Summary
The cathepsin B-like protease-encoding gene AtCathB3 positively influences germination in Arabidopsis seeds, and GBF1 has been identified as its transcriptional repressor that negatively affects the process.
Key words: Arabidopsis thaliana, AtCathB3, bZIP, cathepsin B-like enzymes, GBF1, phylogenetic shadowing, reserve mobilization, seed germination, transcriptional regulation.
Abstract
Protein hydrolysis plays an important role during seed germination and post-germination seedling establishment. In Arabidopsis thaliana, cathepsin B-like proteases are encoded by a gene family of three members, but only the AtCathB3 gene is highly induced upon seed germination and at the early post-germination stage. Seeds of a homozygous T-DNA insertion mutant in the AtCathB3 gene have, besides a reduced cathepsin B activity, a slower germination than the wild type. To explore the transcriptional regulation of this gene, we used a combined phylogenetic shadowing approach together with a yeast one-hybrid screening of an arrayed library of approximately 1200 transcription factor open reading frames from Arabidopsis thaliana. We identified a conserved CathB3-element in the promoters of orthologous CathB3 genes within the Brassicaceae species analysed, and, as its DNA-interacting protein, the G-Box Binding Factor1 (GBF1). Transient overexpression of GBF1 together with a PAtCathB3::uidA (β-glucuronidase) construct in tobacco plants revealed a negative effect of GBF1 on expression driven by the AtCathB3 promoter. In stable P35S::GBF1 lines, not only was the expression of the AtCathB3 gene drastically reduced, but a significant slower germination was also observed. In the homozygous knockout mutant for the GBF1 gene, the opposite effect was found. These data indicate that GBF1 is a transcriptional repressor of the AtCathB3 gene and affects the germination kinetics of Arabidopsis thaliana seeds. As AtCathB3 is also expressed during post-germination in the cotyledons, a role for the AtCathB3-like protease in reserve mobilization is also inferred.
Introduction
Seed germination begins with water uptake and is completed when the radicle protrudes through the layers surrounding the embryo (germination sensu stricto). The protrusion also marks the onset of post-germination events, where seedling growth is mainly supported by the hydrolysis of nutritional reserves until it becomes a photosynthetic competent entity. Seed germination and storage reserve mobilization are two distinct, highly controlled processes, independently regulated and influenced by genetic and environmental factors (Bewley, 1997; Pritchard et al., 2002; Vicente-Carbajosa and Carbonero, 2005; Finch-Savage and Leubner-Metzger, 2006; Nonogaki et al., 2007; Holdsworth et al., 2008; Piskurewicz et al., 2009; Iglesias-Fernández et al., 2011). Brassicaceae mature seeds, such as those of Arabidopsis thaliana, mainly accumulate reserves as lipids and seed storage proteins (SSPs). SSPs are deposited in protein storage vacuoles making up the protein bodies of the cotyledons and the embryonic axis (Jiang et al., 2001; Hills, 2004; Tan-Wilson and Wilson, 2012). During imbibition of these seeds, protein degradation begins at the radicle tip and continues at the pre-vascular strands and subepidermal cell layers where cell expansion and differentiation are being initiated. In the post-germination period, the hydrolysis of SSPs occurs mainly in the cotyledons when proteins of the embryonic axis are nearly exhausted (Müntz, 2007).
Cathepsin B-like cysteine proteases (EC 3.4.22.1; CysProt) belong to the Papain-like class of CysProt (MEROPS Peptidase Database; Rawlings et al., 2012) and are endopeptidases preferentially associated with SSP degradation and mobilization during seed germination and post-germination (García-Lorenzo et al., 2006; Tan-Wilson and Wilson, 2012). These peptidases are synthesized as inactive precursors (zymogens) that are converted into active enzymes by the proteolysis of an N-end peptide (Khan and James, 1998). Upon seed imbibition, SSP mobilization occurs in part by zymogen activation, but during post-germination the major SSP hydrolysis occurs due to the action of de novo-synthesized proteases in the cotyledons. However, transcriptomic analyses of Arabidopsis seeds have shown that during the early stages of germination, genes involved in protein synthesis and degradation are over-represented, reflecting the importance of protein turnover throughout this process (Nakabayashi et al., 2005).
Phytohormones, mainly gibberellins (GA) and abscisic acid (ABA) orchestrate the progression of seed germination (Nambara et al., 2010). The endogenous ABA content in dry seeds rapidly declines upon imbibition and, by contrast, GA synthesis increases rapidly during the early and the late phases of germination (Holdsworth et al., 2008; Weitbrecht et al., 2011). In Arabidopsis, germination sensu stricto is inhibited by ABA, but this hormone seems not to affect lipid mobilization (Pritchard et al., 2002). In cereal aleurone layers, the synthesis of hydrolases, such as CysProt, takes place mainly during post-germination and is activated in response to GA diffused from the embryo. These enzymes are secreted into the endosperm where they mobilize the bulk of the stored reserves (Sun and Gubler, 2004; Eastmond and Jones, 2005). Functional analyses of the promoters of hydrolase genes induced by GA in the barley aleurone (Cejudo et al., 1992; Gubler et al., 1999) have identified a conserved tripartite cis-acting GA-responsive complex. This complex comprises the GARE motif (GA-responsive element, 5′-TAACAAA-3′), the pyrimidine box (5′-CCTTTT-3′), and the 5′-TATCCAC-3′ box, which are recognized by transcription factors (TFs) belonging to the R2R3MYB, DOF, and R1MYB (SHAQKYF) families, respectively (Gubler et al., 1995; Mena et al., 2002; Isabel-Lamoneda et al., 2003; Rubio-Somoza et al., 2006; Moreno-Risueño et al., 2007). However, the GA-responsive complex has not been described in promoters of hydrolase genes induced upon germination of dicotyledoneous seeds (Holdsworth et al., 2008; Weitbrecht et al., 2011; Iglesias-Fernández et al., 2013).
In this paper, we explore the physiological significance and transcriptional regulation of AtCathB3, a gene that encodes a cathepsin B-like protease, during Arabidopsis thaliana seed germination and post-germination periods. AtCathB3 transcripts are the most abundant CathB-like mRNAs throughout both periods. Homozygous germinating seeds for a knockout (KO) in this AtCathB3 gene present a significantly slower germination besides having a decreased cathepsin B-like protease activity compared with that of the wild type (Wt). The transcriptional regulation of this gene has been further explored and the G-box Binding Factor 1 (GBF1) has been identified as a transcriptional repressor of the AtCathB3 gene. The regulatory function of GBF1 upon AtCathB3 gene expression is validated by trans-activation assays in tobacco leaves, real-time quantitative PCR (RT-qPCR) analyses, and mRNA fluorescence in situ hybridization (FISH) experiments, and also by the germination kinetics of both overexpressor (oex) and T-DNA KO insertion lines in the GBF1 gene. The oex-GBF1 lines not only have a significant reduction of the AtCathB3 gene expression levels upon seed germination, but these seeds also germinate slower than the Wt seeds. The opposite behaviour is observed in the KO gbf1 mutant seeds.
Materials and methods
Plant material and germination assays
Plant growth conditions were as follows: ethanol sterilization of Arabidopsis seeds was done for 20min before germination in Petri dishes containing 0.5× MS medium including vitamins (Duchefa-Biochemie, Haarlem, The Netherlands) and solidified with 0.8% agar. Immediately after plating, seeds were stratified for 3 d at 4 °C and then incubated at 21 °C under long-day conditions (16h/8h light/dark; light intensity: 155 μmol photons m–2 s–1) for 2 weeks and then transferred to pots in the greenhouse. Mature seeds for Wt and homozygous for T-DNA insertion and oex mutants were stored at 21 °C and 30% relative humidity, until used for germination assays.
Three replicates of 50 after-ripening seeds were germinated in 90mm Petri dishes on two layers of filter paper (Whatman No. 1) moistened with 3ml of sterile water. Germination was done at 21 °C in growth chambers under long-day conditions as described above, and seeds were always sown in the morning. Seeds were neither surface sterilized nor stratified at 4 °C in order to avoid influencing their dormancy status; nevertheless, fungal infections were not detected by light microscopy during the studied period. Seeds were scored as germinated when the radicle emergence through the seed coat was visible under a magnifying lens. Germination tests were performed in three biological samples using three technical replicates. Statistical analyses were done using the GERMINATOR package program (http://www.pph.wur.nl/UK/seedlab/resources/germinator; Joosen et al., 2010).
Generation of transgenic lines
All material used in this study was derived from Arabidopsis thaliana ecotype Col-0. The T-DNA insertion mutant in AtCathB3 exon 8 (SALK_019630) was obtained from the Nottingham Arabidopsis Stock Centre, and homozygous plants were selected by PCR using a gene-specific primer and a primer derived from the left border of the T-DNA (Supplementary Table S1 available at JXB online; http://www.signal.salk.du/tdnaprimers.2.html). The homozygous T-DNA insertion line in GBF1 exon 9 (SALK_144534) was kindly provided by Dr Zengraft from University of Tubingen, Germany (Smykowski et al., 2010).
