Summary
Twenty-eight BnaMAPKKK genes were cloned. Phylogenetic and expression profiling analyses indicated their relationship and roles in stress and hormone signalling. Two novel BnaMAPKKK genes were identified to mediate cell death independent of pathogens.
Key words: Abiotic stress, Brassica napus, cell death, MAPKKK, MKK, Sclerotinia sclerotiorum.
Abstract
Mitogen-activated protein kinase (MAPK) signalling cascades, consisting of three types of reversibly phosphorylated kinases (MAPKKK, MAPKK, and MAPK), are involved in important processes including plant immunity and hormone responses. The MAPKKKs comprise the largest family in the MAPK cascades, yet only a few of these genes have been associated with physiological functions, even in the model plant Arabidopsis thaliana. Canola (Brassica napus L.) is one of the most important oilseed crops in China and worldwide. To explore MAPKKK functions in biotic and abiotic stress responses in canola, 66 MAPKKK genes were identified and 28 of them were cloned. Phylogenetic analysis of these canola MAPKKKs with homologous genes from representative species classified them into three groups (A–C), comprising four MAPKKKs, seven ZIKs, and 17 Raf genes. A further 15 interaction pairs between these MAPKKKs and the downstream BnaMKKs were identified through a yeast two-hybrid assay. The interactions were further validated through bimolecular fluorescence complementation (BiFC) analysis. In addition, by quantitative real-time reverse transcription–PCR, it was further observed that some of these BnaMAPKKK genes were regulated by different hormone stimuli, abiotic stresses, or fungal pathogen treatments. Interestingly, two novel BnaMAPKKK genes, BnaMAPKKK18 and BnaMAPKKK19, which could elicit hypersensitive response (HR)-like cell death when transiently expressed in Nicotiana benthamiana leaves, were successfully identified. Moreover, it was found that BnaMAPKKK19 probably mediated cell death through BnaMKK9. Overall, the present work has laid the foundation for further characterization of this important MAPKKK gene family in canola.
Introduction
To survive harsh conditions, plants have developed sophisticated mechanisms to sense environmental cues and transmit these signals to regulate plant development and defence. Plant mitogen-activated protein kinase (MAPK) cascades are vital to these functions (Asai et al., 2002; Ichimura et al., 2002; Rodriguez et al., 2010; Meng and Zhang, 2013). MAPK cascades are conserved in eukaryotes through evolution and are composed of MAPKK kinases (MAPKKKs, MAP3K, or MEKK), MAPK kinases (MAPKKs, MAP2Ks, MKKs, or MEKs), and MAPKs (MPKs). Basically, extracellular stimuli sensed by receptors are sequentially transmitted through phosphorylation by MAPKKKs to MKKs, and then to MPKs to regulate intracellular responses including the transcriptional and metabolic responses (Hamel et al., 2006). The activated MAPKKKs phosphorylate either the serine (S) or threonine (T) residues of MKKs and the activated MKKs phosphorylate both T and tyrosine (Y) residues of the MPKs (Asai et al., 2002).
Several MKK and MPK genes have been well characterized, including a description of their downstream components and the physiological processes they mediate (Frye et al., 2001; Asai et al., 2002; Jin et al., 2002; del Pozo et al., 2004; Liu et al., 2004; Nakagami et al., 2004; Colcombet and Hirt, 2008; Melech-Bonfil and Sessa, 2010; Ning et al., 2010; Oh et al., 2010). However, only a limited number of MAPKKK genes have been functionally characterized (Rodriguez et al., 2010; Meng and Zhang, 2013). A possible reason underlying this is that MAPKKKs form a large gene family, making functional redundancy inevitable. In Arabidopsis, MAPKKK is the largest group of the MAPK cascade components, with 80 members classified into three subfamilies, MEKK, Raf, and ZIK, harbouring 21, 11, and 48 genes, respectively (Jonak et al., 2002). The MEKK subfamily is the best characterized and includes tobacco NPK1 (Jin et al., 2002; Liu et al., 2004), Arabidopsis MEKK1 (Asai et al., 2002), alfalfa OMTK1 (oxidative stress activated MAPK triple-kinase 1, MEKK1) (Nakagami et al., 2004), tobacco NbMAPKKKα, NbMAPKKKγ, and NbMAPKKKε, and tomato SlMAPKKKα and SlMAPKKKε (del Pozo et al., 2004; Melech-Bonfil and Sessa, 2010; Oh et al., 2010). Characterized genes of the second subfamily of MAPKKKs include Arabidopsis CTR1/Raf1 (Kieber et al., 1993; Clark et al., 1998), EDR1/Raf2 (Frye et al., 2001), and rice (Oryza sativa) DSM1 (Ning et al., 2010). The ZIK subfamily, also called WNK (With No lysine Kinase), is reported to regulate flowering time and circadian rhythms in rice and Arabidopsis (Wang et al., 2008; Kumar et al., 2011).
Once signals are transmitted from MAPKKKs to terminal MPKs through sequential phosphorylation, activated MPKs then phosphorylate a wide range of substrates, such as WRKY transcription factors or enzymes (Rodriguez et al., 2010; Liang et al., 2013). The first identified complete signalling module is the FLS2–MEKK1–MKK4/5–MPK3/6–WRKY22/29 pathway in Arabidopsis, which is effective against different pathogens including bacteria and the fungal pathogen Botrytis cinerea (Asai et al., 2002; Galletti et al., 2011). Another module, MEKK1–MKK1/2–MPK4, of Arabidopsis was shown to regulate defence responses against biotrophic pathogens negatively while positively regulating defences against necrotrophic fungi (Petersen et al., 2000; Ichimura et al., 2006; Qiu et al., 2008). The induction of salicylic acid (SA) as well as systemic acquired resistance (SAR) in mpk4, mekk1, or mkk1mkk2 double mutants is a result of release and activation of WRKY25 and -33 in nuclei by MAPK substrate 1 (MKS1) in Arabidopsis (Andreasson et al., 2005; Petersen et al., 2010). Further study found that AtMPK4 could induce camelexin biosynthesis upon challenge by a bacterial pathogen (Andreasson et al., 2005; Ichimura et al., 2006; Qiu et al., 2008) but not by a fungal pathogen (Mao et al., 2011). Recently, it was found that AtMEKK1 and AtMKK1/MKK2 negatively regulate MEKK2-mediated immune responses as well as programmed cell death (PCD) (Kong et al., 2012). In tobacco, NPK1–MEK1–Ntf6 mediate resistance to Tobacco mosaic virus (TMV) triggered by the R protein N (Jin et al., 2002; Liu et al., 2004). Moreover, AtEDR1, a Raf-like MAPKKK, could function at the top of a MAPK cascade to regulate SA-inducible defence responses negatively (Frye et al., 2001). However, the functions of most other MAPKKK genes in Arabidopsis are as yet unknown.
Though investigations of the MAPKKK gene family in Arabidopsis, rice, and maize have been reported in recent years (Jouannic et al., 1999; Singh et al., 2012; Kong et al., 2013), no similar study has yet been conducted in canola. Canola is a very important oil crop worldwide and its yield is frequently limited by environmental factors including drought, salinity, cold, and biotic factors, such as stem rot caused by Sclerotinia sclerotiorum. Sclerotinia sclerotiorum is a necrotrophic pathogen and no efficient way has been identified to control this disease. An oxidative burst, or accumulation of reactive oxygen species (ROS), has been associated with many abiotic stresses and pathogen infections (Jaspers and Kangasjarvi, 2010; Heller and Tudzynski, 2011), especially S. sclerotiorum (Rietz et al., 2012). Research with MAPK cascades in Arabidopsis has shown that they are involved in signalling multiple defence responses, including the biosynthesis and signalling of plant stress/defence hormones, ROS production, stomatal closure, defence gene activation, phytoalexin biosynthesis, cell wall strengthening, and hypersensitive response (HR) cell death (Meng and Zhang, 2013). However, the identity and role of MAPKKK genes in canola responses to abiotic and biotic stresses are unknown. It is therefore necessary to characterize the MAPKKK gene family in canola before stress/disease-tolerant canola species can be developed. In previous transcriptomic profiling studies in canola, a few components of the MAPK module were identified, including several MAPKKK genes elicited by S. sclerotiorum (Yang et al., 2007). A few novel BnaMKK (Bna for Brassica napus) and BnaMPK genes and modules were further characterized (Liang et al., 2013). The functions of some of the MKK, MPK, and WRKY genes are under investigation through loss-of-function and gain-of-function strategies. However, the upstream components of MKK–MAPK modules, which are BnaMAPKKKs, have not yet been characterized. Hence, to explore the role of MAPKKK genes in immune and abiotic stress responses in canola, the publicly available expressed sequence tags (ESTs) were mined to identify MAPKKK genes in canola. The cDNA sequences of 28 BnaMAPKKK genes were then cloned, followed by a yeast two-hybrid (Y2H)-based analysis of interactions between canola MAPKKKs and MKKs, and part of the interactions were confirmed in planta. The responses of selected genes under a range of abiotic and biotic stress conditions were also examined. Interestingly, two novel BnaMAPKKK genes that could elicit cell death when transiently expressed in tobacco leaves were successfully identified. These may mediate cell death by regulating specific downstream MKKs. To the authors’ knowledge, this is the first report of canola MAPKKK genes, and the data presented here will lay the foundation for further characterization of this important MAPKKK gene family in canola responses to abiotic and biotic stresses.
Materials and methods
Database search and identification of MAPKKK ESTs in canola
The identification of canola ESTs representing MAPKKK genes was performed as described previously (Liang et al., 2013). In brief, ESTs representing MAPKKKs were retrieved through BLAST search in the NCBI dbEST (http://www.ncbi.nlm.nih.gov/dbEST/index.html, release 01012012) and DFCI (http://compbio.dfci.harvard.edu/tgi/cgi-bin/tgi/gimain.pl?gudb=oilseed_rape,release5.0) using the 80 public AtMAPKKK cDNA sequences downloaded from TAIR9.0 (www.arabidopsis.org) as the queries with an e-value cut-off <10–4. After manual curation, these ESTs were clustered and assembled by DNASTAR (DNASTAR Inc., USA). Subsequently, contigs and singletons were run in a reciprocal BLAST search against the Arabidopsis database to assign a putative orthologue based on the best hit (Supplementary Table S1 available at JXB online).
Plant growth and gene cloning
Canola (double haploid DH12075) plants were grown as described previously (Liang et al., 2013). RNA was isolated using the Plant RNA kit (Omega bio-tek, USA). First-strand cDNA synthesis and high-fidelity PCR amplification using PrimeSTAR HS DNA polymerase (TaKaRa, Japan) were performed as previously described (Liang et al., 2013) with the primers listed in Supplementary Table S2 available at JXB online. PCR products were purified and cloned into the pJET1.2 vector supplied in the CloneJET PCR cloning kit (Fermentas, USA), sequenced, and analysed by DNASTAR. The cDNA sequences of genes cloned in this study were deposited in the GenBank database under the accession numbers KC190095–KC190110 and KF129395–KF129406.
Phylogenetic tree reconstruction, multiple alignment analysis, and conserved signature detection
The MAPKKK genes of rice were downloaded from the rice genome annotation project (http://rice.plantbiology.msu.edu), while those of other species were either identified from Phytozome v9.1 (http://www.phytozome.net/) or retrieved from the NCBI by a keyword search (Supplementary Table S3 available at JXB online). To investigate the evolutionary relationship among MAPKKKs, the predicted amino acid sequences of the products of the MAPKKK genes of canola and other species were aligned and a phylogenetic tree was reconstructed as described previously (Liang et al., 2013). Motif analysis of BnaMAPKKKs was determined by using the Prosite program (http://prosite.expasy.org/prosite.html), and a schematic diagram of amino acid motifs of each BnaMAPKKK was drawn accordingly. The respective domains of MAPKKK proteins were aligned using ClutsalX1.83 and analysed in MEME 4.9.0 (release date 3 October 3, 11:07:26 EST 2012) or illustrated by Boxshade (http://www.ch.embnet.org/software/BOX_form.html). The percentage identity and similarity of sequences was calculated using the program MatGAT v2.02 (Campanella et al., 2003).
