Abstract
The covalent conjugation of bovine serum albumin (BSA) to disulfide cross-linked polymeric nanogels is reported. Polymeric nanogel precursors were synthesized via a reversible addition-fragmentation chain transfer (RAFT) random copolymerization of poly(ethylene glycol) methyl ether methacrylate (PEGMA) and pyridyl disulfide methacrylate (PDSMA). Reaction of the p(PEGMA-co-PDSMA) with dithiothreitol resulted in the formation of nanogels. PDSMA serves as both a crosslinking agent and a reactive handle for the surface modification of the nanogels. Lipophilic dye, DiI, was sequestered within the nanogels by performing the crosslinking reaction in the presence of the hydrophobic molecule. Thiol-enriched BSA was conjugated to nanogels loaded with DiI via a disulfide reaction between the BSA and the surface exposed nanogel pyridyl disulfides. Conjugation was confirmed by fast protein liquid chromatography, dynamic light scattering, and agarose and polyacrylamide gel electrophoresis. We expect that this methodology is generally applicable to the preparation of nanogel-protein therapeutics.
1. Introduction
The development of therapeutic encapsulated nanomaterials is critical for the delivery of therapeutic agents.1, 2 “Smart” or stimuli-responsive materials, which encapsulate lipophilic small molecules, have been synthesized to provide a vast array of possibilities for release of such molecules.3 Furthermore, incorporation of multiple molecules into a single carrier is of great interest in areas such as theranostics and combinatorial therapy.4, 5 Conjugation of therapeutic proteins is important, because the resulting nanomaterials often exhibit improved therapeutic activity compared to the unmodified counterparts.6
Due to their size, physical properties, and high degree of functionality, polymeric nanogels are ideal materials for drug delivery.7-10 Synthesis of nanogels and similar polymeric nanomaterials has been achieved by a variety of methods including micromolding techniques,11 emulsion methods,12, 13 dispersion polymerizations,14, 15 and crosslinking of polymeric starting materials.16-20 We have previously reported polymeric nanogels, which demonstrated excellent stability and encapsulation capabilities.21-24 These nanogels can be assembled in aqueous solution by a simple self-crosslinking reaction from random copolymer precursors in which the template structure contains a hydrophilic monomer, poly(ethylene glycol) methyl ether methacrylate (PEGMA), and a hydrophobic monomer, pyridyl disulfide methacrylate (PDSMA), that is also used as the cross-linker. This self-crosslinking strategy avoids the harsh conditions of microemulsion polymerization. Manipulation of the polymeric structure affords control over encapsulation of hydrophilic/hydrophobic molecules, as well as ligand incorporation.25 This system provides for well defined nanogels with sizes ranging from 10 to 200 nm, such that nanogels could be designed to take advantage of the enhanced permeation and retention (EPR) effect.26, 27 These nanogels have been shown to successfully encapsulate lipophilic small molecules such as 1,1′-dioctadecyl-3,3,3′3′-tetramethylindocarbocyanine perchlorate (DiI), a lipophilic carbocyanine dye, and doxorubicin. The gels were also easily surface-modified by thiol-containing ligands such as cysteine-modified folic acid.21-24 Recently, we have reported a delivery system based on this nanogel technology that concurrently encapsulates lipophilic small molecules within the interior of a nanoassembly and complexes negatively charged proteins at the positively charged nanogel surface.24 Although this non-covalent electrostatic interaction is a simple way to modify hydrogels, it is less robust and therefore may not be suitable for all applications. Covalent conjugation of proteins to the nanogel surface is an attractive alternative to physical adsorption. By covalently conjugating proteins to the nanogel surface, the proteins will likely remain bound to the nanogel in complex biological environments, such as blood, for longer periods of time than if the protein and nanogel were bound by electrostatic interactions.
Bioconjugation of polymers to proteins depends on a variety of factors, such as the availability of reactive amino acid residues, mild reaction conditions, and solubility of reagents. For direct-covalent attachment, amino acid residues such as lysine, and cysteine are commonly exploited.28 If a protein does not contain the required amino acid residue for conjugation, the desired residue can be genetically engineered into the protein structure.29, 30 A simpler alternative to manipulation of the protein amino acid sequence is the use of engineered linkers or ligands to install the required functionality. Such linkers have shown great success in expanding the reactivity of a protein, while retaining protein structure and function. These linkers broaden the possibilities of protein conjugation to supramolecular, microsized or nanosized materials.
