Summary
The transcription factor Foxj1 is expressed by cells destined to differentiate into epithelial cells projecting motile cilia into fluid- or air- filled cavities. Here we report the generation of an inducible knock-in Foxj1CreERT2::GFP mouse which we show reliably induces Cre-mediated recombination for genetic studies in epithelial cells with motile cilia throughout embryonic and postnatal development. Induction during embryonic stages revealed efficient recombination in the epithelial component of the choroid plexus in the developing brain as early as E12.5. Induction during late embryonic stages showed confined recombination not only in the choroid plexus, but also in the ventricular walls of the brain. Recombination induced during postnatal periods expanded to include epithelia of the lungs, testis, oviduct, and brain. Using these mice, we confirmed our recent discovery of a perinatally derived neuronal population in the mouse olfactory bulbs which is derived from the Foxj1 lineage. Our Foxj1CreERT2::GFP knock-in mouse will be a powerful tool for studying molecular mechanisms associated with the continuum of cells that form the Foxj1 lineage, and for assessing their physiological significance during development and aging.
Keywords: Foxj1, CreERT2::GFP, knock-in, ependymal cells, motile cilia, epithelial cells
Results and Discussion
We previously established that the transcription factor forkhead box J1 (Foxj1) is required for the progression of ependymal cell differentiation during perinatal stages (Jacquet et al., 2009). Moreover, Foxj1-dependent differentiation is necessary for maintenance of postnatal neurogenesis in the olfactory bulbs (Jacquet et al., 2011; Paez-Gonzalez et al., 2011). Foxj1 is a master transcriptional regulator of the motile ciliogenic program in the embryonic node and various organs including the oviducts, testis, lungs, and the central nervous system (Blatt et al., 1999; Brody et al., 2000; Chen et al., 1998). As motile cilia in the embryonic node assist in the establishment of left-right asymmetry, germline deletion of Foxj1 can result in situs inversus and loss of asymmetry in the body (Brody et al., 2000; Chen et al., 1998). Moreover, since the ciliogenic program is critical in the context of various ciliopathies (Fliegauf et al., 2007), tools that enhance the study of development and function of motile cilia are important toward better understanding the etiology of various ciliopathic conditions. Here we report the first knock-in Foxj1CreERT2::GFP mouse for studying the Foxj1 lineage throughout embryonic and postnatal development in the central nervous system (CNS) as well as in peripheral organs whose functions depend on motile cilia.
Generation and Characterization of the Foxj1CreERT2::GFP knock-in allele
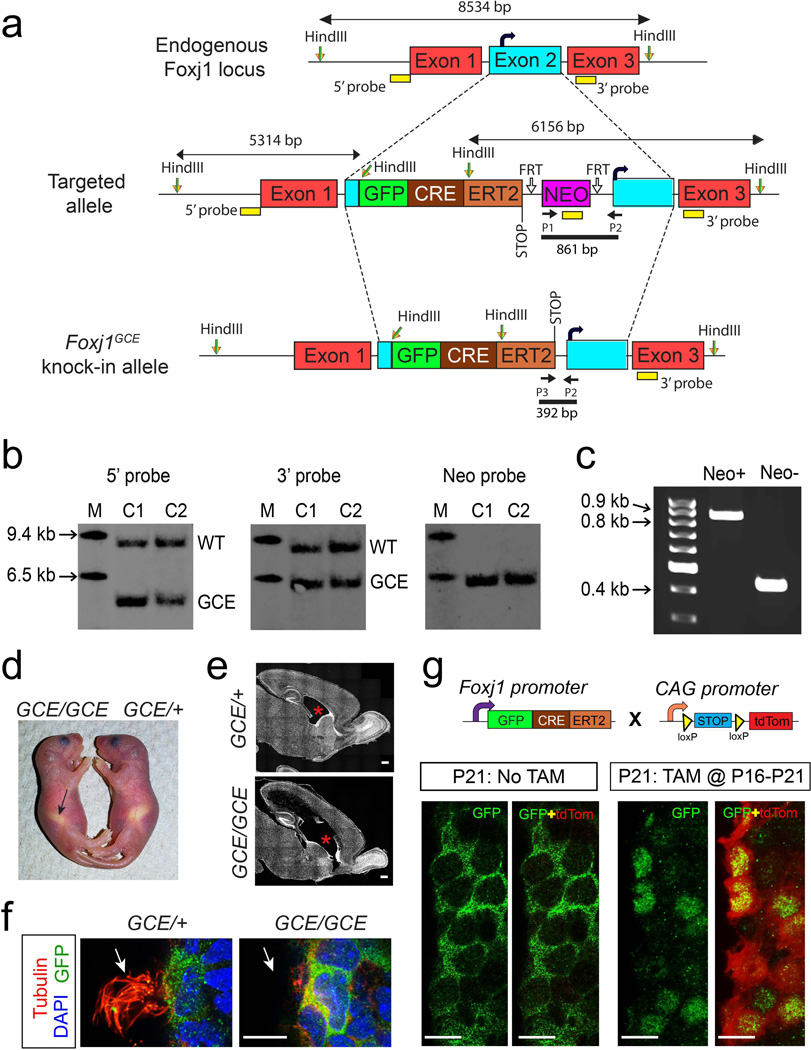
A targeting vector was generated using BAC recombineering to insert the coding regions of GFP fused CreERT2 (GFP::CreERT2) before the endogenous start codon in the mouse Foxj1 Exon2 on chromosome 11 (Fig. 1a). The cassette included a poly A signal to terminate GFP::CreERT2 transcription. The vector was electroporated into mouse embryonic stem (ES) cells and positive clones were injected into mouse blastocysts and Foxj1CreERT2::GFP chimeras were obtained (Fig. 1b). Male chimeras were crossed to wild-type C57BL6/J female mice to obtain Foxj1CreERT2::GFP mice (Fig. 1c).