Subcellular localization and confocal microscopy
To examine the localization of selected BnaMAPKKKs in planta, the coding regions were amplified using Pfu polymerase (Bioer, China) with the primers listed in Supplementary Table S2 available at JXB online. These were digested by the corresponding restriction enzymes and fused upstream of the green fluorescent protein gene (GFP) in the pYJGFP vector. These constructs and p19 protein of Tomato bushy stunt virus were transformed into Agrobacterium tumefaciens GV3101 individually, and overnight cell cultures were resuspended in infiltration media before being infiltrated into 5-week-old leaves of Nicotiana benthamiana (Liang et al., 2013). Two days later, leaf discs were plasmolysed with 500mM mannitol or not treated, and observation of GFP was conducted under an A1R confocal microscope (Nikon, Japan).
Quantitative reverse transcription–PCR (qRT–PCR) assay
Eighteen-day-old canola grown in a greenhouse with a photoperiod of 16h light/8h dark were treated with S. sclerotiorum inoculation, 5mM oxalic acid (OA; Sigma-Aldrich) inoculation, and agar inoculation as a control for S. sclerotiorum and OA treatment. Different chemical treatments include 200mM NaCl, 100 μM jasmonic acid (JA; Sigma-Aldrich, USA), 2mM SA (Sigma-Aldrich), 50 μM abscisic acid (±-ABA; Invitrogen, USA), 25 μM 1-aminocyclopropane-1-carboxylic acid (ACC; Sigma-Aldrich), 10 μM methyl viologen (MV; Sigma-Aldrich); alternatively, plants were treated at 4 ºC (cold) and 37 ºC (heat), and mock-treated plants were used as the control (Liang et al., 2013). Leaves were collected at 1h (except S. sclerotiorum and OA, which were collected after 3h) and 24h post-treatments, flash frozen in liquid nitrogen, and stored at –80 °C. Total RNA samples were isolated and the first-strand cDNAs were synthesized from 2.5 μg of total RNA as described previously (Liang et al., 2013). Three independent biological replicates of each sample were prepared at different times.
qRT–PCR was performed using 10-fold diluted cDNA and a SYBR Green I kit (CWBIO, China) on a CFX96 real-time PCR machine (Bio-Rad, USA). Primers used for qRT–PCR were designed using the PrimerSelect program (DNASTAR Inc.), which was targeted mainly at the 3’-untranslated region (UTR) with an amplicon size of 75–250bp (Supplementary Table S2 available at JXB online). The specificity and amplification efficiency of each pair of primers were examined through both a BLASTn search in the NCBI database and by running standard curves with melting curves. Three independent biological replicates and two technical replicates for each biological replicate were run and the significance was determined through t-test of SPSS statistical software (P<0.05).
Yeast two-hybrid assay
The coding regions of canola MAPKKK and MKK genes were subcloned into pGBKT7 (BD) and pGADT7 (AD) vectors, respectively, using the primers listed in Supplementary Table S2 available at JXB online. Then recombinant plasmids were transformed sequentially into yeast AH109 competent cells according to the Yeast Protocols Handbook (Clontech, USA). The interactions between BnaMAPKKKs and BnaMKKs were tested by streaking both on non-selective SD-LW (synthetic dropout without leucine and tryptophan), and on selective SD-LWH (SD–Leu–Trp–His+5mM 3’AT) and SD-LWHA (-SD-Leu-Trp-His-Ade) media. Plates were incubated at 30 ºC for up to 7 d before being photographed. The titration and colony-lift filter assays were conducted as described previously (Liang et al., 2013).
Bimolecular fluorescence complementation (BiFC)
To verify interaction partners in planta, yellow fluorescent protein (YFP)-based BiFC analysis was performed. The coding regions of BnaMAPKKK and BnaMKK genes were subcloned into pSPYNE(R)173 and pSPYCE(M) vectors, respectively (Waadt et al., 2008). Primers used are listed in Supplementary Table S2 available at JXB online. The recombinant plasmids were transformed into Agrobacterium GV3101 competent cells before being used to infiltrate the leaves of 5-week-old N. benthamiana plants as described previously (Liang et al., 2013). Three to four days later, YFP signals in at least three slides were examined under an A1R confocal microscope (Nikon, Japan).
Site-directed mutagenesis
The coding regions of relevant BnaMAPKKK and BnaMKK genes were PCR amplified using Pfu polymerase and Gateway-compatible gene-specific primers (Supplementary Table S2 available at JXB online) before being introduced into a Gateway entry vector pDONOR/Zeo (Invitrogen, USA). For substitution of one amino acid residue with another, two overlapping primers harbouring mutated nucleotides in the middle were used to run PrimeSTAR-mediated PCR (Li and Wilkinson, 1997). The BnaM3K18-K32G-F/BnaM3K18-K32G-R and BnaM3K19-K37G-F/BnaM3K19-K37G-R primers were used to change the lysine (K) at residues 32 and 37 to glycine (G), respectively (Melech-Bonfil and Sessa, 2010; Hashimoto et al., 2012). Primers BnaMKK9-K74R-F and BnaMKK9-K74R-R were designed to mutate lysine (K) at residue 74 to arginine (R), while primers of BnaMKK9-S193D/S199E-F and BnaMKK9-S193D/S199E-R were designed to mutate serine (S) on residues 193 and 199 to aspartic acid (D) and glutamic acid (E) individually. BnaMKK9K74R and BnaMKK9S193D/S199E are constitutively inactive and active forms of BnaMKK9, respectively (Popescu et al., 2009). PCR products were purified, followed by DpnI (Fermentas, USA) digestion overnight. After purification, the restricted PCR product was transformed into Escherichia coli DH5α competent cells and was selected on low-salt LB medium supplemented with 50mg l–1 zeocin, with the plasmid isolated and sequenced to confirm that the mutated regions were correct.
Transient expression and physiological assay
The coding regions of the respective genes and their mutated derivates were isolated by restriction digestion of the aforementioned pJET or pDONR/Zeo recombinant plasmids, which was sometimes preceded by PCR amplification using high-fidelity Pfu polymerase and primers containing corresponding restriction sites, as listed in Supplementary Table S2 available at JXB online. After digestion, the products were inserted downstream of a double Cauliflower mosaic virus (CaMV) 35S promoter in the binary vector pYBHA or pYBMyc, which was modified from the pYJGFP vector (Liang et al., 2013). Recombinant plasmids were transformed into A. tumefaciens GV3101 and overnight cell cultures were resuspended in infiltration media containing 10mM MES-KOH (pH 5.6), 10mM MgCl2, and 0.15mM acetosyringone, adjusted to an OD600 of 0.5 before equal volumes of each cell culture were infiltrated into the lower epidermal side of 4-week-old leaves of N. benthamiana plants. For each construct, 21 independent leaves of seven independent plants (three leaves per plant) were used for each time point tested. After that, infiltrated plants were kept under normal growth conditions with the phenotype observed and recorded daily beginning 2 d after infiltration and continuing until day 7. To quantify the degree of PCD, electrolyte leakage was measured according to Oh and Martin (2011) with modifications. In brief, five leaf discs (10mm in diameter) were taken from each agro-infiltrated area and kept in deionized water under vacuum for 10min, followed by incubation for 30 min at 25 °C. Ion conductivity was measured using a DDS-307 ion conductivity meter (Leici, China). After boiling for 5min and cooling to room temperature, the ion conductivity was measured again. Distribution of hydrogen peroxide (H2O2) was detected by 3,3’-diaminobenzidine (DAB) staining according to Daudi et al. (2012).
Results and Discussion
Identification and cloning of MAPKKK genes from canola
Although the functions of a few MAPKKK genes in Arabidopsis and a few other plant species have been reported, little is known about this gene family in the important oilseed crop, canola (B. napus). A previous transcriptomic study identified several pathogen- or defence hormone-responsive MAPKKK genes, including BnaMAPKKK17 and BnaMAPKKK18 (Yang et al., 2007), suggesting that MAPKKKs may play a role in canola defence against fungal pathogens. More recently, as a follow-up work, MKK and MAPK genes, as well as the downstream WRKY transcription factor genes in canola were systemically studied and part of them were characterized (Yang et al., 2009; Liang et al., 2013). Ongoing work with selected components of MKK–MPK–WRKY cascades in canola demonstrated interesting phenotypes of gain- and loss-of-function plants (unpublished data). However, the upper components of MAPK cascades, namely MAPKKKs, have not yet been described in canola. All of these prompted the authors to clone and study them in the context of abiotic and biotic stress conditions. To this end, public EST databases were mined, since whole-genome sequencing of B. napus is not yet complete. Eighty cDNA sequences of AtMAPKKK genes were used to search for ESTs of canola that showed high similarity to AtMAPKKK genes. Altogether 839 unique ESTs were obtained including 80 singletons and 145 contigs representing putative MAPKKK genes in canola (Supplementary Table S1 available at JXB online). To facilitate comparisons between species, the established nomenclature of AtMAPKKK genes was followed when naming the BnaMAPKKK (Brassica napus MAPKKK) genes. As a result, 66 MAPKKK genes from canola were identified, which are composed of 18 MAPKKK genes, nine ZIK genes, and 39 Raf genes. It was noted that among all the BnaMAPKKK genes identified, BnaRaf28 has the largest number (73) of ESTs, followed by BnRaf22 and BnRaf21 with a total of 63 and 51 ESTs, respectively; while BnaMAPKKK2, BnaMAPKKK19, ZIK3, ZIK7, Raf2, and Raf7 have only one EST each (Table 1; Supplementary Table S1 available at JXB online). To facilitate the following work, primers were designed based on the identified ESTs to obtain full-length cDNA sequences, at least for the coding regions, employing RT–PCR together with rapid amplification of cDNA ends (RACE). As a result, the cloning of cDNA sequences of 28 BnaMAPKKK genes was achieved (Table 1). Conceptual translation of these cDNA sequences and reciprocal BLAST searches against the Arabidopsis genome indicated that they bore domains and motifs that were typical of MAPKKK proteins. The number of amino acids in each BnaMAPKKK protein ranged from 337 to 1062, with a pI of 4.62–9.42. A previous report identified three MAPKKK genes (MAP3Kα1, MAP3Kβ1, and MAP3Kε1) from B. napus (Jouannic et al., 1999), which corresponds to BnaMAPKKK3, -6, and -8, respectively (Supplementary Table S1 available at JXB online), and they were added to the analysis. Further comparison of these 31 BnaMAPKKK genes with their 31 respective orthologous AtMAPKKK genes demonstrated that they show an identity of 14.2–91%, with a similarity of 14.2–91.5% at the nucleotide level. At the protein level, the maximum identity is 95.2% with a minimum of 6.6%, whereas the similarity ranges from 10.5% to 97.7% (Supplementary Table S4 available at JXB online). However, a comparison of BnaMAPKKKs with all the 75 OsMAPKKKs showed identity of 6.6–71.7%, with a similarity of 6.6–72.9% at the nucleotide level. At the protein level, the maximum identity is 74.6% with the minimum 3.8%, whereas the similarity ranges from 5.6% to 87.8% (Supplementary Table S4 available at JXB online). At this step, it as possible to identify putative orthologues of these BnaMAPKKK genes in both Arabidopsis and rice using the program InParanoid (Table 1).
Table 1.
