Abstract
Palatogenesis, the formation of the palate, is a dynamic process that is regulated by a complex series of context-dependent morphogenetic signaling events. Many genes involved in palatogenesis have been discovered through the use of genetically-manipulated mouse models as well as from human genetic studies, but the roles of these genes and their products in signaling networks regulating palatogenesis are still poorly known. In this review, we give a brief overview on palatogenesis and introduce key signaling cascades leading to formation of the intact palate. Moreover, we review conceptual differences between pathway biology and network biology and discuss how some of the recent technological advances in conjunction with mouse genetic models have contributed to our understanding of signaling networks regulating palate growth and fusion.
INTRODUCTION
Mid-facial fusion defects, such as cleft lip with/or without cleft palate, are among the most common congenital birth defects in humans1. These defects result from a failure of facial/palatal processes to grow and/or fuse appropriately during the first trimester of human development2. Etiology of cleft lip with or without cleft (secondary) palate differs from that of cleft (secondary) palate only3. In this review we concentrate on developmental mechanisms that result in formation of the secondary palate.
The secondary palate, which separates the oral cavity from the nasal cavity, consists of an anterior hard palate (bony) and posterior soft palate (muscular)4. It plays a critical role in breathing, feeding, swallowing and speech. The secondary palate starts to develop from the maxillary process of the first pharyngeal arch as paired processes called palatal shelves. The key cell types in palate development are the neural crest-derived palatal mesenchyme, the ectoderm-derived epithelial lining, the most apical layer composed of periderm cells and the cranial paraxial mesoderm-derived myogenic cells in the soft palate. The palatal shelves first grow bilaterally down along the sides of the tongue. Then they rapidly elevate (as the tongue descends to the floor of the mouth), form a contact in the midline and fuse (Fig 1). All of this takes place between weeks 7 and 11 in human gestation (embryonal days E11.5 – E16 in mice; See Fig 1). Failure in any of these processes, i.e., growth, elevation or fusion, and even a post fusion rupture, results in cleft palate.
Fig. 1.
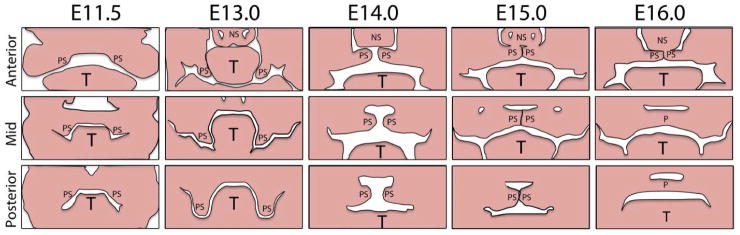
Schematic representation of palatal growth and fusion. Palatal shelves (PS) can be first seen as outgrowths of the maxillary processes of the first pharyngeal arch (E11.5). They then grow vertically down along the sides of the tongue (T; E13.0), rapidly elevate (E14.0), form a contact with each other and with the nasal septum (NS) in the anterior palate (P) (E15.0) and eventually fuse (E16.0).
During the last 20 years, methodological advances in human genetics, transcriptomics, proteomics, epigenetics and particularly in mouse genetics, i.e, in our ability to manipulate the mouse genome, have provided a wealth of information about roles of individual genes in palatogenesis. Altogether more than 300 genes, and all the important morphogenetic signaling mechanisms, have been implicated in palatal fusion either in humans or in experimental animal models5. The most recent studies have started to address mechanisms by which these genes and their product form regulatory networks to regulate palatal shelf growth, patterning and fusion6, 7.
EPITHELIAL-MESENCHYMAL INTERACTIONS CONTROL PALATAL SHELF GROWTH AND PATTERNING
Palatal shelf growth and patterning are controlled by epithelial-mesenchymal interactions. Since outstanding reviews summarizing key pathways involved in palatogenesis have recently been published4, 5, 8, we will provide here only a concise outline of these previous data, and concentrate on the most recent studies elucidating signaling processes that control palatogenesis, and discuss how systems biology can change our view of complex molecular events taking place during palatal fusion.
Growth and patterning of the secondary palate
Many studies have shown that gene expression patterns and gene functions are remarkably different in the developing anterior and posterior palate, which give rise to the hard- and soft palate, respectively (Fig. 2). The anterior-posterior (AP) boundary in the developing palate is on the level of the most posterior ruga.
Fig. 2.
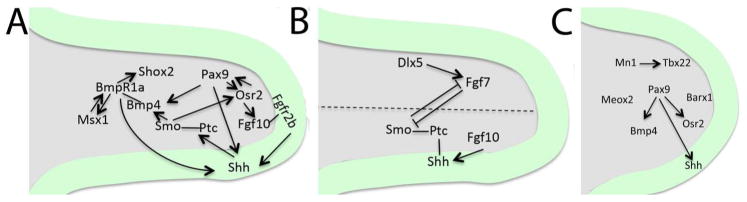
Signaling circuits governing palatal shelf growth and patterning. A, Epithelial-mesenchymal interactions via Pax9-regulated Shh-Bmp and Shh-Fgf feed-back loops control growth and patterning of the anterior palate. B, Oro-nasal patterning of the anterior secondary palate is regulated via Fgf7-mediated Shh repression. C, Pax9-regulated expression of Bmp4 and Osr2 in the posterior palatal mesenchyme and Shh in the posterior palatal epithelium is required for appropriate posterior growth of the secondary palate. In addition, Meox2, Barx1 and Mn1-Tbx22 signaling module regulate posterior palatal growth. Grey, palatal mesenchyme; Green, palatal epithelium.