Figure 1. Generation of Foxj1CreERT2::GFP knock-in mice.
(a) A targeting vector was designed with a GFP::CreERT2 and Neomycin resistance (NEO) cassettes inserted into Exon2 in the Foxj1 locus (curved black arrow denotes endogenous start codon). NEO cassette, flanked by FRT sites, in the targeted allele was excised by crossing F1 progeny with FLP1 mice resulting in the Foxj1CreERT2::GFP knock-in allele. (b) Representative southern blots indicating site-specific insertion of the knock-in cassette in geneticin resistant ES cell clones. Hybridization sites of 5’, 3’, and Neo probes are indicated as yellow rectangles in (a). M – Molecular weight marker; C1 – Clone 1; C2 – Clone 2; WT - wild type band; GCE - Foxj1GCE knock-in allele band. (c) PCR was performed to detect deletion of Neo cassette from the Foxj1CreERT2::GFP knock-in locus after crossing to FLP1 mice. Annealing sites for primers P1, P2, and P3 used in PCR are indicated in (a). (d) Foxj1CreERT2::GFP homozygous (GCE/ GCE) pups display situs inversus where the stomach (arrow) is positioned on the right hand side of the animal. (e) Hydrocephalus indicated by enlargement of the lateral ventricles (red asterisks) was prevalent in GCE/GCE mice. (f) Apical surface of ependymal cells in GCE/GCE mice lacks cilia (red, tubulin staining, arrows). (g) Foxj1+ ependymal cells were genetically labeled by carrying the GCE/+ allele onto a ROSA26CAG-tdTomato reporter background. Tamoxifen (TAM) induced Cre-mediated excision of a stop codon results in constitutive tdTomato (tdTom, red) expression. In the absence of TAM, the GFP::CreERT2 fusion protein is localized to the cytoplasm of ependymal cells resulting in no tdTomato expression. Intraperitoneal injection of TAM at P16 for five consecutive days resulted in translocation of GFP::CreERT2 to the nucleus of ependymal cells at P21 (P21:TAM@P16-P20). Nuclear localization of GFP::CreERT2 overlapped with a high fraction of tdTom+ ependymal cells indicating efficient recombination rate upon TAM exposure. Scale bars: 500µm in (e); 10µm in (f) and (g).
Past studies have shown that Foxj1 is expressed in the embryonic node around embryonic day 7.5 (E7.5) in mice, where it is necessary for development of motile cilia and establishment of left-right asymmetry. We anticipated deletion of functional Foxj1 expression in mice homozygous for targeted insertion of GFP::CreERT2 cassette into the Foxj1 locus (GCE/GCE). However, since the Foxj1 coding sequence on Exon 2 including the endogenous translational START was intact downstream of the knock-in cassette, there was some chance that Foxj1 was transcribed biallelically in homozygous mice. To test this phenotypically, we screened GCE/GCE mice for known Foxj1-null phenotypes (Brody et al., 2000; Jacquet et al., 2009). We indeed found three GCE/GCE mice with situs inversus at birth indicated by reversal of stomach position (Fig. 1d; n=32 mice from 5 litters). In total, development of hydrocephalus was noted in all six GCE/GCE mice that survived up to postnatal day 10 (P10; Fig. 1e) concomitant with absence of cilia on the apical surface of ependymal cells (Fig. 1f; n=32 mice from 5 litters). Thus, the knock-in alleles recapitulate Foxj1-null phenotypes, suggesting disruption in endogenous Foxj1 expression in GCE/GCE mice.
To test the efficiency of our newly generated knock-in allele, we compared the recombination patterns induced in Foxj1CreERT2::GFP mice carrying one knock-in allele (GCE/+) to patterns from a transgenic FOXJ1-Cre line which we previously mapped in the brain (Jacquet et al., 2011). To accomplish this, we crossed the Foxj1CreERT2::GFP mice to ROSA26CAG-tdTomato reporter mice, whereby in resultant mice tamoxifen (TAM) induced nuclear translocation of Cre recombinase mediates the excision of the stop codon flanked by loxP sites for tdTomato (tdTom) expression (Fig. 1g). This recombination results in permanent labeling of Foxj1+ cells as well as any cells derived from putative Foxj1+ progenitors during development (Jacquet et al., 2011). The additional useful element in our new strain is that the GFP fused CreERT2 directly reports on Foxj1 promoter activity. In brain sections from control Foxj1CreERT2::GFP mice at P21 without TAM administration, we found GFP::CreER expression was limited to the cytoplasm of ependymal cells (Fig. 1g). We rarely found tdTom+ cells on the walls of the lateral ventricles (LV) in the absence of TAM (average 0.67 cells per hemisphere; analyzed brain sections every 200µm; n=3 mice at P21), indicating a largely unleaky CreERT2 activity from the Foxj1 locus.
