Abstract
Cellular mechanical properties have been observed to have important implications for pathogenesis and pathophysiology. These observations have led to the recent development of a unique class of biomarkers: mechanical biomarkers. Compared with the traditional biochemical-based biomarkers (e.g., antibodies), mechanical biomarkers have many advantages such as label-free, low cost, convenient maintenance, and reduced assay time. In the past few years, there has been an increasing effort to exploit cellular mechanical biomarkers in microfluidic devices. This trend makes sense because microfluidic devices often feature structures that have characteristic lengths similar to those of cells, which renders them uniquely capable of probing and utilizing mechanical biomarkers. In this Focus article, we discuss a few examples of mechanical biomarker-based microfluidic applications. We believe that these examples are just the tip of the iceberg and that the full potential of mechanical biomarkers in microfluidic-based diagnostics and therapeutics has yet to be revealed.
Introduction
It has long been suggested that intracellular structural rearrangements caused by diseases often lead to changes in cellular mechanical properties.1 One classic example is sickle cell anemia, in which a single-point genetic mutation causes the change of deformability and significant reduction of oxygen-carrying capability of red blood cells (RBCs). Despite the increasing evidence that cellular mechanical properties, such as deformability, are strongly correlated to human disease states, current practices in medical diagnostics are still dominated by biochemical markers such as antibodies. For example, let us consider flow cytometry, a commonly used, high-throughput, single-cell analytical tool. In flow cytometry, the detection of cells is often based on fluorescently labeled antibodies. Cells labeled with fluorescently tagged antibodies are aligned and forced to pass through a light-detection region where they are examined with highly focused laser beams. Cell properties and states can then be extracted by analyzing their fluorescence and/or scattering properties.2
In recent years, efforts to utilize cellular mechanical properties, or mechanical biomarkers, to provide new avenues for detecting and treating diseases have increased.3 Compared with biochemical biomarkers, mechanical biomarkers are label-free—measuring cellular mechanical properties can eliminate many costly sample labeling and preparation steps and significantly reduce assay time and complexity. Since microfluidic devices4–7 often have features with characteristic lengths similar to that of cells, they are excellent platforms for probing cellular mechanical biomarkers to enable novel diagnostic and therapeutic applications. By observing the mechanical behavior of cells in these cell-sized microfluidic structures and exploiting interactions between individual cells and the microfluidic structures, many new and exciting studies and applications become possible. In this Focus article we will discuss several interesting works along this front. We expect that as we improve our understanding of the implication of cell mechanics in various types of diseases, mechanical biomarker-enabled microfluidic approaches will have a more significant impact on many biological studies and clinical applications.
Interrogating disease states via analysis of mechanical biomarkers
Microfluidic devices are unique in their capability to probe cellular mechanical properties such as deformability. Tapping into this capability, one could develop a new class of diagnostic methods. Recently Bow et al. took advantage of the deformability of RBCs and demonstrated a deformability-based, microfluidic flow cytometer with application to malaria, a life-threatening infectious disease.8 Malaria is a result of infection from various types of protists, with Plasmodium falciparum being the most virulent. Plasmodium falciparum acts as a parasite to RBCs, causing symptoms such as fever, coma, or even death. Infected RBCs experience several stages including ring form, trophozoites, and schizonts. During this process, cell deformability is dramatically reduced, with cells becoming several times stiffer in earlier ring form to dozens of times stiffer in late trophozoites and schizonts stages. RBC deformability has strong implications for understanding the pathophysiology of malaria, especially its infection mechanism. Bow et al. developed a high-throughput, deformability-based flow cytometry device for rapid examination of the deformability of infected RBCs. Using this device, the authors measured the dynamic mechanical deformability of cells in addition to conducting typical fluorescence measurements.
Figure 1A shows the schematic of the deformability cytometry device. It contains an array of triangle-shaped pillars. The gaps between pillars are designed to add constraints for RBCs to pass through. In a pressure-driven flow, uninfected and infected RBCs travel through the gaps at different velocities, due to their difference in deformability. Figure 1B shows the identification of infected and uninfected RBCs in the channels, indicated by red and blue arrows, respectively. Using such a deformability-based cytometry device, the difference between the uninfected and infected RBCs can be well characterized. Figure 1C is a two-dimensional (2D) plot of velocity vs. fluorescence intensity of RBCs tested (infected RBCs are fluorescently dyed). Clearly the infected RBCs show much lower speed travelling through the pillar array due to their reduced deformability. This device shows great promise in deformability-based, microfluidic cell analysis for diagnosis and drug development. The authors envision the use of this device for screening drug compounds that alter the rigidity of early-stage infected RBCs to help the spleen remove the infected cells or to reduce the rigidity of late-stage infected RBCs to prevent capillary blockage.