A wide variety of materials have been conjugated to proteins such as small molecules, polymers, and nanoparticles.31-33 Protein PEGylation, pioneered by Abuchowski and coworkers,34, 35 has proven to effectively stabilize proteins in vivo;36 this strategy is currently used in several protein-based therapeutics.37 Controlled radical polymerization techniques (CRP) have been employed for the preparation of protein-polymer conjugates due to the wide monomer tolerance, control of molecular weight distributions, and the ease of introducing protein reactive end-group functionality.33, 38, 39 End-groups such as amine-reactive aldehydes,40 and cysteine-reactive maleimides,41, 42 and pyridyl disulfides,29, 43 are typically used in the formation of protein-polymer conjugates. Proteins can be conjugated to polymeric nanomaterials such as polymersomes,44 and micelles.45 For example, Lu and co-workers have conjugated BSA to the exterior of self-assembled glycopolymer micelles.45 The nanogel system is a simple and effective platform for the construction of multi-functional drug delivery vehicles. Herein we report the conjugation of bovine serum albumin (BSA) to the surface of DiI encapsulated nanogels.
2. Experimental
2.1 Materials
All chemicals, 2,2′-dithiodipyridine, PEGMA (MW 450), d,l-dithiothreitol (DTT), azobisisobutyronitrile (AIBN), DiI, BSA, UltraPure agarose and solvents were purchased from commercial sources (Fischer, Sigma-Aldrich or Invitrogen) and were used as received, except AIBN which was purified by recrystallization. The kD Mini-PROTEAN-TGX gels for SDS-PAGE studies were purchased from Bio-Rad. All buffers for protein conjugations were filtered through 0.22 μm syringe filters and degassed by bubbling with argon for 30 min. N-Succinimidyl-S-acetylthiopropionate (SATP) was prepared according to literature procedures.46
2.2 Measurements
1H-NMR spectra were recorded on a 400 MHz Bruker NMR spectrometer using the residual proton resonance of the solvent as the internal standard. Chemical shifts are reported in parts per million (ppm). Molecular weights of the polymers were estimated by gel permeation chromatography (GPC) in THF using PMMA standards with a refractive index detector. Dynamic light scattering (DLS) measurements were performed using a Malvern Nanozetasizer. UV-Visible spectroscopy was conducted on Thermo Scientific NanoDrop 2000 and Biomate 5 spectrophotometers. Fast protein liquid chromatography (FPLC) was performed on a Bio-Rad BioLogic DuoFlow chromatography system equipped with a GE Healthcare Life Sciences Superose 6 10/300 column. A 50 mM sodium phosphate (pH 7.5) buffer containing 15 mM NaCl at 4 °C was used as the solvent (flow rate: 0.3 mL/min). Fluorescent gel images were obtained on a Bio-Rad FX Pro Plus Fluorimager/PhosphorImager.
2.3 Synthesis
Synthesis of p(PEGMA-co-PDSMA)
A mixture of 2-cyano-2-propyl benzodithioate (RAFT reagent) (21 mg, 0.0949 mmol), PDSEMA (860 mg, 3.37 mmol), PEGMA (620 mg, 1.31 mmol) and AIBN (5.0 mg, 0.0305 mmol) were dissolved in THF (3 mL) and degassed by performing three freeze-pump-thaw cycles. This reaction vessel was sealed and then placed in a pre-heated oil bath at 60 °C for 12 hours. To remove unreacted monomers and purify the polymer, the resultant mixture was precipitated in cold ethyl ether (20 mL) to yield the random copolymer as a waxy solid. GPC (THF) Mn: 23 kDa. PDI: 1.38. 1H NMR (400 MHz, CDCl3) δ: 8.45, 7.66, 7.09, 4.20-4.06, 3.90-3.36, 3.01, 2.15-1.62, 1.43-0.86. The molar ratio between two blocks was determined by integrating the methoxy proton in the polyethylene glycol unit and the aromatic proton in the pyridine and found to be 29:71 PEO:PDSEMA.
Synthesis of DiI Loaded Nanogel
The polymer (10 mg) was dissolved in water (1 mL) and 2 wt% of hydrophobic dye (0.2 mg of DiI) in acetone (100 μL) was added. The mixture was stirred for 6 hours at room temperature, open to the atmosphere allowing the organic solvent to evaporate. To this micellar aggregate solution, was added a measured amount of DTT and then the mixture was stirred for 12 hours at room temperature to allow for crosslinking. The resulting nanogels were purified by filtration and then dialyzed using a membrane with a molecular weight cutoff of 7,000 g/mol (Thermo Fisher SnakeSkin).
