ, Halaby MJ, Zhang Y. The identification of an internal ribosomal entry site in the 5′-untranslated region of p53 mRNA provides a novel mechanism for the regulation of its translation following DNA damage. Oncogene. 2006;25:4613–4619. doi: 10.1038/sj.onc.1209483. [DOI] [PubMed] [Google Scholar]
- 103.Warnakulasuriyarachchi D, Cerquozzi S, Cheung HH, Holcik M. Translational induction of the inhibitor of apoptosis protein HIAP2 during endoplasmic reticulum stress attenuates cell death and is mediated via an inducible internal ribosome entry site element. J Biol Chem. 2004;279:17148–17157. doi: 10.1074/jbc.M308737200. [DOI] [PubMed] [Google Scholar]
- 104.Lang KJ, Kappel A, Goodall GJ. Hypoxia-inducible factor-1alpha mRNA contains an internal ribosome entry site that allows efficient translation during normoxia and hypoxia. Mol Biol Cell. 2002;13:1792–1801. doi: 10.1091/mbc.02-02-0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Bornes S, Prado-Lourenco L, Bastide A, Zanibellato C, Iacovoni JS, Lacazette E, Prats AC, Touriol C, Prats H. Translational induction of VEGF internal ribosome entry site elements during the early response to ischemic stress. Circ Res. 2007;100:305–308. doi: 10.1161/01.RES.0000258873.08041.c9. [DOI] [PubMed] [Google Scholar]
- 106.Al-Fageeh MB, Smales CM. Cold-inducible RNA binding protein (CIRP) expression is modulated by alternative mRNAs. RNA. 2009;15:1164–1176. doi: 10.1261/rna.1179109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Coldwell MJ, deSchoolmeester ML, Fraser GA, Pickering BM, Packham G, Willis AE. The p36 isoform of BAG-1 is translated by internal ribosome entry following heat shock. Oncogene. 2001;20:4095–4100. doi: 10.1038/sj.onc.1204547. [DOI] [PubMed] [Google Scholar]
- 108.Wellensiek BP, Larsen AC, Stephens B, Kukurba K, Waern K, Briones N, Liu L, Snyder M, Jacobs BL, Kumar S, et al. Genome-wide profiling of human cap-independent translation-enhancing elements. Nat Methods. 2013 doi: 10.1038/nmeth.2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Martinez-Salas E, Ramos R, Lafuente E, Lopez de Quinto S. Functional interactions in internal translation initiation directed by viral and cellular IRES elements. J Gen Virol. 2001;82:973–984. doi: 10.1099/0022-1317-82-5-973. [DOI] [PubMed] [Google Scholar]
- 110.Wolin SL, Cedervall T. The La protein. Annu Rev Biochem. 2002;71:375–403. doi: 10.1146/annurev.biochem.71.090501.150003. [DOI] [PubMed] [Google Scholar]
- 111.Pickering BM, Mitchell SA, Spriggs KA, Stoneley M, Willis AE. Bag-1 internal ribosome entry segment activity is promoted by structural changes mediated by poly(rC) binding protein 1 and recruitment of polypyrimidine tract binding protein 1. Mol Cell Biol. 2004;24:5595–5605. doi: 10.1128/MCB.24.12.5595-5605.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Grover R, Ray PS, Das S. Polypyrimidine tract binding protein regulates IRES-mediated translation of p53 isoforms. Cell Cycle. 2008;7:2189–2198. doi: 10.4161/cc.7.14.6271. [DOI] [PubMed] [Google Scholar]
- 113.Gilbert WV, Zhou K, Butler TK, Doudna JA. Cap-independent translation is required for starvation-induced differentiation in yeast. Science. 2007;317:1224–1227. doi: 10.1126/science.1144467. [DOI] [PubMed] [Google Scholar]
- 114.Merrick WC. Cap-dependent and cap-independent translation in eukaryotic systems. Gene. 2004;332:1–11. doi: 10.1016/j.gene.2004.02.051. [DOI] [PubMed] [Google Scholar]
- 115.Sun J, Conn CS, Han Y, Yeung V, Qian SB. PI3K-mTORC1 attenuates stress response by inhibiting cap-independent Hsp70 translation. J Biol Chem. 2011;286:6791–6800. doi: 10.1074/jbc.M110.172882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kozak M. Pushing the limits of the scanning mechanism for initiation of translation. Gene. 2002;299:1–34. doi: 10.1016/S0378-1119(02)01056-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Iacono M, Mignone F, Pesole G. uAUG and uORFs in human and rodent 5′untranslated mRNAs. Gene. 2005;349:97–105. doi: 10.1016/j.gene.2004.11.041. [DOI] [PubMed] [Google Scholar]
- 118.Lee S, Liu B, Huang SX, Shen B, Qian SB. Global mapping of translation initiation sites in mammalian cells at single-nucleotide resolution. Proc Natl Acad Sci U S A. 2012;109:E2424–2432. doi: 10.1073/pnas.1207846109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ingolia NT, Lareau LF, Weissman JS. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011;147:789–802. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Descombes P, Schibler U. A liver-enriched transcriptional activator protein, LAP, and a transcriptional inhibitory protein, LIP, are translated from the same mRNA. Cell. 1991;67:569–579. doi: 10.1016/0092-8674(91)90531-3. [DOI] [PubMed] [Google Scholar]