Daily intraperitoneal (i.p.) injections of TAM (once per day for five consecutive days) beginning at P16 resulted in robust recombination within ependymal cells by P21 in Foxj1CreERT2::GFP brains. Moreover, GFP::CreERT2 had clearly translocated to the nucleus of ependymal cells concomitant with tdTom expression in the majority of GFP+ cells (Fig. 1g). These results are in contrast to the only other available FOXJ1:CreERT2 transgenic line (Rawlins et al., 2007) which in our hands and under the same TAM regimen yields recombination in only 23% of ependymal cells (Jacquet et al., 2011). This discrepancy is likely due to either random integration of the CreERT2 cassette due to transgenic targeting, or because the human FOXJ1 promoter was utilized in transgenic mice possibly resulting in non-specific expression. Thus, our newly generated Foxj1CreERT2::GFP mice are efficient for induction of Cre-mediated recombination in ependymal cells.
Foxj1CreERT2::GFP induction during embryonic development
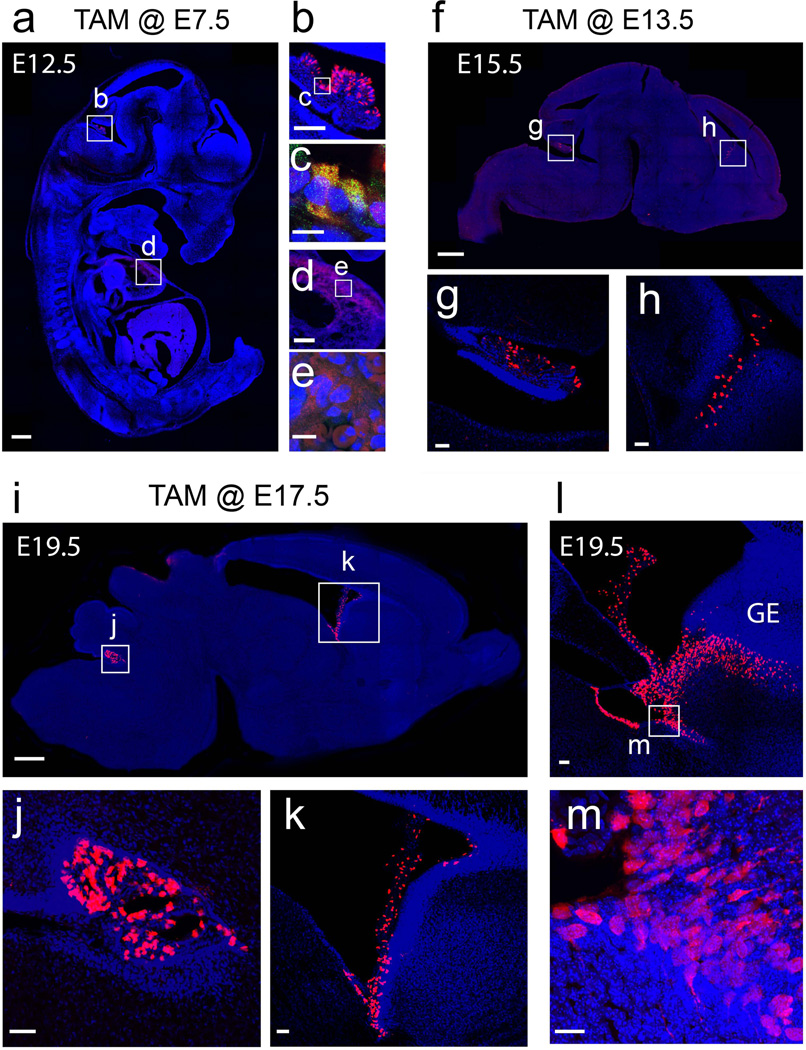
To test the efficiency of recombination during early embryonic development in Foxj1CreERT2::GFP mice, TAM was administered orally to timed-pregnant females harboring embryos co-carrying the ROSA26-CAG-tdTomato reporter allele. Induction in E7.5 embryos and analysis at E12.5 confirmed Foxj1 activity in the choroid plexus (Figs. 2a–c). Low levels of recombination were detected in the dorsal walls of the developing heart (Figs. 2a, 2d, 2e) and throughout the liver (data not shown). However high power imaging revealed that the reporter expression was diffuse in fibroblast-like cells surround the developing cardiac tissue and high levels of autofluorescence were noted in blood cells (Figs. 2d–e). A similarly diffuse signal without detectable GFP::CreERT2 signal was noted in the liver. Moreover, we could not detect any Foxj1 mRNA in the developing heart or liver tissues prior to E15.5 after examining in situ panels in the Allen Brain Atlas (http://developingmouse.brain-map.org/). Thus, it is highly unlikely that the low signals in the heart or liver are due to recombination events in these tissues. Taken together our findings suggest a highly efficient and local recombination in the choroid plexus of the developing brain with early embryonic inductions in Foxj1CreERT2::GFP mice.
Figure 2. Induction of recombination in Foxj1CreERT2::GFP embryos.