Anterior palate development
It has been demonstrated that Sonic Hedgehog (Shh), an important morphogen, plays a crucial organizing role in development and growth of the anterior secondary palate9, 10. Shh is expressed in periodic stripes on the oral side of the developing palate; these stripes correspond to future palatal rugae (transverse ridges on the hard palate)11, 12. Abrogation of Shh in the palatal epithelium (by K14-Cre) or its receptor Smoothened (Smo) in the palatal mesenchyme (using Osr2-Cre) has been shown to result in cleft palate and in reduced expression of a number of genes relevant for palatal shelf growth, e.g., Fgf10, Bmp2, Msx1 and Osr210. Interestingly, mesenchymal Fgf10 expression is also needed for appropriate epithelial Shh signaling via epithelial Fgfr2b activation9, and thus Fgf10-Shh signaling axis provides an important regulatory feedback loop between palatal epithelium and mesenchyme to control palatal anterior growth and patterning9. Similarly, it has been shown that Bmp4 in the palatal mesenchyme is required both for Shox2, which is needed for anterior palatal growth (mesenchymal cell proliferation),13, 14 and for Shh expression in the palatal epithelium15. This provides another signaling loop between the epithelium and mesenchyme to control anterior growth of the secondary palate15. Zhou et al recently demonstrated that deletion of the paired-box gene-9 (Pax9) transcription factor in the palatal mesenchyme resulted in cleft palate that was caused by defective palatal shelf growth and elevation16. In Pax9 mutants, Shh expression in the palatal epithelium, and Bmp4, Fgf10, Msx1 and Osr2 expression in the palatal mesenchyme were dramatically reduced. The authors further showed that a novel knock-in allele, expressing Osr2 in the Pax9 locus (Pax9Osr2KI) was able to rescue the posterior, but not the anterior fusion defects in Pax9 mutants. Consistent with the lack of anterior phenotypic restoration, the expression levels of many key genes, e.g., Bmp4, Msx1 and Shh were not rescued. The authors concluded that Pax9 acts upstream or parallel to Osr2 and controls several different pathways including Shh, Fgf10 and Bmp4 during development of the anterior palate.
Oro-nasal patterning of the anterior secondary palate
As outlined above, Shh is specifically expressed in the oral palatal epithelium, while Shh receptors (Ptch and Smo) and downstream effectors (Msx1, Osr1, Osr2) display a graded expression in the adjacent mesenchyme along the oro-nasal axis17, 18. In contrast, the distal-less homeobox-5 (Dlx5) and Fgf7 genes are specifically expressed in the nasal mesenchyme of the palatal shelf, and the authors suggested that the Dlx5-regulated Fgf7 signaling is critically important in negatively regulating the mesenchymal Shh signaling and palatal oro-nasal patterning17 (Fig. 2).
Posterior palate development
The posterior muscular palate (soft palate) functions during swallowing and speech. In addition to neural crest-derived mesenchymal cells, the posterior palate mesenchyme is populated by myogenic cells derived from the cranial paraxial mesoderm19. The importance of muscle function to palatogenesis was recently demonstrated by Rot-Nikcevic et al, who showed that mouse embryos lacking striated muscles display cleft palate20. Compared to the molecular control of the anterior palate development, much less is known about signaling processes governing the growth and patterning of the posterior secondary palate. Transcription factors Tbx22 and Meox2 are specifically expressed in the posterior palate, while Barx1 and Mn1 are expressed along the entire AP axis, although the expression of these genes is stronger in the posterior than in the anterior mesenchyme21–23, and concordant with the posterior expression domain, mice deficient in Tbx22 suffer from a submucous cleft palate24. Zhou et al recently showed that unlike previously thought, Bmp4 is expressed both in the anterior and posterior palatal mesenchyme, and that its expression is dependent on Pax9 but not on Msx1 in the posterior palate16. Moreover, their results implied that both Osr2 expression in the posterior palatal mesenchyme and Shh expression in the posterior palatal epithelium (in the developing sensory papilla) is regulated by Pax916.
SIGNALING PATHWAYS CONTROLLING PALATAL EPITHELIAL DIFFERENTIATION AND MIDLINE SEAM DISAPPEARANCE
Along with palatal shelf growth, there is growth of maxillary and mandibular processes. This allows the tongue to slide down and forward, which is required for palatal shelf elevation and reorientation25. Pioneering studies of Walker and Fraser and their recent detailed refinement and complementation by Yu and Ornitz showed that mechanisms of the anterior and posterior palatal shelf elevation are different26, 27. While the anterior palatal shelves are elevated by a ‘flipping-up’ mechanism, the posterior palatal shelves undergo tissue-remodeling movement. Molecular mechanisms controlling these events are still poorly known4.