Fig. 1.
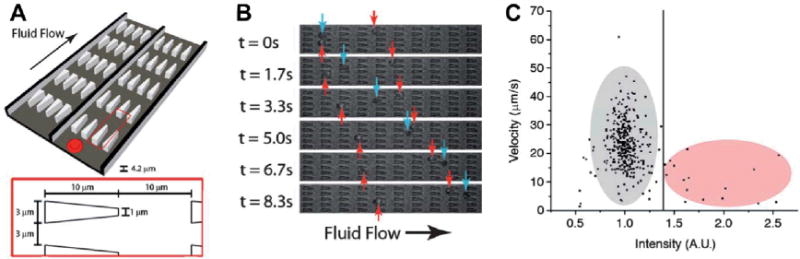
Principles of pillar-based deformability cytometry.8 (A) Schematic of the device design. The device contains an array of obstacles. Physical dimensions of the obstacles are shown in the inset. Each array contains 10 by 200 obstacles. (B) Optical images of infected RBCs (red arrows) and uninfected RBCs (blue arrows) in the device. (C) Velocity vs. intensity for RBCs (infected cells: gray; uninfected cells: red).
The method described above is an excellent example of how cell deformability can be exploited by microfluidic devices to enable novel disease diagnosis and treatment methods. However in the case of malaria, the change of RBC deformability is mainly due to the uptake of rigid protist parasites. Therefore, the question is whether deformability analysis is only helpful in the diagnosis of infectious diseases or might it have a broader impact in the diagnosis of other diseases such as cancer. Recently, Gossett et al. published an interesting work that shed some light onto this question.9 In this work, the authors developed a method for the clinical screening of pleural fluids for malignant cells and the characterization of stem cell differentiation states using a “hydrodynamic stretching” based deformability cytometry device. The device (Fig. 2A) combines the authors’ previously introduced inertial focusing component10 with a “hydrodynamic stretching” component (Fig. 2B). The inertial focusing component ensures the alignment of cells in a single-file fashion along the same streamline, placing them at the same initial condition for better stretching uniformity before they enter the “hydrodynamic stretching” zone (Fig. 2C). The “hydrodynamic stretching” zone is located at the center of an extensional flow. The cells are stretched upon entering this region (Fig. 2D). Stretching-induced changes in cell shape are recorded using a high-speed camera, and images are analyzed to quantify the cell deformability. The “hydrodynamic stretching” based deformability cytometry device provides several advantages, e.g., high throughput, less probability of clogging (especially compared with methods using physical constraints such as pillar arrays), and flexibility for fine-tuning hydrodynamic stress. Using this approach, the authors successfully demonstrated the possibility to predict disease states of cancer and immune activities by measuring the deformability of leukocytes and malignant cells in pleural fluids, and revealed deformability as an early biomarker for pluripotent stem cell differentiation.
Fig. 2.
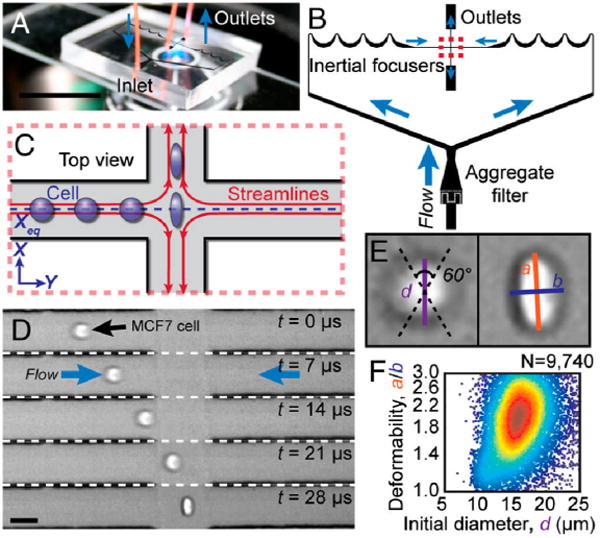
Principles of “hydrodynamic stretching” based deformability cytometry. (A) A photograph of the microfluidic deformability cytometry device. (B) A schematic of the microfluidic device including the “inertial focusing” region and the “hydrodynamic stretching” region. (C) A schematic of the deformation of a cell delivered to the center of an extensional flow via inertial focusing. (D) High-speed microscopic images showing a focused cell entering the extensional flow region. (E) Definition of the shape parameters extracted from images. (F) Density scatter plot of size and deformability measurements. Images reproduced from ref. 9.