The AtCathB3 promoter (–500bp) was amplified from genomic DNA by nested PCR using oligonucleotide pairs containing attB sites (Supplementary Table S1 available at JXB online) for cloning into the pDONOR®207 vector by the Gateway®BP and then transferred by Gateway® LR recombination (Invitrogen, Carlsbad, CA, USA) into the destination vector pMDC163, containing the uidA reporter gene (Curtis & Grossniklaus, 2003). In the T1 generation, plants carrying a pMDC plasmid were identified on the basis of hygromycin resistance. The next generation was tested for single-locus insertion of the transgene based on a 3:1 segregation in MS medium containing hygromycin. Homozygous lines were selected in the T3 generation. The same strategy was used to obtain the oex lines. P35S::GBF1 was cloned into pMDC43, using the plasmid pDEST22® containing the GBF1 open reading frame (ORF), as the entry clone. All constructs were introduced into Agrobacterium tumefaciens strain C58C1 GV3101 by electroporation and these were used to transform Arabidopsis thaliana (Col-0) by the floral dip method (Clough and Bent, 1998).
Histochemical β-glucuronidase (GUS) assays
Qualitative GUS staining assays were performed using the protocol described by Jefferson et al. (1987). Samples were cleared using Hoyer’s light medium as described by Stangeland and Salehian (2002). GUS histochemical staining was visualized under a light stereomicroscope (Leica, Wetzlar, Germany). A representative transgenic line out of 10–12 independent lines was used.
Protease activity assays
Three lots of non-stratified seeds germinated for 12, 24, 36, 48 and 60h were ground under liquid nitrogen and then extracted with protein extraction buffer (340mM sodium acetate, 60mM acetic acid, 4mM EDTA, 8mM dithiothreitol, pH 6). The enzymatic activity of protein extracts was determined using a fluorogenic specific substrate for cathepsin B proteases, Z-Arg-Arg-7-amido-4-methylcoumarin (Z-RR-AMC), following the manufacturer’s instructions (Calbiochem, San Diego, CA, USA). Mixtures of 20 µg of protein extracts and substrate (20 µM) were incubated at 30 °C for 2h and the emitted fluorescence was measured at 20min intervals with a 365nm excitation wavelength and a 465 emission wavelength.
Bioinformatic tools: dendrogram and phylogenetic shadowing
Sequences from the six different species within the Brassicaceae family used in this work were obtained from the Phytozome v8.0 Database (Goodstein et al., 2012; http://www.phytozome.net), except for those of the Arabis alpina genome that were produced by the TRANSNET KBBE consortium (Professor P. Carbonero, Universidad Politécnica de Madrid, Madrid, Spain; personal communication). The major characteristics of the predicted cathepsin B-like proteins are listed in Supplementary Table S2 available at JXB online. The complete deduced amino acid sequences of the 14 CathB-like genes from the six Brassicaceae species were used to reconstruct a phylogenetic dendrogram using the neighbour-joining algorithm (Supplementary Fig. S1 available at JXB online).
The promoter sequences of the six Brassicaceae CathB3 orthologous genes from Arabidopsis thaliana, Arabidopsis lyrata, Capsella rubella, Arabis alpina, Brassica rapa and Eutrema salsugineum are given in Supplementary Table S3 available at JXB online. The mVISTA Shuffle-LAGAN software (http://genome.lbl.gov/vista/index.shtml; Frazer et al., 2004) was used to create pairwise alignments between promoter sequences, and the conserved regions were analysed with MUSCLE (http://www.drive5.com/muscle; Edgar, 2004) and T-Coffee programs (http://www.ebi.ac.uk/Tools/msa/tcoffee; Notredame et al., 2000). Plant cis-acting core regulatory DNA elements were searched through the following data bases: MotifFinder (http://www.genome.jp/tools/motif), BIOBASE (http://www.biobase-international.com), and PlantCARE (Lescot et al., 2002).
Yeast one-hybrid (Y1H) assays
Y1H screenings were performed as described previously (Castrillo et al., 2011). The CathB3-element was amplified by PCR using specific primers that contained XmaI and XbaI restriction sites (Supplementary Table S1 available at JXB online). The engineered restriction sites of the CathB3-element were used to clone it into the XmaI and XbaI sites of the pTUY1H plasmid upstream of a HIS3 reporter gene. The plasmid was introduced into Saccharomyces cerevisiae Y187α (MAT-α) cells. As a prey, a yeast-normalized library of Arabidopsis thaliana TFs containing a collection of approximately 1200 TF ORFs fused to GAL4AD arrayed in a 96-well format into S. cerevisiae YM4271 (MAT-a) was screened. Diploid cells, grown in selection medium lacking leucine (L) and tryptophan (W), were spotted onto plates with the screening medium devoid of L, W and histidine (H) to which increasing concentrations of 3-amino-1,2,4-triazole (3-AT; Sigma, St Louise, MO, USA) were added. Positive colonies were visible after 2–5 d of incubation at 28 °C.
Transient expression assays
For transient expression in tobacco, Nicotiana benthamiana leaves were infiltrated with Agrobacterium tumefaciens (strain C58C1 GV3101) that had previously been transformed with the corresponding reporter (PAtCathB3::uidA) and effector (P35S::GBF1) constructs, or with an empty P35S vector as negative control. Agrobacterium tumefaciens containing pMDC32-P35S::LUC [luciferase (LUC); Gateway Technology, Invitrogen] for normalization and pBIN61-P35S::P19 to avoid silencing (Voinnet et al., 2003) were added. Relative GUS/LUC activities were determined as described by Jefferson et al. (1987) using a Genios Pro 96/384 multifunction microreader (TECAN®; Tecan Group, Männedorf, Switzerland). A minimum of three independent transformation experiments for each construct was done.
RT-qPCR assays
Total RNA was purified, as described previously (Oñate-Sanchez and Vicente-Carbajosa, 2008), from different organs of Arabidopsis 5-week old plants: rosette leaves, roots, stems, and flowers. Siliques at different stages of development and seeds at different germination time points (up to 48h) were also analysed. cDNA was synthesized from 1 µg of total RNA using a First Strand cDNA Synthesis kit for RT-PCR (Roche Applied Science, Mannheim, Germany) following the manufacturer’s instructions. Samples were stored at –20 °C until used.
The specific primers used in the RT-qPCR assays are shown in Supplementary Table S4 available at JXB online, and the ubiquitin gene UBC21 (At5g25760) was used to normalize the data, as it has been demonstrated to be constant throughout the periods studied, as was gene ACT8 (At1g49240; Graeber et al., 2011; Supplementary Fig. S2 available at JXB online). Amplification was performed in an Eco® Real-Time PCR System (Illumina, San Diego, CA, USA). For each 10 µl reaction, 2 µl of the sample’s cDNA was mixed with 5 µl of FastStart SYBR Green Master (Roche Applied Science) and 0.25 µl of each primer (final concentration 500nM), plus sterile water up to the final volume. Samples were subjected to thermal-cycling conditions of 95 °C for 10min, and 40 cycles of 10 s at 95 °C and 30 s at 60 °C for annealing and extension. The melting curve was designed to increase from 55 to 95 °C, and melting temperatures for each amplicon and primer efficiencies were estimated using a calibration dilution curve and slope calculation (Supplementary Table S4 available at JXB online). Three different biological samples for each time point were considered, and expression levels were determined as the number of cycles needed for the amplification to reach a cycle threshold fixed in the exponential phase of the PCR (Pfaffl, 2001).
FISH experiments
The protocol followed was as described by Testillano and Risueño (2009). Digoxygenin-labelled RNA (200–300bp) derived from the 3′ non-coding regions of the Arabidopsis genes analysed was used as a probe (Supplementary Table S1 available at JXB online). After incubation with an alkaline phosphatase-conjugated anti-digoxigenin antibody, the sections were incubated with a secondary antibody, anti-mouse IgG conjugated to Alexa Fluor 488 (Invitrogen). After washing in PBS, the nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI), washed in sterile water, and mounted and examined in a confocal microscope (Leica SP8; Leica).
