Abstract
Weak protein-protein interactions are critical in numerous biological processes. Unfortunately, they are difficult to characterize due to the high concentrations required for the production and detection of the complex population. The inherent sensitivity of nuclear magnetic resonance (NMR) spectroscopy to the chemical environment makes it an excellent tool to tackle this problem. NMR permits the exploration of interactions over a range of affinities, yielding essential insights into dynamic biological processes. The conversion of mRNA to protein is one such process that requires the coordinated association of many low affinity proteins. During start codon recognition, eukaryotic initiation factors assemble into high-order complexes that bind mRNA and bring it to the ribosome for decoding. Many of the structures of the eukaryotic initiation factors have been determined; however, only little is known regarding the weak binary complexes formed and their structure-function mechanisms. Herein, we use start codon recognition as a model system to review the relevant NMR methods for the characterization of weak interactions and the development of small molecule inhibitors.
Keywords: NMR, residual dipolar couplings, paramagnetic relaxation enhancement, chemical shift perturbation, SAXS reconstitution assay, cross saturation, fragment based screening
Introduction
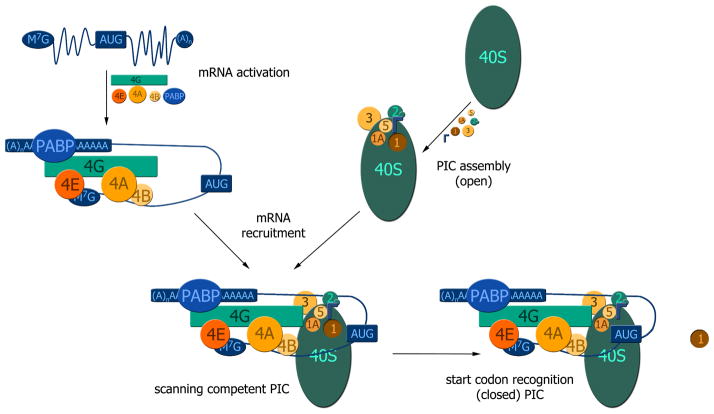
Non-covalent protein interactions exist over a wide range of affinities, from tight complexes with femtomolar affinities to transient interactions with weak, millimolar affinities. These weak protein-protein interactions (PPIs) play fundamental roles in a wide range of biological processes. An excellent example is protein synthesis, where the assembly/disassembly of large, multiprotein complexes is required to occur at fast rates. In eukaryotes, this process is highly regulated at its first step, translation initiation, which requires eIFs to form high-order complexes for regulation of the final step of gene expression. At least a dozen eIFs operate in concert within the decoding center on the 40S ribosome to prime the preinitiation complexes (PICs) with the initiator tRNA hybridized to the mRNA (Figure 1). Two multiprotein complexes assemble independently and then converge to produce an active scanning preinitiation complex (PIC): 1.) eIF4E binds the 5′ cap of the mRNAs along with eIF4A the scaffolding protein eIF4G, along with eIF4B and PABP. 2.) 40S ribosomes serve as platforms for the following eIFs to bind directly: eIF3, eIF2-TC, eIF1, eIF1A and eIF5. Upon mRNA recruitment to PICs, mRNAs are then siphoned through a channel of the 40S ribosome while triplets of nucleotides are inspected for the proper start codon. This dynamic process culminates with the initiator tRNA properly hybridized to the canonical AUG codon of the mRNA [1–3].
Figure 1. Overall schematic of the scanning process that translation preinitiation complexes undergo to reach the start codon.
The messenger RNA is activated by eIF4E binding to the 5′ cap of messenger RNAs (blue) and operates within the larger complex of eIF4F (eIF4E/eIF4G/eIF4A) along with eIF4B and PABP (Top left). eIF1, eIF1A, eIF2-TC, eIF5 and eIF3 bind to the 40S ribosomal particle and prepare it for the recruitment of mRNA. eIF3 binds to the solvent side of the 40S particle, while eIF1A, eIF1, eIF2-TC and eIF5 bind to the subunit interface of the 40S particle, which is assembled into the translation preinitiation complex PIC (open) (Top right). These eIF-driven higher order complexes combine to form a scanning PIC, which inspects triplets in search of the proper start codon (Bottom left). Once the PIC recognizes the start codon, eIF1 is released and the PIC switches from the open to the closed state (Bottom right).
Despite the wealth of structural information for the eukaryotic 40S and the majority of eIFs [4–6], we are only at the cusp of understanding how these actors interact to promote start codon recognition. A major stumbling block to fully understanding the inner workings of the PICs is the weak nature of interactions among eIFs. An arsenal of traditional biochemical techniques have been applied within the field of translation initiation, but it is the implementation of NMR spectroscopy that facilitates the understanding of molecular mechanisms through the characterization of weak PPIs. NMR is extremely sensitive to changes in the chemical environment (i.e. electron environment), which enables the precise mapping of weak protein-protein interfaces that are intractable to other methods. Furthermore, determination of the NMR structure is not a prerequisite to examining molecular interactions. The only requirement is assignment of spectral resonances to the nitrogen, carbon, and hydrogen atoms within a given amino acid sequence. Once this standard task of assigning the polypeptide backbone is complete, a variety of NMR experiments can be performed to map the interface of weak PPIs. NMR can then be applied to screen and rationally develop weak-binding small molecule leads into high affinity drug-like antagonists. This review highlights the most commonly used NMR experiments for the characterization of weak PPIs using start codon recognition as a model system.
Exchange Regimes and Weak Protein-Protein Interactions
Complex formation is governed by the rate constants for association and dissociation, kon and koff, respectively. These two rate constants are combined to produce the exchange rate constant, kex.