Preparation of Thiolated-AlexaFluor 488-BSA
The preparation of thiolated AlexaFluor 488-BSA was undertaken by modifying a literature procedure.47 BSA (5 mg) was dissolved in 0.5 mL of 50 mM sodium phosphate (pH 7.5) containing 1 mM ethylenediaminetetraacetic acid (EDTA). Then, 5 μL of SATP in DMF (16 mg/mL) was added to the BSA solution, and the mixture was incubated for 1 h at 20 °C. To the BSA solution, 1.75 mL of the EDTA in 50 mM sodium phosphate pH 7.5 buffer was added, followed by 250 μL of AlexaFluor 488 carboxylic acid succinimidyl ester in DMF (2 mg/mL). This solution was incubated for 1 h at 20 °C. The SATP and AlexaFluor 488 modified BSA was then purified by ultrafiltration (Amicon Ultra, 30 kDa MWCO). After purification, SATP-AlexaFluor 488-BSA was diluted in 50 mM sodium phosphate buffer (pH 7.5) containing 1 mM EDTA up to a total volume of 0.5 mL and 10 mg/mL concentration. To deprotect the acetylated thiols, 50 μL of a solution containing 0.5 M hydroxyl amine hydrochloride in 50 mM sodium phosphate buffer (pH 7.5) containing 25 mM EDTA was then added to the SATP-AlexaFluor 488-BSA solution and incubated for 2 hours at 20 °C. Following deprotection with hydroxylamine, the thiolated-AlexaFluor 488-BSA was purified by FPLC and concentrated to a final concentration of 2 mg/mL by ultrafiltration. Ellman’s assay was used to determine the extent of the thiol-modification (BSA: free thiol; 1:4.1). UV-Vis spectroscopy was used to determine the extent of AlexaFluor 488 labeling of BSA (BSA: AlexaFluor 488; 1:1.7).
Conjugation of Thiolated-AlexaFluor 488-BSA to DiI Nanogels
A solution of 6% crosslinked, 2 wt% DiI encapsulated nanogels (0.50 mg) in 250 μL 50 mM sodium phosphate (pH 7.5) with 1 mM EDTA was added to a 250 μL solution of thiolated-AlexaFluor 488-BSA (1 mg) in the same buffer. The solution was then agitated for 24 hours at 4 °C on a thermo-shaker (TSZ Scientific). The resulting conjugates were purified by FPLC.
3. Results and Discussion
Synthesis of DiI Loaded Nanogels
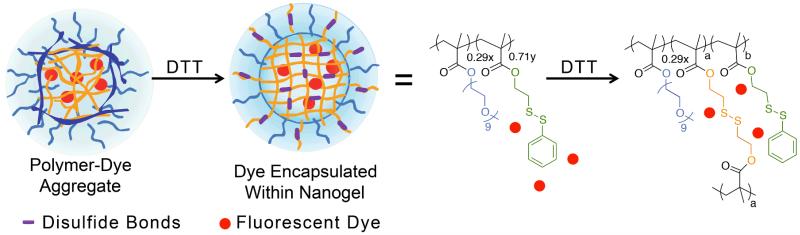
The nanogel was prepared from a random copolymer precursor composed of 29% PEGMA and 71% of a lipophilic PDSMA monomer that also acts as the crosslinkable unit. A 6% crosslinked nanogel was synthesized since we have previously shown that the crosslinked density of these nanogels can be tuned.22, 23 As depicted in Figure 1, self-formation of polymer aggregates with the lipophilic small molecule (DiI) in aqueous media can further lead to the in situ non-covalent loading of the dye within the nanogel interior. The crosslinking event is initiated by the addition of DTT, a reducing agent, and the extent of crosslinking is controlled with the amount of DTT added to the reaction mixture. Although it is possible to use different polymer precursors to achieve crosslinked nanogels, we selected disulfide-based nanogels for convenience and reversibility. Because the crosslink density of the polymeric nanogel is low (6%), it contains a significant number of available pyridyl disulfide functionalities for protein conjugation through the thiol-disulfide exchange reaction.
Fig.1.
Structure of the Nanogel’s polymer precursor and nanogel formation.