Results
The AtCathB3 gene is highly expressed in Arabidopsis thaliana seeds upon germination
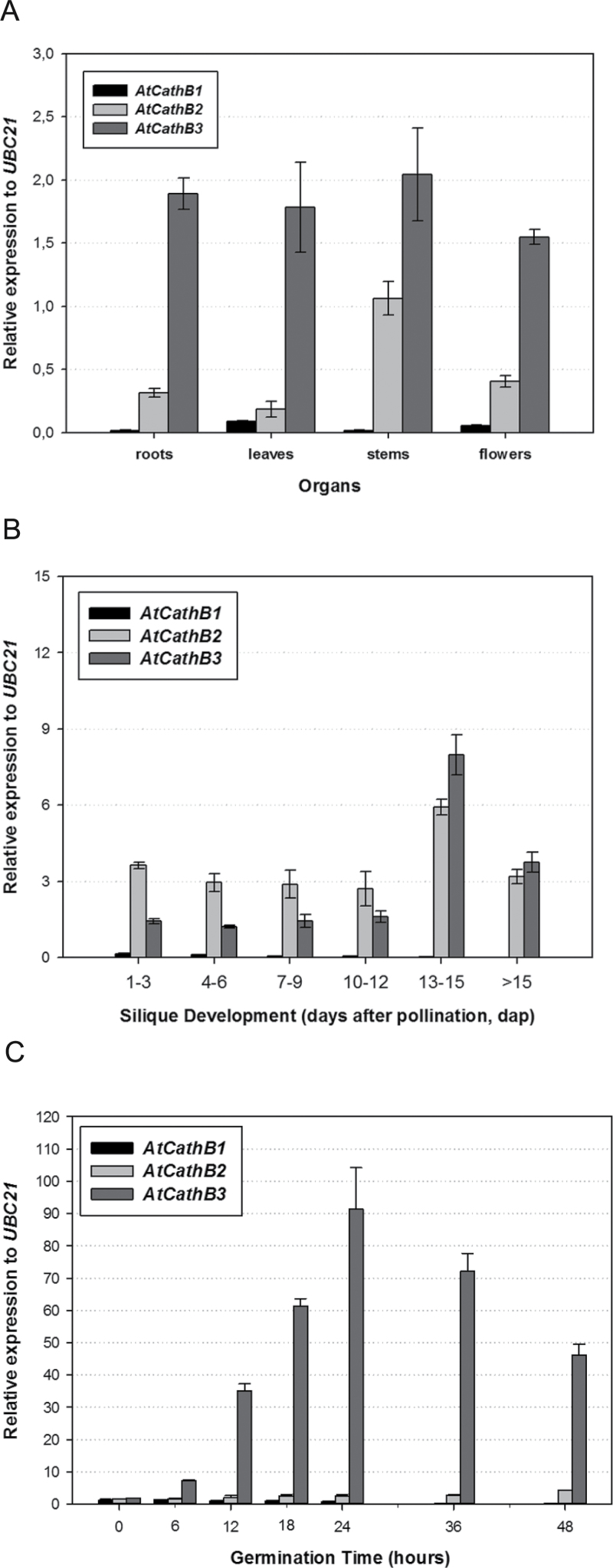
Three CathB-like genes, AtCathB1, AtCathB2, and AtCathB3 (loci At1g02300, At1g02305, and At4g01610, respectively) have been annotated in the Arabidopsis thaliana genome (McLellan et al., 2009). The expression profiles of these AtCathB genes have been analysed by RT-qPCR in different Arabidopsis organs from 5-week old plants: roots, rosette leaves, stems, and flowers. The data in Fig. 1A show that, whereas the AtCathB2 and AtCathB3 transcripts were detected in all the organs analysed, AtCathB1 was only faintly expressed. Siliques at different stages of development (1 to >15 d after pollination, dap) were analysed, and again, while the transcripts of AtCathB1 were almost undetectable, those of AtCathB2 and AtCathB3 appeared throughout fruit development, reaching a peak between 13 and 15 dap (Fig. 1B). The three AtCathB gene transcripts were detected at the same level (<2 relative to UBC21) in dry seeds (Fig. 1C), but AtCathB3 gene expression was highly induced upon germination (>50 times compared with the dry stage), reaching a peak at 24h and still being expressed at 48h when germination was completed (100% radicle emergence).
Fig. 1.
Expression analyses of the AtCathB genes by RT-qPCR. (A) Expression of AtCathB1, AtCathB2, and AtCathB3 in different organs: roots, rosette leaves, stems, and flowers. Plant material was harvested from 5-week old plants. (B) Expression of AtCathB1, AtCathB2, and AtCathB3 throughout silique development: at 1–3, 4–6, 7–9, 10–12, 13–15, and >15 dap. (C) Expression of AtCathB1, AtCathB2, and AtCathB3 during seed germination at 0 (dry seeds), 6, 12, 18, 24, 36 and 48h. Data were normalized to the UBC21 gene and are shown as means±standard error (SE) of three independent experiments.
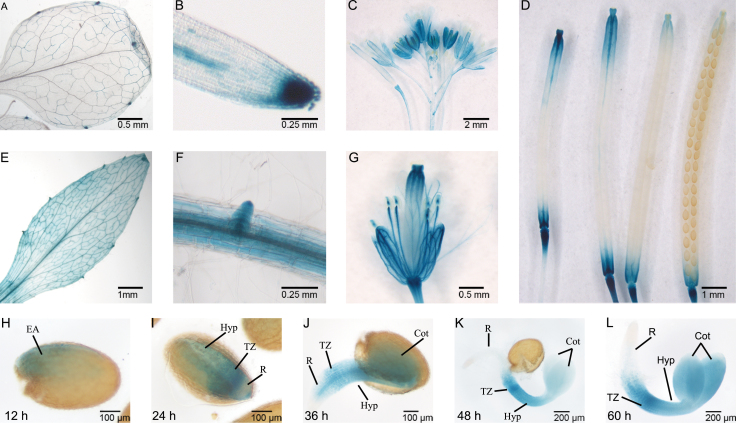
To analyse further the expression of the AtCathB3 gene, stable transgenic lines of Arabidopsis thaliana (Col-0) with a construct of its promoter (–500bp) transcriptionally fused to the reporter uidA gene, which encodes GUS, were produced (PAtCathB3::uidA). GUS histochemical staining of these lines confirmed the ubiquitous expression of the AtCathB3 gene (Fig. 2). In young and adult leaves, GUS expression was particularly intense at the hydathodes and at the vascular tissue (Fig. 2A, E). In roots, GUS activity was detected in the meristematic cells and at the radicle tip, in the incipient vascular elements, and at the initiation of the secondary roots (Figs. 2B, F). GUS was also expressed at the vasculature of the sepals and anthers, and at the stigma of flowers (Fig. 2C, G), and in fruits in the vascular bundles at the base and at the upper part of the silique (Fig. 2D). Upon germination, GUS activity in the PAtCathB3::uidA reporter lines was already detected before radicle protrusion, in the hypocotyl–radicle transition zone of the embryonic axis (12–24h), and later (36–60h) the signal was extended throughout the incipient vascular elements of the radicle and the hypocotyl, as well as to the cotyledons, where GUS expression was enhanced as the cotyledons expanded (Fig. 2H–L).
Fig. 2.
Histochemical localization of GUS expression in Arabidopsis-transformed plants with the PAtCathB3::uidA construct. (A) Young leaf; (B) radicle; (C) inflorescence; (D) siliques; (E) adult leaf; (F) secondary root; (G) flower; (H–L) germinating seeds: at 12 (H), 24 (I), 36 (J), 48 (K), and 60 (L) h. EA, embryonic axis; Hyp, hypocotyl; TZ, transition zone; R, root; Cot, cotyledon.
Seed germination is affected in a homozygous KO mutant of the AtCathB3 gene
To address the question of whether the AtCathB3 gene has a functional role during the germination process of Arabidopsis seeds, a homozygous T-DNA KO line (SALK_019630; atcathb3, T-DNA insertion in exon 8) for this gene was produced and the lack of AtCathB3 transcripts confirmed by RT-qPCR analysis (Fig. 3A, B). Whereas the Wt seeds had completed their germination by 48h, only 80% of the mutants had germinated at 60h (Fig. 3C). Moreover, the t 50 (time to reach 50% of germinated seeds) was significantly delayed in the mutant (40.4±1.9h versus 30.0±0.1h in the Wt; Fig. 3C. inset). Although very low cathepsin B activity was detected in dry seeds, both in the Wt and in the mutant, this enzymatic activity increased upon germination and post-germination (from 24 to 60h) in the Wt seeds. However, in the homozygous atcathb3 seeds, this increase was approximately 50% of that shown in the Wt at 60h. The cathepsin B activity detected in the mutant could be due to the truncated CathB3 protein generated by the T-DNA insertion in exon 8, as the cysteine active site is found in exon 6 (Supplementary Table S2 available at JXB online). These data indicated that the AtCathB3 gene expression affects the germination rate, as well as the protease activity during this process.
Fig. 3.
The mutant line for AtCathB3 (KO atcathb3) is negatively affected in CathB protease activity and germinates slower than the Wt seeds. (A) Position of the T-DNA insertion, indicated with a triangle in exon VIII, in the AtCathB3 mutant analysed (KO atcathb3). (B) Transcript analysis by RT-qPCR of the AtCathB3 gene in Wt and KO atcathb3 lines during seed germination at 0 (dry seeds), 24 and 48h. Data were normalized to the UBC21 gene and are shown as means±SE of three independent experiments. (C) Cathepsin B protease activity (per µg of total protein) in Wt (filled bars) and KO atcathb3 (shaded bars) seeds during germination at 0 (dry seeds), 12, 24, 36, 48 and 60h. Lines indicate the germination kinetics of Wt (filled circles) and KO atcathb3 (open circles). The inset shows t 50 data (time to obtain 50% germinated seeds). KO atcathb3 seeds reached 100% germination at 72h. Data are means±SE of three independent experiments.
Identification of an evolutionarily conserved cis-element in the AtCathB3 gene promoter and of a GBF1 protein recognizing the G-box motif within it
The Arabidopsis thaliana CathB-like family was used to search for orthologous genes in other related Brassicaceae species such as Arabidopsis lyrata, Capsella rubella, Arabis alpina, Eutrema salsugineum, and Brassica rapa (Supplementary Table S2 available at JXB online). The corresponding deduced protein sequences were used to reconstruct an unrooted phylogenetic tree (Supplementary Fig. S1 available at JXB online). The 14 sequences compared were grouped into two major clusters of orthologous genes, supported by bootstrap values >70%. The phylogenetic tree showed that AtCathB1/AtCathB2 and AlCathB1/AlCathB2 were paralogous, probably originating from a recent duplication event occurring after the diversification of the CathB-like genes into two major clusters of orthologous genes. For AtCathB3, only one orthologous gene from each species was found. All the CathB-like proteases annotated contained a peptidase C1A pro-peptide site and a peptidase C1A papain C-terminal domain (Supplementary Table S2 available at JXB online), predicted by the InterProScan program (Quevillon et al., 2005).