(a–e) TAM administration during early embryonic stages E7.5 to E12.5 reveals clear recombination in discrete areas in a sagittal section of the whole embryo (a). (b) Higher zoom of boxed region in (a) showing tdTom+ cells in the choroid plexus. (c) High power image of boxed region in (b) reveals tdTom+ cells in the choroid colocalized with the GFP::Cre::ERT2 signal. (d) Higher zoom of boxed region in (a) showing that the dim tdTom+ signal in the thorax of the embryos is diffuse and surrounded by autofluorescing red blood cells within the dorsal cardiac tissue. (e) GFP::Cre::ERT2 signal was not detectable in the tdTom+ cells in the heart. (f–h) TAM inductions at E13.5 to E15.5 reveal tdTom+ cells in the choroid plexus as shown in a sagittal section of the brain (f). (g–h) Boxed regions in (f) showing tdTom+ cells in the choroid plexus of the fourth (g) and lateral (h) ventricles. (i–m) TAM inductions at E17.5 and analyzed at E19.5 reveals tdTom+ cells in the choroid plexus as well as in developing ependymal cells in the ventricular walls in a sagittal section of the brain (i). (j–k) Boxed areas in (i) exhibiting tdTom+ cells in the fourth (j) and lateral (k) ventricles. (l) Zoomed view of the lateral ventricles surrounding the ganglionic eminence (GE) in a sagittal section close to midline. (m) High power image boxed area in (l) shows tdTom+ cells clearly within the ventricular zone of the GE. Blue: DAPI nuclear staining in all images. Scale bars: 500µm in (a), (f), and (i); 200 µm in (d); 100µm in (b); 50µm in (g), (h), (j), (k), and (l); 20µm in (m); 10µm in (c) and (e).
TAM induction at E13.5 and analysis at E15.5 revealed tdTom+ cells in the choroid plexus of both the lateral (LV) and fourth (4V) ventricles in the brain (Fig. 2f–h). However, with late embryonic inductions (E17.5 induction and analysis at E19.5; Fig. 2i) reporter expression in the brain was no longer confined to the choroid plexus (Figs. 2j–k), but also along the ventricular wall of the lateral ganglionic eminence (Fig. 2l–m) matching our past findings (Jacquet et al., 2011). Taken together, the reporter data from our Foxj1CreERT2::GFP inductions is in line with previously published findings on Foxj1 expression pattern during embryonic development in mice (Jacquet et al., 2009, 2011; Lim et al., 1997).
Postnatal recombination in peripheral organs of Foxj1CreERT2::GFP mice
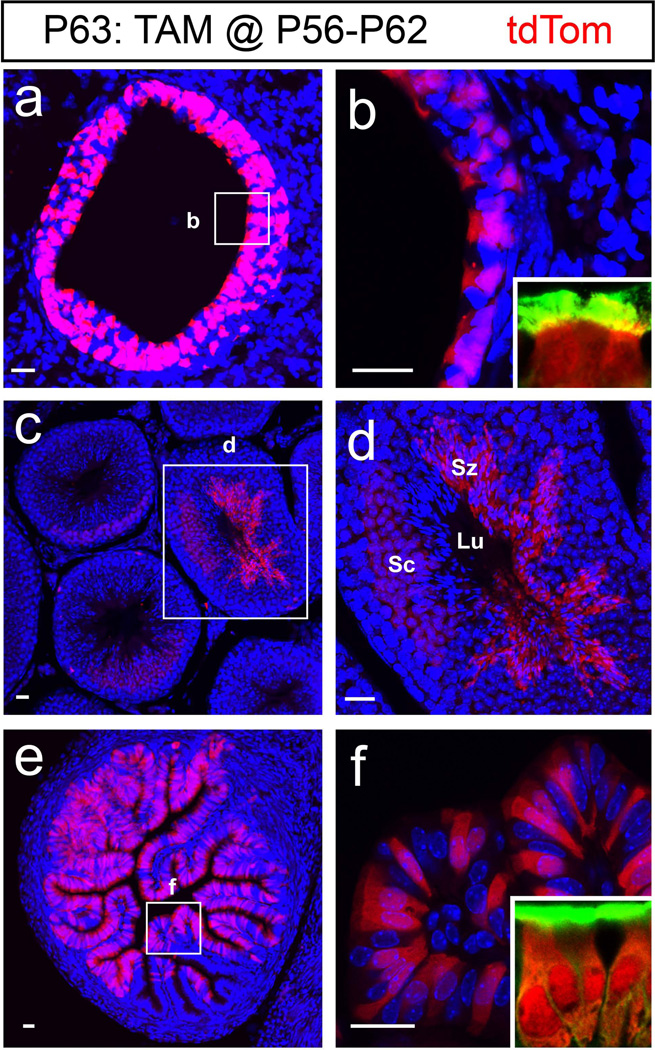
Expression of Foxj1 is well established in organs containing epithelial cells with motile cilia including the lungs, testes, and oviducts (Blatt et al., 1999; Brody et al., 2000; Zhang et al., 2007). To determine if TAM induction in Foxj1CreERT2::GFP mice led to faithful recombination, 8 week old mice were induced (daily for 7 days) and their peripheral organs were collected 24 hours after the last TAM injection. Analysis of sections prepared from the lungs illustrated tdTom+ epithelial cells in the primary bronchioles (Fig. 3a–b). Cross sections of the testes revealed exclusive recombination in the seminiferous tubules, where both the spermatocytes (Sc) as well as the spermatozoa (Sz) expressed tdTom (Fig. 3c–d). In the ampullary segment of the oviducts, tdTom+ cells were found only in the folded columnar epithelium (Fig. 3e–f) whereas the ovaries did not contain tdTom+ cells (data not shown). In addition we found that the tdTom+ cells in the lungs and oviducts were ciliated cells as revealed by α-tubulin staining (green in insets, Figs. 3b and 3f). Similar to the CNS, no tdTom+ cells were found in the lungs, testis, or oviduct of Foxj1CreERT2::GFP mice without TAM administration (data not shown), highlighting the unleaky nature of the GFP::CreERT2 element expressed in our mice.