MAPKKK genes identified and cloned from canola
| Gene | Subfamily | GenBank accession no.a | EST count | No. of amino acids | pI value | Arabidopsis orthologueb/AGI no. | Rice orthologueb locus |
|---|---|---|---|---|---|---|---|
| BnaMAPKKK17 | MEKK | KC190095 | 2 | 368 | 4.92 | AtMAPKKK17/ At2g32510 | OsMAPKKK71/Os02g21700 |
| BnaMAPKKK18 | KC190096 | 2 | 337 | 5.03 | AtMAPKKK18/ At1g05100 | OsMAPKKK57Os05g46750 | |
| BnaMAPKKK19 | KC190097 | 1 | 346 | 5.02 | AtMAPKKK19/ At5g67080 | OsMAPKKK71/Os02g21700 | |
| BnaMAPKKK20 | KC190098 | 3 | 342 | 4.74 | AtMAPKKK20/ At3g50310 | OsMAPKKK71/Os02g21700 | |
| BnaZIK2 | ZIK | KC190099 | 11 | 562 | 5.73 | AtZIK2/At5g58350 | OsMAPKKK20/Os07g38530 |
| BnaZIK3 | KC190100 | 1 | 567 | 4.84 | AtZIK3/At3g22420 | OsMAPKKK20/Os07g38530 | |
| BnaZIK4 | KC190101 | 35 | 679 | 4.95 | AtZIK4/At3g04910 | OsMAPKKK20/Os07g38530 | |
| BnaZIK5 | KF129404 | 26 | 570 | 5.08 | AtZIK5/At3g18750 | OsMAPKKK29/OsWNK4/ Os02g45130 | |
| BnaZIK6 | KF129405 | 17 | 555 | 5.3 | AtZIK6/At5g41990 | OsMAPKKK29/Os02g45130 | |
| BnaZIK8 | KC190102 | 15 | 312 | 5.74 | AtZIK8/At5g55560 | OsMAPKKK64/Os07g39520 | |
| BnaZIK9 | KF129406 | 8 | 475 | 4.87 | AtZIK9/At5g28080 | OsMAPKKK20/Os07g38530 | |
| BnaCTR1 | Raf | KF129395 | 30 | 803 | 5.43 | AtCTR1/At5g03730 | OsMAPKKK12/Os09g39320 |
| BnaRaf17 | KC190103 | 17 | 439 | 7.6 | AtRaf17/At1g14000 | OsMAPKKK65/Os07g43900 | |
| BnaRaf21 | KF129396 | 51 | 546 | 5.57 | AtRaf21/At2g17700 | OsMAPKKK17/Os09g37230 | |
| BnaRaf22 | KF129397 | 63 | 409 | 8.14 | AtRaf22/At2g24360 | OsMAPKKK32/Os08g12750 | |
| BnaRaf23 | KF129398 | 24 | 476 | 9.05 | AtRaf23/At2g31800 | OsMAPKKK74/Os01g66860 | |
| BnaRaf27 | KF129399 | 16 | 459 | 5.85 | AtRaf27/At4g18950 | OsMAPKKK72/Os01g54480 | |
| BnaRaf28 | KC190104 | 73 | 410 | 6.72 | AtRaf28/At4g31170 | OsMAPKKK32/Os08g12750 | |
| BnaRaf29 | KC190105 | 16 | 571 | 5.98 | AtRaf29/At4g35780 | OsMAPKKK17/Os09g37230 | |
| BnaRaf30 | KC190106 | 22 | 568 | 6.49 | AtRaf30 /At4g38470 | OsMAPKKK17/Os09g37230 | |
| BnaRaf33 | KF129400 | 9 | 385 | 7.24 | AtRaf33/At5g50000 | OsMAPKKK34/Os05g50190 | |
| BnaRaf34 | KC190107 | 2 | 352 | 7.92 | AtRaf34/At5g50180 | OsMAPKKK25/Os02g38080 | |
| BnaRaf35 | KC190108 | 10 | 1062 | 5.28 | AtRaf35/At5g57610 | OsMAPKKK35/Os02g54510 | |
| BnaRaf36 | KC190109 | 2 | 510 | 9.42 | AtRaf36/At5g58950 | OsMAPKKK61/Os01g10450 | |
| BnaRaf37 | KF129401 | 5 | 398 | 8.16 | AtRaf37/At5g66710 | OsMAPKKK25/Os02g38080 | |
| BnaRaf39 | KC190110 | 29 | 379 | 8.23 | AtaRaf39/At3g22750 | OsMAPKKK27/Os03g43760 | |
| BnaRaf41 | KF129402 | 6 | 356 | 8.2 | AtRaf41/At3g27560 | OsMAPKKK25/Os02g38080 | |
| BnaRaf46 | KF129403 | 6 | 458 | 9.15 | AtRaf46/At3g59830 | OsMAPKKK74/Os01g66860 |
a The full-length cDNA was cloned and deposited in GenBank.
b Putative orthologues were identified by InParanoid (http://inparanoid.sbc.su.se/cgi-bin/index.cgi) with a score of 1 (the maximum score).
Brassica napus is an amphidiploid species with an AACC genome (2n=38), which is presumably derived from interspecific hybridization of Brassica rapa (2n=20, AA) and Brassica oleracea (2n=18, CC) (Rana et al., 2004). Although partial genome sequencing and comparative chromosome painting suggest that ancestral segmental chromosomal duplications led to effective triplication in Brassica diploids (Lysak et al., 2005, 2007), various mechanisms of genome evolution have contributed to many situations where fewer than three paralogous genes, corresponding to single orthologues in Arabidopsis, are present in the Brassica A or C genome (Osborn, 2004; Parkin et al., 2005). MAPKKK sequences from the sequenced genome of B. rapa were therefore analyzed. As a result, 118 MAPKKK genes were identified from B. rapa (Supplementary Table S3 available at JXB online), which supports that there are not necessarily three paralogous genes existing in the AA genome of B. rapa compared with that in Arabidopsis. Since the CC genome of B. oleracea is not yet known, there is no way to determine the exact number of MAPKKK genes in B. napus. However, since a double haploid canola variety was used, it should greatly facilitate the identification and functional characterization of MAPKKK genes.
Phylogenetic tree reconstruction and domain analysis of BnaMAPKKK proteins
To examine the evolutionary relationships of canola MAPKKKs to other representative crops and models, a rooted phylogenetic tree was produced by alignment of full-length amino acid sequences using a maximum parsimony (MP) algorithm. The species used represented the major land plant lineages including several mono- and eudicotyledonous angiosperms, namely: eudicots A. thaliana (At), B. rapa (Bra), N. benthamiana (Nb), and Solanum lycopersicum (Sl); monocot O. sativa (Os); bryophyte Physcomitrella patens (Pp); lycophyte Selaginella moellendorffii (Sm), as well as the pico-eukaryotic prasinophyte green alga Ostreococcus lucimarinus, which has one of the highest gene densities known in eukaryotes (Lanier et al., 2008). Furthermore, an MAPKKK from a marine green alga Ostreococcus tauri (Ot), the world’s smallest free-living eukaryote, has also been identified and was used to root the tree (Fig. 1; Supplementary Fig. S1, Supplementary Table S3 available at JXB online). It can be seen that most of the characterized MAPKKKs from other species including NbMAPKKKγ, NbMAPKKKα, LeMAPKKKα, NbMAPKKKε, SlMAPKKKε, OsNK1, and OsCDR1 were clustered within the MEKK subfamily and the others were clustered into the Raf subfamily (Supplementary Fig. S1 available at JXB online). Moreover, the presence of a smaller set of MAPKKK members in the green alga O. lucimarinus and primitive land plants including P. patens and S. moellendorffii compared with eudicots and monocots indicated that recent genome duplications may have caused expansion of the MAPKKK gene family during the evolution of the angiosperms, which is especially evident within the Raf subfamily (Rao et al., 2010). This is further supported by the fact that the lower plants S. moellendorffii and P. patens have only 38 and 61 MAPKKK genes, respectively (Supplementary Table S3 available at JXB online), which is much fewer than found in higher plants such as Arabidopsis or rice, which have 80 and 75 members, respectively (Rao et al., 2010).
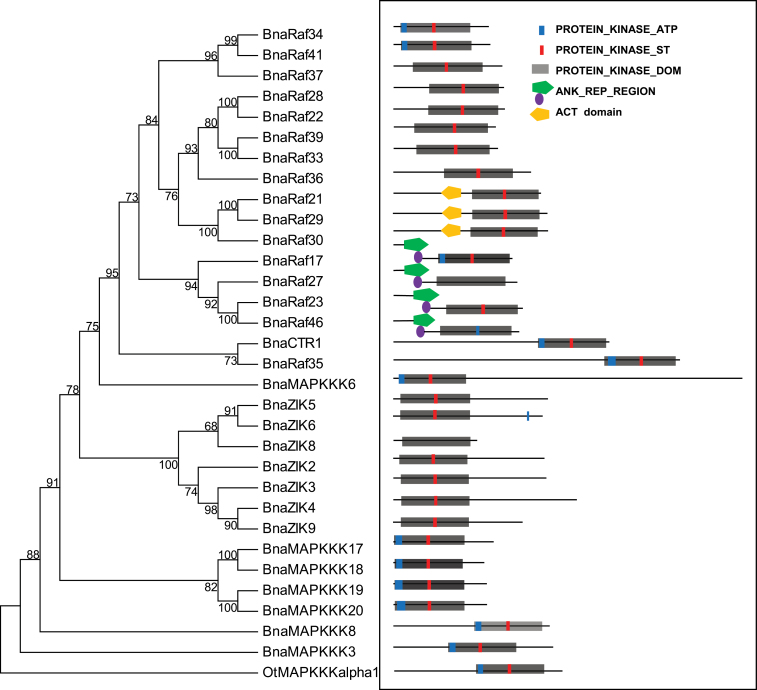
Fig. 1.
Phylogenetic and motif analysis of MAPKKK proteins. (A) Reconstruction of the phylogenetic tree through the maximum parsimony method. The analysis involved 32 amino acid sequences, with BnaMAPKKKs highlighted in green. There are a total of 205 positions in the final data set. The numbers on the nodes are percentages from a bootstrap analysis of 500 replicates. OtMAPKKK alpha1 is used as the outgroup. (B) Schematic diagram of amino acid motifs of BnaMAPKKKs. Motif analysis was determined by using the Prosite program. The subgroup of BnaMAPKKK and BnaRaf has three protein sites. The grey rectangle is the protein kinase domain. The blue rectangular shape indicates the NP_BIND motif, which is the characteristic sequence of IGKGAYGRV, and, of them, ‘K’ is for ATP binding. The ACT_SITE which is ‘D’, is a proton acceptor highlighted in a red rectangular shape. The ANK_REP_REGION is symbolized by a green pentagon and purple circle. The ACT domain is represented by a yellow pentagon.
On the basis of the above phylogenetic analysis, the 31 BnaMAPKKKs could be divided into three major groups, namely groups A(1–4), B(1–4), and C(1–7), which are each supported by significant bootstrap values (Fig. 1; Supplementary Fig. S1 available at JXB online). This is in agreement with the conclusion reached with 60 Arabidopsis MAPKKKs (Ichimura et al., 2002; Jonak et al., 2002). In total, there were four BnaMAPKKKs in the MEKK subfamily, while there were seven in the ZIK subfamily and 17 in the Raf subfamily among the 28 genes cloned (Fig. 1; Supplementary Fig. S1 available at JXB online). It was found that BnaMAPKKK17, -18 -19, and -20 and BnaZIK2, -3, -4, and -8 are in group A, whereas the 17 cloned BnaRaf genes are distributed in the B and C groups. For example, BnaRaf35 and BnaCTR1 are in group B, while BnaRaf17, -21, -22, -23, -27, -28, -29, -30, -33, -34, -36, -37, -39, -41, and -46 belong to group C (Fig. 1). It was also observed that in group A, only A1–A4 have been assigned to AtMAPKKKs, as only 60 AtMAPKKK genes were used to infer the phylogenetic relationship. The remaining 20 novel AtMAPKKK genes, including 11 AtZIK genes (from AtZIK1 to AtZIK11) and nine more AtMAPKKK genes (from AtMAPKKK13 to AtMAPKKK21) were not included in the previous phylogenetic analysis, nor was any specific subgroup assigned. Based on the analysis presented here, it is proposed to assign AtMAPKKK13–AtMAPKKK21 to subgroup A4, since MAPKKKs within each subgroup were clustered together and were supported by high bootstrapping values (Fig. 1; Supplemtenary Fig. S1 available at JXB online). It is noted that orthologous pairs of MAPKKKs between Arabidopsis and canola, and between canola and B. rapa were clustered in the same subclades of the phylogenetic tree, indicating a higher identity/similarity between them (Fig. 1; Supplementary Fig. S1 available at JXB online). Reciprocal Blast analysis and sequence comparison also indicates that the 28 BnaMAPKKK genes cloned are paralogous and are not homeologous genes.
Conserved domains and motifs within BnaMAPKKK proteins were further examined. As reported with Arabidopsis MAPKKK proteins, the characteristic of the MEKK subfamily in canola includes a conserved kinase domain of G(T/S)Px(W/Y/F)MAPEV. The BnaZIK subfamily has GTPEFMAPE(L/V)Y while the BnaRaf-like subfamily has GTxx(W/Y)MAPE (Supplementary Figs S2–S4 available at JXB online) (Jonak et al., 2002). Analysis of the domain architectures of BnaMAPKKKs revealed that most of the 17 BnaRafs have a kinase domain in the C-terminus and a long regulatory domain in the N-terminus. In contrast, eight BnaZIKs have a kinase domain at the N-terminus. As for the four BnaMAPKKKs reported here, the kinase domain is located in the N-terminus (Fig. 1), which is consistent with observations in Arabidopsis and rice (Jouannic et al., 1999; Ichimura et al., 2002; Rao et al., 2010). It is proposed that the long regulatory domain in the N-terminus of the Raf subfamily may function to interact with other proteins and hence to regulate or specify their kinase activity (Jouannic et al., 1999).