The relationship between kex and the chemical shift difference frequency (Δω = ωPL − ωP) between bound (ωPL) and unbound (ωP) states has an important effect and is used to classify NMR resonances as either fast, intermediate or slow exchange [7, 8]. Slow exchange on the NMR timescale occurs when the exchange rate constant is significantly smaller than chemical shift difference frequency (kex ≪ Δω). A resonance in slow exchange yields two peaks, corresponding to the free and bound form, where the intensities are proportional to concentration of each state in equilibrium.
In contrast, fast exchange (kex ≫ Δω) results in a single peak whose position is the population-weighted average of the two states. Intermediate exchange occurs when the exchange rate is similar to the chemical shift difference frequency (kex ~ Δω) and leads to broadening of the resonance. Qualitatively, slow, intermediate, and fast exchange regimes can be used to identify strong (nanomolar), intermediate (low micromolar), and weak (micromolar to millimolar) interactions, respectively.
Chemical Shift Perturbation Assay
This review describes the use of NMR to probe the weak PPIs of eIFs with known structures or backbone assignments. The resonance frequency, or chemical shift, of a resonance reflects the unique chemical environment of a given nuclei [9–11]. Changes in the chemical shift, also known as chemical shift perturbations (CSPs), are manifested in NMR spectra by movements in the position of each cross-peak (Figure 2A) [8].
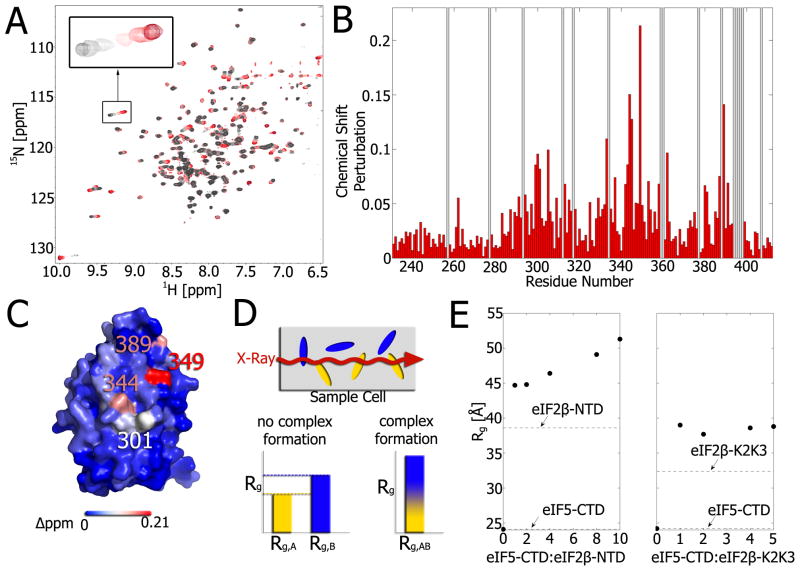
Figure 2. Chemical shift perturbations and SAXS as complementary tools for the study of protein-protein interfaces.
A) Overlay of 1H-15N HSQC spectra of 15N-eIF5-CTD titrated with increasing concentrations of eIF2β–K2K3 from 0–1 molar equivalents (colored from gray to red). The inset shift perturbation exemplifies the expected pattern of chemical shift averaging for fast exchange. B) Plot of chemical shift changes at a 1:1 ratio of eIF5-CTD: eIF2β–K2K3 versus the residue number. Chemical shift perturbations were calculated using the following equation: . Gray bars indicate residues with no assignment. C) Mapping of the perturbations onto the structure of eIF5-CTD (color corresponds to magnitude of shift perturbation). D) Principle of the SAXS reconstitution assay. A mixture of proteins is subjected to the X-ray beam and Rg vales are extracted from the scattering profile. If the two proteins in the mixture don’t interact with each other, the Rg represents a concentration-weighted average of the individual proteins’ Rg values. If the two proteins associate, the Rg is the weighted average of the complex’s Rg and the Rg of the protein in excess. Titration of one protein to another will produce a typical binding curve for the change in Rg values. E) Using the SAXS reconstitution assay, the titration of eIF2β–NTD (left) and eIF2β–K2K3 (right) to eIF5-CTD clearly show that a complex is formed in both cases. In the case of eIF2β–NTD, a second binding event is observed at increasing eIF2β–NTD concentrations.
In the field of translation initiation, CSPs have been used to map the interaction interfaces of many eIFs [12–19]. Typically, 1H-15N heteronuclear single-quantum coherence (HSQC) spectra are collected on 15N-labeled protein in the presence and absence of an unlabeled ligand. The 1H-15N HSQC provides a “fingerprint” of the protein in which each cross-peak corresponds to an individual NH groups and therefore reports on the environment of every amino acid with the exception of proline. Ligand binding causes changes in the chemical environment of nuclei responsible for the interaction with the most perturbed residues qualitatively playing the most critical role (Figure 2B). The amino acid residues with the largest chemical shift changes can then be mapped onto the protein to identify the binding interface (Figure 2C). Since the interactions are studied at equilibrium at high protein concentrations, the assay is suitable for detecting even weak interactions and are limited only by the solubility of the binding partners (reviewed in [7]). Since the amide groups only reflect changes to the protein backbone upon ligand binding, sidechain CSPs can be measured, for example, by collecting 1H-13C HSQC spectra of deuterated proteins labeled with 13C-1H methyl groups.