Conjugation and Purification of Nanogels to Thiolated-BSA
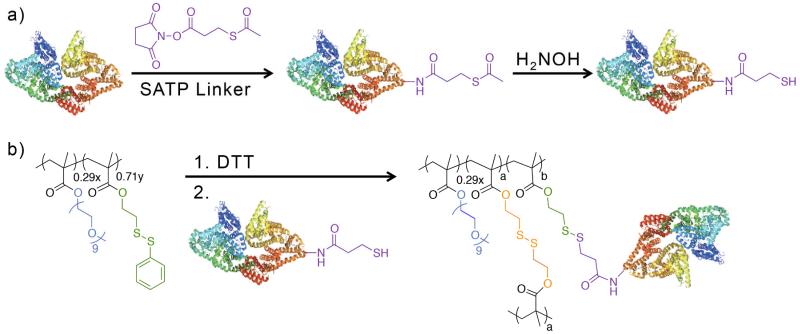
We chose globular BSA protein with molecular weight of 66.5 kDa and pI of 4.7 as the model protein, because it contains a free cysteine on its surface at amino acid 34. Our first attempt to attach the protein to the nanogel was through direct conjugation using the available free thiol of Cys34 of BSA. The conjugation was expected to proceed through a simple thiol-disulfide exchange reaction between the free thiol groups from the BSA and the unreacted PDS moieties of the nanogel. Although the BSA Cys 34 has been successfully used for polymer conjugation,29 direct conjugation to the nanogel surface was not successful. We believe this may be due to the size of the nanogels, which are much larger and have bulkier structure compared to linear polymers. Although this Cys 34 is considered to be surface exposed on BSA, it appears that this amino acid is not accessible to the PDS groups of the nanoassembly.
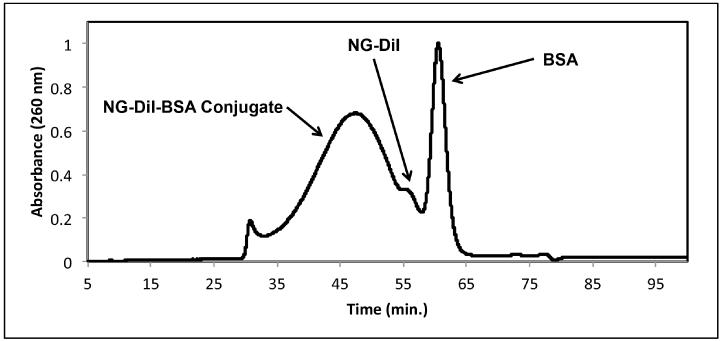
In order to introduce thiol groups that could be more accessible for nanogel conjugation, BSA was modified with the known thiolating agent SATP, resulting in a 1:4 BSA:thiol ratio (Figure 2a.). Bioconjugation was performed in a simple reaction by the addition of the pre-assembled DiI-containing crosslinked nanogel (NG-DiI) to a thiolated-BSA solution in 50 mM sodium phosphate buffer pH 7.5 as a 1:2 ratio (NG-DiI:BSA) (Figure 2b.) followed by purification by fast protein liquid chromatography (FPLC) using a size exclusion column. FPLC was also used for characterization, and Figure 3 shows the FPLC trace of the bioconjugation reaction mixture. It was observed that retention time of 60 and 55 minutes for the unmodified BSA and unmodified NG-DiI, respectively, shifted to ~ 46 min. This indicated that the particles were larger due to the conjugation of the protein, and that the bioconjugation was successfully achieved. A high molecular weight aggregate, with a retention time ~30 min, was also observed; this aggregate was also present in the unmodified nanogels. Fractions containing this aggregate were not collected with the purified nanogel conjugates.
Fig. 2.
(a) BSA modification with SATP linker, (b) NG-DiI-BSA conjugate formation. (BSA structure PDB: 3V03)
Fig. 3.
FPLC trace of nanogel conjugation mixture. Buffer was 50 mM sodium phosphate (pH 7.5) containing 15 mM NaCl, 4 °C, and flow rate: 0.3 mL/min.