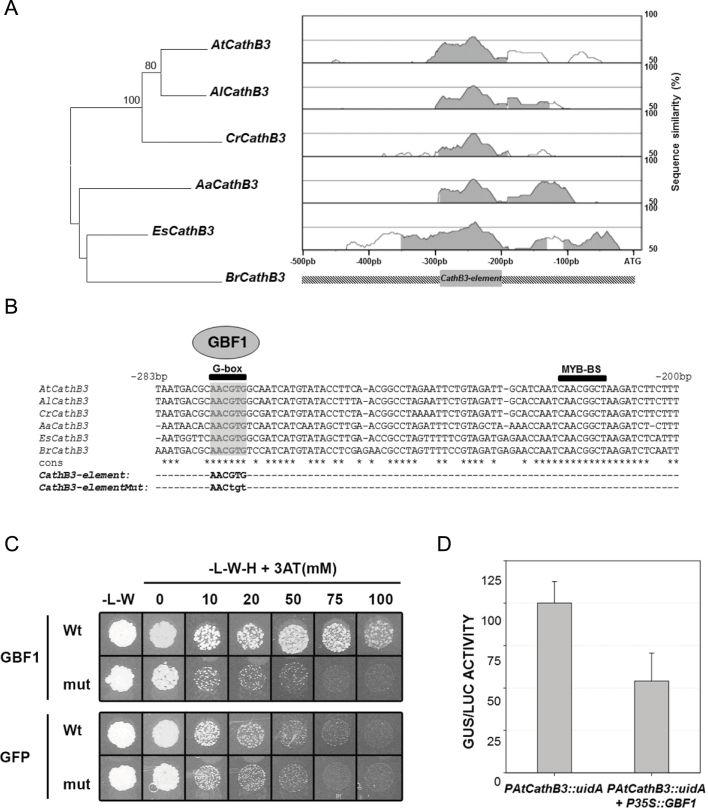
The promoter regions of the CathB3 orthologous genes in the Brassicaceae species studied were searched in silico for conserved sequences through a phylogenomic approach (Vavouri and Elgar, 2005). The promoter regions compared spanned the intergenic distance between the ATG translation initiation codon of AtCathB3 (At4g01610) and its preceding GRAS gene (At4g01600; Supplementary Table S2 available at JXB online). The pairwise alignment among the six promoters analysed using the mVISTA, MUSCLE, and T-Coffee programs (Notredame et al., 2000; Edgar, 2004; Frazer et al., 2004) showed a highly conserved block, with >70% identity, spanning from –283 to –200bp (Fig. 4A, ‘CathB3-element’). This cis-regulatory element contained two highly conserved core sequences (Fig. 4B): a G-box [binding site for basic leucine zipper (bZIP) proteins; Foster et al., 1994] and a MYB-binding site (Prouse and Campbell, 2012).
Fig. 4.
Identification of conserved cis-elements in orthologus CathB3 promoters in the Brassicaceae species: Arabidopsis thaliana (At), Arabidopsis lyrata (Al), Capsella rubella (Cr), Arabis alpina (Aa), Eutrema salsugineum (Es), and Brassica rapa (Br). The GBF1 transcription factor interacted with the CathB3-element in yeast one-hybrid screenings (Y1H) and negatively regulated the expression of the PCathB3::uidA construct in tobacco transient assays. (A) Pairwise alignment of CathB3 promoter sequences from different Brassicaceae species using mVISTA. For the graphical output, a sliding window of 100bp was used and base pair identity ranged from 50 to 100%. Shaded areas show conserved blocks. (B) Sequence alignment of the CathB-element (–283 to –200bp). A G-box and MYB-binding site (MYB-BS) core motives are indicated. The G-box core is shaded and a mutated version of the G-box (CathB3-element Mut) is indicated. (C) GBF1 specifically binds to the G-box sequence of the CathB3-element in a Y1H screening. Growth of diploid cells after mating of two yeast strains containing the CathB3-element–pTUY1H construct and pDEST22-AD:GBF1 or pDEST22-AD:GFP (negative control) on non-selective (–L–W) or selective medium (–L, –W, –H) with increasing concentrations of 3-AT. (D) Tobacco leaves co-infiltrated with Agrobacterium tumefaciens containing PCathB3::uidA as reporter construct and P35S::GBF1 as effector construct. GUS activity was relative to LUC activity, which was used as internal control. Data are means±SE of six biological replicates.
To identify putative TFs able to interact with the CathB3-element, a library of approximately 1200 Arabidopsis thaliana TF ORFs fused to the GAL4 activation domain and arrayed in a 96-well format (RR library; REGIA+REGULATORS library) was used to perform a mating-based Y1H screening. The CathB3-element was cloned in the episomal plasmid (pTUY1H) to be used as a bait for the screening of this RR library (Castrillo et al., 2011).
Following this procedure, the bZIP GBF1 (locus At4g36730) was identified. Diploid yeasts containing the plasmids CathB3-element-pTUY1H and GBF1-pDEST22 were able to grow in an auxotrophic medium lacking histidine even in the presence of 100mM 3-AT, a competitive inhibitor of the product of the HIS3 gene, while growth of the negative control (CathB3-element–pTUY1H+GFP–pDEST22) was inhibited at the lowest concentration of the inhibitor used (10mM 3-AT; Fig. 4C). To determine whether the binding of GBF1 is through the G-box element (CathB3-element: 5′-AACGTG-3′), a mutation was produced within this G-box (CathB3-element mut: 5′-AACtgt-3′; Schindler et al., 1992b ). The growth of diploid yeasts containing plasmids GBF1–pDEST22 and CathB3-element mut–pTUY1H was inhibited at 3-AT concentrations similar to those inhibiting the green fluorescent protein (GFP) negative control (10mM and higher; Fig. 4C). Thus, the G-box-binding site contained in the CathB3-element was specifically recognized by the GBF1 TF.
The interaction between GBF1 and the CathB3-element was investigated in planta (Fig. 4D), using a tobacco transient expression system where PAtCathB3::uidA (GUS) was the reporter construct and a plasmid where GBF1 was under the control of the cauliflower mosaic virus (CaMV) 35S promoter, P35S::GBF1, acted as the effector. GUS activity (of at least six independently transformed plants) was measured in Nicotiana benthamiana leaves co-infiltrated with Agrobacterium tumefaciens containing the PAtCathB3::uidA reporter construct and Agrobacterium tumefaciens containing either a P35S::GBF1 construct or the corresponding empty vector as negative control. A construct containing the CaMV 35S promoter fused to the LUC-encoding ORF was used as an internal control for the transformation experiments. The GUS activity in plants co-transformed with the reporter construct and the empty negative-control vector defined the basal GUS activity (100%). Co-transformation of the reporter construct together with the P35S::GBF1 effector reduced the GUS activity, driven by the AtCathB3 promoter, by about 50% (Fig. 4D).
GBF1 is a transcriptional repressor of the AtCathB3 gene and negatively affects Arabidopsis thaliana seed germination
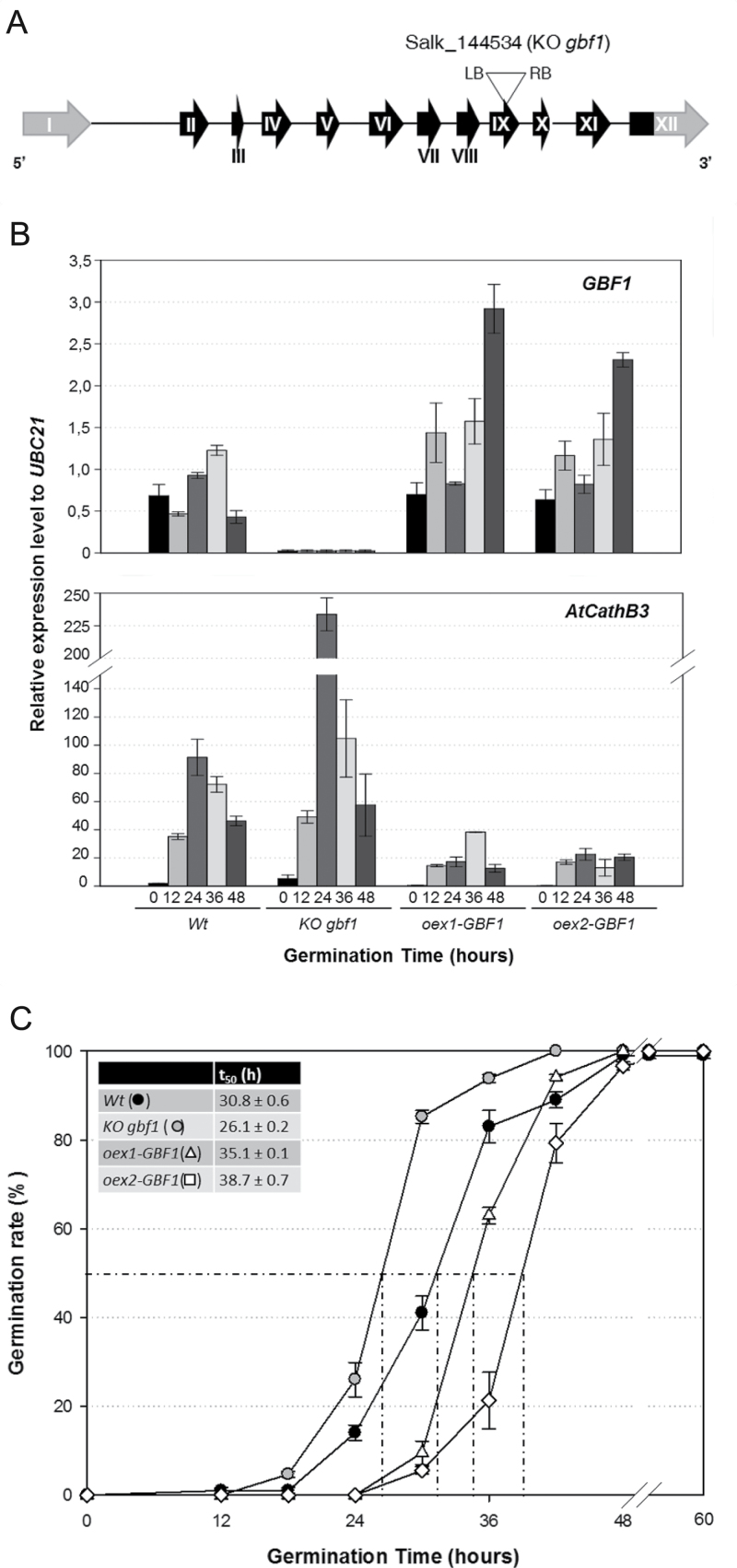
To further verify the effect of GBF1 on AtCathB3 gene expression in Arabidopsis thaliana, the expression profiles of AtCathB3 and GBF1 were analysed in two over-expressor GBF1 lines (oex1-GBF1 and oex2-GBF1) and in the homozygous KO line for GBF1 (gbf1), and compared with those in the Wt. The KO gbf1 homozygous plants as expected, not only presented undetectable levels for the GBF1 transcripts in germinating seeds but also accumulated AtCathB3 transcripts at a level of 2- to 3-fold higher than those in the Wt (Fig. 5A, B). In the two oex-GBF1 lines, the opposite was observed: expression of the AtCathB3 gene was decreased by >75% compared with that in the Wt germinating seeds. The variation observed in GBF1 mRNA levels within a given oex-GBF1 line during seed germination can be explain by the non-homogeneous expression patterns driven by the CaMV 35S promoter during germination, as previously reported (Benfey et al., 1989). However, the GBF1 levels reached were sufficient to negatively influence AtCathB3 expression. The germination in the gbf1 mutant seeds was faster than in those of the Wt (t 50=26.1±0.2 versus t 50=30.8±0.1), whereas in the two oex-GBF1 lines, germination was slower and, as a consequence, the corresponding t 50 values were higher (Fig. 5C, inset). These data indicated that GBF1 represses transcription of the AtCathB3 gene and negatively affects Arabidopsis thaliana seed germination.
