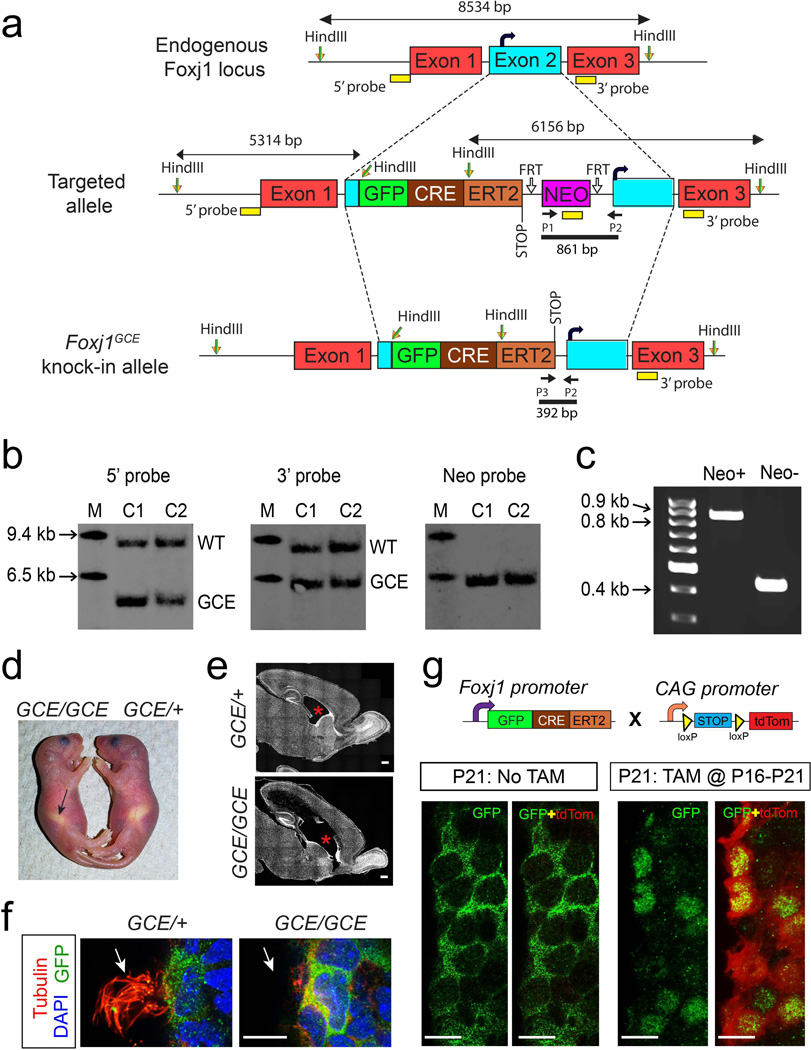
Figure 1. Generation of Foxj1CreERT2::GFP knock-in mice.
(a) A targeting vector was designed with a GFP::CreERT2 and Neomycin resistance (NEO) cassettes inserted into Exon2 in the Foxj1 locus (curved black arrow denotes endogenous start codon). NEO cassette, flanked by FRT sites, in the targeted allele was excised by crossing F1 progeny with FLP1 mice resulting in the Foxj1CreERT2::GFP knock-in allele. (b) Representative southern blots indicating site-specific insertion of the knock-in cassette in geneticin resistant ES cell clones. Hybridization sites of 5’, 3’, and Neo probes are indicated as yellow rectangles in (a). M – Molecular weight marker; C1 – Clone 1; C2 – Clone 2; WT - wild type band; GCE - Foxj1GCE knock-in allele band. (c) PCR was performed to detect deletion of Neo cassette from the Foxj1CreERT2::GFP knock-in locus after crossing to FLP1 mice. Annealing sites for primers P1, P2, and P3 used in PCR are indicated in (a). (d) Foxj1CreERT2::GFP homozygous (GCE/ GCE) pups display situs inversus where the stomach (arrow) is positioned on the right hand side of the animal. (e) Hydrocephalus indicated by enlargement of the lateral ventricles (red asterisks) was prevalent in GCE/GCE mice. (f) Apical surface of ependymal cells in GCE/GCE mice lacks cilia (red, tubulin staining, arrows). (g) Foxj1+ ependymal cells were genetically labeled by carrying the GCE/+ allele onto a ROSA26CAG-tdTomato reporter background. Tamoxifen (TAM) induced Cre-mediated excision of a stop codon results in constitutive tdTomato (tdTom, red) expression. In the absence of TAM, the GFP::CreERT2 fusion protein is localized to the cytoplasm of ependymal cells resulting in no tdTomato expression. Intraperitoneal injection of TAM at P16 for five consecutive days resulted in translocation of GFP::CreERT2 to the nucleus of ependymal cells at P21 (P21:TAM@P16-P20). Nuclear localization of GFP::CreERT2 overlapped with a high fraction of tdTom+ ependymal cells indicating efficient recombination rate upon TAM exposure. Scale bars: 500µm in (e); 10µm in (f) and (g).