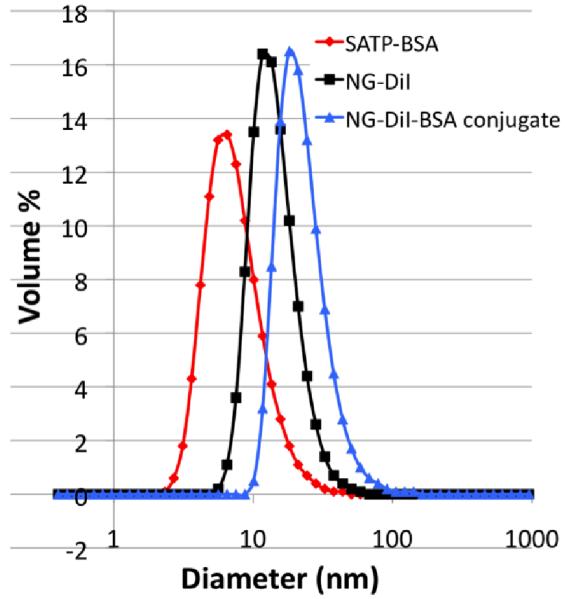
Next, differences in size between free NG-DiI, free BSA and bioconjugates were evaluated by dynamic light scattering (DLS). The hydrodynamic diameter of individual NG-DiI and free thiolated-BSA were found to be 14 nm and 8 nm, respectively. The NG-DiI-BSA bioconjugates sample was larger at 25 nm, also indicative of protein attachment to the surface of the nanogel (Figure 4).
Fig. 4.
DLS measurements of: thiolated BSA (8 nm, red), free NG-DiI (14 nm, black), NG-DiI-BSA conjugates (25 nm, blue).
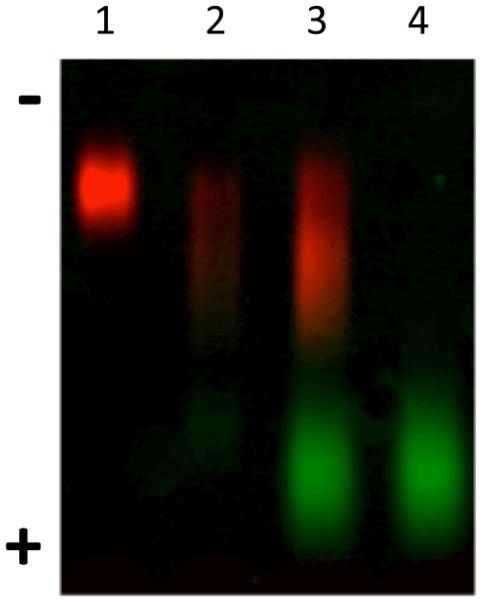
NG-DiI-BSA bioconjugation was verified by agarose electrophoresis, which separates molecules by charge difference (Figure 5). In order to image the results by fluorescence, thiolated-BSA was first labeled with AlexaFluor 488 (AF) dye and was conjugated to the DiI containing nanogel. Samples of free NG-DiI (lane 1, red) and free BSA (lane 4, green) were characterized along with the FPLC purified bioconjugate reaction mixture (lane 2) and the crude bioconjugation mixture (lane 3). The gel clearly shows a shift of the nanogel band towards the cathode in lane 2 and 3, indicating bioconjugation. Lane 3 and 2 are the same conjugate samples before and after purification, respectively. This demonstrates that the majority of the unconjugated BSA was successfully removed by FPLC.
Fig. 5.
Agarose electrophoresis gel of: NG-DiI (lane 1), purified NG-DiI-BSA conjugate (lane 2), crude NG-DiI-BSA conjugates reaction mixture (lane 3) and AlexaFluor 488 labeled BSA (lane 4).
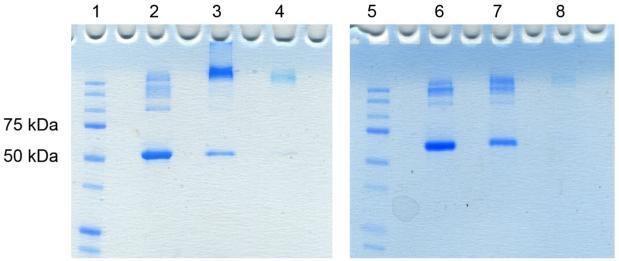
Next, we investigated the formation of NG-DiI-BSA bioconjugates by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) (Figure 6). The nanogels themselves are only slightly stained by coomassie blue. Thus, we anticipated that if conjugation was successful a more intense band should be visible at slightly higher molecular than free nanogel under non-reducing conditions. We also expected that under non-reducing conditions the NG-DiI-BSA bioconjugate band should stay intact and retain binding to the BSA. On the other hand, under reducing conditions the nanogels should disassemble and the conjugated protein should be released. SDS-PAGE confirmed the successful conjugation of NG-DiI-BSA. Under non-reducing conditions, the bioconjugate (lane 3) shows an increase in molecular weight compared to free BSA (lane 2) in addition to a faint band of free protein. Although this band is present in the stained SDS-PAGE gel, it is evident that most of the protein has been conjugated to the nanogel. Under reducing conditions the NG-DiI-BSA conjugate is disrupted (lane 7) by the reduction of disulfide bonds leading to release of the protein, as evident by the band for free BSA in lane 6. In this gel, non-reduced BSA (lane 2) appears to be smaller in size than the reduced BSA (lane 6); this is typical for BSA.