A careful examination of orthologous MAPKKK17, -18, -19, and -20 genes between Arabidopsis and canola showed that they had a GxGxxS/AxV motif instead of the typical sequence GxGxxGxV in subdomain I, which was also the ATP active site (Supplementary Fig. S2A, B available at JXB online). These could be assigned to the A4 subgroups as they existed as an independent subclade within the group A (Supplementary Fig. S1 available at JXB online). In addition, the ATP-binding region signature within the kinase domain of BnaMAPKKKs in subgroup A4 was totally different from that of other members in MEKK, Raf, or even the ZIK subfamilies (Supplementary Fig. S2 available at JXB online).
The ZIK or WNK subfamily genes, with 10 and nine members in Arabidopsis and rice, respectively, are reported to regulate flowering time and circadian rhythms in rice and Arabidopsis (Wang et al., 2008; Kumar et al., 2011). However, AtZIK8 was not considered to belong to any of the groups in WNK. The protein kinase ATP-binding region signature of the ZIK subgroup was therefore inspected; this is (L/I)GXG(A/S)(F/V/S)KXVXX(G/A)X(5–18)AVK of Motif 1. The second lysine (K) in domain II was replaced by an asparagine (N) residue, while it is serine (S) in BnaZIK2 (Supplementary Fig. S3A available at JXB online), which was consistent with previous findings in rice and Arabidopsis (Wang et al., 2008; Kumar et al., 2011). In OsWNK2, -3, -7, and -8, R is present in place of K, while in OsWNK4 and -9, S was present in place of K, and G replaces K in OsWNK6 (Kumar et al., 2011). The missing K residue in domain II in the ZIK subgroup, originally responsible for the coordination of ATP in the active centre, was replaced by a K residue in subdomain I (Xu et al., 2000). It was also noted that there is a 60 amino acid region at the C-terminus of BnaZIKs, which is likely to be the autoinhibitory domain (Supplementary Fig. S3A available at JXB online). This is similar to the Arabidopsis orthologues (Wang et al., 2008). Furthermore, from the phylogenetic study of ZIKs, it was noted that they could be clustered into four clades or subgroups, with each containing ZIKs from both monocots and eudicots (Supplementary Fig. S3C available at JXB online), suggesting that about four ZIK ancestral genes existed before the split of monocots and eudicots. As multiple ZIK genes exist in higher plants such as Arabidopsis, rice, and canola, this suggests a relatively recent duplication of ZIK genes (Wang et al., 2008). In addition, it was obvious that AtZIKs and BnaZIKs were always clustered together, suggesting a higher similarity of orthologous pairs between Arabidopsis and canola, the two representative species of the Brassicaceae family. A previous study identified that plant and animal WNKs form completely different groups, indicating divergence from a common ancestor (Wang et al., 2008). ZIK/WNK genes do not exist in either yeast or bacterial genomes but exist in some other unicellular eukaryotic genomes such as oomycetes and diplomonads, indicating the origin of early eukaryotes (Wang et al., 2008). Moreover, the monophyletic clade for eudicots indicates the same ancestor before splitting of the monocots and eudicots, while a polyphyletic group may suggest that more than one gene existed before the splitting of the monocots and eudicots (Wang et al., 2008).
From a multiple alignment of the Raf subfamily of canola MAPKKKs, it was inferred that BnaRafs contain a conserved [RK][IV]GXG[SF][FY]G[TE]VX[KRH][GA]X[WF][HFN]G sequence, which is the signature sequence of subdomain I and also discriminates them from the other MAPKKKs (Supplementary Fig. S4 available at JXB online) (Jouannic et al., 1999). It was noted that an ACT domain is present only in BnaRaf21, -29, and -30, and this domain is reported to be involved in amino acid metabolism or protein–protein interaction (dimerization) (Feller et al., 2006); however, the significance of it in Raf protein function awaits experimental study. The presence of an ACT domain in some Raf kinases was also observed in rice (Rao et al., 2010). It was also identified that BnaRaf23, -27, and -46 have three ankyrin (ANK) repeats in the N-terminus while BnaRaf17 has only one (Supplementary Fig. S4 available at JXB online). The ANK repeat is one of the most common protein–protein interaction motifs and has been found in proteins with diverse functions, such as transcriptional initiators, cell cycle regulators, the cytoskeleton, ion transporters, and signal transducers (Sedgwick and Smerdon, 1999). In Arabidopsis, there are 105 predicted ANK repeat-containing proteins, and most of them are transmembrane proteins (Becerra et al., 2004). Since no interactions were detected between BnaRaf23, -27, -46, and eight BnaMKKs (see below), they may interact with some of the unidentified BnaMKKs or other unknown proteins. Another feature of the Raf proteins in plants is the existence of subdomain VIII, which is GTXX(W/Y)MAPE or [LIM]X[SD]X[ST]X[AK]GTP[EQ]W (Jouannic et al., 1999) (Supplementary Fig. S4 available at JXB online); however, the conservation of amino acid residues within this subdomain is limited, except at a few sites, as shown by the MEME analysis (Supplementary Fig. S4B available at JXB online).
Through the aforementioned phylogenetic analysis, multiple alignments and domain analysis of BnaMAPKKKs in canola, it was concluded that MAPKKK genes are ancestral and conserved from lower to higher plants. The classification and function of members of this important gene family may be rather conserved between monocots and dicots. Compared with other species, such as O. lucimarinus, P. patens, and S. moellendorffii, the increasing number of MAPKKK genes is probably due to evolutionary events such as genome duplication or expansion. Taken together, during the long history of evolution, the MAPK signalling cascades were relatively conserved, though subfunctionalization or neofunctionalization may have resulted in differences in gene function in specific species.
Subcellular localization of canola MAPKKK proteins
To investigate further the function of the 28 BnaMAPKKK genes cloned, the localization of the encoded proteins was first predicted by using PSORT (http://psort.hgc.jp/), CELLO v2.5 (http://cello.life.nctu.edu.tw), and ESLPred (http://www.imtech.res.in/raghava/eslpred/index.html). It was found that most of the BnaMPKKKs were predicted to be localized to the nucleus, cytoplasm, or plasma membrane, with the exception of BnaRaf17, -28, -29, and BnaZIK2, which were also present in the cytoskeleton and chloroplast, respectively (Supplementary Table S5 available at JXB online). For instance, BnaRaf17 was predicted to be localized to chloroplast stroma, microbody (peroxisome), chloroplast thylakoid membrane, and chloroplast thylakoid space, while BnaRaf28 was localized to the endoplasmic reticulum (membrane), plasma membrane, chloroplast thylakoid membrane, and mitochondrial inner membrane. BnaRaf29 was predicted to sit in the cytoplasm, microbody (peroxisome), chloroplast, stroma and chloroplast thylakoid membrane. Transmembrane helices (TMH) of these BnaMAPKKKs were also predicted using TMHMM (http://www.cbs.dtu.dk/services/TMHMM-2.0/); however, no TMH was identified for any of the 28 BnaMAPKKK proteins (data not shown), suggesting that none of these proteins is a transmembrane protein.
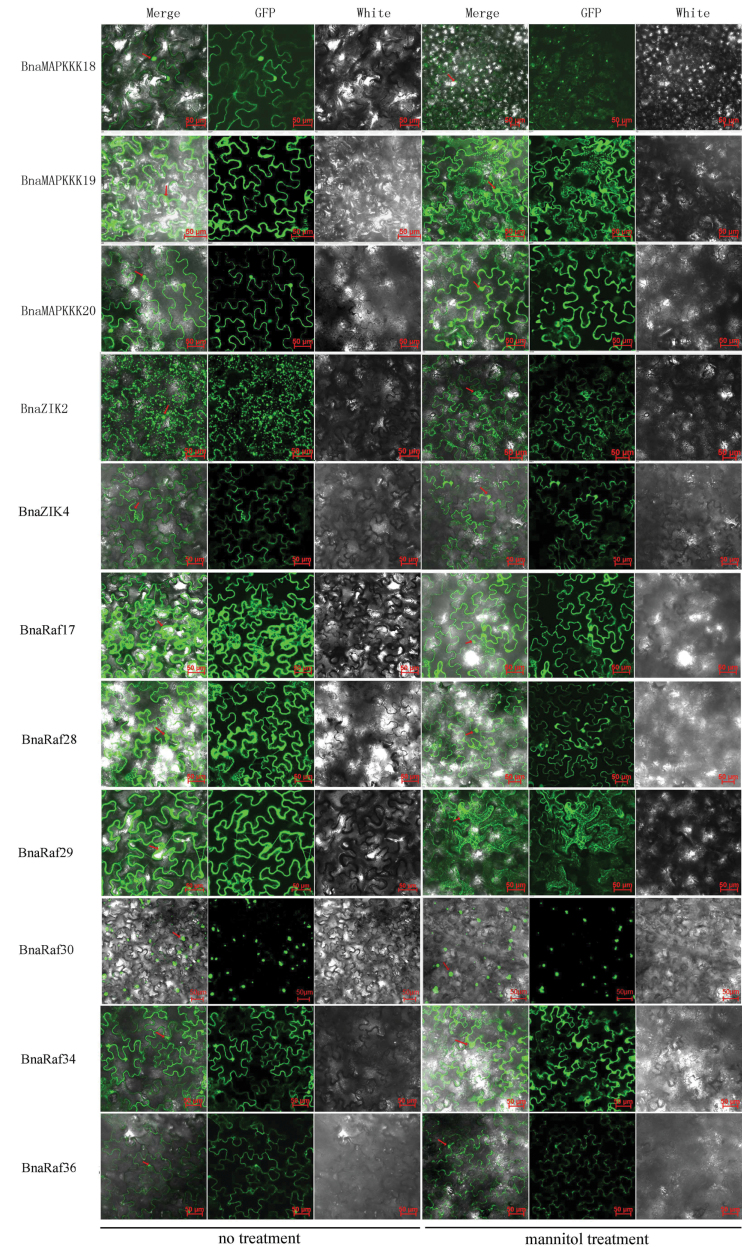
To investigate further the subcellular localization of canola MAPKKK proteins, selected genes representing the three subfamilies of BnaMAPKKKs, namely BnaMAPKKK18, -19, -20, BnaZIK2, and -4, as well as BnaRaf17, -28, -29, -30, -34, and -36 were expressed as fusion proteins with GFP in N. benthamiana. The recombinant plasmids were transformed into agrobacteria and then infiltrated into the lower epidermal leaves of N. benthamiana, with the GFP signals observed 2 d later. It was found that in leaf cells harbouring the fusion proteins of BnaMAPKKK18, -19, and -20, BnaZIK2 and -4, and BnaRaf17, -28, -29, -30, -34, and -36, the GFP signals were present in the cytoplasm and nucleus (Fig. 2). For BnaZIK2, the signals were observed in the chloroplast as well as in the cytoplasm and nucleus, while for BnaRaf30, the GFP signals were only present in nuclei. As a control, the subcellular localization of the GFP protein was also examined in tobacco leaf cells, and green signals were obviously present in both the cytosol and nuclei (data not shown). Leaf discs from the agroinfiltrated plants were further treated with a high osmotic solution (500mM mannitol) for 1h, and the GFP signals were observed again under the same confocal settings. As shown, most of the BnaMAPKKKs tested were still localized in both the cytoplasm and nuclei, except BnaRaf30 (Fig. 2). The in planta demonstration of the subcellular localizations of BnaMAPKKKs is expected to reflect more accurately the natural subcellular localization of these proteins, as compared with the in silico predictions.
Fig. 2.
Subcellular localization of BnaMAPKKK proteins in N. benthamiana cells using green fluorescent protein (GFP). The six panels represented BnaMAPKKK18, -19, -20, and BnaRaf17, -28, and -29, respectively, under normal and mannitol treatment (500mM for 1h). The red arrows indicate the nuclei or dots of cytosol in the focused cells. In each panel, the extreme left is GFP fluorescence, the middle bright field, and the right an overlay of the two images. Scale bar=50 μm.