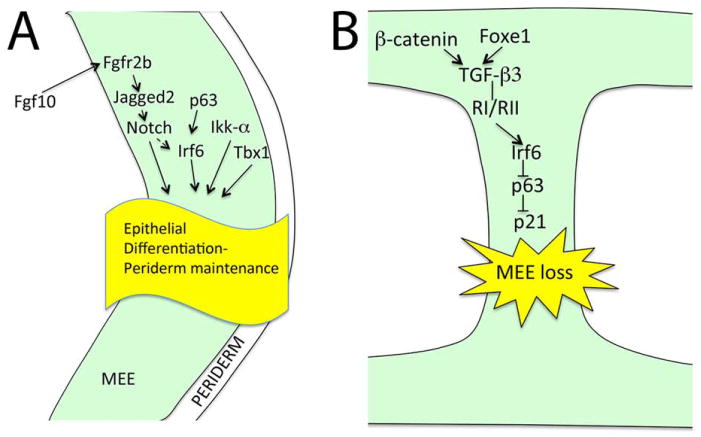
After elevation, the palatal shelves meet in the midline and become adherent28. This event is tightly controlled, since inappropriate adherence prevents palatal shelf elevation resulting in cleft palate29, 30. Periderm cells, joined to each other by tight junctions, have an important role in controlling palatal shelf adherence and epithelial differentiation31, 32. This thin (one cell) layer of flattened cells is thought to function as a protective layer or insulator preventing aberrant adhesions. However, the loss of peridermal cells is required at sites of fusion, e.g., the tips of apposing palatal shelves for appropriate epithelial differentiation and adherence32. Recent studies have shown that mouse embryos lacking Fgf10, Jagged2, Irf6, Ikka (chuk) or Tbx1 show aberrant oral adhesions between the tongue and palatal shelves29, 30, 33, 34, and it has been suggested that Jagged2-Notch and p63-Irf6 signaling regulate maintenance of periderm cells during palatogenesis29, 30, 35(Fig. 3A).
Fig. 3.
A, Molecular control of palatal epithelial (green) differentiation. Fgf-Notch signaling, P63-Irf6 signaling, Tbx1 and Ikk-α regulate differentiation or the prefusion palatal epithelium (green). B, Medial edge epithelial loss is mediated via a signaling cascade involving TGF-β3, Irf6, p63 and p21in the palatal midline epithelial seam (green).
Once the apposing palatal shelves have adhered, the midline epithelial seam (MES) must be removed to obtain a mesenchymal confluence28. This will mostly happen via programmed cell death (apoptosis), although a possibility that one or both of the two other suggested mechanisms, i.e., epithelial-mesenchymal transformation and migration, would also play some role in MES removal cannot be totally excluded31, 36, 37. Nevertheless, it is clear that TGF-β signaling plays a critical role in MES removal. Tgfb3 is strongly and specifically expressed in the MEE38, 39, and mice deficient in Tgfb3 show 100% penetrant cleft palate40, 41. Moreover, palatal epithelial cells defective in genes encoding TGF-β type I or type II receptors fail to undergo apoptosis resulting in a failure of the MES to disappear37, 42. How Tgfb3 expression is regulated in the MEE is poorly known. It was recently shown that epithelial-specific Ctnnb1 (the gene encoding β-catenin) mutants displayed cleft palate and loss of Tgfb3 expression suggesting that canonical Wnt signaling via β-catenin is required for appropriate MEE-specific Tgfb3 expression43. Venza et al showed that mutations in the FOXE1 gene result in Bamforth-Lazarus syndrome characterized with craniofacial defects including cleft palate44, and that Tgfb3 is a direct target of Foxe144.
As outlined above, epithelium-specific deletion of genes encoding TGF-β type I and II receptors resulted in defective palatogenesis37, 42. Therefore, it was rather surprising that epithelium-specific deletion of Smad4, which is a critical intracellular TGF-β signal transducer, did not result in detectable defects in palatal fusion45. Xu et al, further demonstrated that simultaneous deletion of Smad4 and inhibition of p38Mapk resulted in persistent MES in explant cultures suggesting that both Smad-dependent and Smad-independent TGF-β signaling act redundantly for successful palatal epithelial fusion45. Iwata et al recently showed that Irf6 is a direct target of Smad4 in the palatal epithelium, and that overexpression of Irf6 in the palatal epithelium rescued the palatal defect seen in epithelial-specific Tgfbr2 mutants46. Moreover, they showed that Irf6 is needed to suppress dNp63, which is a prerequisite for p21 (Cip1) expression, cell cycle arrest and subsequent MEE loss46(Fig. 3B).