Exploiting mechanical biomarkers for cell separation
Cell separation is another area of microfluidics that could significantly benefit from exploiting mechanical biomarkers. We may take the classic deterministic lateral displacement (DLD)11 as an example. A typical DLD-based separation device (Fig. 3) makes use of the asymmetric bifurcation of laminar flow around obstacles. In such a device, a particle’s path is determined by its size. The flow between two posts can be divided into three lanes (lane 1, 2, and 3, indicated in Fig. 3). If the diameter of the particle is less than the width of lane 1, the particle will flow around the posts and fall repeatedly into the same lane. Thus although the particle displays zigzag motion, overall it always travels in straight lines and the posts will not cause lateral displacements of the particle (Fig. 3A). In contrast, if the diameter of the particle is larger than the width of lane 1, the particle will be displaced laterally due to constantly “bumping” into the posts. The overall effect is that the particle will be forced to travel laterally and be separated from smaller particles (Fig. 3B).
Fig. 3.
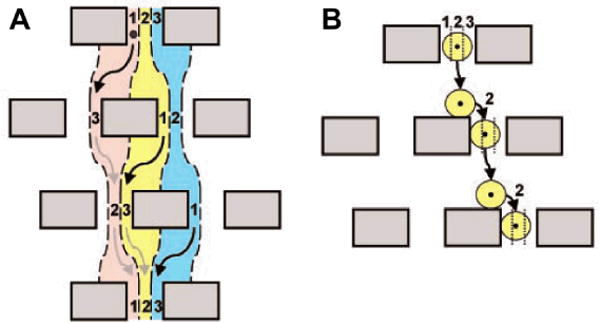
The principle of deterministic lateral displacement (DLD). (A) Three fluid streams (represented by red, yellow, and blue) enter the three lanes (represented by 1, 2, and 3, from left to right, respectively) at the first obstacle row. Small particles will follow the streamlines and fall repeatedly into the same lane. (B) Big particles travel laterally as they enter the next row of obstacles. Image reproduced with permission from ref. 11.
DLD is a very effective and highly precise method for size-based particle/cell separation. For the separation of rigid spherical particles, the outcome is fairly predictable. However, there are often complications for biological cells because their deformation and shape can affect their trajectories and hence potentially change the outcome of the separation. A recent study by Beech et al. demonstrated how shape and deformability can be utilized to develop novel methods for cell separation.12 In this study, the authors employed deformable and disc-shaped RBCs. In a DLD device, the separation of RBCs is determined by the effective diameters of cells (Fig. 4A). The effective diameters of RBCs are dependent on multiple factors including: the state of the cell (Fig. 4B), the extent of cell deformation (Fig. 4C), and the orientation of the cell (Fig. 4D). Beech et al. suggested several simple methods for changing the effective diameters of cells (such as RBCs) in DLD devices. For example, they altered the depth of the DLD device to force cells to be oriented in certain ways (Fig. 4 E, F, and G); they also tuned the fluid shear rates to achieve different cell deformation (Fig. 4C). Both approaches altered effective cell diameters and thus changed the outcome of cell separation. The authors tested their theory using three red blood cell types in devices with different heights and at different shear rates and achieved very promising results. This work indicates great potential in the use of tailored DLD devices to separate cells based on their mechanical properties. One could envision that once we achieve a better understanding of cellular mechanical biomarkers for various diseases, such tailored DLD devices can be invaluable in many applications that require high-throughput, label-free cell separation, e.g., separation of circulating tumor cells (CTCs).
Fig. 4.