Fig. 5.
AtCathB3 gene expression and germination kinetics in two over-expressor lines (oex1-GBF1, oex2-GBF1) and a KO line for GBF1 (gbf1). (A) Position of the T-DNA insertion is indicated with a triangle in exon IX of GBF1 (gbf1 mutant). (B) Transcript analysis by RT-qPCR of the GBF1 and AtCathB3 genes during germination of Wt, KO gbf1, oex1-GBF1, and oex2-GBF1 seeds at 0 (dry seeds), 12, 24, 36, and 48h. Data were normalized to the UBC21 gene and are shown as means±SE of three independent experiments. (C) Germination kinetics of seeds from: Wt (filled circles), KO gbf1 (shaded circles), oex1-GBF1 (triangles), and oex2-GBF1 (squares). Data are means±SE of three independent experiments. The inset shows t 50 data (time to obtain 50% germinated seeds).
AtCathB3 and GBF1 are co-expressed in the same cell types in germinating seeds
If the GBF1 gene encodes a transcriptional regulator of the AtCathB3 gene during seed germination, both genes should be expressed in the same cells at the right time. In order to explore this, FISH analyses of both transcripts were carried out in germinating seeds before radicle protrusion at equivalent physiological stages (24h for gbf1 mutant and 30h for Wt and atcathb3 mutant; Fig. 6). Longitudinal sections hybridized to specific antisense probes, derived from the 3′ non-coding regions of the AtCathB3 and GBF1 genes, showed a strong signal for both transcripts at the epidermis of the embryonic axis and at the incipient vascular elements of the embryo (Fig. 6A, B). As expected, the KO atcathB3 did not affect the expression of GBF1 (Fig. 6D, E); however, when the KO gbf1 line was hybridized with the antisense probe for AtCathB3 gene, the fluorescence signal increased and appeared not only at the epidermis but also throughout the embryonic axis (Fig. 6G). A similar localization of both transcripts in the same tissue types at same time, as well as the increase in the fluorescence signal for AtCathB3 in the gbf1 mutant, is compatible with transcriptional regulation of the AtCathB3 gene by the GBF1 TF. As expected, no signal was detected in the atcathb3 and gbf1 KO mutant seed sections with their corresponding antisense probes (Fig. 6D, H), nor when the Wt seed sections were hybridized with the sense probes as negative controls (Fig. 6J, K). Microscopic analysis of coarse sections (8 µm) of paraffin samples by differential interference contrast (Fig. 6C, F, I, L) showed that the cellular organization of the seed tissues was adequately preserved during sample preparation.
Fig. 6.
FISH analysis of AtCathB3 (A, D, G, J) and GBF1 (B, E, H, K) transcripts in germinating seeds of Wt (A, B) and atcathb3 mutant (D, E) at 30h and gbf1 mutant (G, H) at 24h. Results for control sense probes are shown in (J) and (K). All images are merged for the Alexa Fluor 488-conjugated anti-mouse IgG with DAPI staining to reveal the nuclei. (C, F, I, L) Differential interference constrast images. AL, aleurone; Cot, cotyledon; SC, seed coat; R, radicle; EP, epidermis, VE, vascular elements.
Discussion
In the genome of Arabidopsis thaliana, the CathB-like gene family is represented by three members, AtCathB1, AtCathB2, and AtCathB3. During seed germination, the most highly expressed of these genes is AtCathB3 (Nakabayashi et al., 2005, and this study). The analyses of the GUS promoter reporter lines, PAtCathB3::uidA (–500bp) showed a strong GUS expression in the lower hypocotyl, the hypocotyl–radicle transition zone, and the radicle tip during germination sensu stricto (Fig. 2H, I). Similar results were obtained by mRNA in situ hybridization experiments of imbibed seeds before radicle protrusion (Fig. 6), where transcripts of AtCathB3 were localized to the epidermis of the same regions where GUS was detected in the transformed PAtCathB3::uidA lines. The lower hypocotyl and the hypocotyl–radicle transition zone have been reported as regions of intense activity of cell expansion upon germination of Arabidopsis thaliana seeds. In this region, protein storage vacuoles are rapidly replaced by a central lytic vacuole enabling the elongation of embryonic cells (Sliwinska et al., 2009; Bolte et al., 2011). Interestingly, the protein AtCathB3 has been localized to the vegetative vacuole proteome of Arabidopsis thaliana and it is expected to have a similar vacuolar localization in germinating seeds (Carter et al., 2004).
From a physiological point of view, the KO atcathB3 mutant had a slower germination rate than the Wt, as indicated by their t 50 values (40.4±1.9h vs 30.0±0.1h, respectively; Fig. 3C), indicating that AtCathB3 positively influences germination sensu stricto in Arabidopsis seeds. This germination role is different from that in cereal grains where the proteases encoded by CathB-like genes are expressed in the aleurone layer and participate in the mobilization of SSPs during post-germination but do not seem to play a role in germination sensu stricto (Cejudo et al., 1992; Gubler et al., 1999; Mena et al., 2002; Isabel-Lamoneda et al., 2003).
When analysing the PAtCathB3::uidA reporter lines after radicle protrusion (≥36h), GUS expression was found in the cotyledons, enhancing the intensity of the signal with the cotyledon expansion, as well as being detected in the incipient vascular elements of the hypocotyl (Fig. 2). These findings are in agreement with a dynamic model for reserve mobilization that proposes hydrolysis of the bulk of SSPs at the protein storage vacuoles of the cotyledons when those of the embryonic axis are exhausted (Müntz, 2007). The implication of other proteases has been reported, like serine carboxipeptidase III (CPIII) from wheat, in the programmed cell death of the tracheary element differentiation between the radicle axis and the cotyledons to promote vascular development (Dominguez et al., 2002). Our data indicate a role for AtCathB3 not only in the incipient vasculature development during germination sensu stricto but also in SSP mobilization during post-germination.
To gain insight into the cis–trans transcriptional regulatory code of the AtCathB3 gene, a phylogenomic approach was used to identify a conserved region of 84bp (the CathB3-element) in the promoters of the CathB3 orthologous genes among several Brassicaceae species (Fig. 4). The power of this approach has been demonstrated in Saccharomyces (Cliften et al., 2001), primates (Boffelli et al., 2003), and Arabidopsis and related Brassicaceae (Koch et al., 2001; Hong et al., 2003; Adrian et al., 2010; Iglesias-Fernández et al., 2013). Using as a bait the CathB3-element for screening a yeast library of approximately 1200 TF ORFs of Arabidopsis thaliana (Castrillo et al., 2011), GBF1 (At4g36730) was isolated and the specificity of the binding through the G-box element 5′-(A/T)ACGTG-3′ (Schindler et al., 1992b ) was demonstrated (Fig. 4B, C). GBF1 belongs to the G group of bZIP TFs, which includes five members: GBF1, GBF2, GBF3, AtbZIP16, and AtbZIP68. These TFs share not only their DNA-binding domain but also additional highly conserved motives in the N-terminal region (Jakoby et al., 2002; Supplementary Fig. S3A, B available at JXB online). Transient co-expression of the P35S::GBF1 and the PAtCathB3::uidA constructs in tobacco leaves revealed that GBF1 acts as a negative regulator of the AtCathB3 promoter in planta (Fig. 4D).