Fig. 6.
SDS-PAGE visualized by coomassie blue staining (Lane 1: protein marker; Lane 2: BSA non-reducing conditions; Lane 3: NG-DiI-BSA conjugate non-reducing conditions; Lane 4: NG-DiI non-reducing conditions; Lane 5: protein marker; Lane 6: BSA reducing conditions; Lane 7: NG-DiI-BSA conjugate reducing conditions; Lane 8: NG-DiI reducing conditions).
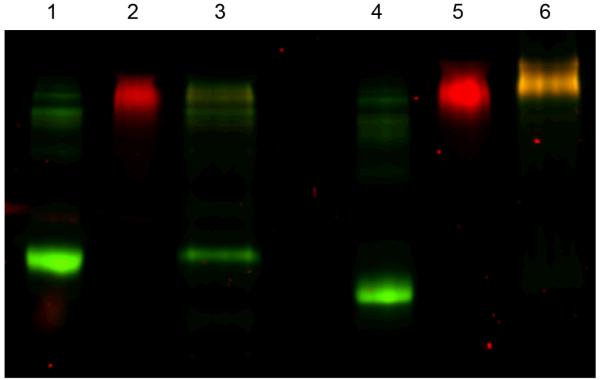
Figure 7 is the SDS-PAGE electrophoresis fluorescent images from the same samples. In these gels, free BSA (lanes 1 and 4) can be seen as a green band and free NG-DiI as a red band (lanes 2 and 5). When the nanogel and protein are conjugated the color is observed as yellow under non-reducing conditions (lane 6), demonstrating co-localization of the two fluorophores. Once the samples are subjected to reducing conditions release of the protein is evidenced by the appearance of green color again (lane 3) and disappearance of the red color from the release of DiI dye upon dissolution of the nanogel. Interestingly, the band for the unconjugated nanogel (lane 2) did show the red fluorescence of the DiI under these reducing conditions; however, the intensity was decreased. This indicates that the unmodified nanogel cleaved to some extent, but did not completely degrade during the short exposure time to DTT. It may be that the presence of the protein facilitates reduction of the nanogel because of its hydrophilic nature or that residual pyridyl disulfides on the unmodified gels cause reoxidation of the material. This is interesting because it suggests that nanogel degradation may be influenced by the surface functionality, and these studies are underway.
Fig. 7.
SDS-PAGE visualized by fluorescence (Lane 1: BSA reducing conditions; Lane 2: NG-DiI reducing conditions; Lane 3: NG-DiI-BSA conjugate reducing conditions; Lane 4: BSA non-reducing conditions; Lane 5: NG-DiI non-reducing conditions; Lane 6: NG-DiI-BSA conjugate non-reducing conditions).
Together, this data demonstrates that proteins can be successfully conjugated to nanogels loaded with hydrophobic molecules. Development of a polymeric nanocarrier like one presented herein provides a tool for the incorporation of imaging agents and therapeutic proteins for theranostic or combinatorial therapy applications. The use of nanocarriers for theranostics provides the opportunity for image-guided therapy. This ability to monitor treatment in real time enhances the probability of providing a more accurate disease treatment by allowing physicians to detect and treat simultaneously. In addition to this, combination of several therapeutics in a single carrier, especially if incorporation of therapeutics with different characteristics is facilitated, may enhance the effectiveness of the therapy by providing a synergistic therapeutic effect. Importantly the data show that proteins can be modified on the protein nanogels, suggesting that targeting proteins or antibodies may be possible, which would increase effectiveness, and this work is underway.
4. Conclusion
We have demonstrated the versatility of our nanogel system to encapsulate lipophilic small molecules on the interior and conjugate proteins at the nanogel exterior by covalent attachment. BSA was modified with a thiol linker, and was readily conjugated to the nanogels via disulfide exchange reactions with pyridyl disulfide moieties at the nanogel exterior. The results suggest that this nanogel system is a simple and effective platform for development of sophisticated drug delivery systems. This work provides the groundwork for successful application of therapeutic proteins and antibodies to the nanogels, which should be important for selective targeting of the nanogels.