NETWORK BIOLOGY AND PALATOGENESIS
Pathway biology vs network biology
As outlined above, genetically manipulated mouse models have helped to identify genes, pathways and signaling modules controlling both palatal shelf growth, patterning and fusion. While providing valuable conceptual models, these molecule-centric ‘pathway biology’ approaches are often highly simplified, ignoring context-dependent interactions and are thus potentially misleading. Completion of genome sequencing projects of several different species, and subsequent technological advances in expression profiling (microarray and RNA-Seq), proteomics (mass spectrometry, protein arrays), analyses of protein-protein interactions (yeast two-hybrid screen, mass spectrometry) and protein-DNA interactions (Chip-Seq) have provided basic tools for the understanding of complex regulatory networks. From these advances, new paradigms have emerged that are collectively called systems biology or network biology (Fig. 4).
Fig. 4.
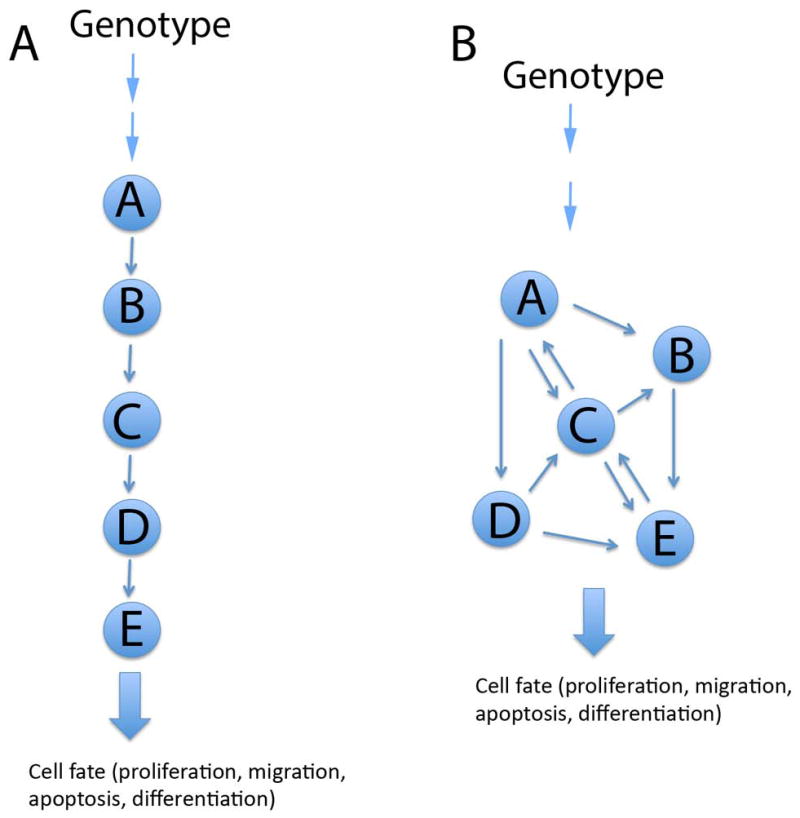
Schematic comparison of a pathway biology model (A) to a network biology model (B). Arrows depict interactions between signaling molecules. In a pathway biology model (A), genomic information regulating a cell fate is mediated by a linear pathway, where every downstream function is directly affected by an upstream signaling molecule. In a network biology model, genomic information controlling a certain cell fate is mediated by a network of interacting molecules.
Based on chosen high-throughput methods, networks can be further divided into transcription regulation-, protein-, signaling-, metabolic- and regulatory networks or different combinations of intergrated networks (as reviewed by Emmert-Streib and Glazko47). A common feature of these interactome networks is that they are highly complex and context dependent placing multidimensional and multidirectional networks between genotypes and phenotypes48. Network biology is rapidly contributing to our understanding of complex biological processes. However, due to the extreme complexity and the volume of the generated data, their interpretations would not have been possible without parallel advances in computational biology. Together these developments have also made it possible to apply machine learning and other virtual biology approaches to develop new hypotheses and to prioritize and to test existing hypotheses in situations where experimental biology is not feasible, for instance, due to the high cost or ethical reasons.
Despite enormous promise, network biology paradigms have not been intensely applied to complex developmental processes, e.g., palatogenesis. This is largely due to the fact that developmental events are highly context dependent, both spatially and temporally. Therefore, it has been challenging to isolate sufficient quantities of highly purified transient cell types, e.g., palatal medial edge epithelial cells, without introducing changes to their endogenous gene expression profiles or interfering with pre-existing protein-protein or protein-DNA interactions.
Initial attempts to understand gene regulatory networks during palatogenesis have focused on epigenetic and expression screens on samples harvested from palatal tissues containing both the epithelium and mesenchyme. Pelikan et al. used conventional microarray approaches on samples harvested on prefusion palatal shelves of control and neural crest-specific Tgfbr2 mutants (Tgfbr2/Wnt1-Cre) during palatogenesis6. Tgfbr2/Wnt1-Cre mutants display cleft palate resulting from reduced growth of prefusion palatal shelves49. Since most of the mesenchyme in palatal shelves is derived from the cranial neural crest50, these studies allowed identification of genes that are differentially expressed between control mice and mice lacking the gene encoding the TGF-β type II receptor in the palatal mesenchyme. The authors performed gene ontology, transcription factor binding prediction, and miRNA enrichment analyses and analyzed predicted functional relationships of differentially expressed genes using pathway analyses. They grouped the differentially expressed genes based on different functions, e.g., cell cycle regulation, micro-tubule function and cholesterol synthesis6, and have used these data to further dissect the role of TGF-β type II receptor-mediated signaling in the palatal mesenchyme51, 52.