It was shown previously that mammalian eIF5-CTD interacts with eIF2β-NTD [20], however, the binding interface was unknown. Titration of eIF2β-NTD into eIF5-CTD resulted in broadening of the resonances, indicative of intermediate exchange on the NMR timescale, and hampered determination of the interface. Repeating the titration with eIF2β-K2K3, a variant of eIF2β-NTD shortened by one of its three K-boxes, pushed the complex into the fast exchange regime and enabled determination of the interface. Figure 2A demonstrates how the chemical shift of each eIF5-CTD cross-peak changes throughout the titration of eIF2β-NTD. The chemical shift changes are plotted as a function of residue number (Figure 2B) and mapped onto the structure of eIF5-CTD to highlight the binding epitope (Figure 2C). Mapping of the interface was critical for elucidating the role of eIF5-CTD in terminating start codon recognition [15].
In some cases, chemical shift perturbation might be very small and inconclusive. For these situations, we previously developed a small-angle X-ray scattering (SAXS) reconstitution assay to analyze changes to the radius of gyration (Rg) upon increasing the concentration of one of the complex components [21]. A general scheme is shown in Figure 2D. If no binding occurs the readout would be a weighted average of the Rg values of the monomeric compounds. Upon complex formation, the Rg value would increase. A sharp increase in Rg value would occur for strong binding affinities and a more gradual increase for weaker interactions [21]. We have applied this method previously to translation initiation complexes important for start codon recognition [15]. Figure 2E shows an example of the SAXS reconstitution assay for the complex formation of eIF5-CTD with eIF2β-K2K3 and eIF2β–NTD. The first two points of the SAXS titration cover the entire concentration range of the NMR titration shown in Figure 2A. Increasing eIF2β-K2K3 concentrations produced no further change in Rg and suggested saturation of binding. In contrast, further increases in the eIF2β-NTD concentration resulted in a more gradually increasing, non-saturating Rg value and suggest a second, lower affinity binding site. Because addition of eIF2β-K2K3 did not show a second, gradual increase this data suggests that all three K-boxes are required for binding to both sites. SAXS is an excellent independent tool to complement the study of weak PPIs. Another orthogonal method to complement NMR data in the study of complexes that require binding of divalent cations such as Mg2+ is XAFS (X-ray absorption fine structure). Similar to SAXS, XAFS can be used as a reconstitution assay by titrating protein into a constant concentration of the Mg-bound ligand (substituting Mn2+ for Mg2+ for technical reasons) [22].
Residual Dipolar Couplings
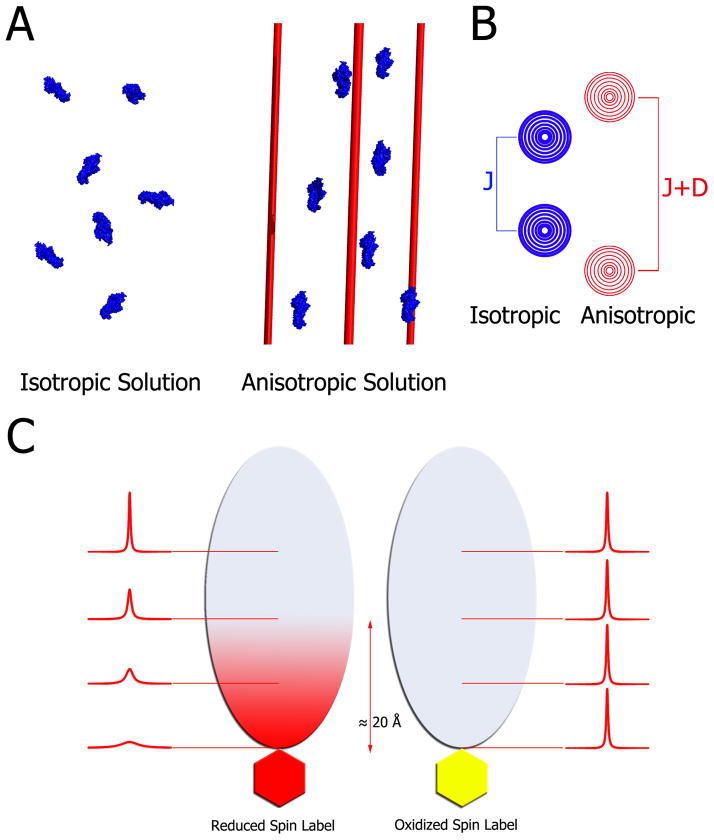
Residual Dipolar Couplings (RDCs) have become a routine constraint in NMR structure determination by providing short- and long-range orientation information (see for example [23]). Figure 3A and B provide a schematic of the principle of RDCs. Under isotropic conditions, dipolar couplings average to zero. Weak partial alignment of the protein in an anisotropic medium (such as phage, nanotubes, bicelles etc.) leads to non-zero averaging of dipolar couplings. Conveniently, RDCs contribute to the observed 1JNH coupling in 1H-15N HSQC experiments and, therefore, can be extracted by taking the difference of the effective 1JNH coupling value in the presence and absence of alignment media [24]. The RDCs obtained are an average of all conformations in solution and would therefore present a mixture of contributions from the free and bound state making the analysis very complicated. Recently, Blackledge et al. developed a protocol for measuring and analyzing RDCs for weak protein-protein complexes by combining differential isotope labeling and linear extrapolation [25]. They successfully applied this protocol to the complexation of CD2AP SH3-C and ubiquitin (KD = 132 μM).
Figure 3. Principles of residual dipolar couplings and paramagnetic relaxation enhancement.