Acknowledgements
HDM thanks the National Institutes of Health (R21 CA 137506-01) for funding. ST thanks the Army Research Office (57858CH) and the National Science Foundation for support through the Center for Hierarchical Manufacturing at UMass Amherst (CMMI-0531171). NMM thanks the National Science Foundation for a Graduate Research Fellowship (DGE-0707424). DCG thanks for NSF for support by the IGERT program through the Institute for Cellular Engineering at UMass Amherst.
Notes and References
- 1.Baldwin SP, Saltzman WM. Adv. Drug Deliv. Rev. 1998;33:71–86. doi: 10.1016/s0169-409x(98)00021-0. [DOI] [PubMed] [Google Scholar]
- 2.Avichezer D, Schechter B, Arnon R. React. Funct. Polym. 1998;36:59–69. [Google Scholar]
- 3.Zha L, Banik B, Alexis F. Soft Matter. 2011;7:5908–5916. [Google Scholar]
- 4.Agrawal V, Paul MK, Mukhopadhyay AK. J. Lipos. Res. 2005;15:141–155. doi: 10.1080/08982100500364081. [DOI] [PubMed] [Google Scholar]
- 5.Hoffmann S, Vystrcilova L, Ulbrich K, Etrych T, Caysa H, Mueller T, Maeder K. Biomacromolecules. 2012;13:652–663. doi: 10.1021/bm2015027. [DOI] [PubMed] [Google Scholar]
- 6.Broyer RM, Grover GN, Maynard HD. Chem. Commun. 2011;47:2212–2226. doi: 10.1039/c0cc04062b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Asadian-Birjand M, Sousa-Herves A, Steinhilber D, Cuggino JC, Calderon M. Curr. Med. Chem. 2012;19:5029–5043. doi: 10.2174/0929867311209025029. [DOI] [PubMed] [Google Scholar]
- 8.Raemdonck K, Demeester J, De Smedt S. Soft Matter. 2009;5:707–715. [Google Scholar]
- 9.Oh JK, Drumright R, Siegwart DJ, Matyjaszewski K. Prog. Polym. Sci. 2008;33:448–477. doi: 10.1016/j.progpolymsci.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yallapu MM, Jaggi M, Chauhan SC. Drug Discov. Today. 2011;16:457–463. doi: 10.1016/j.drudis.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rolland JP, Maynor BW, Euliss LE, Exner AE, Denison GM, DeSimone JM. J. Am. Chem. Soc. 2005;127:10096–10100. doi: 10.1021/ja051977c. [DOI] [PubMed] [Google Scholar]
- 12.Mitra S, Gaur U, Ghosh PC, Maitra AN. J. Control. Release. 2001;74:317–323. doi: 10.1016/s0168-3659(01)00342-x. [DOI] [PubMed] [Google Scholar]
- 13.Lee H, Mok H, Lee S, Oh YK, Park TG. J. Control. Release. 2007;119:245–252. doi: 10.1016/j.jconrel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 14.Rieger J, Grazon C, Charleux B, Alaimo D, Jerome C. J. Polym. Sci. Pol. Chem. 2009;47:2373–2390. [Google Scholar]
- 15.Shen WQ, Chang YL, Liu GY, Wang HF, Cao AN, An ZS. Macromolecules. 2011;44:2524–2530. [Google Scholar]
- 16.Thurmond KB, Kowalewski T, Wooley KL. J. Am. Chem. Soc. 1996;118:7239–7240. [Google Scholar]
- 17.Rijcken CJ, Snel CJ, Schiffelers RM, van Nostrum CF, Hennink WE. Biomaterials. 2007;28:5581–5593. doi: 10.1016/j.biomaterials.2007.08.047. [DOI] [PubMed] [Google Scholar]
- 18.Liu SY, Weaver JVM, Tang YQ, Billingham NC, Armes SP. Macromolecules. 2002;35:6121–6131. [Google Scholar]
- 19.Cheng C, Qi K, Germack DS, Khoshdel E, Wooley KL. Adv. Mater. 2007;19:2830–2835. [Google Scholar]
- 20.Stevens DM, Tempelaar S, Dove AP, Harth E. ACS Macro Letters. 2012;1:915–918. doi: 10.1021/mz300179r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ryu J-H, Bickerton S, Zhuang J, Thayumanavan S. Biomacromolecules. 2012;13:1515–1522. doi: 10.1021/bm300201x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ryu J-H, Chacko RT, Jiwpanich S, Bickerton S, Babu RP, Thayumanavan S. J. Am. Chem. Soc. 2010;132:17227–17235. doi: 10.1021/ja1069932. [DOI] [PubMed] [Google Scholar]