Ozturk et al. combined database analysis and RNA-Seq to identify cleft palate genes in a Tgfb3 knockout model53. The original data analysis revealed a large number of genes that were differentially expressed between control and mutants throughout palatal fusion. By analyzing differentially expressed genes within the set of 322 cleft palate-associated genes, the authors could identify eight unique genes that all followed the Smad-dependent pathway.
Seelan et al analyzed methylated promoters and microRNA expression profiles using array-based platforms, and analyzed their data using computational gene interaction predictions7. The authors discussed the benefits of understanding epigenetic gene regulation during palatal fusion, and pointed out that some teratogenic compounds, e.g., an antiepileptic drug, valproic acid, alter chromatin conformational state and that its use during early pregnancy results in birth defects including cleft palate in humans.
As mentioned above, major limitations in applying modern high-throughput methods to embryological processes are the lack of a temporal control in analysis of dynamic developmental events and the difficulty in obtaining pure populations of cells without introducing a myriad of artifactual variables. The current methods, e.g., fluorescence activated cell sorting (FACS), laser capture or manual microdissection and immunopanning all have obvious limitations: FACS and immunopanning remove cells from their natural environment, which will likely introduce changes to endogenous gene expression programs, while dissection methods are prone to operator errors and when combined with laser capture, often compromise the quality and quantity of the harvested specimen. Novel methods based on in vivo labeling of cell type-specific transcripts, e.g., INTACT, Ribo-tag, TRAP have attempted to address some of these problems54–56. A recently published TU-tagging method is particularly interesting, since in addition to allowing cell-type specific in vivo labeling, it can be used to identify newly synthetized mRNAs and non-coding RNAs, including microRNAs, in intact mouse embryos or in postnatal mice57. Moreover, this can be accomplished in specific cell types and at specific times57.
CONCLUSIONS
Advances made in mouse genetics during the last 25 years have largely contributed to our understanding of molecular mechanisms controlling palatal shelf growth, patterning and fusion. They have helped to identify genes required for palatogenesis, and have provided information about pathways in which these genes are functioning during palate development and fusion. These studies have also revealed some significant limitations in classical molecule- or pathway-centric approaches. At the same time, several methodological advances in high throughput- and computational techniques, have contributed to an emergence of new paradigms of network biology. While these novel integrated approaches are changing the way we think about biological processes, their application to highly dynamic context dependent processes taking place during embryonal development, e.g., palatogenesis, has been challenging. Notwithstanding, initial studies have addressed changes in transcriptomes in particular mouse cleft palate models and contributed to understanding of the role of epigenetics during palate development. As new tools and techniques become available, our understanding of molecular mechanisms regulating palate development will likely advance rapidly, which will also help us to better understand pathogenetic mechanisms of cleft palate syndrome, one of the most common birth defects in humans.
Acknowledgments
Our work is supported by the National Institutes of Health grant no. RO1DE13085.
References
- 1.Murray JC. Gene/environment causes of cleft lip and/or palate. Clin Genet. 2002;61:248–256. doi: 10.1034/j.1399-0004.2002.610402.x. [DOI] [PubMed] [Google Scholar]
- 2.Schutte BC, Murray JC. The many faces and factors of orofacial clefts. Hum Mol Genet. 1999;8:1853–1859. doi: 10.1093/hmg/8.10.1853. [DOI] [PubMed] [Google Scholar]
- 3.Dixon MJ, Marazita ML, Beaty TH, Murray JC. Cleft lip and palate: understanding genetic and environmental influences. Nat Rev Genet. 2011;12:167–178. doi: 10.1038/nrg2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bush JO, Jiang R. Palatogenesis: morphogenetic and molecular mechanisms of secondary palate development. Development. 2012;139:231–243. doi: 10.1242/dev.067082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gritli-Linde A. Molecular control of secondary palate development. Dev Biol. 2007;301:309–326. doi: 10.1016/j.ydbio.2006.07.042. [DOI] [PubMed] [Google Scholar]