Identification and validation of BnaMAPKKK and BnaMKK interactions
To explore the function of MAPKKK genes in canola, it is essential to identify their direct MKK targets. Therefore, a Y2H assay was performed to identify the BnaMKKs interacting with each of the 28 BnaMAPKKKs. To this end, the coding regions of 28 BnaMAPKKK and eight BnaMAPKK genes were first cloned into the GAL4-BD bait and GAL4-AD prey vectors, respectively. The autoactivation activity of each of the 28 BnaMAPKKK proteins in yeast was then tested, and it was found that BnaMAPKKK18, BnaZIK3, BnaZIK4, and BnaRaf35 showed evident autoactivation activity (data not shown). To solve this issue, the coding regions of BnaMAPKKK18, BnaZIK3, BnaZIK4, and BnaRaf35 were fused in-frame with the GAL4-AD domain whereas those of the eight BnaMKK genes were fused with the GAL4-BD domain. Thirdly, individual BnaMAPKKK–BnaMKK pairs were co-transformed into yeast cells, with colonies tested on both selective media, followed by titration and β-galactosidase activity assay to examine the strength and genuineness of interactions. As a result, 22 pairs of interactions were identified, and six of the BnaMKK proteins were found to interact with at least one BnaMAPKKK protein in the Y2H assay, while BnaMKK1 and BnaMKK4 did not interact with any of the 28 BnaMAPKKK proteins assayed (Table 2; Supplementary Fig. S5 available at JXB online), suggesting that these two MKK proteins may be transducer signals from other upstream MAPKKK proteins. There were six BnaMAPKKKs, BnaMAPKKK17, -18, and -20, BnaZIK3 and -4, and BnaRaf35, that interacted with BnaMKK3; six BnaMAPKKKs, BnaMAPKKK17, -19, -20, BnaZIK2, -9, and BnaRaf28, with BnaMKK9; three BnaMAPKKKs, BnaZIK2, -9, and BnaRaf28, with BnaMKK5; and five BnaMAPKKKs, BnaMAPKKK17, -19, -20, BnaZIK2, and BnaRaf28 with BnaMKK8. As regards BnaMKK2 and -6, only BnaRaf28 interacted with both of them (Table 2).
Table 2.
Summary of yeast two-hybrid assay of interactions between BnaMAPKKK and BnaMKK proteins
| pGBKT7 pGADT7 |
BnaMKK1 | BnaMKK2 | BnaMKK3 | BnaMKK4 | BnaMKK5 | BnaMKK6 | BnaMKK8 | BnaMKK9 |
|---|---|---|---|---|---|---|---|---|
| BnaMAPKKK17 | – | – | ++BiFC | – | – | – | ++ | +BiFC |
| BnaMAPKKK18 | – | – | ++ | – | – | – | – | – |
| BnaMAPKKK19 | – | – | – | – | – | – | ++ | +BiFC |
| BnaMAPKKK20 | – | – | ++BiFC | – | – | – | ++ | +BiFC |
| BnaZIK2 | – | – | – | – | + BiFC | – | ++ | +BiFC |
| BnaZIK3 | – | – | + | – | – | – | – | – |
| BnaZIK4 | – | – | ++BiFC | – | – | – | – | – |
| BnaZIK5 | – | – | – | – | – | – | – | – |
| BnaZIK6 | – | – | – | – | – | – | – | – |
| BnaZIK8 | – | – | – | – | – | – | – | – |
| BnaZIK9 | – | – | – | – | ++ | – | – | ++ |
| BnaCTR1 | – | – | – | – | – | – | – | – |
| BnaRaf17 | – | – | – | – | – | – | – | – |
| BnaRaf21 | – | – | – | – | – | – | – | – |
| BnaRaf22 | – | – | – | – | – | – | – | – |
| BnaRaf23 | – | – | – | – | – | – | – | – |
| BnaRaf27 | – | – | – | – | – | – | – | – |
| BnaRaf28 | – | ++ BiFC | – | – | + BiFC | + BiFC | ++ | ++ BiFC |
| BnaRaf29 | – | – | – | – | – | – | – | – |
| BnaRaf30 | – | – | – | – | – | – | – | – |
| BnaRaf33 | – | – | – | – | – | – | – | – |
| BnaRaf34 | – | – | – | – | – | – | – | – |
| BnaRaf35 | – | – | ++ | – | – | – | – | – |
| BnaRaf36 | – | – | – | – | – | – | – | – |
| BnaRaf37 | – | – | – | – | – | – | – | – |
| BnaRaf39 | – | – | – | – | – | – | – | – |
| BnaRaf41 | – | – | – | – | – | – | – | – |
| BnaRaf46 | – | – | – | – | – | – | – | – |
The interaction strength was scored visually, from no interaction (–) to strong interaction (++).
BiFC indicates interactions confirmed through bimolecular fluorescence complementation in planta.
Parts of the protein–protein interactions identified in the Y2H assays were confirmed using the BiFC procedure in plant cells. The presence of yellow fluorescence signals showed that BnaMAPKKK17, -19, -20, BnaZIK2, -9, and BnaRaf28 interacted with BnaMKK9; BnaRaf28 with BnaMKK6; BnaZIK2 and BnaRaf28 with BnaMKK5; BnaMAPKKK17, -20, and BnaZIK4 with BnaMKK3; and BnaRaf28 with BnaMKK2 in the epidermal cells of N. benthamiana (Supplementary Fig. S6 available at JXB online). The respective negative controls (target proteins fused to half of YFP co-expressed with the other half of the YFP) did not yield detectable YFP signals (Supplementary Fig. S6 available at JXB online). Taken together, the identification of BnaMAPKKK–BnaMKK pairs provided useful information to dissect their function in canola response to abiotic and biotic stresses.
Since MAPKKKs usually exert their function through phosphorylation of downstream MKK proteins, co-localization of interacting MAPKK and MKK proteins is normally expected for signal transduction. Therefore, the in vivo localizations of pairs of interacting BnaMAPKKKs and BnaMKKs was compared, including Raf28–MKK2, MAPKKK18–MKK3, MAPKKK20–MKK3, and ZIK4–MKK3. The subcellular localization of MKK2 and MKK3 was previously shown to be the cytoplasm and nucleus (Liang et al., 2013). The co-localization of MKK2 and MKK3 with BnaMAPKKKs probably facilitates their interactions in the cytoplasm as well as the nuclei (Fig. 2; Supplementary Fig. S6 available at JXB online). Hence, the co-localization of BnaMAPKKK–BnaMKK interaction pairs facilitates their interaction and phosphorylation to mediate timely responses to external and internal stimuli. However, it should be noted that even if two proteins are identified to interact in a Y2H assay or after co-expression in plants, they may not necessarily do so under natural conditions, as a result of differences in spatiotemporal expression patterns.
Expression analysis of BnaMAPKKK genes in response to stress treatments
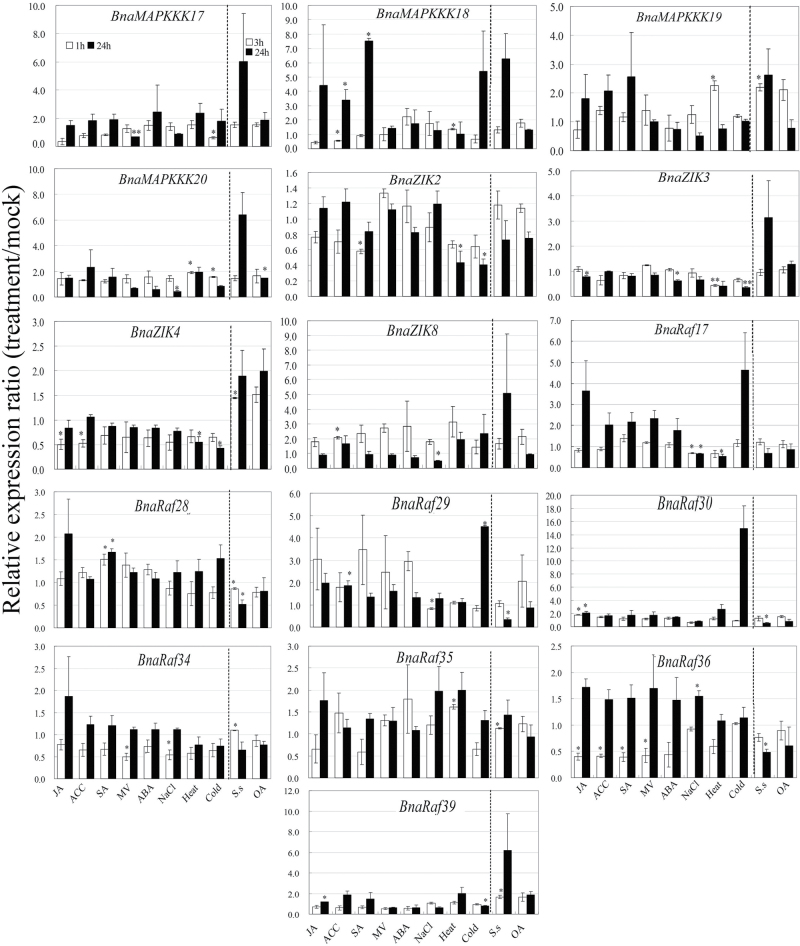
To elucidate of the functions of BnaMAPKKK genes in the context of abiotic and biotic stresses, the expression patterns of 16 selected BnaMAPKKK genes, including four MAPKKK genes, four ZIK genes, and eight Raf genes, were studied using qRT–PCR. Among the stress treatments applied, ABA is a well-known abiotic stress hormone, and JA, ethylene, and SA are well-known defence hormones, while MV can trigger ROS burst in plant cells. OA, on the other hand, is a pathogenicity factor produced by S. sclerotiorum, and can elicit PCD and suppress the oxidative burst of host plants (Cessna et al., 2000; Guimaraes and Stotz, 2004; Kim et al., 2008). Canola seedlings were subjected to moderate stress treatments at two time points to better monitor the transcript changes of the investigated BnaMAPKKK genes. Data of three independent biological replicates were subjected to statistical analysis to identify BnaMAPKKK genes responsive to one or a combination of stress conditions (Fig. 3; Supplementary Fig. S7A available at JXB online). It was found that JA modulated expression of five genes, among which BnaRaf30 was induced while BnaZIK3, BnaZIK4, BnaRaf36, and BnaRaf39 were repressed at one or both time points tested. Five genes, BnaMAPKKK18, BnaZIK3, BnaZIK8, BnaRaf29, and BnaRaf36, were responsive to ACC, among which BnaZIK8 and BnaRaf29 were up-regulated while BnaMAPKKK18, BnaZIK3, and BnaRaf36 were down-regulated. SA significantly induced BnaMAPKKK18 and BnaRaf28 while it repressed BnaZIK2, BnaRaf34, and BnaRaf36 at specific time points. Expression of BnaMAPKKK17, BnaRaf34, and BnaRaf36 was down-regulated by MV treatment at one time point tested. ABA seemed only to repress BnaZIK3 expression and exerted no effect on the transcription of the other genes. On the other hand, salinity induced BnaMAPKKK20 and BnaRaf36 at the 1h and 24h time points, respectively, whereas it down-regulated BnaMAPKKK20 (24h), BnaZIK3, BnaZIK8, and BnaRaf17 (1h and 24h), as well as BnaRaf29 and BnaRaf34 (1h). Seven genes were affected by heat treatment, among which the transcript levels of BnaMAPKKK18, -19, and BnaRaf35 were increased at the 1h time point, while those of BnaZIK2, -3, -4, and BanRaf17 were decreased at 24h or at both time points by heat stress. In addition, moderate cold stress increased the transcript abundance of BnaMAPKKK20 and BnaRaf29 at the 1h time point, but decreased that of BnaMAPKKK17, ZIK2, -3, -4, and BanRaf39 mostly at the 24h time point. Upon challenge by the fungal pathogen S. sclerotiorum, BnaMAPKKK19, ZIK4, Raf34, -35, and -39 were significantly induced at the 3h time point, while BnaRaf28, -29, -30, and -36 were repressed at the 24h time point. Lastly, only BnaMAPKKK20 was induced by OA treatment (Fig. 3; Supplementary Fig. S7A available at JXB online), although to a rather small extent. Taken together, these data indicate that on one hand, some BnaMAPKKK genes participate in transduction of multiple stresses; and, on the other hand, a specific stress activates transcription of more than one BnaMAPKKK gene, providing evidence that BnaMAPKKK plays a role in the cross-talk of multiple stresses, including both abiotic and biotic stresses.
Fig. 3.
Expression analyses of BnaMAPKKK genes in response to various treatments, including 20 μM JA, 1mM ACC, 2mM SA, 10 μM paraquat (MV), 50 μM ABA, 200mM NaCl, heat (37 °C), cold (4 °C), S. sclerotiorum infection, and 5mM oxalic acid (OA). Data are the mean of three biological replicates ±SE. Asterisks denote significant differences (compared with 1) by Student t-test analysis (*P≤0.05; **P≤0.01).