Figure 3. Foxj1 expression in peripheral organs.
Recombination induced in peripheral organs after 7 days of TAM administration beginning at P56 were analyzed 24 hours after the last TAM injection (P63: TAM@P56-P62). (a) Cross section of the lung epithelium showing tdTom+ cells in the primary bronchioles. (b) High magnification of primary bronchioles expressing tdTom in the boxed area. Inset reveals the tdTom+ cells as ciliated cells. (c) Cross section through the testis shows tdTom+ cells in the seminiferous tubule. (d) High magnification image of boxed area in (c) reveals the tdTom+ population in the seminiferous tubules includes the spermatocytes (Sc) as well as the spermatozoa (Sz) with elongated nucleus adjacent to the lumen (Lu). (e) Cross section of a female Foxj1CreERT2::GFP ampullary segment of the oviduct shows tdTom+ cells in the ciliated epithelium. (f) High magnification of boxed area in (e) highlights the tdTom+ ciliated cells in the epithelial folds. Insets in b and f show that tdTom+ cells are ciliated indicated by α-tubulin staining (green). Blue: DAPI nuclear staining in all images. Scale bars: 20µm in all images.
Cellular profiling of the Foxj1CreERT2::GFP lineage in the postnatal brain
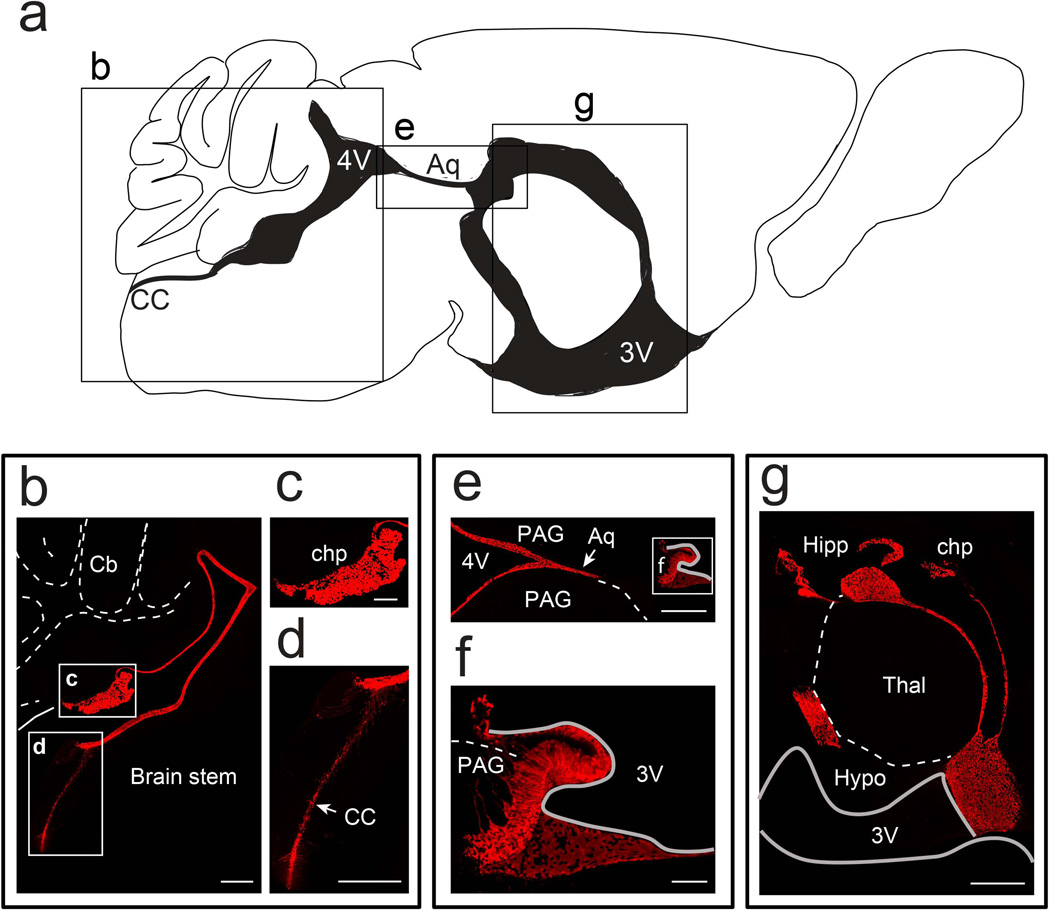
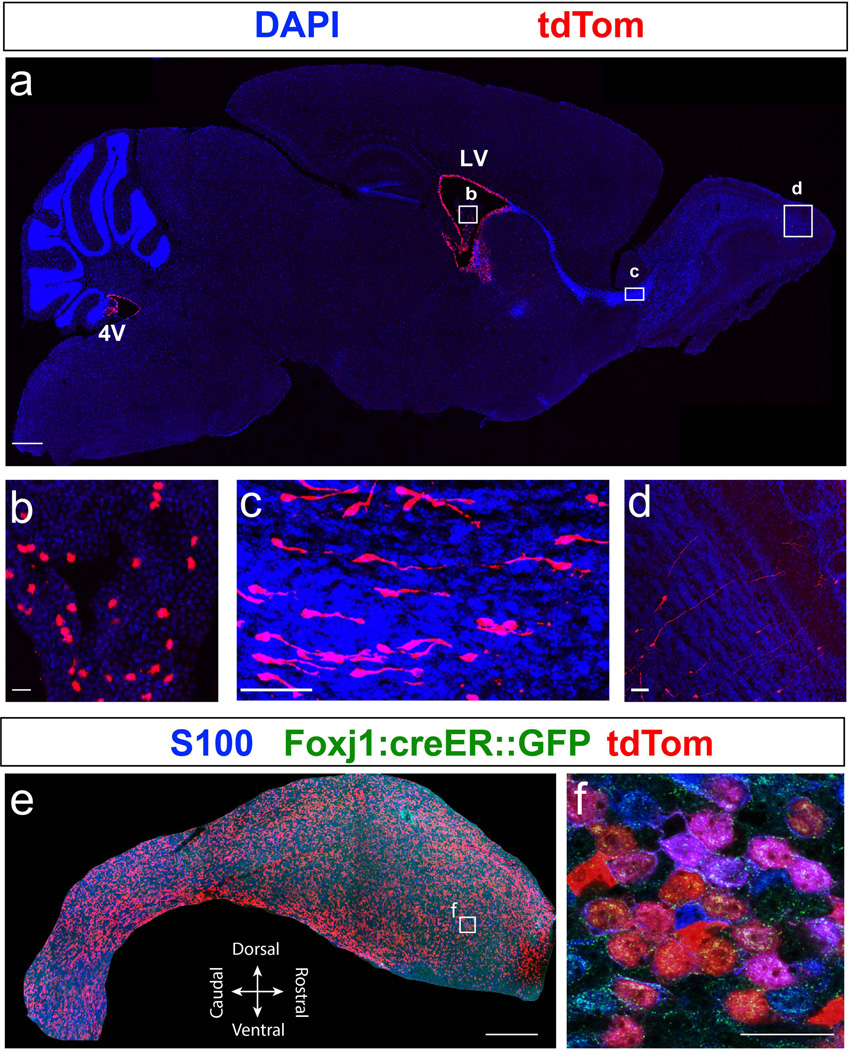
Recent findings from our lab established that Foxj1 is required for early postnatal development of the adult neural stem cell niche in the lateral ventricles through its direct participation in differentiation of ependymal cells (Jacquet et al., 2009, 2011). To test the fidelity of our new Foxj1CreERT2::GFP allele and compare its targeted populations with the transgenic line used in our lineage tracing studies in the olfactory bulbs (Jacquet et al., 2011), TAM was administered i.p. to females with new born pups to induce recombination in the suckling mice at P0. Analysis of induced brains at P21 revealed tdTom+ ependymal and choroid plexus (chp) cells along the entire ventricular system (Figs. 4–5). Wholemount view of the ventricular wall showed high degree of colocalization between tdTom+ cells and ependymal cells marked by S100 immunostaining (Fig. 5e–f). In addition, we noticed cytoplasmic localization of CreERT2::GFP signal in some ependymal cells indicating that Foxj1 promoter is active in a fraction of this population. Thus, our GFP reporter when combined with the permanent Cre-responsive tdTomato reporter is highly useful for identification of cells that express Foxj1.