- 6.Pelikan RC, Iwata J, Suzuki A, Chai Y, Hacia JG. Identification of candidate downstream targets of TGFbeta signaling during palate development by genome-wide transcript profiling. J Cell Biochem. 2013;114:796–807. doi: 10.1002/jcb.24417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seelan RS, Mukhopadhyay P, Pisano MM, Greene RM. Developmental epigenetics of the murine secondary palate. ILAR J. 2012;53:240–252. doi: 10.1093/ilar.53.3-4.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meng L, Bian Z, Torensma R, Von den Hoff JW. Biological mechanisms in palatogenesis and cleft palate. J Dent Res. 2009;88:22–33. doi: 10.1177/0022034508327868. [DOI] [PubMed] [Google Scholar]
- 9.Rice R, Spencer-Dene B, Connor EC, Gritli-Linde A, McMahon AP, Dickson C, Thesleff I, Rice DP. Disruption of Fgf10/Fgfr2b-coordinated epithelial-mesenchymal interactions causes cleft palate. J Clin Invest. 2004;113:1692–1700. doi: 10.1172/JCI20384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lan Y, Jiang R. Sonic hedgehog signaling regulates reciprocal epithelial-mesenchymal interactions controlling palatal outgrowth. Development. 2009;136:1387–1396. doi: 10.1242/dev.028167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Economou AD, Ohazama A, Porntaveetus T, Sharpe PT, Kondo S, Basson MA, Gritli-Linde A, Cobourne MT, Green JB. Periodic stripe formation by a Turing mechanism operating at growth zones in the mammalian palate. Nat Genet. 2012;44:348–351. doi: 10.1038/ng.1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baek JA, Lan Y, Liu H, Maltby KM, Mishina Y, Jiang R. Bmpr1a signaling plays critical roles in palatal shelf growth and palatal bone formation. Dev Biol. 2011;350:520–531. doi: 10.1016/j.ydbio.2010.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hilliard SA, Yu L, Gu S, Zhang Z, Chen YP. Regional regulation of palatal growth and patterning along the anterior-posterior axis in mice. J Anat. 2005;207:655–667. doi: 10.1111/j.1469-7580.2005.00474.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li L, Lin M, Wang Y, Cserjesi P, Chen Z, Chen Y. BmprIa is required in mesenchymal tissue and has limited redundant function with BmprIb in tooth and palate development. Dev Biol. 2011;349:451–461. doi: 10.1016/j.ydbio.2010.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Z, Song Y, Zhao X, Zhang X, Fermin C, Chen Y. Rescue of cleft palate in Msx1-deficient mice by transgenic Bmp4 reveals a network of BMP and Shh signaling in the regulation of mammalian palatogenesis. Development. 2002;129:4135–4146. doi: 10.1242/dev.129.17.4135. [DOI] [PubMed] [Google Scholar]
- 16.Zhou J, Gao Y, Lan Y, Jia S, Jiang R. Pax9 regulates a molecular network involving Bmp4, Fgf10, Shh signaling and the Osr2 transcription factor to control palate morphogenesis. Development. 2013 doi: 10.1242/dev.099028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Han J, Mayo J, Xu X, Li J, Bringas P, Jr, Maas RL, Rubenstein JL, Chai Y. Indirect modulation of Shh signaling by Dlx5 affects the oral-nasal patterning of palate and rescues cleft palate in Msx1-null mice. Development. 2009;136:4225–4233. doi: 10.1242/dev.036723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lan Y, Ovitt CE, Cho ES, Maltby KM, Wang Q, Jiang R. Odd-skipped related 2 (Osr2) encodes a key intrinsic regulator of secondary palate growth and morphogenesis. Development. 2004;131:3207–3216. doi: 10.1242/dev.01175. [DOI] [PubMed] [Google Scholar]
- 19.Zhang L, Yoshimura Y, Hatta T, Otani H. Myogenic determination and differentiation of the mouse palatal muscle in relation to the developing mandibular nerve. J Dent Res. 1999;78:1417–1425. doi: 10.1177/00220345990780080701. [DOI] [PubMed] [Google Scholar]
- 20.Rot-Nikcevic I, Reddy T, Downing KJ, Belliveau AC, Hallgrimsson B, Hall BK, Kablar B. Myf5−/−: MyoD−/− amyogenic fetuses reveal the importance of early contraction and static loading by striated muscle in mouse skeletogenesis. Dev Genes Evol. 2006;216:1–9. doi: 10.1007/s00427-005-0024-9. [DOI] [PubMed] [Google Scholar]
- 21.Li Q, Ding J. Gene expression analysis reveals that formation of the mouse anterior secondary palate involves recruitment of cells from the posterior side. Int J Dev Biol. 2007;51:167–172. doi: 10.1387/ijdb.062212ql. [DOI] [PubMed] [Google Scholar]