A) In an isotropic medium (left) proteins tumble freely averaging out any dipolar couplings. Addition of filamentous phage, nanotubes, bicelles, compressed gels etc. produces a modest anisotropy of the proteins (right). For illustration purposes all molecules here are aligned; however, in practice, the media are tuned to produce about 0.1% alignment. This partial alignment leads to an incomplete averaging of anisotropic magnetic interactions manifested in residual dipolar couplings. B) In isotropic conditions, the spectrum of a coupled resonance yields two peaks separated by the J-coupling. In anisotropic conditions, the peaks are separated by the sum of the J-coupling and the residual dipolar coupling. Therefore, RDCs can be easily extracted by comparing the effective couplings under isotropic and anisotropic conditions. C) For paramagnetic relaxation enhancement, a paramagnetic spin label, which is a stable free radical, is covalently linked to the thiol of a lone cysteine residue (left side). This spin-labeled protein is then added to 15N-labeled cognate partner, which subsequently broadens the interface resonances on its 1H-15N HSQC spectrum. Subsequently, the radical is quenched by reduction with ascorbic acid, and the spectrum is collected under the same conditions. Upon reduction, the previously broadened resonances reappear, which facilitates mapping of the interaction interface within ~20 Å of the spin-labeled probe.
Paramagnetic Relaxation Enhancement Experiments
Paramagnetic relaxation enhancement (PRE) is a method that can not only qualitatively identify the complex interface, but also provide distance restraints between one protein and its cognate partner. Hence in combination with the CSP assay, PRE experiments serve as a complimentary method to map an interaction interface. As opposed to NOEs which require a relatively long time to develop and are only observable up to 5 Å, PREs can monitor transient, long-range interactions up to 20–25 Å. Attachment of a paramagnetic prosthetic group, usually via thiol linkage, to an otherwise unlabeled protein leads to the broadening of cross-peaks present at the interface of an isotopically-labeled binding partner (Figure 3C, left). A reducing agent, such as ascorbic acid, is then added to return resonances to their normal peak height and intensity for assignment (Figure 3C, right). For a comprehensive overview over this method see [26]. Often PREs are more sensitive than RDCs for measuring low populated conformations. The Ubbin k group has utilized PREs extensively in the study of the transient complex between cytochrome c and cytochrome c peroxidase [27, 28]. In the field of translation initiation, PRE experiments were performed on eIF4E to collect distance information for refining structures with limited NOE data [29].
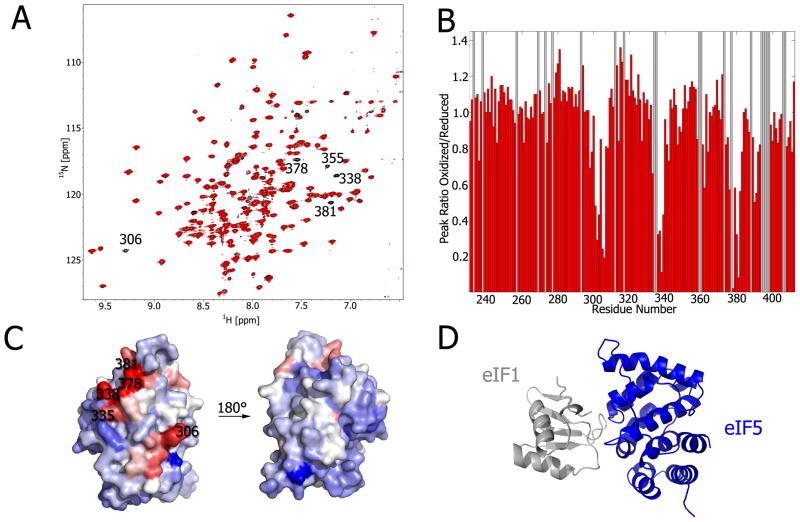
Regarding start codon recognition, PRE experiments were used to map the distances of eIF interfaces, which validated the CSP experiments and confirmed that eIF1 binds to one face of the eIF5 CTD [15]. The reagent (1-oxyl-2,2,5,5-tetramethyl-3-pyrroline-Δ3-methyl) methanethiosulfonate (MTSL) was incubated with the single cysteine mutant eIF1-K57C. Following MTSL attachment and purification via column chromatography, we combined eIF1-K57C-MTSL with 15N-isotopically labeled eIF5-CTD. The paramagnetic property of the hydroxyl radical on the eIF1-K57C mutant broadens the resonances of eIF5-CTD (Figure 4A; red), while the addition of ascorbic acid (vitamin C) reduces the spin label (Figure 4A; black). The paramagnetic broadening effect can be readily observed in the 1H-15N HSQC overlay of the of oxidized and reduced forms (Figure 4A). Since the backbone resonance assignments of eIF5-CTD have been previously determined [15], the magnitude of resonance broadening can be quantified from the HSQC overlay (Figure 4B). In addition, eIF1-K57C mutant’s interacting interface on eIF5-CTD was mapped using this PRE data (Figure 3C). Since the structures of both eIF1 and eIF5-CTD have been solved previously, we combined our CSP mapping data along with the distance restraints obtained from the PRE data to model a protein complex (Figure 4D).
Figure 4. Modeling weak protein complexes with PREs.
A) Overlay of 1H-15N HSQC spectra of 0.2 mM 15N-labeled eIF5-CTD in the presence of 0.2 mM eIF1-K57C-MTSL (oxidized, red; reduced, black). B) This histogram corresponds with the 1H-15N HSQC spectra in A; oxidized and reduced, eIF1-K57C-MTSL. The ratio of oxidized/reduced cross-peak intensities is plotted as a function of eIF5-CTD residue number (adapted from [15]). Grey bars represent backbone resonances that are not assigned. C) eIF5-CTD residues most effected by eIF1-K57C-MTSL are mapped onto the protein surface. D) CSP and PRE experiments were used to restrain a model for the eIF1:eIF5-CTD complex using the software HADDOCK (Adapted from [15]).