During seed germination (between 0 and 36h), GBF1 mRNA expression increased up to 2-fold, being the most abundantly expressed bZIP gene within the G group (Fig. 5B, Supplementary Fig. S3C available at JXB online). Our results showed that an increment in the AtCathB3 transcript levels was observed in the germinating gbf1 mutant seeds, and these mutants displayed a faster germination rate than the Wt seeds. Moreover, the two oex-GBF1 lines analysed presented not only lower AtCathB3 expression but also a significantly slower germination rate (Fig. 5). Taken together, these results indicate that GBF1 is a negative transcriptional regulator of the AtCathB3 gene during seed germination.
Transcriptional changes occur that are both temporally and spatially separated in the endosperm and embryo of the germinating seeds of Arabidopsis (Penfield et al., 2006; Dekkers et al., 2013). GBF1 is expressed specifically at the epidermis of the embryonic axis upon seed imbibition, and when this gene is knocked out, a delay of radicle protrusion and enhancement of AtCathB3 expression occurs in the germinating seeds (Figs 5 and 6). Other bZIPs have been reported to be repressors of seed germination, such as ABI5, which provokes inhibition of the endosperm rupture, and its transcripts are localized at the embryonic axis and at the micropylar endosperm in ABA-treated imbibed seeds (Finkelstein and Lynch, 2000; López-Molina et al., 2002; Piskurewicz et al., 2008). Recently, the positive influence on seed germination sensu stricto of AtbZIP44 (At1g75390; S-group) has been reported, with its mRNAs localized to the endosperm and embryonic axis of germinating Arabidopsis thaliana seeds. AtbZIP44 is a positive transcriptional activator of the endo-β-mannanase-encoding gene AtMAN7 (At5g66460). Endo-β-mannanases are important hydrolytic enzymes involved in loosening of the cell walls of mannan-rich endosperm cells during germination of dicotyledonous seeds (Iglesias-Fernández et al., 2011; Rodríguez-Gacio et al., 2012). KO AtbZIP44 mutants have a statistically significant slower germination rate than the Wt, being affected in two phases of the germination process: rupture of the seed coat and radicle protrusion through the micropylar endosperm (Iglesias-Fernández et al., 2013).
GBF1 transcripts accumulate during Arabidopsis seed germination at the embryonic axis and vascular elements of the embryo at the same time as those of AtCathB3 (Figs 5 and 6). However, the transcriptional effect of GBF1 upon its target genes can be modified by phosphorylation by casein kinase II, which stimulates its DNA-binding activity (Klimczak et al., 1992; Menkens et al., 1995), and can be also modulated by its interaction with other TFs of the same or different families. It has been reported previously that GBF1 can dimerize with all the members of the G group (GBF1, GBF2, GBF3, bZIP16, and bZIP68) and with HY5 and HYH from the H group, which are master positive regulators of photomorphogenesis, generating positive and negative regulatory responses in light-regulated gene expression and seedling development (Schindler et al., 1992a ; Singh et al., 2012). During photomorphogenesis, when shortening of the hypocotyls, cotyledon expansion, and development of mature chloroplasts occur, GBF1 plays a regulatory role in cryptochrome-mediated blue-light signalling at early seedling development, acting as a negative regulator of the inhibition of hypocotyl elongation, and as a positive regulator of cotyledon expansion. Moreover, GBF1 may function either as a transcriptional activator or repressor of its target genes, as it negatively regulates the gene encoding ribulose-bisphosphate-carboxylase small subunit (RBCS), whereas it is a positive regulator of the chlorophyll a/b-binding protein-encoding gene (CAB1 gene, synonymous LHCB1.3; Mallappa et al., 2006). This antagonistic role for a given TF in the regulation of different seed genes has been reported not only with bZIPs but also with DOF proteins. In barley seeds, the bZIPs BLZ1 and BLZ2 activate the Hor2 gene encoding a B-hordein storage protein during seed maturation (Vicente-Carbajosa et al., 1998; Oñate et al., 1999), while they are transcriptional repressors of the α-amylase-encoding Amy6.4 gene during seed germination (Fuentes, 2008). Similarly, the DOF BPBF (DOF24) activates expression of the Hor2 gene upon maturation while repressing the Al21 gene encoding a CathB-like protease induced in the aleurone layers in response to GA during seed germination (Mena et al., 1998; 2002). The interaction between GBF1 and COP1 has also been described, a well-characterized negative regulator of light signalling during photomorphogenesis, and this physical interaction is required for the optimum accumulation of GBF1 in light-grown seedlings (Mallappa et al., 2008). All these data indicate the importance of the combinatorial interaction of GBF1 with other TFs in transcriptional regulation. Whether GBF1 negatively regulates AtCathB3 expression at the embryonic axis in germinating seeds through its interaction with other bZIPs or other TFs belonging to other families remains to be elucidated.
GBF1 has been also described as being involved in leaf senescence, repressing the expression of genes as RUBISCO (RBCS) and CATALASE2 involved in the redox response (Smykowski et al., 2010), and activating a LHCB2.4 gene encoding a chlorophyll a/b-binding protein belonging to the chloroplast antennal system (Shaikhali et al., 2012), different from that encoded by LHCB1.3, mentioned previously. Interestingly, AtCathB3 and GBF1 are expressed at the incipient vascular elements during seed germination and post-germination where these cells undergo a programmed cell death that implicates redox reactions and vacuolization. Our group has previously reported that DOF6 (At3g45610) negatively affects seed germination and is also localized at the incipient vascular elements of the embryo (Rueda-Romero et al., 2011). However, DOF6 and GBF1 do not interact in yeast two-hybrid assays (data not shown). The gene encoding the DOF DAG1 is also a negative regulator of seed germination, while its paralogous DAG2 is a transcriptional activator being expressed in the incipient vascular elements of the embryo (Gualberti et al., 2002; Gabriele et al., 2010).
In summary, this work describes a new role for the AtCathB3 gene, not only during germination sensu stricto when the mobilization of proteins occurs at the embryonic axis but also in the post-germination process when SSPs are hydrolysed in the cotyledons, and demonstrates that GBF1 is a bZIP transcriptional regulator negatively affecting AtCathB3 expression during seed germination. As GBF1 functions in cryptochrome-mediated blue-light signalling during seedling establishment, further investigation is needed to explore the involvement of GBF1 blue-light regulation in protein mobilization during germination.
Supplementary data
Supplementary data are available at JXB online.
Supplementary Fig. S1. Phylogenetic tree with the deduced protein sequences of the cathepsin B-like genes of the Brassicaceae species.
Supplementary Fig. S2. Transcription levels of the housekeeping gene (UBC21) used in the RT-qPCR analyses, presented as C t mean values. Expression of the ACT8 gene is used for comparison.
Supplementary Fig. S3. Arabidopsis thaliana bZIP transcription factors of the G subfamily.
Supplementary Table S1. List of primers used for cloning, for probe synthesis in FISH, and for T-DNA genotyping of mutant insertion lines.
Supplementary Table S2. Major characteristics of the Brassicaceae predicted CathB-like protein sequences.
Supplementary Table S3. Genomic sequences of the CathB3 gene promoters of the Brassicaceae species used in the phylogenetic shadowing analysis.
Supplementary Table S4. Sequences and characteristic of the primers used in the RT-qPCR analyses.
Acknowledgements
We thank Dr Javier Paz-Ares (Centro Nacional de Biotecnología, CSIC-Madrid, Spain) for providing promoter sequences for orthologous CathB3 genes from several Brassicaceae species, and Dr Ulrike Zentgraf (University of Tubingen, Germany) for providing the KO gbf1 homozygous mutant seeds. This work was financially supported by the Spanish Grants CONSOLIDER CSD 2007-00057 and BFU2009-11809, from the Ministerio de Ciencia e Innovación (MICINN, Spain). RI-F is the recipient of a post-doctoral contract JCI2010-07909 (MICINN, Spain). Unpublished data shared from the Arabis alpina genome project (PLANT-KBBE 2008M EUI2008-03716) is acknowledged. We thank Professor A. J. Matilla (Universidad de Santiago de Compostela, USC, Spain) for critical reading of the manuscript.
Glossary
Abbreviations:
- 3-AT
3-amino-1,2,4-triazole
- ABA
abscisic acid
- bZIP
basic leucine zipper
- CaMV
cauliflower mosaic virus
- DAPI
4′,6-diamidino-2-phenylindole
- FISH
fluorescence in situ hybridization
- GA
gibberellin
- GFP
green fluorescent protein
- GUS
β-glucuronidase
- KO
knockout
- LUC
luciferase
- oex
overexpressor
- RT-qPCR
real-time quantitative PCR
- SE
standard error
- SSP
seed storage protein
- TF
transcription factor
- Wt
wild type
- Y1H
yeast one-hybrid.