- 121.Stern-Ginossar N, Weisburd B, Michalski A, Le VT, Hein MY, Huang SX, Ma M, Shen B, Qian SB, Hengel H, et al. Decoding human cytomegalovirus. Science. 2012;338:1088–1093. doi: 10.1126/science.1227919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Calvo SE, Pagliarini DJ, Mootha VK. Upstream open reading frames cause widespread reduction of protein expression and are polymorphic among humans. Proc Natl Acad Sci U S A. 2009;106:7507–7512. doi: 10.1073/pnas.0810916106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Brar GA, Yassour M, Friedman N, Regev A, Ingolia NT, Weissman JS. High-resolution view of the yeast meiotic program revealed by ribosome profiling. Science. 2012;335:552–557. doi: 10.1126/science.1215110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Mueller PP, Hinnebusch AG. Multiple upstream AUG codons mediate translational control of GCN4. Cell. 1986;45:201–207. doi: 10.1016/0092-8674(86)90384-3. [DOI] [PubMed] [Google Scholar]
- 125.Dever TE, Feng L, Wek RC, Cigan AM, Donahue TF, Hinnebusch AG. Phosphorylation of initiation factor 2 alpha by protein kinase GCN2 mediates gene-specific translational control of GCN4 in yeast. Cell. 1992;68:585–596. doi: 10.1016/0092-8674(92)90193-g. [DOI] [PubMed] [Google Scholar]
- 126.Powley IR, Kondrashov A, Young LA, Dobbyn HC, Hill K, Cannell IG, Stoneley M, Kong YW, Cotes JA, Smith GC, et al. Translational reprogramming following UVB irradiation is mediated by DNA-PKcs and allows selective recruitment to the polysomes of mRNAs encoding DNA repair enzymes. Genes Dev. 2009;23:1207–1220. doi: 10.1101/gad.516509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kondo T, Plaza S, Zanet J, Benrabah E, Valenti P, Hashimoto Y, Kobayashi S, Payre F, Kageyama Y. Small peptides switch the transcriptional activity of Shavenbaby during Drosophila embryogenesis. Science. 2010;329:336–339. doi: 10.1126/science.1188158. [DOI] [PubMed] [Google Scholar]
- 128.Arribere JA, Gilbert WV. Roles for transcript leaders in translation and mRNA decay revealed by transcript leader sequencing. Genome Res. 2013;23:977–987. doi: 10.1101/gr.150342.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Pelechano V, Wei W, Steinmetz LM. Extensive transcriptional heterogeneity revealed by isoform profiling. Nature. 2013;497:127–131. doi: 10.1038/nature12121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Xue S, Barna M. Specialized ribosomes: a new frontier in gene regulation and organismal biology. Nat Rev Mol Cell Biol. 2012;13:355–369. doi: 10.1038/nrm3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Vesper O, Amitai S, Belitsky M, Byrgazov K, Kaberdina AC, Engelberg-Kulka H, Moll I. Selective translation of leaderless mRNAs by specialized ribosomes generated by MazF in Escherichia coli. Cell. 2011;147:147–157. doi: 10.1016/j.cell.2011.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Gilbert WV. Functional specialization of ribosomes? Trends Biochem Sci. 2011;36:127–132. doi: 10.1016/j.tibs.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Landry DM, Hertz MI, Thompson SR. RPS25 is essential for translation initiation by the Dicistroviridae and hepatitis C viral IRESs. Genes Dev. 2009;23:2753–2764. doi: 10.1101/gad.1832209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Park HS, Himmelbach A, Browning KS, Hohn T, Ryabova LA. A plant viral “reinitiation” factor interacts with the host translational machinery. Cell. 2001;106:723–733. doi: 10.1016/s0092-8674(01)00487-1. [DOI] [PubMed] [Google Scholar]
- 135.Parenteau J, Durand M, Morin G, Gagnon J, Lucier JF, Wellinger RJ, Chabot B, Elela SA. Introns within ribosomal protein genes regulate the production and function of yeast ribosomes. Cell. 2011;147:320–331. doi: 10.1016/j.cell.2011.08.044. [DOI] [PubMed] [Google Scholar]
- 136.Kondrashov N, Pusic A, Stumpf CR, Shimizu K, Hsieh AC, Xue S, Ishijima J, Shiroishi T, Barna M. Ribosome-mediated specificity in Hox mRNA translation and vertebrate tissue patterning. Cell. 2011;145:383–397. doi: 10.1016/j.cell.2011.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Nguyen-Lefebvre AT, Leprun G, Morin V, Vinuelas J, Coute Y, Madjar JJ, Gandrillon O, Gonin-Giraud S. V-erbA generates ribosomes devoid of RPL11 and regulates translational activity in avian erythroid progenitors. Oncogene. 2013 doi: 10.1038/onc.2013.93. [DOI] [PubMed] [Google Scholar]
- 138.Sonenberg N, Hinnebusch AG. New modes of translational control in development, behavior, and disease. Mol Cell. 2007;28:721–729. doi: 10.1016/j.molcel.2007.11.018. [DOI] [PubMed] [Google Scholar]
- 139.Gkogkas CG, Khoutorsky A, Ran I, Rampakakis E, Nevarko T, Weatherill DB, Vasuta C, Yee S, Truitt M, Dallaire P, et al. Autism-related deficits via dysregulated eIF4E-dependent translational control. Nature. 2013;493:371–377. doi: 10.1038/nature11628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Santagata S, Mendillo ML, Tang YC, Subramanian A, Perley CC, Roche SP, Wong B, Narayan R, Kwon H, Koeva M, et al. Tight coordination of protein translation and HSF1 activation supports the anabolic malignant state. Science. 2013;341:1238303. doi: 10.1126/science.1238303. [DOI] [PMC free article] [PubMed] [Google Scholar]