Cross Saturation Transfer as an Alternative Method to Determine Binding Interfaces
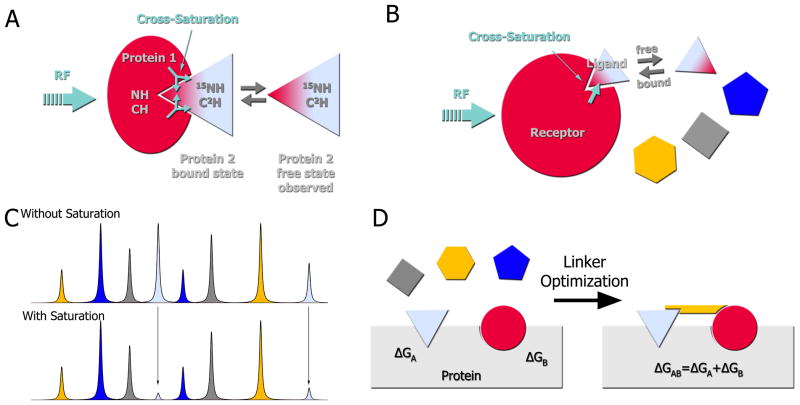
Chemical shift perturbations do not exclusively occur in resonances at the complex interface but may also manifest in nuclei distal to the binding site (for example, as a result of changes in hydrogen bonding or conformational changes). Cross saturation transfer, and related methods, provide an alternative for the identification of large protein-protein interfaces and can be applied to complexes with strong or weak affinity [30, 31]. Figure 5A illustrates the principle of cross saturation and cross saturation transfer. The non-labeled receptor (protein 1) binds the ligand (protein 2) which is 15N,2H labeled. The aliphatic frequencies of protein 1 are then selectively irradiated leading to saturation of the aliphatic, aromatic and amide resonances through spin diffusion. The magnetization is transferred to the 1H-15N pairs of protein 2 located at the binding site, which results in diminished 1H-15N HSQC peak intensities. If the complex has high affinity, the chemical shift values in the HSQC spectrum represent the bound form of the ligand. However, if the complex is weak, the peak positions represent the free ligand. Regardless, the saturation has a longer lifetime than the exchange rate between the free and bound forms and the free form still “remembers” the saturation.
Figure 5. Cross-saturation transfer experiments for protein-protein complexes and drug discovery.
A) Principle of cross saturation transfer: The unlabeled receptor (protein 1) binds the 15N,2H-ligand (protein 2). The aliphatic frequencies of the receptor are then selectively irradiated, which then saturates the aliphatic, aromatic and amide resonances of the receptor through spin diffusion. This saturation is also transferred to ligand resonances located at the binding site and leads to a decrease in the respective HSQC peak intensities. If the complex has a high affinity, the HSQC spectrum would show the peaks of the bound form of the ligand. If the complex is weak, the HSQC would show the spectrum of the free ligand; nonetheless, the saturation has a longer lifetime than the exchange rate between free and bound form and the free form still “remembers” the saturation. B) Application of the cross saturation transfer principle to a mixture of fragments. The same principle as described in A) applies to any ligand binding to the receptor. C) In a mixture of compounds, only fragments that bind to the receptor (gray compound) would exhibit a change in peak intensities. D) Illustration of the SAR by NMR method [35]. First, a fragment library is screened against a protein of interest until two compounds, A and B, are identified that can simultaneously bind to proximal sites. These compounds are then linked together and again screened against the protein. Theoretically, these compounds can be linked into a single molecule whose binding affinity is the summation of the individual binding energies.
Use of NMR for Fragment-Based Drug Design and Rational Drug Discovery
Following the identification and characterization of weak interfaces, such as occur during start codon recognition, our goal as biomedical researchers is to modulate protein function by developing therapeutics. Traditional drug discovery focuses on screening large compound matrices by characterizing their effect in functional tests. Unfortunately, the number of successful inhibitors or ‘hits’ typically comprises less than 0.1% the compound array [32]. The low success rate is attributed to the high affinities that are required to antagonize functional tests. The hits are further disadvantaged in that they possess higher molecular mass and suboptimal physiochemical properties. In contrast to their larger counterparts, smaller compounds, or fragments, serve as optimal starting places for developing high affinity molecules. NMR fills a niche in drug discovery by being well suited to characterize weak-affinity molecular fragments with the added benefit of providing atomic level information on the binding site. As thorough reviews exist elsewhere for the design, development, and screening of fragment libraries, we will only touch briefly on these topics [33, 34]. Here, we will review how NMR’s power to characterize weak binding interfaces can be turned towards the development of small molecule antagonists.