- 22.Liu W, Lan Y, Pauws E, Meester-Smoor MA, Stanier P, Zwarthoff EC, Jiang R. The Mn1 transcription factor acts upstream of Tbx22 and preferentially regulates posterior palate growth in mice. Development. 2008;135:3959–3968. doi: 10.1242/dev.025304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bush JO, Lan Y, Maltby KM, Jiang R. Isolation and developmental expression analysis of Tbx22, the mouse homolog of the human X-linked cleft palate gene. Dev Dyn. 2002;225:322–326. doi: 10.1002/dvdy.10154. [DOI] [PubMed] [Google Scholar]
- 24.Pauws E, Hoshino A, Bentley L, Prajapati S, Keller C, Hammond P, Martinez-Barbera JP, Moore GE, Stanier P. Tbx22null mice have a submucous cleft palate due to reduced palatal bone formation and also display ankyloglossia and choanal atresia phenotypes. Hum Mol Genet. 2009;18:4171–4179. doi: 10.1093/hmg/ddp368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ferguson MW. Palate development: mechanisms and malformations. Ir J Med Sci. 1987;156:309–315. doi: 10.1007/BF02951261. [DOI] [PubMed] [Google Scholar]
- 26.Walker BD, Fraser FC. Closure of the secondary palate in three strains of mice. J Embryol Exp Morphol. 1956;4:176–189. [Google Scholar]
- 27.Yu K, Ornitz DM. Histomorphological study of palatal shelf elevation during murine secondary palate formation. Dev Dyn. 2011;240:1737–1744. doi: 10.1002/dvdy.22670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ferguson MW. Palate development. Development. 1988;103 (Suppl):41–60. doi: 10.1242/dev.103.Supplement.41. [DOI] [PubMed] [Google Scholar]
- 29.Ingraham CR, Kinoshita A, Kondo S, Yang B, Sajan S, Trout KJ, Malik MI, Dunnwald M, Goudy SL, Lovett M, et al. Abnormal skin, limb and craniofacial morphogenesis in mice deficient for interferon regulatory factor 6 (Irf6) Nat Genet. 2006;38:1335–1340. doi: 10.1083/ng1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Richardson RJ, Dixon J, Jiang R, Dixon MJ. Integration of IRF6 and Jagged2 signalling is essential for controlling palatal adhesion and fusion competence. Hum Mol Genet. 2009;18:2632–2642. doi: 10.1093/hmg/ddp201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vaziri Sani F, Hallberg K, Harfe BD, McMahon AP, Linde A, Gritli-Linde A. Fate-mapping of the epithelial seam during palatal fusion rules out epithelial-mesenchymal transformation. Dev Biol. 2005;285:490–495. doi: 10.1016/j.ydbio.2005.07.027. [DOI] [PubMed] [Google Scholar]
- 32.Yoshida M, Shimono Y, Togashi H, Matsuzaki K, Miyoshi J, Mizoguchi A, Komori T, Takai Y. Periderm cells covering palatal shelves have tight junctions and their desquamation reduces the polarity of palatal shelf epithelial cells in palatogenesis. Genes Cells. 2012;17:455–472. doi: 10.1111/j.1365-2443.2012.01601.x. [DOI] [PubMed] [Google Scholar]
- 33.Funato N, Nakamura M, Richardson JA, Srivastava D, Yanagisawa H. Tbx1 regulates oral epithelial adhesion and palatal development. Hum Mol Genet. 2012;21:2524–2537. doi: 10.1093/hmg/dds071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Humphreys R, Zheng W, Prince LS, Qu X, Brown C, Loomes K, Huppert SS, Baldwin S, Goudy S. Cranial neural crest ablation of Jagged1 recapitulates the craniofacial phenotype of Alagille syndrome patients. Hum Mol Genet. 2012;21:1374–1383. doi: 10.1093/hmg/ddr575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Casey LM, Lan Y, Cho ES, Maltby KM, Gridley T, Jiang R. Jag2-Notch1 signaling regulates oral epithelial differentiation and palate development. Dev Dyn. 2006;235:1830–1844. doi: 10.1002/dvdy.20821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dudas M, Li WY, Kim J, Yang A, Kaartinen V. Palatal fusion - Where do the midline cells go? A review on cleft palate, a major human birth defect. Acta Histochem. 2007;109:1–14. doi: 10.1016/j.acthis.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 37.Xu X, Han J, Ito Y, Bringas P, Jr, Urata MM, Chai Y. Cell autonomous requirement for Tgfbr2 in the disappearance of medial edge epithelium during palatal fusion. Dev Biol. 2006;297:238–248. doi: 10.1016/j.ydbio.2006.05.014. [DOI] [PubMed] [Google Scholar]
- 38.Pelton RW, Dickinson ME, Moses HL, Hogan BL. In situ hybridization analysis of TGF beta 3 RNA expression during mouse development: comparative studies with TGF beta 1 and beta 2. Development. 1990;110:609–620. doi: 10.1242/dev.110.2.609. [DOI] [PubMed] [Google Scholar]
- 39.Millan FA, Denhez F, Kondaiah P, Akhurst RJ. Embryonic gene expression patterns of TGF beta 1, beta 2 and beta 3 suggest different developmental functions in vivo. Development. 1991;111:131–143. doi: 10.1242/dev.111.1.131. [DOI] [PubMed] [Google Scholar]