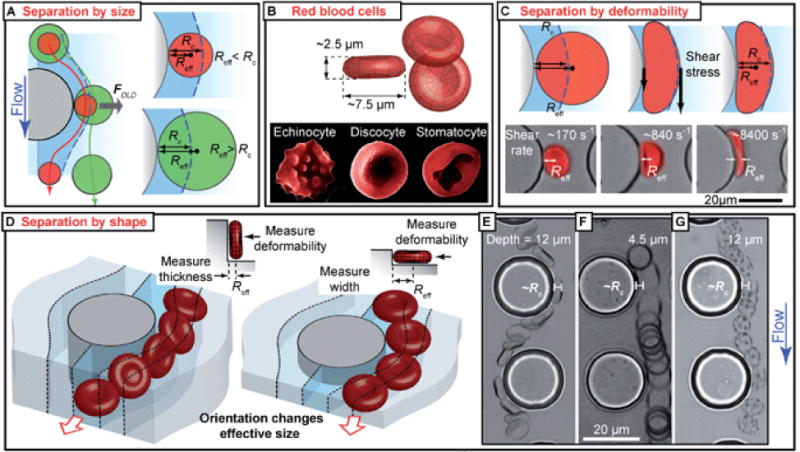
DLD-based separation by cell shape and deformability.12 (A) DLD separation based on effective cell diameter. (B) Different forms of RBCs in normal and abnormal states. (C) Shear-induced deformation of RBCs affects effective cell diameter. (D) Orientation affects effective cell diameter. (E, F, G) Channel depth affects RBC orientation, hence effective cell diameter and cell trajectory in a DLD device.
Conclusion
Because the critical lengths of many microfluidic structures are similar to those of biological cells, microfluidic devices provides an excellent platform for taking advantage of cellular mechanical biomarkers and enabling a new class of mechanical biomarker-based, diagnostic and therapeutic tools. The recent advances outlined in this Focus article are intriguing and will continue to inspire others to pursue the potential of cellular mechanical biomarkers in microfluidic applications. We envision that by marrying cell mechanics and microfluidics, and by exploiting the merits offered by both disciplines, the field of mechanical biomarker based microfluidics will not only lead to more effective, less expensive, and faster cell-based analytical methods, but also help devise more efficient strategies for disease treatment.
Acknowledgments
This research was supported by National Institutes of Health (NIH) Director’s New Innovator Award (1DP2OD007209-01). We thank Joseph Rufo and Alex Ahmadi for helpful discussion.
References
- 1.Suresh S, Spatz J, Mills JP, Micoulet A, Dao M, Lim CT, Beil M, Seufferlein T. Acta Biomaterialia. 2005;1:15–30. doi: 10.1016/j.actbio.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 2.Mao X, Nawaz AA, Lin S-CS, Lapsley MI, Zhao Y, McCoy JP, El-Deiry WS, Huang TJ. Biomicrofluidics. 2012;6:024113. doi: 10.1063/1.3701566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mills JP, Diez-Silva M, Quinn DJ, Dao M, Lang MJ, Tan KSW, Lim CT, Milon G, David PH, Mercereau-Puijalon O, Bonnefoy S, Suresh S. Proc Nat Acad Sci. 2007;104:9213–9217. doi: 10.1073/pnas.0703433104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shi J, Ahmed D, Mao X, Lin S-CS, Lawit A, Huang TJ. Lab on a Chip. 2009;9:2890–2895. doi: 10.1039/b910595f. [DOI] [PubMed] [Google Scholar]
- 5.Moraes C, Sun Y, Simmons CA. Integrative Biology. 2011;3:959–971. doi: 10.1039/c1ib00056j. [DOI] [PubMed] [Google Scholar]
- 6.Zheng Y, Shojaei-Baghini E, Azad A, Wang C, Sun Y. Lab Chip, Lab Chip. 2012;12:2560–2567. doi: 10.1039/c2lc21210b. [DOI] [PubMed] [Google Scholar]
- 7.Ding X, Lin S-CS, Kiraly B, Yue H, Li S, Chiang I-K, Shi J, Benkovic SJ, Huang TJ. Proc Nat Acad Sci. 2012;109:11105–11109. doi: 10.1073/pnas.1209288109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bow H, Pivkin IV, Diez-Silva M, Goldfless SJ, Dao M, Niles JC, Suresh S, Han J. Lab Chip. 2011;11:1065–1073. doi: 10.1039/c0lc00472c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gossett DR, Tsea HTK, Leec SA, Yinge Y, Lindgren AG, Yang OO, Rao J, Clark AT, Di Carlo D. Proc Nat Acad Sci. 2012 doi: 10.1073/pnas.1200107109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Di Carlo D, Irimia D, Tompkins RG, Toner M. Proc Nat Acad Sci. 2007;104:18892–18897. doi: 10.1073/pnas.0704958104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang LR, Cox EC, Austin RH, Sturm JC. Science. 2004;304:987–990. doi: 10.1126/science.1094567. [DOI] [PubMed] [Google Scholar]
- 12.Beech JP, Holm SH, Adolfsson K, Tegenfeldt JO. Lab Chip. 2012;12:1048–1051. doi: 10.1039/c2lc21083e. [DOI] [PubMed] [Google Scholar]