Exploring low molecular weight molecules by NMR yields several advantages (Figure 5C). Foremost, the diversity of chemical space is smaller in low molecular weight compounds and therefore coverage can be achieved with a smaller library [35]. Additionally, starting with small scaffolds affords chemists’ the opportunity to improve binding affinity and activity by modifying a scaffold, which almost ubiquitously results from adding functionalities, while still maintaining a molecular weight consistent with good ADME properties. The work of Shuker and colleagues on the calcineurin-inhibitor, FKBP, is the prime example of the fragment-based drug discovery method [36]. Known as structure-activity relationships by NMR (SAR by NMR), the method relies on identifying and linking molecules that bind to an adjacent site (Fig. 5D). A compound library is built around small chemical scaffolds that are easily modified by chemistry. Molecules are typically 50–250 Da and correspondingly bind weakly (Kd ~ 10−3–10−4 M). As illustrated in Figure 5D, fragments are screened for a single site. After a molecule is identified, that site is saturated and more fragments are screened with the goal of identifying adjacent binding pockets. Although far from a trivial task, subsequent linking of these compounds together produces an additive free energy of binding. SAR by NMR is usually performed by recording the protein chemical shift changes in 2D 1H-15N HSQC spectra. Fragments can be easily characterized using routine NMR experiments as outlined in previous sections (for example, chemical shift perturbations and cross saturation), but various other experiments have been devised to improve the screening throughput.
The majority of NMR-based screening experiments are considered ligand-based because they focus on the NMR signal originating from the small molecules. The throughput of these techniques can be increased by judiciously combining compounds, which possess unique chemical shifts that are easily discriminated. Ligand-based methods also conserve the quantity of precious isotopically-labeled protein samples. Saturation-transfer difference (STD) experiments, a ligand-based analog of cross saturation, are primarily used for screening large fragment libraries. Water ligand observation via gradient spectroscopy (WaterLOGSY) is a similar ligand-based technique that relies on the transfer of magnetization from bulk water to the protein binding site and to the bound ligand [37]. These are the two most common ligand-screening techniques, but there are a variety of others such as diffusion filters [38], NOE-pumping [39], line-broadening, and target immobilized NMR screening (TINS) [40]. In addition to high sensitivity for identification of weak protein-protein interactions, NMR has the additional advantage of providing structural information on the binding site at the atomic level.
Knowledge of the protein-ligand interface provides a unique opportunity to rationally develop small molecule therapies. The protein structure is first inspected for druggable epitopes, such as residues critical for complex formation. Until recently, NMR structures were considered too low-resolution for structure-based drug development. However, the realization that the inherent conformational flexibility of NMR ensembles may improve compound identification (similar to docking models that promote flexible side chains) has been proven successful. Virtual compound libraries, that contain molecules on the order of tens of millions, are screened in silico in several days to weeks. The docking quality is then scored and the top ranking molecules are prioritized for NMR compound screening – such as 1H-15N HSQC or STD experiments. Molecules with the highest affinity and binding site specificity should then be confirmed in an independent assay (such as fluorescence polarization). If affinities are sufficiently strong a functional assay may be utilized. A series of similar compounds should then be screened by NMR to explore structure-activity relationships and refine the pharmacophore. This information can be simultaneously used to rescreen for molecules in silico as well as chemically optimize the hits into lead-like compounds. The ability of NMR spectroscopy to examine both weak and strong interactions at atomic resolution is a powerful tool for the rational design of small molecule inhibitors.
Concluding Remarks
The importance of weak interactions cannot be understated, since it is the cumulative effect of weak interactions in combination with stronger interactions that allow for dynamic biological processes to occur. NMR is a unique tool capable of monitoring both weak- and strong-affinity complexes at an atomic level without necessitating structure determination. Furthermore, NMR spectroscopy is also an excellent tool for rational drug design of small molecules, wherein one can precisely determine the amino acids involved in the ligand interface. Although we utilized start codon recognition as a model system, the utility of NMR spectroscopy can be readily applied to elucidating the biological mechanisms of other dynamic processes involving weak PPIs.
Acknowledgments
REL is funded by NCI R01CA068262 and NIGMS P01GM047467. SRA is funded by NIGMS P01GM047467. JJZ is funded by Individual Ruth L. Kirschstein NRSA Postdoctoral Fellowship F32GM103005. GW thanks NCI and NIGMS for funding this work: NCI R01CA068262 and NIGMS P01GM047467.