References
- Adrian J, Farrona S, Reimer JJ, Albani MC, Coupland G, Turk F. 2010. Cis-regulatory elements and chromatin state co-ordinately control temporal and spatial expression of FLOWERING LOCUS T in Arabidopsis. Plant Cell 22, 1425–1440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Bodèn M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. 2009. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Research 37, W202–W208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benfey PN, Ren L, Chua NH. 1989. The CaMV 35S enhancer contains at least two domains which can confer different developmental and tissue-specific expression patterns. EMBO Journal 8, 2195–2202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bewley JD. 1997. Seed germination and dormancy. Plant Cell 9, 1055–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boffelli D, McAuliffe J, Ovcharenko D, Lewis KD, Ovacharenko I, Pachter L, Rubin EM. 2003. Phylogenetic shadowing of primate sequences to find functional regions of the human genome. Science 299, 1391–1393 [DOI] [PubMed] [Google Scholar]
- Bolte S, Lanquar V, Soler M-N, Beebo A, Satiat-Jeunemaitre B, Bouhidel K, Thomine S. 2011. Distinct lytic vacuolar compartments are embedded inside the protein storage vacuole of dry and germinating Arabidopsis thaliana seeds. Plant Cell Physiology 52, 1142–1152 [DOI] [PubMed] [Google Scholar]
- Carter C, Pan S, Zouhar J, Avila EL, Girke T, Raikhel NV. 2004. The vegetative vacuole proteome of Arabidopsis thaliana reveals predicted and unexpected proteins. Plant Cell 16, 3285–3303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castrillo G, Turck F, Leveugle M, Lecharny A, Carbonero P, Coupland G, Paz-Ares J, Oñate-Sanchez L. 2011. Speeding cis-trans regulation discovery by phylogenomic analyses coupled with screenings of an arrayed library of Arabidopsis transcription factors. PLoS ONE 6, e21524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cejudo FJ, Ghose TK, Stabel P, Baulcombe DC. 1992. Analysis of the gibberellin-responsive promoter of a cathepsin B-like gene from wheat. Plant Molecular Biology 20, 849–856 [DOI] [PubMed] [Google Scholar]
- Cliften PF, Hillier LW, Fulton L, Graves T, Miner T, Gish WR, Waterston RH, Jonhnston M. 2001. Surveying Saccharomyces genomes to identify functional elements by comparative DNA sequence analyses. Genome Research 11, 1175–1186 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana . The Plant Journal 16, 735–743 [DOI] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U. 2003. A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiology 133, 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekkers BJ, Pearce S, van Bolderen-Veldkamp RP, et al. 2013. Transcriptional dynamics of two seed compartments with opposing roles in Arabidopsis seed germination. Plant Physiology 163, 205–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez F, González MC, Cejudo FJ. 2002. A germination-related gene encoding a serine carboxypeptidase is expressed during the differentiation of the vascular tissue in wheat grains and seedlings. Planta 215, 727–734 [DOI] [PubMed] [Google Scholar]
- Eastmond PJ, Jones RL. 2005. Hormonal regulation of gluconeogenesis in cereal aleurone is strongly cultivar-dependent and gibberellin action involves SLENDER1 but not GAMYB. The Plant Journal 44, 483–493 [DOI] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research 32, 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finch-Savage WE, Leubner-Metzger G. 2006. Seed dormancy and the control of germination. New Phytologist 171, 501–523 [DOI] [PubMed] [Google Scholar]
- Finkelstein RR, Lynch TJ. 2000. Abscisic acid inhibition of radicle emergence but not seedling growth is suppressed by sugars. Plant Physiology 122, 1179–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster R, Izawa T, Chua NH. 1994. Plant bZIP proteins gather at ACGT elements. Journal of the Federation of American Societies for Experimental Botany 8, 192–200 [DOI] [PubMed] [Google Scholar]
- Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I. 2004. VISTA: computational tools for comparative genomics. Nucleic Acids Research 32, W273–W279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuentes R. 2008. Molecular characterization of new bZIP genes of the Opaque-2 subfamily in barley. Doctoral thesis, Universidad Politécnica de Madrid, Spain.
- Gabriele S, Rizza A, Martone J, Circelli P, Costantino P, Vittorioso P. 2010. The Dof protein DAG1 mediates PIL5 activity on seed germination by negatively regulating GA biosynthetic gene AtGA3ox1 . The Plant Journal 61, 312–323 [DOI] [PubMed] [Google Scholar]
- García-Lorenzo M, Sjodin A, Jansson S, Funk C. 2006. Protease gene families in Populus and Arabidopsis. BMC Plant Biology 6, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodstein DM, Shu S, Howson R, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N, Rokhsar DS. 2012. Phytozome: a comparative platform for green plant genomics. Nucleic Acid Research 40, D1178–D1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graeber K, Linkies A, Wood AT, Leubner-Metzger G. 2011. A guide line to family-wide comparative state-of-the-art quantitative RT-PCR analysis exemplified with a Brassicaceae cross-species seed germination case study. Plant Cell 23, 2045–2063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gualberti G, Papi M, Bellucci L, Ricci I, Bouchez D, Camilleri C, Costantino P, Vittorioso P. 2002. Mutations in the Dof zinc finger genes DAG2 and DAG1 influence with opposite effects the germination of Arabidopsis seeds. Plant Cell 14, 1253–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler F, Kalla R, Roberts JK, Jacobsen JV. 1995. Gibberellin-regulated expression of a myb gene in barley aleurone cells: evidence for Myb transactivation of a high-pI α-amylase gene promoter. Plant Cell 7, 1879–1891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler F, Raventos D, Deys M, Watts R, Mundy J, Jacobsen JV. 1999. Target genes and regulatory domains of the GAMYB transcriptional activator in cereal aleurone. The Plant Journal 17, 1–9 [DOI] [PubMed] [Google Scholar]
- Hills JM. 2004. Control of storage-product synthesis in seeds. Current Opinion in Plant Biology 7, 302–308 [DOI] [PubMed] [Google Scholar]
- Holdsworth MJ, Bentsink L, Soppe WJ. 2008. Molecular networks regulating Arabidopsis seed maturation, after-ripening, dormancy and germination. New Phytologist 179, 33–54 [DOI] [PubMed] [Google Scholar]
- Hong RL, Hamaguchi L, Busch MA, Weigel D. 2003. Regulatory elements of the floral homeotic gene AGAMOUS identified by phylogenetic footprinting and shadowing. Plant Cell 15, 1296–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias-Fernández R, Barrero-Sicilia C, Carrillo-Barral N, Oñate-Sanchez L, Carbonero P. 2013. Arabidopsis thaliana bZIP44: a transcription factor affecting seed germination and expression of the mannanase-encoding gene AtMAN7 . The Plant Journal 74, 767–780 [DOI] [PubMed] [Google Scholar]
- Iglesias-Fernández R, Rodriguez-Gacio MC, Barrero-Sicilia C, Carbonero P, Matilla A. 2011. Three endo-β-mannanase genes expressed in the micropylar endosperm and in the radicle influence germination of Arabidopsis thaliana seeds. Planta 233, 25–36 [DOI] [PubMed] [Google Scholar]
- Isabel-Lamoneda I, Diaz I, Martinez M, Mena M, Carbonero P. 2003. SAD: a new DOF protein from barley that activates transcription of a cathepsin B-like thiol protease gene in the aleurone of germinating seeds. The Plant Journal 33 329–340 [DOI] [PubMed] [Google Scholar]
- Jakoby M, Weisshaar B, Dröge-Laser W, Vicente-Carbajosa J, Tiedemann J, Kroj T, Parcy F. 2002. bZIP transcription factors in Arabidopsis. Trends in Plant Science 7, 106–111 [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. 1987. GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO Journal 6, 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Phillips TE, Hamm CA, Drozdowicz YM, Rea PA, Maeshima M, Rogers SW, Rogers JC. 2001. The protein storage vacuole: a unique compound organelle. Journal of Cell Biology 155, 991–1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joosen RV, Kodde J, Willems LA, Ligterink W, van der Plas LH, Hilhorst HW. 2010. GERMINATOR: a software package for high-throughput scoring and curve fitting of Arabidopsis seed germination. The Plant Journal 62, 148–159 [DOI] [PubMed] [Google Scholar]
- Khan AR, James MN. 1998. Molecular mechanisms for the conversion of zymogens to active proteolytic enzymes. Protein Science 7, 815–836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimczak LJ, Schindler U, Cashmore AR. 1992. DNA-binding activity of the Arabidopsis G-box binding factor GBF1 is stimulated by phosphorylation by casein kinase from broccoli. Plant Cell 4, 87–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch M, Weisshaar B, Kroymann J, Haubold B, Mitchell-Olds T. 2001. Comparative genomics and regulatory evolution: conservation and function of the Chs and Apetala3 promoters. Molecular Biology and Evolution 18, 1882–1891 [DOI] [PubMed] [Google Scholar]
- Lescot M, Dehais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouze P, Rombauts S. 2002. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Research 30, 325–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Molina L, Mongrand S, McLachlin DT, Chait BT, Chua NH. 2002. ABI5 acts downstream of ABI3 to execute an ABA-dependent growth arrest during germination. The Plant Journal 32, 317–328 [DOI] [PubMed] [Google Scholar]