Since public data on Arabidopsis gene expression are available and Arabidopsis is a close relative of canola, comparative analysis of orthologous MAPKKK genes between these two species was performed to examine to what extent the behaviours of AtMAPKKK and BnaMAPKKK genes in response to stress are correlated. To this end, expression profiles of AtMAPKKK genes in response to different hormone and stress treatments were investigated using ATH1 GeneChip data. It should be noted that the stress conditions applied to canola seedlings for the qRT–PCR assay are very similar to those for the Arabidopsis data set. As shown in Supplementay Fig. S7B available at JXB online, AtAMPKKK17 was induced in seedlings 3h after treatment with 10 μM ABA and 6h after heat (38 °C) treatment. AtMAPKKK18 was up-regulated by 10 μM ABA at 1h and 3h after treatment. AtMPKKK19 was induced by B. cinerea, 10 μM MeJA at 30min, 1h, and 3h, and 10 μM SA, while it was repressed after treatment with 150mM NaCl. Botrytis cinerea is a grey mould fungus and it is taxonomically closely related to the white mould fungus S. sclerotiorum (Amselem et al., 2011). Both pathogens are necrotrophic by producing similar virulence factors such as OA, cell wall-degrading enzymes, etc. (Gentile, 1954; Choquer et al., 2007; Kim et al., 2008; Amselem et al., 2011). AtMAPKKK20 was up-regulated by 10 μM MeJA after 1h treatment. It was also identified that both AtMAPKKK17 and -18 are also induced in the root samples after treatment with 150mM NaCl for 30min to 24h. MAPKKK19 orthologues in both canola and Arabidopsis are induced by necrotrophic pathogens (Fig. 3; Supplementary Fig. S7 available at JXB online). However, differences in the expression profiles of BnaMAPKKK and AtMAPKKK after some treatments are also evident (Supplementary Fig. S7 available at JXB online). For instance, AtZIK4 was repressed by SA and heat, whereas it was induced by the biotrophic bacterial pathogen P. syringae. BnaZIK4 was also repressed by JA, heat, cold, and ACC, but was induced by S. sclerotiorum (Fig. 3; Supplementary Fig. S7A available at JXB online). AtZIK8 was induced by B. cinerea, SA, and drought (Supplementary Fig. S7B available at JXB online), whereas BnaZIK8 was induced by ACC only (Fig. 3). AtRaf17 was induced by SA treatment, whereas BnaRaf17 was repressed by NaCl and heat. AtRaf18 was induced by SA while it was repressed by salt treatment (Supplementary Fig. S7 available at JXB online). AtRaf29 was induced by ABA; AtRaf30 was induced by MeJA, SA, and cold, while it was repressed by salt. AtRaf34 was repressed by cold and heat, and AtRaf35 was induced by ABA, heat, and salt treatments (Supplementary Fig. S7B available at JXB online).
In summary, comparison of transcript expression profiles of some presumed orthologous MAPKKK genes in Arabidopsis and canola show similarities in response to abiotic stress, hormone, and pathogen treatments. For instance, Raf28 was induced by SA, Raf30 was induced by JA, and MAPKKK19 was induced by necrotrophic fungi. However, significant differences were also observed between these two species and this may be attributed to evolutionary differences in gene expression that have occurred, and also experimental differences, especially sampling time.
Characterization of BnaMAPKKK gene functions in cell death
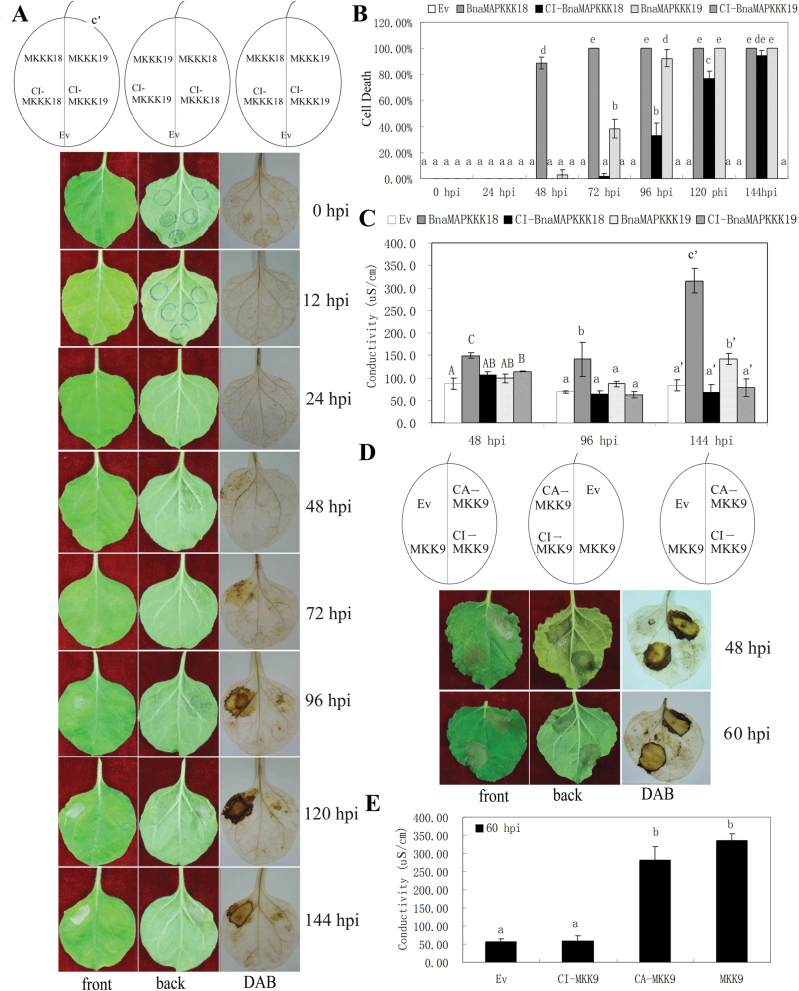
In the above Y2H and qRT–PCR studies, certain BnaMAPKKK genes were identified whose encoded proteins interacted with specific BnaMKKs, and that also increased in transcript abundance in response to multiple stress conditions. To explore their functions further, it was selected to express multiple genes transiently, including BnaMAPKKK18, -19, and BnaMKK9 as well as their constitutively inactive (CI) mutant forms individually under the CaMV 35S promoter through agroinfiltration into N. benthamiana leaves. The inactive form of the MAPKKKs or MKKs was achieved by replacing the ATP-binding site (lysine, K) with an arginine (R) or with a methionine (M) (Melech-Bonfil and Sessa, 2010; Hashimoto et al., 2012). Interestingly, ectopic expression of either BnaMAPKKK18 or -19 caused pathogen-independent cell death compared with the CI version of their respective genes or empty vector control, beginning 48h or 72h post-infiltration (hpi), and this lasted till 144 hpi (Fig. 4A). It was also observed that the symptom of a water-soaked area became apparent as early as 24 hpi (Fig. 4A). To explore the role of hydrogen peroxide (H2O2) during cell death, DAB staining was performed and the results showed that there was strong staining in sites expressing only BnaMAPKKK18 or -19 beginning at 48 hpi and continued till 144 hpi, but not in any control sites (Fig. 4A, B). Nitroblue tetrazolium (NBT) staining of superoxide showed similar changes (data not shown). Moreover, the electrolyte leakage of leaf discs taken from leaves expressing BnaMAPKKK18, BnaMAPKKK18 K32G, BnaMAPKKK19, BnaMAPKKK19 K37G, and the empty vector was examined. The results showed that a significant increase in ion leakage was visible 3 d after agroinfiltration of BnaMAPKKK18 or -19, in contrast to that of leaves expressing BnaMAPKKK18 K32R or the control plasmid (Fig. 4C), which further demonstrates that the hypersensitive response (HR)-like cell death associated with hydrogen peroxide production is triggered by high expression of BnaMAPKKK18 and -19. From the phylogenetic analysis, BnaMAPKKK18 and -19 were classified into subgroup A4 (Supplementary Fig. S1 available at JXB online). Interestingly, a literature search and phylogenetic analysis identified that SlMAPKKKε and NbMAPKKKε in the A4 group positively regulate cell death signalling in plant immunity (Supplementary Fig. S1 available at JXB online) (Melech-Bonfil and Sessa, 2010; Hashimoto et al., 2012). Epistasis experiments showed that SlMAPKKKε-mediated cell death is negatively regulated by SIPKK (Melech-Bonfil and Sessa, 2010). The results therefore implied that BnaMAPKKK18 and -19 are two novel kinases mediating cell death in plants.
Fig. 4.
Overexpression of BnaMAPKKK18, -19, and MKK9 induced pathogen-independent cell death in N. benthamiana leaves. Leaves were infiltrated with agrobacteria carrying wild-type genes or mutated versions. All experiments were performed three times with similar results obtained. (A) Symptoms of N. benthamiana leaf areas expressing BnaMAPKKK18 and BnaMAPKKK19 genes and their mutated forms 24, 48, 72, 96, 120 and 144h post-infiltration (hpi). The left, middle, and right panels represent the front and back, and DAB staining, respectively. (B) Quantification of cell death in N. benthamiana leaves expressing BnaMAPKKK18 or BnaMAPKKK19 and their mutated derivates by examining the percentages of leaf sites with water-soaked symptoms at various time points. (C) Measurement of electrolyte leakage in leaf discs expressing BnaMAPKKK18 or BnaMAPKKK19 and their mutated derivates at 48, 96, and 144 hpi. (D) Symptoms of N. benthamiana leaf areas expressing BnaMKK9 or its mutated versions at 48 and 60 hpi. (E) Measurement of electrolyte leakage in leaf discs transiently expressing BnaMKK9 and its mutated derivates at 60 hpi. Values represent the means of three independent assays for each time point ±SE. Identical and different letters represent non-significant and significant differences (P≤0.05), respectively.
From our Y2H results, BnaMAPKKK18 interacted with BnaMKK3. However, expression of BnaMKK3 in N. benthamiana did not induce significant cell death (data not shown). Though the function of BnaMKK3 has not been identified yet, its orthologue AtMKK3 is known to regulate the JA signal transduction pathway together with AtMPK6 (Takahashi et al., 2007) and also plays an important role in plant immune and stress responses possibly through interacting with group C MPKs, including MPK1, -2, -7, and -14, and the downstream pathogenesis-related 1 (PR1) (Doczi et al., 2007).
In the above Y2H assay, it was found that BnaMAPKKK19 interacted with BnaMKK9. Interestingly, it was also found that BnaMKK9 also induced cell death when transiently expressed in N. benthamiana leaves (Fig. 4D). Interestingly, expression of the constitutively active BnaMKK9 (CA-MKK9) also induced strong cell death, whereas the CI BnaMKK9 (CI-MKK9) or empty plasmid did not (Fig. 4D). DAB and NBT staining demonstrated that there was evident accumulation of ROS at sites expressing BnaMKK9 or its constitutive form, but not in the sites expressing the inactive form or empty vector (Fig. 4D). To quantity further the extent of cell death, electrolyte leakages of leaf discs taken from leaves expressing the wild-type or mutant form of the BnaMKK9 gene and the empty vector were monitored. The results showed that a significant increase in ion leakage was visible 3 d after agroinfiltration of canola MKK9 or CA-MKK9 in contrast to that of leaves expressing CI-MKK9 or the control plasmid (Fig. 4E). A literature search indicated that Arabidopsis MKK9 can similarly accelerate cell death in N. benthamiana through Sgt1, a known regulator of cell death (Popescu et al., 2009), which suggests that orthologous MKK9 between canola and Arabidopsis may have a conserved function.
To understand further the role of the MAPKKK–MKK signaling cascade in cell death and ROS accumulation, co-overexpression analysis of BnaMAPKKK19 and BnaMKK9 in N. benthamiana was performed, considering the fact that BnaMAPKKK19 and BnaMKK9 interacted in both Y2H and BiFC. As shown in Supplementary Fig. S8 available at JXB online, BnaMKK9 alone is already active, and co-expression of MKK9 and MAPKKK19 does not have a significant additive or synergistic effect, as shown from statistical analysis of the cell death index and electrolyte leakage (Supplementary Fig. S8B, C available at JXB online). Meanwhile, cell death induced by expression of BnaMKK9 was not significantly influenced in the absence of upstream BnaMAPKKK19, neither was BnaMAPKKK19 in the absence of downstream BnaMKK9. It would be interesting to identify the substrates of MAPKKK19 and MAPKKK18 in N. benthamiana, and to explore the relationship between these two MAPKKKs and ROS-generating enzymes Rbohs (respiratory burst oxidase homologues) (Torres and Dangl, 2005). Moreover, whether the other cloned BnaMAPKKK and BnaMKK genes could regulate cell death and ROS signalling is still under investigation in our lab.