Abbreviations
- ADME
absorption, distribution, metabolism, and excretion
- CTD
C-terminal domain
- eIF
eukaryotic initiation factor
- CSP
chemical shift perturbation
- HSQC
heteronuclear single-quantum coherence
- MTSL
(1-oxyl-2,2,5,5-tetramethyl-3-pyrroline-Δ3-methyl) methanethiosulfonate
- NMR
nuclear magnetic resonance
- NTD
N-terminal domain
- PIC
preinitiation complex
- PPI
protein-protein interaction
- RDC
residual dipolar coupling
- PRE
paramagnetic relaxation enhancement
- SAR
structure-activity relationship
- SAXS
small-angle X-ray scattering
- STD
saturation transfer difference
- TC
ternary complex
- WaterLOGSY
water ligand observation via gradient spectroscopy
- XAFS
X-ray absorption fine structure
References
- 1.Aitken CE, Lorsch JR. A mechanistic overview of translation initiation in eukaryotes. Nature structural & molecular biology. 2012;19:568–576. doi: 10.1038/nsmb.2303. [DOI] [PubMed] [Google Scholar]
- 2.Hinnebusch AG. Molecular mechanism of scanning and start codon selection in eukaryotes. Microbiology and molecular biology reviews: MMBR. 2011;75:434–467. doi: 10.1128/MMBR.00008-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Voigts-Hoffmann F, Klinge S, Ban N. Structural insights into eukaryotic ribosomes and the initiation of translation. Curr Opin Struct Biol. 2012 doi: 10.1016/j.sbi.2012.07.010. [DOI] [PubMed] [Google Scholar]
- 4.Ben-Shem A, Garreau de Loubresse N, Melnikov S, Jenner L, Yusupova G, Yusupov M. The structure of the eukaryotic ribosome at 3.0 A resolution. Science. 2011;334:1524–1529. doi: 10.1126/science.1212642. [DOI] [PubMed] [Google Scholar]
- 5.Ben-Shem A, Jenner L, Yusupova G, Yusupov M. Crystal structure of the eukaryotic ribosome. Science. 2010;330:1203–1209. doi: 10.1126/science.1194294. [DOI] [PubMed] [Google Scholar]
- 6.Rabl J, Leibundgut M, Ataide SF, Haag A, Ban N. Crystal structure of the eukaryotic 40S ribosomal subunit in complex with initiation factor 1. Science. 2011;331:730–736. doi: 10.1126/science.1198308. [DOI] [PubMed] [Google Scholar]
- 7.Marintchev A, Frueh D, Wagner G. NMR methods for studying protein-protein interactions involved in translation initiation. Methods Enzymol. 2007;430:283–331. doi: 10.1016/S0076-6879(07)30012-8. [DOI] [PubMed] [Google Scholar]
- 8.Williamson MP. Using chemical shift perturbation to characterise ligand binding. Progress in nuclear magnetic resonance spectroscopy. 2013;73:1–16. doi: 10.1016/j.pnmrs.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 9.Chen Y, Reizer J, Saier MH, Jr, Fairbrother WJ, Wright PE. Mapping of the binding interfaces of the proteins of the bacterial phosphotransferase system, HPr and IIAglc. Biochemistry. 1993;32:32–37. doi: 10.1021/bi00052a006. [DOI] [PubMed] [Google Scholar]
- 10.Gronenborn AM, Clore GM. Identification of the contact surface of a streptococcal protein G domain complexed with a human Fc fragment. J Mol Biol. 1993;233:331–335. doi: 10.1006/jmbi.1993.1514. [DOI] [PubMed] [Google Scholar]
- 11.van Nuland NA, Kroon GJ, Dijkstra K, Wolters GK, Scheek RM, Robillard GT. The NMR determination of the IIA(mtl) binding site on HPr of the Escherichia coli phosphoenol pyruvate-dependent phosphotransferase system. FEBS letters. 1993;315:11–15. doi: 10.1016/0014-5793(93)81122-g. [DOI] [PubMed] [Google Scholar]
- 12.ElAntak L, Tzakos AG, Locker N, Lukavsky PJ. Structure of eIF3b RNA recognition motif and its interaction with eIF3j: structural insights into the recruitment of eIF3b to the 40 S ribosomal subunit. J Biol Chem. 2007;282:8165–8174. doi: 10.1074/jbc.M610860200. [DOI] [PubMed] [Google Scholar]
- 13.Elantak L, Wagner S, Herrmannova A, Karaskova M, Rutkai E, Lukavsky PJ, Valasek L. The indispensable N-terminal half of eIF3j/HCR1 cooperates with its structurally conserved binding partner eIF3b/PRT1-RRM and with eIF1A in stringent AUG selection. J Mol Biol. 2010;396:1097–1116. doi: 10.1016/j.jmb.2009.12.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gelev V, Aktas H, Marintchev A, Ito T, Frueh D, Hemond M, Rovnyak D, Debus M, Hyberts S, Usheva A, et al. Mapping of the auto-inhibitory interactions of protein kinase R by nuclear magnetic resonance. J Mol Biol. 2006;364:352–363. doi: 10.1016/j.jmb.2006.08.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luna RE, Arthanari H, Hiraishi H, Nanda J, Martin-Marcos P, Markus MA, Akabayov B, Milbradt AG, Luna LE, Seo HC, et al. The C-Terminal Domain of Eukaryotic Initiation Factor 5 Promotes Start Codon Recognition by Its Dynamic Interplay with eIF1 and eIF2beta. Cell Rep. 2012;1:689–702. doi: 10.1016/j.celrep.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marintchev A, Edmonds KA, Marintcheva B, Hendrickson E, Oberer M, Suzuki C, Herdy B, Sonenberg N, Wagner G. Topology and regulation of the human eIF4A/4G/4H helicase complex in translation initiation. Cell. 2009;136:447–460. doi: 10.1016/j.cell.2009.01.014. S0092-8674(09)00020-8 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marintchev A, Kolupaeva VG, Pestova TV, Wagner G. Mapping the binding interface between human eukaryotic initiation factors 1A and 5B: a new interaction between old partners. Proc Natl Acad Sci U S A. 2003;100:1535–1540. doi: 10.1073/pnas.0437845100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reibarkh M, Yamamoto Y, Singh CR, del Rio F, Fahmy A, Lee B, Luna RE, Ii M, Wagner G, Asano K. Eukaryotic initiation factor (eIF) 1 carries two distinct eIF5-binding faces important for multifactor assembly and AUG selection. J Biol Chem. 2008;283:1094–1103. doi: 10.1074/jbc.M708155200. [DOI] [PubMed] [Google Scholar]