- Mallappa C, Singh A, Ram H, Chattopadhyay S. 2008. GBF1, a transcription factor of blue light signaling in Arabidopsis, is degraded in the dark by a proteasome-mediated pathway independent of COP1 and SPA1. Journal of Biological Chemistry 283, 35772–35782 [DOI] [PubMed] [Google Scholar]
- Mallappa C, Yadav V, Negi P, Chattopadhyay S. 2006. A basic leucine zipper transcription factor, G-box-binding factor 1, regulates blue light-mediated photomorphogenic growth in Arabidopsis. Journal of Biological Chemistry 281, 22190–22199 [DOI] [PubMed] [Google Scholar]
- McLellan H, Gilroy EM, Yun BW, Birch PR, Loake GJ. 2009. Functional redundancy in the Arabidopsis Cathepsin B gene family contributes to basal defence, the hypersensitive response and senescence. New Phytologist 183, 408–418 [DOI] [PubMed] [Google Scholar]
- Mena M, Cejudo FJ, Isabel-Lamoneda I, Carbonero P. 2002. A role for the DOF transcription factor BPBF in the regulation of gibberellin-responsive genes in barley aleurone. Plant Physiology 130, 11–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mena M, Vicente-Carbajosa J, Schmidt RJ, Carbonero P. 1998. An endosperm-specific DOF protein from barley, highly conserved in wheat, binds to and activates transcription from the prolamin-box of a native B-hordein promoter in barley endosperm. The Plant Journal 16, 53–62 [DOI] [PubMed] [Google Scholar]
- Menkens AE, Schindler U, Cashmore AR. 1995. The G-box: a ubiquitous regulatory DNA element in plants bound by the GBF family of bZIP proteins. Trends in Biochemical Sciences 20, 506–510 [DOI] [PubMed] [Google Scholar]
- Moreno-Risueño MA, Díaz I, Carrillo L, Fuentes R, Carbonero P. 2007. The HvDOF19 transcription factor mediates the abscisic acid-dependent repression of hydrolase genes in germinating barley aleurone. The Plant Journal 51, 352–365 [DOI] [PubMed] [Google Scholar]
- Müntz K. 2007. Protein dynamics and proteolysis in plant vacuoles. Journal of Experimental Botany 58, 2391–2407 [DOI] [PubMed] [Google Scholar]
- Nakabayashi K, Okamoto M, Koshiba T, Kamiya Y, Nambara E. 2005. Genome-wide profiling of stored mRNA in Arabidopsis thaliana seed germination: epigenetic and genetic regulation of transcription in seed. The Plant Journal 41, 697–709 [DOI] [PubMed] [Google Scholar]
- Nambara E, Okamoto M, Tatematsu K, Yano R, Seo M, Kamiya Y. 2010. Abscisic acid and the control of seed dormancy and germination. Seed Science Research 20, 55–67 [Google Scholar]
- Nonogaki H, Chen F, Bradford K. 2007. Mecanisms and genes involved in germination sensu stricto . In: Bradford K, Nonogaki H, eds. Seed development, dormancy and germination. Oxford: Blackwell Publishing, 264–304 [Google Scholar]
- Notredame C, Higgins DG, Heringa J. 2000. T-Coffee: a novel method for fast and accurate multiple sequence alignment. Journal of Molecular Biology 302, 205–217 [DOI] [PubMed] [Google Scholar]
- Oñate L, Vicente-Carbajosa J, Lara P, Díaz I, Carbonero P. 1999. Barley BLZ2, a seed-specific bZIP protein that interacts with BLZ1 in vivo and activates transcription from the GCN4-like motif of B-hordein promoters in barley endosperm. Journal of Biological Chemistry 274, 9175–9182 [DOI] [PubMed] [Google Scholar]
- Oñate-Sanchez L, Vicente-Carbajosa J. 2008. DNA-free RNA isolation protocols for Arabidopsis thaliana, including seeds and siliques. BMC Research Notes 1, 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penfield S, Li Y, Gilday AD, Graham S, Graham IA. 2006. Arabidopsis ABA INSENSITIVE4 regulates lipid mobilization in the embryo and reveals repression of seed germination by the endosperm. Plant Cell 18, 1887–1899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Research 29, e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piskurewicz U, Jikumaru Y, Kinoshita N, Nambara E, Kamiya Y, Lopez-Molina L. 2008. The gibberellic acid signaling repressor RGL2 inhibits Arabidopsis seed germination by stimulating abscisic acid synthesis and ABI5 activity. Plant Cell 20, 2729–2745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piskurewicz U, Tureckova V, Lacombe E, Lopez-Molina L. 2009. Far-red light inhibits germination through DELLA-dependent stimulation of ABA synthesis and ABI3 activity. EMBO Journal 28, 2259–2271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard SL, Charlton WL, Baker A, Graham IA. 2002. Germination and storage reserve mobilization are regulated independently in Arabidopsis. The Plant Journal 31, 639–647 [DOI] [PubMed] [Google Scholar]
- Prouse MB, Campbell MM. 2012. The interaction between MYB proteins and their target DNA binding sites. Biochemica et Biophysica Acta 1819, 67–77 [DOI] [PubMed] [Google Scholar]
- Quevillon E, Silventoinen V, Pillai S, Harte N, Mulder N, Apweiler R, Lopez R. 2005. InterproScan: protein domains identifier. Nucleic Acids Research 33, W116–W120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlings ND, Barrett AJ, Bateman A. 2012. MEROPS: the database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Research 40, D343–D350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Gacio MC, Iglesias-Fernández R, Carbonero P, Matilla AJ. 2012. Softening-up mannan-rich cell walls. Journal of Experimental Botany 63, 3975–3988 [DOI] [PubMed] [Google Scholar]
- Rubio-Somoza I, Martinez M, Abraham Z, Díaz I, Carbonero P. 2006. Ternary complex formation between HvMYBS3 and other factors involved in transcriptional control in barley seeds. The Plant Journal 47, 269–281 [DOI] [PubMed] [Google Scholar]
- Rueda-Romero P, Barrero-Sicilia C, Gómez-Cadenas A, Carbonero P, Oñate-Sánchez L. 2011. Arabidopsis thaliana DOF6 negatively affects germination in non-after-ripened seeds and interacts with TCP14. Journal of Experimental Botany 63, 1937–1949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler U, Menkens AE, Beckmann H, Ecker JR, Cashmore AR. 1992a. Heterodimerization between light-regulated and ubiquitously expressed Arabidopsis GBF bZIP proteins. EMBO Journal 11, 1261–1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler U, Terzaghi W, Beckmann H, Kadesch T, Cashmore AR. 1992b. DNA binding site preferences and transcriptional activation properties of the Arabidopsis transcription factor GBF1. EMBO Journal 11, 1275–1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaikhali J, Norén L, de Dios Barajas-López J, Srivastava V, König J, Sauer UH, Wingsle G, Dietz KJ, Strand Å. 2012. Redox-mediated mechanisms regulate DNA binding activity of the G-group of basic region leucine zipper (bZIP) transcription factors in Arabidopsis. Journal of Biological Chemistry 287, 27510–27525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, Ram H, Abbas N, Chattopadhyay S. 2012. Molecular interactions of GBF1 with HY5 and HYH proteins during light-mediated seedling development in Arabidopsis thaliana . Journal of Biological Chemistry 287, 25995–26009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sliwinska E, Bassel GW, Bewley JD. 2009. Germination of Arabidopsis thaliana seeds is not completed as a result of elongation of the radicle but of the adjacent transition zone and lower hypocotyl. Journal of Experimental Botany 60, 3587–3594 [DOI] [PubMed] [Google Scholar]
- Smykowski A, Zimmermann P, Zentgraf U. 2010. G-Box binding factor1 reduces CATALASE2 expression and regulates the onset of leaf senescence in Arabidopsis. Plant Physiology 153, 1321–1331 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Stangeland B, Salehian Z. 2002. An improved clearing method for GUS assay in Arabidopsis endosperm and seeds. Plant Molecular Biology Reporter 20, 107–114 [Google Scholar]
- Sun TP, Gubler F. 2004. Molecular mechanism of gibberellin signalling in plants. Annual Review of Plant Biology 55, 197–223 [DOI] [PubMed] [Google Scholar]
- Tan-Wilson AL, Wilson KA. 2012. Mobilization of seed protein reserves. Physiologia Plantarum 145, 140–153 [DOI] [PubMed] [Google Scholar]
- Testillano P, Risueño MC. 2009. Tracking gene and protein expression during microspore embryogenesis by confocal laser scanning microscopy. In: Touraev A, Forster BP, Jain SM, eds. Advances in haploid production in higher plants. Heidelberg: Springer-Verlag, 339–347 [Google Scholar]
- Vavouri T, Elgar G. 2005. Prediction of cis-regulatory elements using binding site matrices—the successes, the failures and the reasons for both. Current Opinion in Genetics and Development 15, 395–402 [DOI] [PubMed] [Google Scholar]
- Vicente-Carbajosa J, Oñate L, Lara P, Diaz I, Carbonero P. 1998. Barley BLZ1: a bZIP transcriptional activator that interacts with endosperm-specific gene promoters. The Plant Journal 13, 629–640 [DOI] [PubMed] [Google Scholar]
- Vicente-Carbajosa V, Carbonero P. 2005. Seed maturation: developing an intrusive phase to accomplish a quiescent state. International Journal of Developmental Biology 49, 645–651 [DOI] [PubMed] [Google Scholar]
- Voinnet O, Rivas S, Mestre P, Baulcombe D. 2003. An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. The Plant Journal 33, 949–956 [DOI] [PubMed] [Google Scholar]
- Weitbrecht K, Müller K, Leubner-Metzger G. 2011. First off the mark: early seed germination. Journal of Experimental Botany 62, 3289–3309 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.