A complete module, which includes BnaMKK9–BnaMPK1/2–BnaWRKY53 was previously identified (Liang et al., 2013), which links the BnaMKK9–MPK1/2 cascade to downstream WRKY transcription factor(s), and they may together regulate cell death and/or leaf senescence, since AtWRKY53 was identified to regulate leaf senescence (Miao et al., 2007). Whether BnaMAPKKK19 mediates cell death through MKK9–BnaMAPK1/2–BnaWRKY53 and how the ethylene signalling pathway is integrated by this module need to be further elucidated. Besides BnaMAPKKK19, four more BnaMAPKKKs, namely BnaMAPKKK17, -20, BnaZIK2, and BnaRaf28, were also identified to interact with BnaMKK9 (Table 2; Supplementary Figs S5, S6 available at JXB online); however, none of these BnaMAPKKKs was shown to elicit cell death (data not shown), indicating that they are possibly involved in other biological processes.
The activation of AtMKK5 can lead to HR-like cell death through ethylene signalling perception (Liu et al., 2008). Arabidopsis MKK4 and MKK5 belong to group C MKKs with D sites K/R-K/R-X(1–5)-L/I-X-L/I at the N-termini (Bardwell et al., 2001; Grewal et al., 2006). More recently, it is reported that the D site in SlMKK2 of S. lycopersicum is critical for interacting with SlMPK3 and triggering PCD (Oh et al., 2013). Though Arabidopsis YODA–MKK4/MKK5–MPK3/MPK6 modules regulate stomatal development and patterning (Wang et al., 2007), the direct upstream component(s) of AtMKK5 has not been reported yet. In the Y2H assay, two BnaMAPKKKs, namely BnaZIK2 and BnaRaf28, interacted with BnaMKK5 (Table 2; Supplementary Fig. S5 available at JXB online), and the interactions were confirmed through BiFC (Supplementary Fig. S6 available at JXB online). However, no cell death induced by BnaZIK2 and BnaRaf28 overexpression in N. benthamiana was observed (data not shown). Whether there are other canola MAPKKK genes mediating cell death or ROS signalling awaits further investigation. Also, whether and how BnaMAPKKK18 and -19 modulate plant immunity against fungal pathogens such as S. sclerotiorum needs to be experimentally determined.
Conclusion
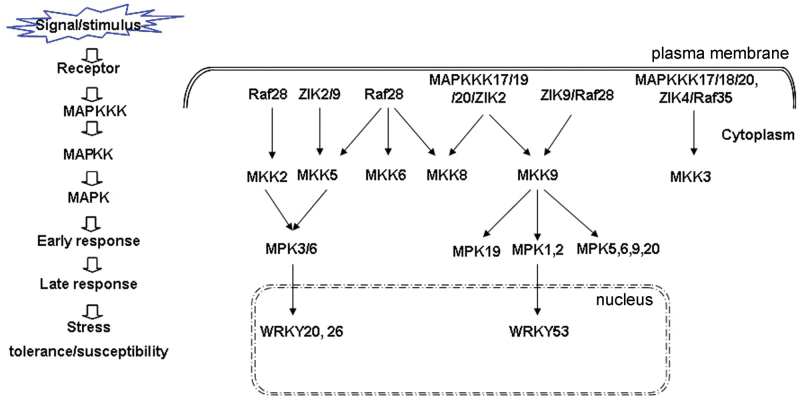
MAPK signalling pathways are very important in plant development, abiotic stress, and defence responses. So far, the function of only a few MAPKKK genes in Arabidopsis, rice, tomato, and tobacco have been reported (Rodriguez et al., 2010; Meng and Zhang, 2013). In the present study, the identification of the MAPKKK gene family in the important oilseed crop, canola, was described. A total of 28 novel MAPKKK genes in canola were cloned and characterized, and two novel MAPKKK genes, BnaMAPKKK18 and -19, mediating cell death were identified. A few complete MAPK modules were identified through linking MAPKKKs to recently identified downstream components (Liang et al., 2013). For example, BnaZIK2/BnaRaf28–BnaMKK5–BnaMPK3/6–BnaWRKY20/26, BnaRaf28–BnaMKK2/5–BnaMPK3/6–BnaWRKY20/26, and BnaMAPKKK17/19/20/ZIK2/BnaRaf28–BnaMKK9–BnaMPK5/-9/-19/-20 (Fig. 5). Many of these have not been reported even in Arabidopsis, indicating that different combinations of BnaMAPKKK–BnaMKK–BnaMPKs are very likely to be involved in the responses to different external and internal stimuli in canola. In the present and previous studies, it was found that there are differences in expression patterns of orthologous MAPKKK, MKK, and MPK genes in response to different stimuli between Arabidopsis and canola, and it was also identified that even the interaction pairs are not conserved between these two species, which highlights the limitations of applying conclusions from the model species Arabidopsis to canola (Liang et al., 2013).
Fig. 5.
A proposed model summarizing MAPK cascades in canola. Dotted lines indicate the possibility of the MAPK cascades acting either in a parallel or a sequentially co-dependent manner. Solid lines indicate confirmed interactions and dashed lines with arrows mean possible regulation at the transcriptional level.
In a previous study, canola MAPKKK17, MAPKKK18, as well as MPK3, MPK4, MPK6, and MPK17 were identified as being responsive to S. sclerotiorum infection (Yang et al., 2007). Seven MKK and 12 MPK genes from canola were recently identified, and their function was analysed in the context of abiotic and biotic stresses (Liang et al., 2013). Here the identification and characterization of their upstream regulators, MAPKKKs, were described. Although the present 28 BnaMAPKKK genes comprise only 44% of the 66 identified genes for canola, 15 BnaMAPKKK–BnaMKK interaction pairs were identified, which provide clues to dissect the function of individual BnaMAPKKKs in canola. For instance, BnaRaf28 could interact with BnaMKK2, -5, -6, -8, and -9, while six different BnaMAPKKKs could interact with BnaMKK9, and six BnaMAPKKKs interacted with BnaMKK3 (Table 2; Fig. 5). It is proposed that the different combinations of BnaMKKK–BnaMKK pairs may fulfil their function in response to diverse abiotic stress signalling pathways and plant immune responses. However, the possibility cannot be excluded that even if two proteins are found to interact in Y2H assay or after co-expression in N. benthamiana, they may not necessarily do that in the natural situation. Moreover, it was identified that two novel MAPKKK genes, BnaMAPKKK18 and -19, could induce cell death when transiently expressed in leaves of N. benthamiana, possibly through accumulation of ROS. Moreover, BnaMKK9 and its constitutively active form also induced strong cell death and H2O2 accumulation. However, a more complete picture of MAPKKK genes and the related signalling cascade in canola will not be available before the remaining MAPKKK genes are cloned from canola and characterized. Overall, the present study of MAPKKK genes in canola lays the foundation for further exploration of their roles in abiotic stress signalling and in plant immunity against S. sclerotiorum. It also provides important information for genetially manipulating the abundance and/or activity of related MAPKKKs and their components in the MAPK pathway to improve stress tolerance of canola.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Phylogenetic analysis of MAPKKKs from representative species.
Figure S2. Multiple alignment of MEKK subfamily MAPKKK proteins in representative species.
Figure S3. Multiple alignment of ZIK subfamily MAPKKK proteins in representative species.
Figure S4. Multiple alignment of Raf subfamily MAPKKK proteins in representative species.
Figure S5. Yeast two-hybrid (Y2H) assay of interactions between MAPKKK and MKK proteins in canola.
Figure S6. Analysis of BnaMAPKKK and BnaMKK interactions in N. benthamiana through bimolecular fluorescence complementation (BiFC).
Figure S7. The heat maps of the expression profiles of MAPKKK genes of canola and Arabidopsis in responses to abiotic and biotic stresses.
Figure S8. Co-expression analysis of BnaMAPKKK19 and MKK9 in eliciting pathogen-independent cell death in N. benthamiana leaves.
Table S1. BnaMAPKKK EST summary.
Table S2. Primers used in this study.
Table S3. MAPKKK sequences from different species used for phylogenetic analysis.
Table S4. Similarity and identity analysis of MAPKKK genes/proteins between Arabidopsis, rice, and canola.
Table S5. Computational prediction of subcellular localizations of MAPKKK proteins in canola.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (no. 31301648 to BY and 31270293 to Y-QJ) and the Chinese Ministry of Education Program for New Teachers in Universities (no. 20110204120005 to BY). We would like to acknowledge Professor Lili Huang (Northwest A&F University) for providing the Sclerotinia scleortiorum strain SX09-904, and Professor Jörg Kudla (Universität Münster, Germany) for the BiFC vectors. We would also thank the anonymous reviewers for constructive advice on improving the manuscript.
References
- Amselem J, Cuomo CA, van Kan JA, et al. 2011. Genomic analysis of the necrotrophic fungal pathogens Sclerotinia sclerotiorum and Botrytis cinerea . PLoS Genetics 7, e1002230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreasson E, Jenkins T, Brodersen P, et al. 2005. The MAP kinase substrate MKS1 is a regulator of plant defense responses. EMBO Journal 24, 2579–2589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai T, Tena G, Plotnikova J, Willmann MR, Chiu WL, Gomez-Gomez L, Boller T, Ausubel FM, Sheen J. 2002. MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415, 977–983 [DOI] [PubMed] [Google Scholar]
- Bardwell AJ, Flatauer LJ, Matsukuma K, Thorner J, Bardwell L. 2001. A conserved docking site in MEKs mediates high-affinity binding to MAP kinases and cooperates with a scaffold protein to enhance signal transmission. Journal of Biological Chemistry 276, 10374–10386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becerra C, Jahrmann T, Puigdomenech P, Vicient CM. 2004. Ankyrin repeat-containing proteins in Arabidopsis: characterization of a novel and abundant group of genes coding ankyrin-transmembrane proteins. Gene 340, 111–121 [DOI] [PubMed] [Google Scholar]
- Campanella JJ, Bitincka L, Smalley J. 2003. MatGAT: an application that generates similarity/identity matrices using protein or DNA sequences. BMC Bioinformatics 4, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cessna SG, Sears VE, Dickman MB, Low PS. 2000. Oxalic acid, a pathogenicity factor for Sclerotinia sclerotiorum, suppresses the oxidative burst of the host plant. The Plant Cell 12, 2191–2200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choquer M, Fournier E, Kunz C, Levis C, Pradier JM, Simon A, Viaud M. 2007. Botrytis cinerea virulence factors: new insights into a necrotrophic and polyphageous pathogen. FEMS Microbiology Letters 277, 1–10 [DOI] [PubMed] [Google Scholar]
- Clark KL, Larsen PB, Wang X, Chang C. 1998. Association of the Arabidopsis CTR1 Raf-like kinase with the ETR1 and ERS ethylene receptors. Proceedings of the National Academy of Sciences, USA 95, 5401–5406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colcombet J, Hirt H. 2008. Arabidopsis MAPKs: a complex signalling network involved in multiple biological processes. Biochemical Journal 413, 217–226 [DOI] [PubMed] [Google Scholar]
- Daudi A, Cheng Z, O’Brien JA, Mammarella N, Khan S, Ausubel FM, Bolwell GP. 2012. The apoplastic oxidative burst peroxidase in Arabidopsis is a major component of pattern-triggered immunity. The Plant Cell 24, 275–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Pozo O, Pedley KF, Martin GB. 2004. MAPKKKalpha is a positive regulator of cell death associated with both plant immunity and disease. EMBO Journal 23, 3072–3082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doczi R, Brader G, Pettko-Szandtner A, Rajh I, Djamei A, Pitzschke A, Teige M, Hirt H. 2007. The Arabidopsis mitogen-activated protein kinase kinase MKK3 is upstream of group C mitogen-activated protein kinases and participates in pathogen signaling. The Plant Cell 19, 3266–3279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feller A, Hernandez JM, Grotewold E. 2006. An ACT-like domain participates in the dimerization of several plant basic-helix–loop–helix transcription factors. Journal of Biological Chemistry 281, 28964–28974 [DOI] [PubMed] [Google Scholar]