- 19.Suzuki C, Garces RG, Edmonds KA, Hiller S, Hyberts SG, Marintchev A, Wagner G. PDCD4 inhibits translation initiation by binding to eIF4A using both its MA3 domains. Proc Natl Acad Sci U S A. 2008;105:3274–3279. doi: 10.1073/pnas.0712235105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Das S, Maiti T, Das K, Maitra U. Specific interaction of eukaryotic translation initiation factor 5 (eIF5) with the beta-subunit of eIF2. J Biol Chem. 1997;272:31712–31718. doi: 10.1074/jbc.272.50.31712. [DOI] [PubMed] [Google Scholar]
- 21.Akabayov B, Akabayov SR, Lee SJ, Wagner G, Richardson CC. Impact of macromolecular crowding on DNA replication. Nat Commun. 2013;4:1615. doi: 10.1038/ncomms2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Akabayov B, Richardson CC. Binding of Mn-deoxyribonucleoside triphosphates to the active site of the DNA polymerase of bacteriophage T7. Powder Diffr. 2011;26:159–162. doi: 10.1154/1.3583156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lipsitz RS, Tjandra N. Residual dipolar couplings in NMR structure analysis. Annu Rev Biophys Biomol Struct. 2004;33:387–413. doi: 10.1146/annurev.biophys.33.110502.140306. [DOI] [PubMed] [Google Scholar]
- 24.Ottiger M, Delaglio F, Bax A. Measurement of J and dipolar couplings from simplified two-dimensional NMR spectra. J Magn Reson. 1998;131:373–378. doi: 10.1006/jmre.1998.1361. [DOI] [PubMed] [Google Scholar]
- 25.Ortega-Roldan JL, Jensen MR, Brutscher B, Azuaga AI, Blackledge M, van Nuland NA. Accurate characterization of weak macromolecular interactions by titration of NMR residual dipolar couplings: application to the CD2AP SH3-C:ubiquitin complex. Nucleic Acids Res. 2009;37:e70. doi: 10.1093/nar/gkp211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clore GM, Iwahara J. Theory, Practice, and Applications of Paramagnetic Relaxation Enhancement for the Characterization of Transient Low-Population States of Biological Macromolecules and Their Complexes. Chemical Reviews. 2009;109:4108–4139. doi: 10.1021/cr900033p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ubbink M. Dynamics in transient complexes of redox proteins. Biochem Soc Trans. 2012;40:415–418. doi: 10.1042/BST20110698. BST20110698 [pii] [DOI] [PubMed] [Google Scholar]
- 28.Volkov AN, Ubbink M, van Nuland NA. Mapping the encounter state of a transient protein complex by PRE NMR spectroscopy. J Biomol NMR. 2010;48:225–236. doi: 10.1007/s10858-010-9452-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Battiste JL, Wagner G. Utilization of site-directed spin labeling and high-resolution heteronuclear nuclear magnetic resonance for global fold determination of large proteins with limited nuclear overhauser effect data. Biochemistry. 2000;39:5355–5365. doi: 10.1021/bi000060h. [DOI] [PubMed] [Google Scholar]
- 30.Nakanishi T, Miyazawa M, Sakakura M, Terasawa H, Takahashi H, Shimada I. Determination of the interface of a large protein complex by transferred cross-saturation measurements. J Mol Biol. 2002;318:245–249. doi: 10.1016/S0022-2836(02)00018-9. [DOI] [PubMed] [Google Scholar]
- 31.Takahashi H, Nakanishi T, Kami K, Arata Y, Shimada I. A novel NMR method for determining the interfaces of large protein-protein complexes. Nat Struct Biol. 2000;7:220–223. doi: 10.1038/73331. [DOI] [PubMed] [Google Scholar]
- 32.Doman TN, McGovern SL, Witherbee BJ, Kasten TP, Kurumbail R, Stallings WC, Connolly DT, Shoichet BK. Molecular docking and high-throughput screening for novel inhibitors of protein tyrosine phosphatase-1B. J Med Chem. 2002;45:2213–2221. doi: 10.1021/jm010548w. [DOI] [PubMed] [Google Scholar]
- 33.Methods in Enzymology. 1. Vol. 493. Elsevier; 2011. [Google Scholar]
- 34.Pellecchia M, Bertini I, Cowburn D, Dalvit C, Giralt E, Jahnke W, James TL, Homans SW, Kessler H, Luchinat C, et al. Perspectives on NMR in drug discovery: a technique comes of age. Nature Reviews Drug Discovery. 2008;7:738–745. doi: 10.1038/nrd2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hajduk PJ. SAR by NMR: putting the pieces together. Mol Interv. 2006;6:266–272. doi: 10.1124/mi.6.5.8. [DOI] [PubMed] [Google Scholar]
- 36.Shuker SB, Hajduk PJ, Meadows RP, Fesik SW. Discovering high-affinity ligands for proteins: SAR by NMR. Science. 1996;274:1531–1534. doi: 10.1126/science.274.5292.1531. [DOI] [PubMed] [Google Scholar]
- 37.Dalvit C, Pevarello P, Tato M, Veronesi M, Vulpetti A, Sundstrom M. Identification of compounds with binding affinity to proteins via magnetization transfer from bulk water. J Biomol NMR. 2000;18:65–68. doi: 10.1023/a:1008354229396. [DOI] [PubMed] [Google Scholar]
- 38.Hajduk PJ, Olejniczak ET, Fesik SW. One-dimensional relaxation- and diffusion-edited NMR methods for screening compounds that bind to macromolecules. J Am Chem Soc. 1997;119:12257–12261. doi: 10.1021/ja9715962. [DOI] [Google Scholar]
- 39.Chen A, Shapiro MJ. NOE pumping: A novel NMR technique for identification of compounds with binding affinity to macromolecules. J Am Chem Soc. 1998;120:10258–10259. doi: 10.1021/ja982152o. [DOI] [Google Scholar]
- 40.Vanwetswinkel S, Heetebrij RJ, van Duynhoven J, Hollander JG, Filippov DV, Hajduk PJ, Siegal G. TINS, target immobilized NMR screening: An efficient and sensitive method for ligand discovery. Chemistry & Biology. 2005;12:207–216. doi: 10.1016/j.chembiol.2004.12.004. [DOI] [PubMed] [Google Scholar]