Figure 4. Labeling of the Foxj1 lineage in the ventricular system of the postnatal brain.
(a) Diagram detailing the ventricular system (shaded black) of adult mouse brain in a sagittal section adjacent to the midline. (b) Sagittal section of Foxj1CreERT2::GFP mouse brain reveals Foxj1 lineage (tdTom+, red) in the fourth ventricle (4V) as well as the central canal (cc). Cb – cerebellum. (c) High magnification view of choroid plexus (chp) in the 4V. (d) High magnification image of tdTom+ cells in the central canal of the spinal cord. (e–f) Cerebral aqueduct (Aq) contains tdTom+ cells that extend to the subcommissural organ (f). PAG – Periaqueductal gray. (g) Foxj1 lineage in the third ventricle (3V) contains ependymal cells lining the ventricles and the choroid plexus. Hipp – hippocampus, Thal – thalamus, and Hypo – hypothalamus. Scale bars: 500µm in (b), (d), (e) and (g); 200µm in (c); 100µm in (f).
Figure 5. Labeling of the Foxj1 lineage in the postnatal forebrain.
(a) Sagittal section of a P21 brain from Foxj1CreERT2::GFP mice, induced with TAM at P0, shows tdTom+ cells (red) in the fourth ventricle (4V), lateral ventricle (LV), choroid plexus (b), rostral migratory stream (RMS; c), and olfactory bulbs (OB; d). (b) High magnification image of boxed area in (a) shows sporadic expression of tdTom+ cells in the choroid plexus of the LV. (c) High magnification of boxed area in (a) reveals numerous tdTom+ neuroblasts migrating in the RMS towards the OB, as reported in a previous study (Jacquet et al., 2011). (d) High magnification view of boxed area in (a) shows tdTom+ granule neurons in the OB. (e) S100 (blue) stained wholemount excised from the lateral ventricle from P21 Foxj1CreERT2::GFP mice brain reveals robust colocalization with tdTom+ ependymal cells. (f) High magnification image of boxed area in (e) reveals high degree of colocalization between S100 stained ependyma and tdTom+ cells. Scale bars: 500µm in (a) and (e); 50µm in (b), (c), and (d); 20µm in (f).