- 40.Proetzel G, Pawlowski SA, Wiles MV, Yin M, Boivin GP, Howles PN, Ding J, Ferguson MW, Doetschman T. Transforming growth factor-beta 3 is required for secondary palate fusion. Nat Genet. 1995;11:409–414. doi: 10.1038/ng1295-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kaartinen V, Voncken JW, Shuler C, Warburton D, Bu D, Heisterkamp N, Groffen J. Abnormal lung development and cleft palate in mice lacking TGF-beta 3 indicates defects of epithelial-mesenchymal interaction. Nat Genet. 1995;11:415–421. doi: 10.1038/ng1295-415. [DOI] [PubMed] [Google Scholar]
- 42.Dudas M, Kim J, Li WY, Nagy A, Larsson J, Karlsson S, Chai Y, Kaartinen V. Epithelial and ectomesenchymal role of the type I TGF-beta receptor ALK5 during facial morphogenesis and palatal fusion. Dev Biol. 2006;296:298–314. doi: 10.1016/j.ydbio.2006.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.He F, Xiong W, Wang Y, Li L, Liu C, Yamagami T, Taketo MM, Zhou C, Chen Y. Epithelial Wnt/beta-catenin signaling regulates palatal shelf fusion through regulation of Tgfbeta3 expression. Dev Biol. 2011;350:511–519. doi: 10.1016/j.ydbio.2010.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Venza I, Visalli M, Parrillo L, De Felice M, Teti D, Venza M. MSX1 and TGF-beta3 are novel target genes functionally regulated by FOXE1. Hum Mol Genet. 2011;20:1016–1025. doi: 10.1093/hmg/ddq547. [DOI] [PubMed] [Google Scholar]
- 45.Xu X, Han J, Ito Y, Bringas P, Jr, Deng C, Chai Y. Ectodermal Smad4 and p38 MAPK are functionally redundant in mediating TGF-beta/BMP signaling during tooth and palate development. Dev Cell. 2008;15:322–329. doi: 10.1016/j.devcel.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Iwata J, Suzuki A, Pelikan RC, Ho TV, Sanchez-Lara PA, Urata M, Dixon MJ, Chai Y. Smad4-Irf6 genetic interaction and TGFbeta-mediated IRF6 signaling cascade are crucial for palatal fusion in mice. Development. 2013;140:1220–1230. doi: 10.1242/dev.089615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Emmert-Streib F, Glazko GV. Network biology: a direct approach to study biological function. Wiley Interdiscip Rev Syst Biol Med. 2011;3:379–391. doi: 10.1002/wsbm.134. [DOI] [PubMed] [Google Scholar]
- 48.Bensimon A, Heck AJ, Aebersold R. Mass spectrometry-based proteomics and network biology. Annu Rev Biochem. 2012;81:379–405. doi: 10.1146/annurev-biochem-072909-100424. [DOI] [PubMed] [Google Scholar]
- 49.Ito Y, Yeo JY, Chytil A, Han J, Bringas P, Jr, Nakajima A, Shuler CF, Moses HL, Chai Y. Conditional inactivation of Tgfbr2 in cranial neural crest causes cleft palate and calvaria defects. Development. 2003;130:5269–5280. doi: 10.1242/dev.00708. [DOI] [PubMed] [Google Scholar]
- 50.Chai Y, Jiang X, Ito Y, Bringas P, Jr, Han J, Rowitch DH, Soriano P, McMahon AP, Sucov HM. Fate of the mammalian cranial neural crest during tooth and mandibular morphogenesis. Development. 2000;127:1671–1679. doi: 10.1242/dev.127.8.1671. [DOI] [PubMed] [Google Scholar]
- 51.Iwata J, Hacia JG, Suzuki A, Sanchez-Lara PA, Urata M, Chai Y. Modulation of noncanonical TGF-beta signaling prevents cleft palate in Tgfbr2 mutant mice. J Clin Invest. 2012;122:873–885. doi: 10.1172/JCI61498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Iwata J, Suzuki A, Pelikan RC, Ho TV, Sanchez-Lara PA, Chai Y. Modulation of lipid metabolic defects rescues cleft palate in Tgfbr2 mutant mice. Hum Mol Genet. 2013 doi: 10.1093/hmg/ddt410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ozturk F, Li Y, Zhu X, Guda C, Nawshad A. Systematic analysis of palatal transcriptome to identify cleft palate genes within TGFbeta3-knockout mice alleles: RNA-Seq analysis of TGFbeta3 Mice. BMC Genomics. 2013;14:113. doi: 10.1186/1471-2164-14-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Doyle JP, Dougherty JD, Heiman M, Schmidt EF, Stevens TR, Ma G, Bupp S, Shrestha P, Shah RD, Doughty ML, et al. Application of a translational profiling approach for the comparative analysis of CNS cell types. Cell. 2008;135:749–762. doi: 10.1016/j.cell.2008.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sanz E, Yang L, Su T, Morris DR, McKnight GS, Amieux PS. Cell-type-specific isolation of ribosome-associated mRNA from complex tissues. Proc Natl Acad Sci U S A. 2009;106:13939–13944. doi: 10.1073/pnas.0907143106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Deal RB, Henikoff S. The INTACT method for cell type-specific gene expression and chromatin profiling in Arabidopsis thaliana. Nat Protoc. 2011;6:56–68. doi: 10.1038/nprot.2010.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gay L, Miller MR, Ventura PB, Devasthali V, Vue Z, Thompson HL, Temple S, Zong H, Cleary MD, Stankunas K, et al. Mouse TU tagging: a chemical/genetic intersectional method for purifying cell type-specific nascent. RNA Genes Dev. 2013;27:98–115. doi: 10.1101/gad.205278.112. [DOI] [PMC free article] [PubMed] [Google Scholar]