- Frye CA, Tang D, Innes RW. 2001. Negative regulation of defense responses in plants by a conserved MAPKK kinase. Proceedings of the National Academy of Sciences, USA 98, 373–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galletti R, Ferrari S, De Lorenzo G. 2011. Arabidopsis MPK3 and MPK6 play different roles in basal and oligogalacturonide- or flagellin-induced resistance against Botrytis cinerea. Plant Physiology 157, 804–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentile AC. 1954. Carbohydrate metabolism and oxalic acid synthesis by Botrytis cinerea. Plant Physiology 29, 257–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal S, Molina DM, Bardwell L. 2006. Mitogen-activated protein kinase (MAPK)-docking sites in MAPK kinases function as tethers that are crucial for MAPK regulation in vivo . Cell Signaling 18, 123–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guimaraes RL, Stotz HU. 2004. Oxalate production by Sclerotinia sclerotiorum deregulates guard cells during infection. Plant Physiology 136, 3703–3711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamel LP, Nicole MC, Sritubtim S, et al. 2006. Ancient signals: comparative genomics of plant MAPK and MAPKK gene families. Trends in Plant Science 11, 192–198 [DOI] [PubMed] [Google Scholar]
- Hashimoto M, Komatsu K, Maejima K, Okano Y, Shiraishi T, Ishikawa K, Takinami Y, Yamaji Y, Namba S. 2012. Identification of three MAPKKKs forming a linear signaling pathway leading to programmed cell death in Nicotiana benthamiana . BMC Plant Biology 12, 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller J, Tudzynski P. 2011. Reactive oxygen species in phytopathogenic fungi: signaling, development, and disease. Annual Review of Phytopathology 49, 369–390 [DOI] [PubMed] [Google Scholar]
- Ichimura K, Casais C, Peck S, Shinozaki K, Shirasu K. 2006. MEKK1 is required for MPK4 activation and regulates tissue-specific and temperature-dependent cell death in Arabidopsis. Journal of Biological Chemistry 281, 36969–36976 [DOI] [PubMed] [Google Scholar]
- Ichimura K, Shinozaki K, Tena G, et al. 2002. Mitogen-activated protein kinase cascades in plants: a new nomenclature. Trends in Plant Science 7, 301–308 [DOI] [PubMed] [Google Scholar]
- Jaspers P, Kangasjarvi J. 2010. Reactive oxygen species in abiotic stress signaling. Physiologia Plantarum 138, 405–413 [DOI] [PubMed] [Google Scholar]
- Jin H, Axtell MJ, Dahlbeck D, Ekwenna O, Zhang S, Staskawicz B, Baker B. 2002. NPK1, an MEKK1-like mitogen-activated protein kinase kinase kinase, regulates innate immunity and development in plants. Developmeental Cell 3, 291–297 [DOI] [PubMed] [Google Scholar]
- Jonak C, Okresz L, Bogre L, Hirt H. 2002. Complexity, cross talk and integration of plant MAP kinase signalling. Current Opinion in Plant Biology 5, 415–424 [DOI] [PubMed] [Google Scholar]
- Jouannic S, Hamal A, Leprince AS, Tregear JW, Kreis M, Henry Y. 1999. Plant MAP kinase kinase kinases structure, classification and evolution. Gene 233, 1–11 [DOI] [PubMed] [Google Scholar]
- Kieber JJ, Rothenberg M, Roman G, Feldmann KA, Ecker JR. 1993. CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the raf family of protein kinases. Cell 72, 427–441 [DOI] [PubMed] [Google Scholar]
- Kim KS, Min JY, Dickman MB. 2008. Oxalic acid is an elicitor of plant programmed cell death during Sclerotinia sclerotiorum disease development. Molecular Plant-Microbe Interactions 21, 605–612 [DOI] [PubMed] [Google Scholar]
- Kong Q, Qu N, Gao M, et al. 2012. The MEKK1–MKK1/MKK2–MPK4 kinase cascade negatively regulates immunity mediated by a mitogen-activated protein kinase kinase kinase in Arabidopsis. The Plant Cell 24, 2225–2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong X, Lv W, Zhang D, Jiang S, Zhang S, Li D. 2013. Genome-wide identification and analysis of expression profiles of maize mitogen-activated protein kinase kinase kinase. PLoS One 8, e57714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar K, Rao KP, Biswas DK, Sinha AK. 2011. Rice WNK1 is regulated by abiotic stress and involved in internal circadian rhythm. Plant Signaling and Behavior 6, 316–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanier W, Moustafa A, Bhattacharya D, Comeron JM. 2008. EST analysis of Ostreococcus lucimarinus, the most compact eukaryotic genome, shows an excess of introns in highly expressed genes. PLoS One 3, e2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Wilkinson MF. 1997. Site-directed mutagenesis: a two-step method using PCR and DpnI. Biotechniques 23, 588–590 [DOI] [PubMed] [Google Scholar]
- Liang W, Yang B, Yu B, et al. 2013. Identification and analysis of MKK and MPK gene families in canola (Brassica napus L.). BMC Genomics 14, 392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Wang Y, Xu J, Su T, Liu G, Ren D. 2008. Ethylene signaling is required for the acceleration of cell death induced by the activation of AtMEK5 in Arabidopsis. Cell Research 18, 422–432 [DOI] [PubMed] [Google Scholar]
- Liu Y, Schiff M, Dinesh-Kumar SP. 2004. Involvement of MEK1 MAPKK, NTF6 MAPK, WRKY/MYB transcription factors, COI1 and CTR1 in N-mediated resistance to tobacco mosaic virus. The Plant Journal 38, 800–809 [DOI] [PubMed] [Google Scholar]
- Lysak MA, Cheung K, Kitschke M, Bures P. 2007. Ancestral chromosomal blocks are triplicated in Brassiceae species with varying chromosome number and genome size. Plant Physiology 145, 402–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lysak MA, Koch MA, Pecinka A, Schubert I. 2005. Chromosome triplication found across the tribe Brassiceae. Genome Research 15, 516–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao G, Meng X, Liu Y, Zheng Z, Chen Z, Zhang S. 2011. Phosphorylation of a WRKY transcription factor by two pathogen-responsive MAPKs drives phytoalexin biosynthesis in Arabidopsis. The Plant Cell 23, 1639–1653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melech-Bonfil S, Sessa G. 2010. Tomato MAPKKKepsilon is a positive regulator of cell-death signaling networks associated with plant immunity. The Plant Journal 64, 379–391 [DOI] [PubMed] [Google Scholar]
- Meng X, Zhang S. 2013. MAPK cascades in plant disease resistance signaling. Annual Review of Phytopathology 51, 245–266 [DOI] [PubMed] [Google Scholar]
- Miao Y, Laun TM, Smykowski A, Zentgraf U. 2007. Arabidopsis MEKK1 can take a short cut: it can directly interact with senescence-related WRKY53 transcription factor on the protein level and can bind to its promoter. Plant Molecular Biology 65, 63–76 [DOI] [PubMed] [Google Scholar]
- Nakagami H, Kiegerl S, Hirt H. 2004. OMTK1, a novel MAPKKK, channels oxidative stress signaling through direct MAPK interaction. Journal of Biological Chemistry 279, 26959–26966 [DOI] [PubMed] [Google Scholar]
- Ning J, Li X, Hicks L, Xiong L. 2010. A Raf-like MAPKKK gene DSM1 mediates drought resistance through reactive oxygen species scavenging in rice. Plant Physiology 152, 876–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh CS, Hwang J, Choi MS, Kang BC, Martin GB. 2013. Two leucines in the N-terminal MAPK-docking site of tomato SlMKK2 are critical for interaction with a downstream MAPK to elicit programmed cell death associated with plant immunity. FEBS Letters 587, 1460–1465 [DOI] [PubMed] [Google Scholar]
- Oh CS, Martin GB. 2011. Tomato 14-3-3 protein TFT7 interacts with a MAP kinase kinase to regulate immunity-associated programmed cell death mediated by diverse disease resistance proteins. Journal of Biological Chemistry 286, 14129–14136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh CS, Pedley KF, Martin GB. 2010. Tomato 14-3-3 protein 7 positively regulates immunity-associated programmed cell death by enhancing protein abundance and signaling ability of MAPKKK α. The Plant Cell 22, 260–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborn TC. 2004. The contribution of polyploidy to variation in Brassica species. Physiologia Plantarum 121, 531–536 [Google Scholar]
- Parkin IA, Gulden SM, Sharpe AG, Lukens L, Trick M, Osborn TC, Lydiate DJ. 2005. Segmental structure of the Brassica napus genome based on comparative analysis with Arabidopsis thaliana. Genetics 171, 765–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen K, Qiu JL, Lutje J, Fiil BK, Hansen S, Mundy J, Petersen M. 2010. Arabidopsis MKS1 is involved in basal immunity and requires an intact N-terminal domain for proper function. PLoS One 5, e14364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen M, Brodersen P, Naested H, et al. 2000. Arabidopsis MAP kinase 4 negatively regulates systemic acquired resistance. Cell 103, 1111–1120 [DOI] [PubMed] [Google Scholar]
- Popescu S, Popescu G, Bachan S, Zhang Z, Gerstein M, Snyder M, Dinesh-Kumar S. 2009. MAPK target networks in Arabidopsis thaliana revealed using functional protein microarrays. Genes and Development 23, 80–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu JL, Fiil BK, Petersen K, et al. 2008. Arabidopsis MAP kinase 4 regulates gene expression through transcription factor release in the nucleus. EMBO Journal 27, 2214–2221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rana D, van den Boogaart T, O’Neill CM, Hynes L, Bent E, Macpherson L, Park JY, Lim YP, Bancroft I. 2004. Conservation of the microstructure of genome segments in Brassica napus and its diploid relatives. The Plant Journal 40, 725–733 [DOI] [PubMed] [Google Scholar]
- Rao KP, Richa T, Kumar K, Raghuram B, Sinha AK. 2010. In silico analysis reveals 75 members of mitogen-activated protein kinase kinase kinase gene family in rice. DNA Research 17, 139–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rietz S, Bernsdorff FE, Cai D. 2012. Members of the germin-like protein family in Brassica napus are candidates for the initiation of an oxidative burst that impedes pathogenesis of Sclerotinia sclerotiorum . Journal of Experimental Botany 63, 5507–5519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez MC, Petersen M, Mundy J. 2010. Mitogen-activated protein kinase signaling in plants. Annual Review of Plant Biology 61, 621–649 [DOI] [PubMed] [Google Scholar]
- Sedgwick SG, Smerdon SJ. 1999. The ankyrin repeat: a diversity of interactions on a common structural framework. Trends in Biochemical Science 24, 311–316 [DOI] [PubMed] [Google Scholar]
- Singh R, Lee MO, Lee JE, et al. 2012. Rice mitogen-activated protein kinase interactome analysis using the yeast two-hybrid system. Plant Physiology 160, 477–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi F, Yoshida R, Ichimura K, Mizoguchi T, Seo S, Yonezawa M, Maruyama K, Yamaguchi-Shinozaki K, Shinozaki K. 2007. The mitogen-activated protein kinase cascade MKK3–MPK6 is an important part of the jasmonate signal transduction pathway in Arabidopsis. The Plant Cell 19, 805–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres MA, Dangl JL. 2005. Functions of the respiratory burst oxidase in biotic interactions, abiotic stress and development. Current Opinion in Plant Biology 8, 397–403 [DOI] [PubMed] [Google Scholar]
- Waadt R, Schmidt LK, Lohse M, Hashimoto K, Bock R, Kudla J. 2008. Multicolor bimolecular fluorescence complementation reveals simultaneous formation of alternative CBL/CIPK complexes in planta. The Plant Journal 56, 505–516 [DOI] [PubMed] [Google Scholar]
- Wang H, Ngwenyama N, Liu Y, Walker JC, Zhang S. 2007. Stomatal development and patterning are regulated by environmentally responsive mitogen-activated protein kinases in Arabidopsis. The Plant Cell 19, 63–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Liu K, Liao H, Zhuang C, Ma H, Yan X. 2008. The plant WNK gene family and regulation of flowering time in Arabidopsis. Plant Biology (Stuttgart) 10, 548–562 [DOI] [PubMed] [Google Scholar]
- Xu B, English JM, Wilsbacher JL, Stippec S, Goldsmith EJ, Cobb MH. 2000. WNK1, a novel mammalian serine/threonine protein kinase lacking the catalytic lysine in subdomain II. Journal of Biological Chemistry 275, 16795–16801 [DOI] [PubMed] [Google Scholar]
- Yang B, Jiang Y, Rahman MH, Deyholos MK, Kav NN. 2009. Identification and expression analysis of WRKY transcription factor genes in canola (Brassica napus L.) in response to fungal pathogens and hormone treatments. BMC Plant Biology 9, 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B, Srivastava S, Deyholos MK, Kav NNV. 2007. Transcriptional profiling of canola (Brassica napus L.) responses to the fungal pathogen Sclerotinia sclerotiorum . Plant Science 173, 156–171 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.