We recently reported that the cellular lineage derived from Foxj1promoter- active cells in the forebrain not only gives rise to ependymal cells but also consists of a small population of neurons that occupy the olfactory bulbs (Jacquet et al., 2011). Our past lineage tracing indicated that these neurons are only derived during early postnatal development, and their development comes to a halt during early adult periods. Analysis of Foxj1CreERT2::GFP forebrains at P21, following TAM-inductions at P0, confirmed the presence of tdTom+ neuroblasts migrating through the rostral migratory stream (Fig. 5c). Additionally, we noticed several tdTom+ neurons in the granule cell layer of the olfactory bulb (OB) which possessed long apical dendrites that branched at their contact with the mitral cell layer (Fig. 5d). Thus, with our new knock-in mouse we can conclusively confirm the presence of a unique set of OB neurons that are derived from a progenitor pool shared with the ependymal lineage. The properties and functional significance of this unique population of neurons remain to be determined. Taken together, the inducible Foxj1CreERT2::GFP knock-in mice reported here will be highly suitable to study Foxj1 transcriptional activity in different tissues, mechanisms of differentiation in Foxj1+ cells, and the biology of motile ciliogenesis.
Material and Methods
Animals
Mice used in this study were bred and housed in the College of Veterinary Medicine vivarium according to Institutional Animal Care and Use Committee (IACUC) and North Carolina State University regulations. Chimeras carrying the Foxj1CreERT2::GFP knock-in allele (GCE) were obtained from injection of E14Tg2a.4 cells (derived from mouse strain 129/P2OlaHsd) after electroporation with the targeting vector and standard selection procedures (Fig. 1a). GCE mice (B6;129-Foxj1tmHtg) were derived by intercrossing F1 generation in C57BL/6J background. FLP1 mice (The Jackson Laboratory stock #003946) were used for Neo cassette removal to derive Foxj1CreERT2::GFP knock-in allele. Mice carrying the knock-in allele were genotyped using purified DNA extracted by Phenol-Chloroform-Isoamylalcohol method from harvested tail tissue. Extracted DNA was amplified by PCR (primers: GCE Forward – 5’ CTC CCA CAT CAG GCA CAT GAG TAA 3’and GCE Reverse – 5’ GCA AAC AAC AGA TGG CTG GCA ACT 3’) using an annealing temperature of 60°C which yields a 392bp product (Fig. 1c). ROSA26-CAG-tdTomato reporter mice (The Jackson Laboratory stock #007908) were used for lineage tracing. Foxj1CreERT2::GFP knock-in mice will be made available for distribution to the research community.
Southern blotting
Genomic DNA was extracted (Phenol:Chloroform:Isoamyl alcohol, 25:24:1; Fisher Scientific Cat.# BP1752) and digested with restriction enzyme HindIII (New England Biolabs) for 3 hours at 37°C. Digested genomic DNA was run in 0.7% agarose gel, prepared in 1xTAE buffer, at 50V for 4 hours and stained using ethidium bromide solution. Following depurination using 250 mM HCl for 10 minutes at room temperature (RT), the gel was immersed in denaturation buffer (0.5M NaOH in 1.5M NaCl) for 15 minutes at RT. Gel was then immersed in neutralization buffer (0.5M Tris-HCl pH 7.5 in 1.5M NaCl) for 15 minutes at RT and then equilibrated in 20xSSC buffer. Neutral transfer of digested genomic DNA to Nytran SuPerCharge nylon membrane was performed using TurboBlotter system (Whatman) by following manufacturer’s instructions. Following crosslinking, the membrane and DIG labeled probes were prepared for hybridization using DIG easy hyb reagent (Roche) and the probe was let hybridize overnight (5’ and 3’ probe at 51.5°C; Neo probe at 45°C). After ON hybridization the membrane was prepared for detection following low and high stringency buffer washes following manufacturer’s instructions using DIG wash and block buffer set (Roche). Digested genomic DNA fragments were detected on nylon membranes using alkaline phosphatase conjugated anti-Digoxigenin Fab fragments and CDP-Star chemiluminescent substrate (Roche).
Embryonic and postnatal Tamoxifen administration
The day vaginal plug was recorded as embryonic day 0.5. Tamoxifen (TAM) stock (10mg/ mL) was prepared by dissolving dry TAM (sigma catalogue# T5648) in corn oil and stored at −20°C until use. For embryonic inductions TAM was delivered once to pregnant females at 50 mg/kg body weight orally via feeding tubes. For P0 inductions, TAM was administered (75mg/kg body weight) intraperitoneally for five consecutive days to female with new born pups, which then passed on the TAM to their feeding pups through milk. In our experience P0 pups cannot tolerate even low doses of TAM when administered directly via intradermal or intraperitoneal routes. TAM was administered at 100mg/kg body weight directly to mice by intraperitoneal injections after P14.