- 23.Ryu JH, Jiwpanich S, Chacko R, Bickerton S, Thayumanavan S. J. Am. Chem. Soc. 2010;132:8246–8247. doi: 10.1021/ja102316a. [DOI] [PubMed] [Google Scholar]
- 24.Gonzalez-Toro DC, Ryu J-H, Chacko RT, Zhuang J, Thayumanavan S. J. Am. Chem. Soc. 2012;134:6964–6967. doi: 10.1021/ja3019143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bickerton S, Jiwpanich S, Thayumanavan S. Mol. Pharm. 2012;9:3569–3578. doi: 10.1021/mp3004226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matsumura Y, Maeda H. Cancer Res. 1986;46:6387–6392. [PubMed] [Google Scholar]
- 27.Duncan R. Nat. Rev. Drug Discov. 2003;2:347–360. doi: 10.1038/nrd1088. [DOI] [PubMed] [Google Scholar]
- 28.Sletten EM, Bertozzi CR. Angewandte Chemie International Edition. 2009;48:6974–6998. doi: 10.1002/anie.200900942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bontempo D, Heredia KL, Fish BA, Maynard HD. J. Am. Chem. Soc. 2004;126:15372–15373. doi: 10.1021/ja045063m. [DOI] [PubMed] [Google Scholar]
- 30.Lundblad RL. Techniques in Protein Modification. CRC Press; Boca Raton, FL: 1995. [Google Scholar]
- 31.Rana S, Yeh Y-C, Rotello VM. Curr. Opin. Chem. Biol. 2010;14:828–834. doi: 10.1016/j.cbpa.2010.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thompson S. In: Walker JM, editor. Humana Press; 2009. pp. 1715–1726. [Google Scholar]
- 33.Grover GN, Maynard HD. Curr. Opin. Chem. Biol. 2010;14:818–827. doi: 10.1016/j.cbpa.2010.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abuchowski A, Vanes T, Palczuk NC, Davis FF. J. Biol. Chem. 1977;252:3578–3581. [PubMed] [Google Scholar]
- 35.Pasut G, Veronese F. In: Polymer Therapeutics I. Satchi-Fainaro R, Duncan R, editors. Springer; Berlin Heidelberg: 2006. pp. 95–134. [Google Scholar]
- 36.Joralemon MJ, McRae S, Emrick T. Chem. Commun. 2010;46:1377–1393. doi: 10.1039/b920570p. [DOI] [PubMed] [Google Scholar]
- 37.Alconcel SNS, Baas AS, Maynard HD. Polym. Chem. 2011;2:1442–1448. [Google Scholar]
- 38.Le Droumaguet B, Nicolas J. Polym. Chem. 2010;1:563–598. [Google Scholar]
- 39.Sumerlin BS. ACS Macro Letters. 2011;1:141–145. doi: 10.1021/mz200176g. [DOI] [PubMed] [Google Scholar]
- 40.Tao L, Mantovani G, Lecolley F, Haddleton DM. J. Am. Chem. Soc. 2004;126:13220–13221. doi: 10.1021/ja0456454. [DOI] [PubMed] [Google Scholar]
- 41.Velonia K, Rowan AE, Nolte RJM. J. Am. Chem. Soc. 2002;124:4224–4225. doi: 10.1021/ja017809b. [DOI] [PubMed] [Google Scholar]
- 42.De P, Li M, Gondi SR, Sumerlin BS. J. Am. Chem. Soc. 2008;130:11288–11289. doi: 10.1021/ja804495v. [DOI] [PubMed] [Google Scholar]
- 43.Liu JQ, Bulmus V, Herlambang DL, Barner-Kowollik C, Stenzel MH, Davis TP. Angew. Chem. Int. Edit. 2007;46:3099–3103. doi: 10.1002/anie.200604922. [DOI] [PubMed] [Google Scholar]
- 44.van Dongen SFM, Nallani M, Cornelissen JJLM, Nolte RJM, van Hest JCM. Chemistry – A European Journal. 2009;15:1107–1114. doi: 10.1002/chem.200802114. [DOI] [PubMed] [Google Scholar]
- 45.Xiao N-Y, Li A-L, Liang H, Lu J. Macromolecules. 2008;41:2374–2380. [Google Scholar]
- 46.Liu L, Rozenman M, Breslow R. J. Am. Chem. Soc. 2002;124:12660–12661. doi: 10.1021/ja028151k. [DOI] [PubMed] [Google Scholar]
- 47.Hermanson GT. Bioconjugate Techniques. Elsevier; San Diego: 2008. [Google Scholar]