Immunohistochemistry
Mice were transcardially perfused using 4% paraformaldehyde (PFA) prepared in phosphate buffered saline (PBS) and tissues harvested were postfixed in 4% PFA overnight. Brains were sagittally sectioned to 50um thick sections using vibratome (Leica VT 1000S) and stored in a solution of 1xPBS with 0.05% sodium azide. Similarly lungs, testis, and oviducts were sectioned coronally. For immunohistochemical analysis tissue sections were immersed in blocking buffer (10% goat serum and 1% Triton-X-100 in 1xPBS) for an hour at room temperature (RT). Following blocking, sections were incubated with appropriate primary antibodies [chicken anti-GFP (Abcam) at 1:2000; rabbit anti-RFP (Abcam) at 1:1000; mouse anti-Tubulin (Sigma) at 1:1000; rabbit S100 (DAKO) at 1:1000] in antibody diluent (1% goat serum and 0.03% Triton-X-100 in 1xPBS) for overnight at 4°C. Sections were then washed three times, 5 minutes each, using 1xPBS and incubated with appropriate secondary antibodies in antibody diluent with DAPI [(Sigma) at 1µg/ mL] for an hour at RT. Following PBS washes, sections were mounted to plus-sided slides and coverslip placed on sections with mounting media to avoid air bubbles. Confocal images of stained sections were captured using either Nikon Eclipse C1 or Olympus fluoview FV1000 confocal microscopes.
Acknowledgments
We are grateful to Dr. Andy McMahon (Harvard and USC) for kindly provided us with the GFP::CreERT2 cassette. All ES cell work and generation of chimeric mice were conducted by TransViragen/UNC Animal Models Core (Chapel Hill, NC). We thank Raul Salinas, Sooyoung Franck, and Jermaine McGill for technical assistance at different stages of the development and characterization of the Foxj1CreERT2::GFP ES cells and chimeric mice. We appreciate members of the Ghashghaei lab for critical reading of the manuscript. This work was supported by National Institutes of Health Grant R01NS062182, a grant from the American Federation for Aging Research, and institutional funds to H.T.G.
References
- Blatt EN, Yan XH, Wuerffel MK, Hamilos DL, Brody SL. Forkhead transcription factor HFH-4 expression is temporally related to ciliogenesis. Am. J. Respir. Cell Mol. Biol. 1999;21:168–176. doi: 10.1165/ajrcmb.21.2.3691. [DOI] [PubMed] [Google Scholar]
- Brody SL, Yan XH, Wuerffel MK, Song SK, Shapiro SD. Ciliogenesis and left-right axis defects in forkhead factor HFH-4-null mice. Am. J. Respir. Cell Mol. Biol. 2000;23:45–51. doi: 10.1165/ajrcmb.23.1.4070. [DOI] [PubMed] [Google Scholar]
- Chen J, Knowles HJ, Hebert JL, Hackett BP. Mutation of the mouse hepatocyte nuclear factor/forkhead homologue 4 gene results in an absence of cilia and random left-right asymmetry. J. Clin. Invest. 1998;102:1077–1082. doi: 10.1172/JCI4786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fliegauf M, Benzing T, Omran H. When cilia go bad: cilia defects and ciliopathies. Nat. Rev. Mol. Cell Biol. 2007;8:880–893. doi: 10.1038/nrm2278. [DOI] [PubMed] [Google Scholar]
- Jacquet BV, Salinas-Mondragon R, Liang H, Therit B, Buie JD, Dykstra M, Campbell K, Ostrowski LE, Brody SL, Ghashghaei HT. FoxJ1-dependent gene expression is required for differentiation of radial glia into ependymal cells and a subset of astrocytes in the postnatal brain. Development. 2009;136:4021–4031. doi: 10.1242/dev.041129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquet BV, Muthusamy N, Sommerville LJ, Xiao G, Liang H, Zhang Y, Holtzman MJ, Ghashghaei HT. Specification of a Foxj1-dependent lineage in the forebrain is required for embryonic-to-postnatal transition of neurogenesis in the olfactory bulb. J. Neurosci. 2011;31:9368–9382. doi: 10.1523/JNEUROSCI.0171-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim L, Zhou H, Costa RH. The winged helix transcription factor HFH-4 is expressed during choroid plexus epithelial development in the mouse embryo. Proc. Natl. Acad. Sci. U.S.A. 1997;94:3094–3099. doi: 10.1073/pnas.94.7.3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paez-Gonzalez P, Abdi K, Luciano D, Liu Y, Soriano-Navarro M, Rawlins E, Bennett V, Garcia-Verdugo JM, Kuo CT. Ank3-dependent SVZ niche assembly is required for the continued production of new neurons. Neuron. 2011;71:61–75. doi: 10.1016/j.neuron.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlins EL, Ostrowski LE, Randell SH, Hogan BLM. Lung development and repair: contribution of the ciliated lineage. Proc. Natl. Acad. Sci. U.S.A. 2007;104:410–417. doi: 10.1073/pnas.0610770104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Huang G, Shornick LP, Roswit WT, Shipley JM, Brody SL, Holtzman MJ. A transgenic FOXJ1-Cre system for gene inactivation in ciliated epithelial cells. Am. J. Respir. Cell Mol. Biol. 2007;36:515–519. doi: 10.1165/rcmb.2006-0475RC. [DOI] [PMC free article] [PubMed] [Google Scholar]